Abstract
Due to their cytotoxic activities, many anticancer drugs cause extensive damage to the intestinal mucosa and have antibiotic activities. Here we show that cisplatin induces significant changes in the repertoire of intestinal commensal bacteria that exacerbate mucosal damage. Restoration of the microbiota through fecal-pellet gavage drives healing of cisplatin-induced intestinal damage. Bacterial translocation to the blood stream is correspondingly abrogated, resulting in a significant reduction in systemic inflammation, as evidenced by decreased serum IL-6 and reduced mobilization of granulocytes. Mechanistically, reversal of dysbiosis in response to fecal gavage results in the production of protective mucins and mobilization of CD11b+ myeloid cells to the intestinal mucosa, which promotes angiogenesis. Administration of Ruminococcus gnavus, a bacterial strain selectively depleted by cisplatin treatment, could only partially restore the integrity of the intestinal mucosa and reduce systemic inflammation, without measurable increases in the accumulation of mucin proteins. Together, our results indicate that reconstitution of the full repertoire of intestinal bacteria altered by cisplatin treatment accelerates healing of the intestinal epithelium and ameliorates systemic inflammation. Therefore, fecal microbiota transplant could paradoxically prevent life-threatening bacteremia in cancer patients treated with chemotherapy.
Keywords: Immunotherapy, ovarian cancer, cisplatin, microbiome
INTRODUCTION
Platinum derivatives are one of the most commonly used chemotherapies for cancer. Cisplatin is used alone or in combination as a first-line treatment for advanced-stage ovarian, cervical, pancreatic, non-small-cell lung cancer or head and neck cancer. Due to inhibition of DNA replication, cisplatin is known to have antibiotic effects on both Gram-negative and Gram-positive bacterial strains, including some Bacillus and E. coli [1].
Despite its broad applications, cisplatin induces multiple adverse effects that severely deteriorate the quality of life of cancer patients and even precludes therapeutic continuation. One of such adverse effects, shared with other chemotherapeutic agents, is damage of the intestinal epithelium. The intestinal epithelium is subject to a rapid cell turn-over and proliferation. Cisplatin binds to DNA and forms crosslinks that impair replication. This damage results in loss of integrity in the intestinal mucosa that prevents chemotherapy continuation and causes bacteremia, which can be life-threatening. Damage to the intestinal mucosa is a major cause of infection in cancer patients treated with chemotherapy [2]. Systemic bloodstream infection due to intestinal bacterial translocation is associated with alterations in the repertoire of commensal microorganisms [3].
Thus far there are no effective measures to prevent chemotherapy-induced intestinal damage. Preclinical attempts using systemic TGF-β [4] or IL-11 [5] administration with different chemotherapies have shown promise. However, the tumor promoting effect of these cytokines has precluded clinical translation [6, 7].
Here we describe how cisplatin therapy alters the intestinal microbiota, and that restoration of the pre-chemotherapy microbiota through fecal gavage accelerates intestinal healing after cisplatin-associated intestinal damage.
MATERIALS AND METHODS
Animals and cell lines
Wild-type C57BL/6 (8–12 week old) were purchased from Charles River.
Parental ID8 cells were provided by Katherine Roby (Department of Anatomy and Cell Biology, University of Kansas Medical Center, Kansas City, KS) [8] and retrovirally transduced to express Defb29 and Vegf-a [9]. Ruminococcus gnavus (29149) was purchased from ATCC.
Animal experiments were approved by the Institutional Animal Care and Use Committee at The Wistar Institute and at the University of South Florida.
Cisplatin treatment and microbiota gavage
We treated mice with i.p. cisplatin at 10mg/kg diluted in PBS at day 21 of tumor progression. We resuspended one fecal pellet in 1ml of PBS. Pellets from non-cisplatin treated tumor bearing mice from the same cohort were used for fecal gavage after resuspension in PBS. Anaerobic cultures of Ruminococcus gnavus in Brain Heart Infusion Broth (Sigma 53286) supplemented with Yeast extract (Fluka 70161) and Hemin (Sigma H9039) (24) were resuspended in PBS at saturated concentration (OD600 >1; >109 bacteria/mL). Each mouse was gavaged with 200 μL of the microbiota mix 2 days after cisplatin treatment.
Microbiota determination
Fecal DNA was purified using PowerFecal DNA Isolation Kit (Mo Bio). We performed 16S Tag sequencing using MiSeq 500-cycle chemistry (Illumina). Groups of 5 mice were analyzed.
The sequenced 16S reads were analyzed using the QIIME software package [10] and STAR (https://stamps.mbl.edu/index.php/Picrust_stamp_lab). Reads were removed from the analysis if they did not match the barcode (up to two mismatches were allowed) and primer sequence. Operational taxonomic units (OTUs) were created by clustering the reads at 97% identity using UCLUST [11]. Representative sequences from each OTU were aligned using PyNAST [12], and a phylogenetic tree was inferred using FastTree v. 2.1.3 [13] after applying the standard Lane mask for 16S sequences [14]. STAR software was used for the comparisons. Taxonomic assignments were generated by the UCLUST consensus method of QIIME 1.8, using the GreenGenes 16S database v. 13_8 [15].
Antibodies, flow cytometry, ELISA analysis and immunohistochemistry
We used fluorochrome-conjugated anti-mouse antibodies, as follows: anti-CD3e (17A2), CD45 (30-F11), CD11b (M1/70), Ly6G (1A8), Ly6C (HK1.4) (all from Biolegend). Live/dead exclusion was done with Zombie Yellow viability probe (Biolegend).
Samples were run using a BD LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo.
Ileum and cecum samples were paraffin embedded. For staining, they were subjected to antigen retrieval and deparaffinized. Slides were then stained with hematoxylin and eosin or fixed with acetone and washed with PBS, and sections blocked using normal goat serum followed by staining with CD11b (LS Bio/LS-C141892, Run on Ventana Discovery XT, Ventana OmniMap 760-149) and Muc3 antibodies (ABIN2426703), followed by a biotinylated rabbit anti-mouse and completion of immunohistochemical procedure according to manufacturer instructions (Vector Labs). Slides were viewed using Nikon ECLIPSE 80i microscope and the NIS-Element Imaging software.
Infiltration of CD11b+ cells and density of MUC3 were analyzed by Aperio™ (Vista, CA) AT2 with a 200x/0.8NA objective lens at a rate of 3 minutes per slide via Basler tri-linear-array detection. Each slide was then analyzed using the Default Nuclear v9 Algorithm in the Spectrum database. The image algorithm used the following thresholds: Weak =210, Moderate = 188, Strong = 162 to segment positive staining of various intensities. The algorithm was applied to the entire digital slide image to determine the percentage of positive biomarker staining by applicable area.
Staining for CD31 (Novus Biologicals NB600-1475) was then analyzed using the Microvessel Analysis v1 in the Spectrum database. The algorithm was applied to the entire digital slide image to determine the total number of vessels.
Western-blot analysis of Muc3 expression was performed using anti-Muc3A ab138510 (Abcam) on proteins extracted from snap-frozen mouse ilea.
To deplete immature myeloid cells, tumor-bearing mice received 350 μg of anti-Gr1 (RB6-8C5; BioXCell) vs. control isotype antibodies i.p. daily, starting at the time of cisplatin treatment.
We analyzed IL-6 by ELISA (eBioscience), following manufacturer’s instructions.
Quantitative real-time PCR
Tissue RNA was isolated from snap-frozen samples by mechanical disruption and extracted using RNeasy kits (QIAGEN) according to manufacturer’s instruction. RNA was reverse transcribed using High Capacity Reverse Transcription kits (Applied-Biosystems). Quantification of bacterial 16S was performed using SYBR green reagents and primers (Forward 5′-TCCTACGGGAGGCAGCAGT-3′; Reverse: 5′-GGACTACCAGGGTATCTAATCCTGTT-3′). Quantification of mouse Muc2 and Muc3 were performed using SYBR green reagents and primers (Muc2: Forward: 5′-AAACTCAGCTGGGAAGAACTG G-3′; and Reverse: 5′-TTGGGAGTGGAAGTCTCAATGAT-3′; Muc3: Forward: 5′-CACCCCAGCACCTACCACTACT-3′; and Reverse: 5′-ATAGAA GAGGCTGGTGCACTGAC-3′). mRNA expression was normalized by GAPDH levels (primers Forward: 5′-CCTGCACCACCAACTGCTTA-3′; and Reverse: 5′-AGTGATGGCATGGACTGTGGT-3′). The average of three independent analyses for gene and sample was calculated using the ΔΔ threshold cycle (Ct) method and was normalized to the endogenous reference control gene Gapdh.
Primers for Eubacteria: Forward: 5′-GTAGTCCACGCCGTAAACGATG-3′; and Reverse: 5′-ACACGAGCTGACGACAACCATG-3′; Actinobacteria: Forward: 5′-TACGGCCGCAAGGCTA-3′; and Reverse: 5′-TCRTCCCCACCTTCCTCCG-3′; Lactobacillus: Forward: 5′-AGCAGTAGGGAATCTTCCA-3′; and Reverse 5′-CGCCACTGGTGTTCYTCCATATA-3′; Prevotella: Forward: 5′-CACRGTAAACGATGGATGCC-3′; and Reverse: 5′-GGTCGGGTTGCAGACC-3′; γProteobacteria: Forward: 5′-TCGTCAGCTCGTGTYGTGA-3′; and Reverse: 5′-CGTAAGGGCCATGATG-3′; αProteobacteria: Forward: 5′-CIAGTGTAGAGGTGAAATT-3′; and Reverse: 5′-CCCCGTCAATTCCTTTGAGTT-3′ and R. gnavus: Forward: 5′-GAAAGCGTGGGGAGCAAACAGG-3′; and Reverse: 5′-GACGACAACCATGCACCACCTG-3′
Statistical analysis
Unless noted otherwise, all experiments were repeated at least twice and with similar results. Differences between the means of experimental groups were calculated using Kruskal–Wallis test. Error bars represent standard error of the mean. All statistical analyses were done using Graph Pad Prism 5.0. p< 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
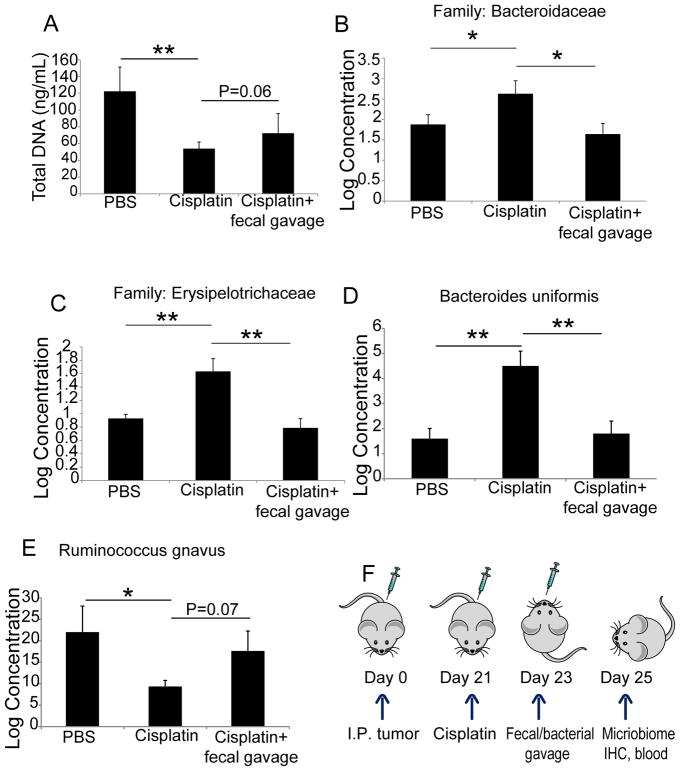
To determine the effect of cisplatin administration on the composition of intestinal commensal bacteria, we first treated different cohorts of mice bearing peritoneal ID8-Defb29/Vegf-a ovarian carcinomatosis [16–20] with different doses of cisplatin. Doses of 20 mg/mL i.p. resulted in >60% mortality (not shown), while 10 mg/mL delayed malignant progression without fatal toxicities and was therefore selected for the rest of the study. Under these conditions, the amount of fecal DNA retrieved from cisplatin treated mice was significantly lower compared with that of untreated mice after 48 hrs. (Figure 1A). Furthermore, sequencing of 16S rRNA from fecal DNA showed that cisplatin treatment causes measurable dysbiosis. This was evidenced by significant increases in bacteria of the Bacteroidaceae and Erysipelotrichaceae families, as well as in Bacteroides uniformis (Figure 1B–D and Supplemental File 1). In contrast, cisplatin caused a significant decrease of Ruminococcus gnavus, a trans-sialidase expressing bacterial strain that acquires nutritional competitive advantage by degrading mucins (Figure 1E) [21, 22].
Figure 1. Cisplatin-induced dysbiosis is reversed by fecal gavage.
(A) Quantity of DNA extracted from fecal pellets of the different group of mice. (B–E) Levels of the families of Bacteroidaceae and Erysipelotrichaceae, and the bacterial strains Bacterioides uniformis and Ruminococcus gnavus, as determined by 16S ribosomal RNA sequencing and subsequent bioinformatical analysis. (F) Schematic depiction of the experimental approach. Kruskal–Wallis test; *p<0.05; *p<0.01.
To determine whether the administration of intestinal microbiota from untreated tumor-bearing mice could prevent dysbiosis, we gavaged cisplatin-treated mice with either a fecal pellet suspension from pre-treated tumor-bearing mice or PBS, 2 days after cisplatin treatment (experimental scheme shown in Figure 1F). As shown in Figure 1B–E, gavage of fecal pellets abrogated cisplatin-induced increases in the relative amounts of Bacteroidaceae and Erysipelotrichaceae family bacteria constituting the gut microbiome. As expected, the alterations in Bacteroides uniformis and Ruminococcus gnavus strains were also partially reversed within 48 hrs. Therefore, cisplatin has a selective antibiotic effect that reduces the amount of specific bacterial species, such as Ruminococcus gnavus, thus permitting overgrowth of other bacterial families. More importantly, pre-treatment composition of the intestinal microbiome can be partially restored through oral gavage of fecal materials.
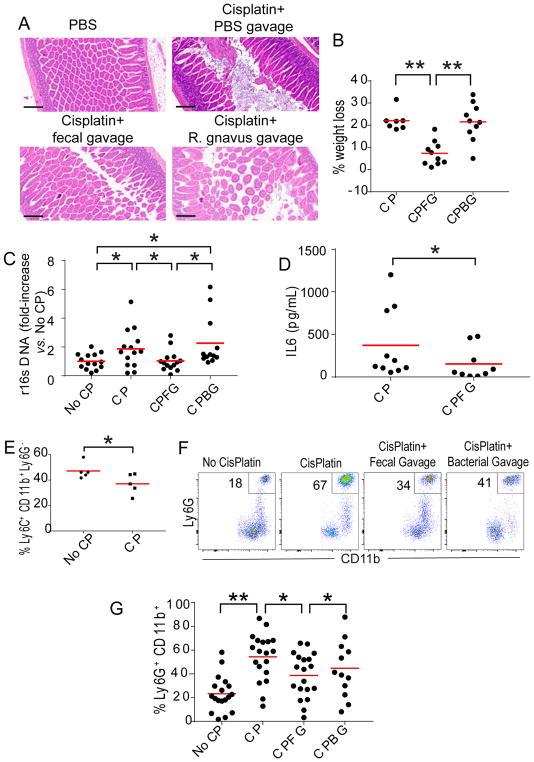
Cisplatin and other chemotherapeutic agents are known to damage the intestinal epithelium. Accordingly, we found significant disruption of the intestinal mucosa within 96 hrs of cisplatin treatment (Figure 2A). Alterations were more pronounced in the ileum, but were also detectable in the colonic mucosa. More importantly, gavage of fecal pellets resulted in decreased damage of the intestinal lining after cisplatin administration in 4 independent experiments (Figure 2A), suggesting that the restoration of the pre-treatment intestinal microbiota is able to facilitate healing of the intestinal epithelium. Gavage of Ruminococcus gnavus cultures also resulted in a trend for beneficial effects on the intestinal mucosa, although not significant (Figure 2A and Supplemental Figure 1A).
Figure 2. Transplantation of commensal microbiota accelerates healing of cisplatin-induced intestinal damage in tumor-bearing mice.
(A) Hematoxylin-eosin staining of ileum samples from control mice receiving PBS, or mice treated with cisplatin followed by gavage with PBS, fecal pellets or Ruminococcus gnavus (OD600 >1). Representative of 4 independent experiments, with similar results. (B) Percentage of weight loss 4 days after indicated interventions. CP, Cisplatin; CPFG, Cisplatin plus fecal pellet gavage; CPBG, Cisplatin plus Ruminococcus gnavus gavage. Pooled from 3 independent experiments (n=7–10 mice/group, total; Kruskal–Wallis test; *p<0.05; *p<0.01.). (C) Q-PCR quantification of the fold-increase of 16S ribosomal subunit DNA in serum, referred to the signal in PBS-treated mice (No CP). Pooled from 3 independent experiments (n=12–14 mice/group, total; Kruskal–Wallis test; *p<0.05). (D) Quantification of IL-6 in the serum of mice receiving CisPlatin, CP, or CisPlatin plus fecal gavage, CPFG (t-test; p<0.05). Pooled from 2 independent experiments (n=9–10 mice/group, total). (E) Ly6C+CD11b+Ly6G− myelomonocytic cells in the peripheral blood of tumor-bearing-mice 24 hrs after receiving PBS vs. cisplatin (t-test; p<0.05). (F&G) Ly6GhighCD11b+ granulocytes mobilized in the peripheral blood of tumor-bearing-mice treated with PBS or cisplatin, followed by PBS or fecal vs. Ruminococcus gnavus gavage (gated on CD11b+ cells). Pooled from 4 independent experiments (n=12–19 mice/group, total). Bar, 200 μm. Kruskal–Wallis test; *p<0.05; *p<0.01.
To understand the systemic effects of cisplatin-induced intestinal damage and disruption of the microbiome, we treated different cohorts of mice bearing established ID8-Defb29/Vegf-a peritoneal carcinomatosis with cisplatin, followed by gavage of pre-chemotherapy fecal pellets, Ruminococcus gnavus, or PBS. As shown in Figure 2B, cisplatin treatment induced severe weight loss, which was ameliorated by bacteriotherapy with fecal pellets but not Ruminococcus gnavus alone. Correspondingly, loss of intestinal integrity resulted in increased translocation of bacterial products to the blood stream in cisplatin-treated mice. In contrast, reversing dysbiosis through fecal gavage (but not through gavage of Ruminococcus gnavus alone) prevented bacterial translocation from the gastrointestinal tract (Figure 2C). Using genus- and strain-specific primers, we detected the presence of Eubacteria, Lactobacillus, Prevotella and Ruminococcus gnavus in the blood of all treated groups, without significant differences associated with the gavage of different products (not shown).
The passage of bacterial products from the gut to the blood stream was associated with corresponding elevations in serum IL-6 levels, which were again significantly ameliorated by fecal gavage (Figure 2D). Accordingly, although cisplatin administration induced significant decreases in the myelomonocytic compartment in peripheral blood 24 hrs after treatment, we also found increased levels of (CD45+CD11b+Ly6Ghigh) granulocytes 4 days after cisplatin treatment (Figure 2E&F). More importantly, the administration of fecal pellets through oral gavage drove significant reductions in inflammation-associated mobilization of these granulocytes (Figure 2FE&G). Higher doses of cisplatin in our model resulted in similar neutrophilia at the same temporal point (Supplemental Figure 1B). Altogether, these data show that cisplatin-induced damage to the intestinal epithelium causes increased bacterial translocation and neutrophilia within 4 days of treatment. However, restoration of the pre-cisplatin microbiome is associated with a healthier intestinal epithelium and less neutrophilia.
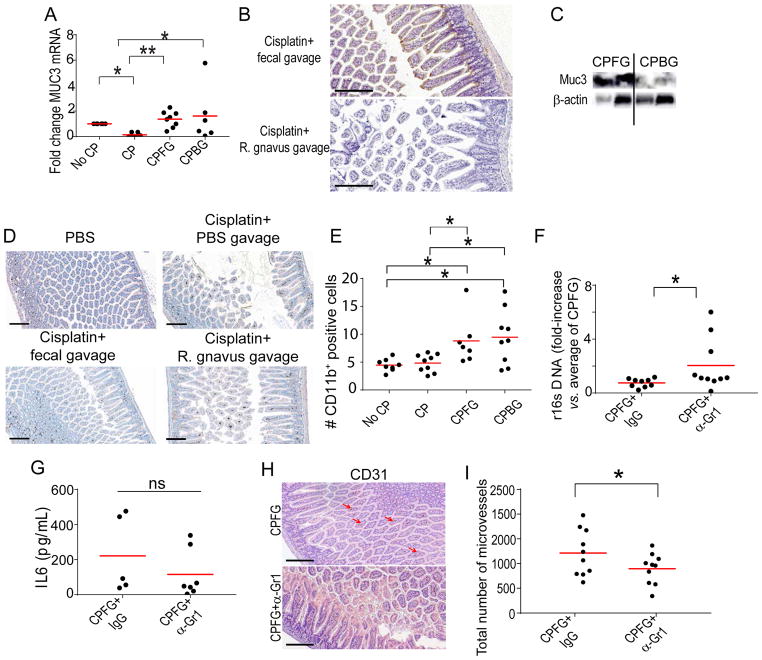
To understand the mechanisms whereby commensal bacteria promote the healing of the intestinal mucosa, we focused on the role of the microbiota in the maintenance of the mucus layer, a protective barrier comprised of glycoproteins, trefoil factors and mucins [3]. Supporting that restoration of the pre-treatment repertoire of commensal bacteria after cisplatin treatment promotes mucus secretion, we found that gavage with fecal pellets promoted higher expression of Muc3, both at the mRNA and protein levels (Figure 3A–C). The administration of Ruminococcus gnavus was also associated with stimulation of Muc3 production at the mRNA level (Figure 3A). However, the administration of this bacterial strain, which has mucin-degrading activity [21], did not result in the accumulation of Muc3 at the protein level in the gut (Figure 3B&C, and Supplemental Figure 1C).
Figure 3. Reversing dysbiosis through fecal gavage promotes mucus secretion and accumulation of myeloid cells.
(A) Normalized Q-PCR of Muc3 expression in ilea of control or cisplatin treated mice followed by administration of PBS, fecal gavage or gavage of Ruminococcus gnavus (≥5 samples/group pooled from 2 independent experiments). (B) Representative staining of Muc3 protein in the ilea of tumor-bearing mice receiving fecal pellets or Ruminococcus gnavus after cisplatin in 2 independent experiments (10 mice/group, total). (C) Representative Western-blot analysis of Muc3 in the ilea of the 2 mice described in B. (D) Representative staining of infiltration of CD11b+ myeloid cells in the ilea of mice treated with cisplatin followed by gavage with PBS, fecal pellets or Ruminococcus gnavus in 2 independent experiments (10 mice/group, total). (E) Quantification of CD11b+ spots in the experiments shown in D. (F) Fold-increase in bacterial r16s quantified by Q-PCR in the peripheral blood of receiving gavage with fecal pellets plus Gr1 depleting antibodies vs. control IgGs (iIgG). Pooled from 2 independent experiments (9–10 mice/group in each). (G) ELISA quantification of IL-6 in the serum of the mice in one of these experiments. (H) Representative CD31 staining of the ilea of cisplatin-treated mice receiving gavage with fecal pellets plus Gr1 depleting antibodies vs. control IgGs in 2 independent experiments (10 mice/group, total). (I) Quantification of the total number of microvessels, as determined through CD31 staining, in the same samples. No CP, PBS; CP, cisplatin; CPFG, cisplatin followed by fecal gavage; CPBG, cisplatin followed by Ruminococcus gnavus gavage. Kruskal–Wallis test; *p<0.05; *p<0.01. Bar, 200 μm.
Mucus production has been shown to be regulated by myeloid cells in other tissues [23]. To further understand how replacement of commensal bacteria promotes mucus secretion, we next analyzed the inflammatory infiltrates in the intestinal mucosa after different treatments. We found that gavage of both fecal pellets and Ruminococcus gnavus increased the accumulation of CD11b+ myeloid cells in the ileum (Figure 3D&E). To understand the contribution of these intestinal myeloid cells to the production of mucus and, subsequently, mucosal healing, we again performed cisplatin treatments followed by oral gavage of fecal materials in the presence vs. the absence of Gr1 depleting antibodies. As shown in Figure 3F, effective depletion of Gr1+ myeloid cells (Supplementary Figure 1D) abrogated protection against bacterial translocation in response to fecal pellet gavage. However, IL-6 was not elevated in serum, indicating that Gr1+ myeloid cells are the major source of IL-6 production after cisplatin chemotherapy (Figure 3G). Consistent with increased bacterial translocation upon myeloid cell depletion, intestinal healing also disappeared (Figure 3H). Importantly, this was associated with a significant decrease in the total number of microvessels, as determined by CD31 staining (Figure 3H&I).
Together, these results indicate that the administration of intestinal microbiota from an untreated tumor-bearing mouse is associated with increased mobilization of intestinal myeloid cells. This promotes the production of elements that comprise the mucus layer and the mobilization of myeloid cells to damaged areas, which is associated with enhanced angiogenesis and faster intestinal healing. Importantly, mucus production is independent from myeloid cell mobilization, as MUC3 production was not affected by Gr1 depletion (not shown). Administration of Ruminococcus gnavus, which is selectively depleted by cisplatin, induces comparable myeloid cell mobilization and Muc3 mRNA up-regulation. However, this mucin-degrading bacterium does not allow the accumulation of Muc3 at the protein level, resulting in decreased protection against mucosal damage.
Overall, our study demonstrates the effectiveness of administering healthy gut microbiota to accelerate healing from cisplatin-associated epithelial damage and unveils a novel intervention to improve patient wellbeing and chemotherapy completion. We found a significant decrease in R. gnavus in cisplatin treated mice, which was restored with fecal gavage. However, although reconstitution of R. gnavus induced a similar myeloid mobilization and Muc3 mRNA upregulation as fecal gavage, it was not sufficient to explain the full effect of fecal gavage in promoting the healing of the intestinal mucosa. Interestingly, increases in Ruminococcaceae in fecal microbiomes of melanoma patients have been recently associated with better response to immunotherapy, while increases in Bacteroidales had the opposite effect [24]. Cisplatin, therefore, appears to promote a microbiome associated with resistance to immunotherapy, although further studies in humans need to clarify this issue.
Cisplatin-associated intestinal damage is an important determinant of chemotherapy dose reduction, delay in treatment or even cessation of the cancer treatment [2, 25]. For the past few years, the importance of the gut microbiota has taken off, and its implications on human health and disease are starting to be understood. Bacteriotherapy is currently being used to treat recurrent C. difficile colitis [26]. For a clinical translation of our results, feces could be collected pre-chemotherapy for fecal autotransplantation and reconstitution of the microbiota after treatment with cisplatin. Fecal transplant could, therefore, become a feasible and safe approach in the treatment of chemotherapy-associated intestinal damage.
Supplementary Material
Summary sentence.
Fecal microbiota transplant accelerates intestinal healing and prevents bacteriemia after cisplatin treatment.
Acknowledgments
Support for Shared Resources was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute and CA076292 to H. Lee Moffitt Cancer Center & Research Institute. This study was supported by R01CA157664, R01CA124515, R01CA178687, The Jayne Koskinas&Ted Giovanis Breast Cancer Research Consortium at Wistar and Ovarian Cancer Research Fund Program Project Development awards. MJA and NS were supported by T32CA009171. KKP was supported by was supported by T32CA009140. APP was supported by the Ann Schreiber Mentored Investigator Award (OCRF).
ABBREVIATIONS
- DNA
Deoxyribonucleic acid
- E. coli
Escherichia Coli
- IL-6
Interleukin 6
- IL-11
Interleukin 11
- mRNA
Messenger Ribonucleic acid
- PBS
Phosphate Buffered Saline
- TGF-β
Transforming Growth Factor beta
Footnotes
AUTHORSHIP
A.P.P. and J.P.S. designed and performed most experiments and co-wrote the manuscript; M.R.R. provided intellectual and technical support; N.S., M.J.A. and K.K.P. contributed to the design of in vivo experiments and performed in vitro experiments; J.W. and C.A. performed the bioinformatical analysis of the microbiome. T.L.C. and J.A.M. processed and stored specimens and oversaw mouse experiments; R.A.C. and J.C. performed Q-PCR experiments and provided critical insight; L.N. cultured Ruminococcus gnavus and provided technical support; J.R.C.G. oversaw and designed the study and experiments, analyzed data, and co-wrote the manuscript.
COI: The authors declare no conflict of interest.
References
- 1.Joyce K, Saxena S, Williams A, Damurjian C, Auricchio N, Aluotto S, Tynan H, Demain AL. Antimicrobial spectrum of the antitumor agent, cisplatin. J Antibiot (Tokyo) 2010;63:530–2. doi: 10.1038/ja.2010.64. [DOI] [PubMed] [Google Scholar]
- 2.Taur Y, Pamer EG. Microbiome mediation of infections in the cancer setting. Genome Med. 2016;8:40. doi: 10.1186/s13073-016-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Vliet MJ, Harmsen HJ, de Bont ES, Tissing WJ. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 2010;6:e1000879. doi: 10.1371/journal.ppat.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonis ST, Lindquist L, Van Vugt A, Stewart AA, Stam K, Qu GY, Iwata KK, Haley JD. Prevention of chemotherapy-induced ulcerative mucositis by transforming growth factor beta 3. Cancer Res. 1994;54:1135–8. [PubMed] [Google Scholar]
- 5.Du XX, Doerschuk CM, Orazi A, Williams DA. A bone marrow stromal-derived growth factor, interleukin-11, stimulates recovery of small intestinal mucosal cells after cytoablative therapy. Blood. 1994;83:33–7. [PubMed] [Google Scholar]
- 6.Putoczki T, Ernst M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J Leukoc Biol. 2010;88:1109–17. doi: 10.1189/jlb.0410226. [DOI] [PubMed] [Google Scholar]
- 7.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, Byrom D, Riera A, Rossell D, Mangues R, Massague J, Sancho E, Batlle E. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–84. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roby KF, Taylor CC, Sweetwood JP, Cheng Y, Pace JL, Tawfik O, Persons DL, Smith PG, Terranova PF. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–91. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- 9.Conejo-Garcia JR, Benencia F, Courreges MC, Kang E, Mohamed-Hadley A, Buckanovich RJ, Holtz DO, Jenkins A, Na H, Zhang L, Wagner DS, Katsaros D, Caroll R, Coukos G. Tumor-infiltrating dendritic cell precursors recruited by a beta-defensin contribute to vasculogenesis under the influence of Vegf-A. Nat Med. 2004;10:950–8. doi: 10.1038/nm1097. [DOI] [PubMed] [Google Scholar]
- 10.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PloS one. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephen TL, Payne KK, Chaurio RA, Allegrezza MJ, Zhu H, Perez-Sanz J, Perales-Puchalt A, Nguyen JM, Vara-Ailor AE, Eruslanov EB, Borowsky ME, Zhang R, Laufer TM, Conejo-Garcia JR. SATB1 Expression Governs Epigenetic Repression of PD-1 in Tumor-Reactive T Cells. Immunity. 2017;46:51–64. doi: 10.1016/j.immuni.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephen TL, Rutkowski MR, Allegrezza MJ, Perales-Puchalt A, Tesone AJ, Svoronos N, Nguyen JM, Sarmin F, Borowsky ME, Tchou J, Conejo-Garcia JR. Transforming Growth Factor beta-Mediated Suppression of Antitumor T Cells Requires FoxP1 Transcription Factor Expression. Immunity. 2014;41:427–39. doi: 10.1016/j.immuni.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svoronos N, Perales-Puchalt A, Allegrezza MJ, Rutkowski MR, Payne KK, Tesone AJ, Nguyen JM, Curiel TJ, Cadungog MG, Singhal S, Eruslanov EB, Zhang P, Tchou J, Zhang R, Conejo-Garcia JR. Tumor Cell-Independent Estrogen Signaling Drives Disease Progression through Mobilization of Myeloid-Derived Suppressor Cells. Cancer Discov. 2017;7:72–85. doi: 10.1158/2159-8290.CD-16-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tesone AJ, Rutkowski MR, Brencicova E, Svoronos N, Perales-Puchalt A, Stephen TL, Allegrezza MJ, Payne KK, Nguyen JM, Wickramasinghe J, Tchou J, Borowsky ME, Rabinovich GA, Kossenkov AV, Conejo-Garcia JR. Satb1 Overexpression Drives Tumor-Promoting Activities in Cancer-Associated Dendritic Cells. Cell Rep. 2016;14:1774–86. doi: 10.1016/j.celrep.2016.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perales-Puchalt A, Svoronos N, Rutkowski MR, Allegrezza MJ, Tesone AJ, Payne KK, Wickramasinghe J, Nguyen JM, O’Brien SW, Gumireddy K, Huang Q, Cadungog MG, Connolly DC, Tchou J, Curiel TJ, Conejo-Garcia JR. Follicle-Stimulating Hormone Receptor Is Expressed by Most Ovarian Cancer Subtypes and Is a Safe and Effective Immunotherapeutic Target. Clin Cancer Res. 2017;23:441–453. doi: 10.1158/1078-0432.CCR-16-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crost EH, Tailford LE, Monestier M, Swarbreck D, Henrissat B, Crossman LC, Juge N. The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes. 2016;7:302–312. doi: 10.1080/19490976.2016.1186334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graziani F, Pujol A, Nicoletti C, Dou S, Maresca M, Giardina T, Fons M, Perrier J. Ruminococcus gnavus E1 modulates mucin expression and intestinal glycosylation. J Appl Microbiol. 2016;120:1403–17. doi: 10.1111/jam.13095. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Nadel JA. Role of neutrophils in mucus hypersecretion in COPD and implications for therapy. Treat Respir Med. 2004;3:147–59. doi: 10.2165/00151829-200403030-00003. [DOI] [PubMed] [Google Scholar]
- 24.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boussios S, Pentheroudakis G, Katsanos K, Pavlidis N. Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann Gastroenterol. 2012;25:106–118. [PMC free article] [PubMed] [Google Scholar]
- 26.Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol. 2016;14:609–20. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.