Abstract
Nicotinic acetylcholine α4β2* receptors (nAChRs) are implicated in various neurodegenerative diseases and smoking addiction. Imaging of brain high-affinity α4β2* nAChRs at the cellular and subcellular levels would greatly enhance our understanding of their functional role. Since better resolution could be achieved with fluorescent probes, using our previously developed positron emission tomography (PET) imaging agent [18F]nifrolidine, we report here design, synthesis and evaluation of two fluorescent probes, nifrodansyl and nifrofam for imaging α4β2* nAChRs. The nifrodansyl and nifrofam exhibited nanomolar affinities for the α4β2* nAChRs in [3H]cytisine-radiolabeled rat brain slices. Nifrofam labeling was observed in α4β2* nAChR-expressing HEK cells and was upregulated by nicotine exposure. Nifrofam co-labeled cell-surface α4β2* nAChRs, labeled with antibodies specific for a β2 subunit extracellular epitope indicating that nifrofam labels α4β2* nAChR high-affinity binding sites. Mouse brain slices exhibited discrete binding of nifrofam in the auditory cortex showing promise for examining cellular distribution of α4β2* nAChRs in brain regions.
Keywords: Fluorescence imaging, Nicotine, Nifene, Nifrolidine, Nifordansyl, Nifrofam
Graphical abstract
Fluorescent probes for imaging α4β2* nAChRs were synthesized which exhibited nanomolar affinities in [3H]cytisine-radiolabeled rat brain slices. Nifrofam labeling was observed in α4β2* nAChR-expressing HEK cells and was co-labeled with antibodies specific for a β2 subunit extracellular epitope indicating that nifrofam labels α4β2* nAChR.

Neuronal α4β2* nicotinic cholinergic receptors (nAChRs) are involved in learning and memory,1 and functions such as control of movement, pain, reward as well as nicotine addiction. Dysfunction of these receptors has been implicated in numerous human conditions including Alzheimer’s disease, Parkinson’s disease, and others.2 Due to the clinical importance of nAChRs, evaluating the activity of these receptors can potentially assist in understanding human illness. We have developed several positron emission tomography (PET) imaging agents for non-invasive imaging of high-affinity sites on α4β2* receptors including partial agonist [18F]nifene 1 which is currently being used in PET imaging of various species3–5 and humans.6 Fluoroalkyl derivatives such as [18F]nifrolidine 2 were also prepared as potential antagonists of α4β2* receptors.7–9 Both [18F]nifene and [18F]nifrolidine exhibit similar distribution in rat brain slices (Fig-1A–C). We have related [18F]nifene binding in rat forebrain regions to each animal’s performance on an auditory-cued, active avoidance task.10 Our results support the use of 18F-nifene for relating mature nAChRs to behavior, and potentially for tracking the dynamics of brain networks during learning acquisition and memory consolidation.
Figure 1. Lead PET α4β2 nAChRs probes for design of fluorescent probes.
1. Chemical structure of [18F]nifene; 2. Chemical structure of [18F]nifrolidine showing (blue arrow) the possibility of replacing fluorine-18 (in red circle) with a fluorophore; A. Rat brain slice showing different brain regions (FC: frontal cortex; ST: striata; TH: thalamus; HP: hippocampus; SB: subiculum; CB: cerebellum); B. [18F]Nifene binding to the brain regions shown in brain slice A; C. [18F]Nifrolidine binding to the brain regions shown in brain slice A.
Imaging resolution using in vivo PET is in the range of 1.4 mm for small animals and 4.5 mm for humans. In vitro autoradiography using PET radioligands (as seen in Fig-1 B, C) provides better resolution, but does not approach cellular level imaging. Thus, in order to study cellular and subcellular distribution of the α4β2* nAChRs, fluorescent probes are required. Efforts have been reported to derivatize epibatidine to provide fluorescent agonists for neuronal and muscle-type nAChRs.11 Successful efforts have also been made in the development of fluorescent probes for α7 nAChRs based on the structure of α-bungaratoxin and the more selective α-conotoxin.12 Knock-in mouse with yellow fluorescent protein tagged nAChR subunits (including α4YFP) have been used to study chronic nicotine administration and subcellular trafficking.13,14 However, the fluorescent protein tag is present on all α4 subunits and cannot distinguish unassembled subunits and receptors lacking high-affinity binding sites from nAChRs that contain high-affinity binding sites. Indirect fluorescence methods using calcium imaging have been used to map presynaptic functional nAChRs.15 Currently there are no fluorescent small molecules available that bind selectively to high-affinity neuronal α4β2* nAChRs.
Since [18F]nifene lacks appropriate functionality where a fluorophore may be appended, we chose [18F]nifrolidine7 in order to design a suitable fluorescent probe. The fluorine atom in the alkyl chain of nifrolidine can be replaced with the fluorophore while maintaining the primary pharmacophore of the molecule. The 3-carbon chain at the 5-position in nifrolidine does not adversely affect their binding to the α4β2* nAChRs.7–9 Similar findings of retention of affinity for α4β2* nAChRs have been reported for other pyridylethers with alkyl chains at the 5-position.16
However, the effect of the large fluorophores such as dansyl or fluorescein groups on the binding to α4β2* nAChRs is not known. Figure 2 shows energy minimized structures of nifrodansyl 4 (fluorine atom in nifrolidine 3 replaced with dansylamide) and nifrofam 5 (fluorine atom in nifrolidine 3 replaced with carboxyfluorescein). As can be seen in Fig-2A–C, the backbone structure of the nifrolidine is not affected by these large bulky groups. However, the spatial requirements of these large fluorophores may cause steric effects at the nAChR site. Carboxyfluorescein is larger than dansyl group which further increases the spatial requirements for nifrofam compared to that for nifrodansyl.
Figure 2. Potential Fluorescence Imaging Agents for α4β2 nAChRs.
A. Minimized energy structure of nifrolidine, 3 with reported high affinity for α4β2* receptors; B. Energy minimized structure of nifrodansyl, 4, in which the fluorine atom in 3 is replaced with the fluorescent moiety, dansylamide (marked in green bracket in 4). C. Energy minimized structure of nifrofam, 5, in which the fluorine atom in 3 is replaced with the fluorescent moiety, carboxyfluorescein (FAM, marked in green bracket in 5).
Thus, in this paper we report the following: 1. Synthesis and characterization of 5-(3′-propyldansylamide)-3-(2-(S)-pyrrolidinylmethoxy)pyridine (nifrodansyl, Fig-2, 4) and 5-(3′-propylfluoresciencarboxyamide)-3-(2-(S)-pyrrolidinylmethoxy)pyridine (nifrofam, Fig-2, 5); 2. Measurement of in vitro binding affinities of nifrodansyl and nifrofam in rat brain slices using [3H]cytisine to label the α4β2* nAChR sites; 3. In vitro fluorescence binding studies of nifrofam to HEK cells that stably express α4β2 nAChRs; and 4. In vitro fluorescence binding studies of nifrofam in mouse brain sections.
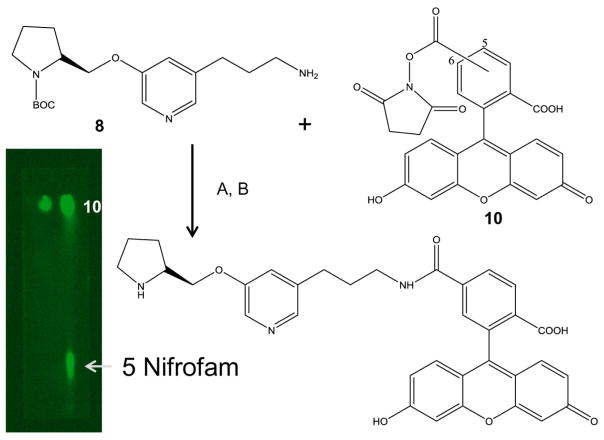
Synthesis of nifrodansyl was carried out by using modifications of the previously described procedures for nifrolidine and is shown in Fig-3.7 The key intermediate necessary for derivatization was the N-BOC substituted amine 8. The N-BOC tosylate 6, synthesized previously7, was first converted to the corresponding azide 7 in 60% yield by refluxing with sodium azide in THF.17 After purification, the azide 7 was subsequently reduced using lithium borohydride to the corresponding N-BOC amine 8 in 40% yield. Dansyl chloride was then reacted with the purified N-BOC amine 8 to provide the substituted N-BOC dansylamide 9 in 80% yield. Removal of the N-BOC protecting group using trifluoroacetic acid resulted in nifrodansyl 4 in 90% yield. Compared to dansylamide, nifrodansyl was found to be more polar due to the presence of the secondary nitrogen in the pyrrolidine ring as seen in the thin layer chromatogram in Fig-3. The purified nifrodansyl 4 trifluoroacetate salt was used for in vitro studies.
Figure 3. Synthesis of Nifrodansyl.
A. Sodium azide, DMF, trace water, reflux 24hrs. B. LiBH4, THF, rt. 24 hrs. C. Dansyl chloride, CH2Cl2, Pyridine, rt, 24 hrs. D. Trifluoroacetic acid (TFA), CH2Cl2, rt, 24 hrs. Thin layer chromatography (9:1 CH2Cl2: CH3OH) showing dansylamide (Rf= 0.8) on the left lane and the more polar nifrodansyl (Rf=0.2) on the right lane.
Synthesis of nifrofam was carried out by reacting the aforementioned N-BOC amine 8 with 5(6)carboxyfluorescein succinimidyl ester 10 to provide the corresponding N-BOC carboxamide in >70% yield (Fig-4).18 Removal of the N-BOC protecting group using trifluoroacetic acid resulted in nifrofam 5 in 75–80% yield. Nifrofam required more polar solvent to elute and was more polar than nifrodansyl due to the presence of the secondary nitrogen in the pyrrolidine ring and the polar phenolic groups as seen in the thin layer chromatogram in Fig-4. the purified nifrofam 5 was used for in vitro studies as a trifluoroacetate salt. It must be noted that the nifrofam 5 shows only the 5-carboxamido derivative but may contain the 6-carboxamido regioisomer as well. In this initial study the 5- versus 6-regioisomeric content of nifrofam was not established.
Figure 4. Synthesis of Nifrofam.
A. FAM 5,6-NHS ester 10, DMSO, rt, 24 hrs. D. Trifluoroacetic acid (TFA), CH2Cl2, rt, 24 hrs. Thin layer chromatography (1:1 CH2Cl2: CH3OH) showing carboxyfluorescien NHS ester 10 on the left lane (Rf= 0.95) and the product mixture containing nifrofam 5 in the right lane (Rf=0.2).
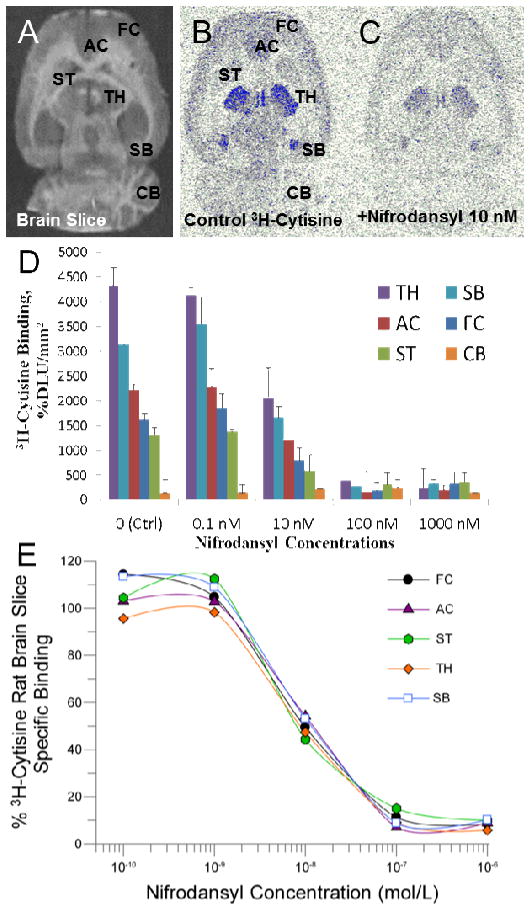
In vitro binding affinity studies of nifrodansyl and nifrofam were carried out on rat brain slices labeled with [3H]cytisine.19 Figure 5B shows [3H]cytisine labeling of rat brain regions of thalamus, frontal cortex, anterior cingulate, striatum, subiculum and cerebellum as previously reported.20,21 With increasing concentration of nifrodansyl, binding of [3H]cytisine was reduced from all brain regions (Fig. 5D). Measured inhibitory constants (IC50) of nifrodansyl in the various brain regions were: frontal cortex= 1.91 nM; anterior cingulate= 5.48 nM; striata= 0.68 nM; thalamus= 5.09 nM; subiculum= 5.36 nM.
Figure 5. Binding affinity curves for Nifrodansyl.
(A) Scan of rat brain slice; (B) total binding of [3H]cytisine in different brain regions (AC, anterior cingulate; TH, thalamus; SB, subiculum; FC, frontal cortex; ST, striatum; CB, cerebellum); (C) binding of [3H]cytisine in the presence of 10 nM nifrodansyl; (D) Binding of [3H]cytisine in the presence of different concentrations of nifrodansyl; (E) Competition binding curves of nifrodansyl with [3H]cytisine binding in rat brain regions shown in (B).
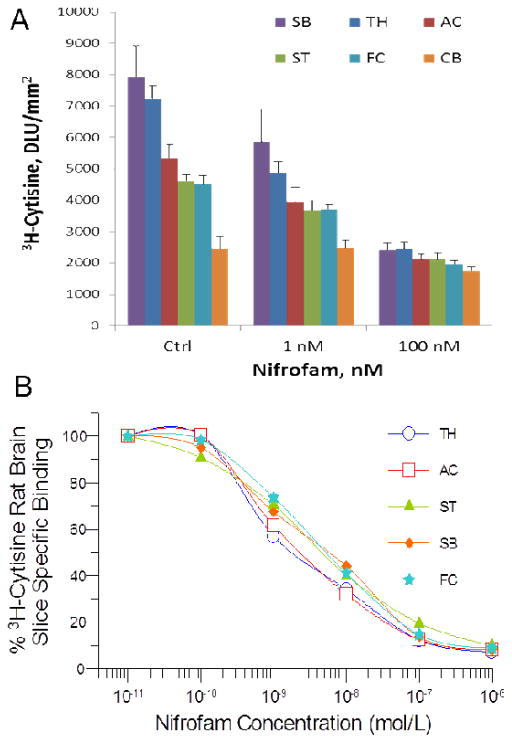
Nifrofam displaced [3H]cytisine from the various rat brain regions of thalamus, frontal cortex, anterior cingulate, striatum, subiculum and cerebellum similar to nifrodansyl (Fig 6A). With increasing concentration of nifrofam, binding of [3H]cytisine was reduced from all brain regions (Fig. 6B). Measured inhibitory constants (IC50) of nifrofam in the various brain regions were: frontal cortex= 10.8 nM; anterior cingulate= 5.91 nM; striata= 16.8 nM; thalamus= 4.04 nM; subiculum= 10.2 nM.
Figure 6. Binding affinity curves for Nifrofam.
(A). Binding of [3H]cytisine in the presence of nifrofam (AC, Anterior cingulate; TH, thalamus; SB, subiculum; FC, frontal cortex; ST, striatum; CB, cerebellum); (B). Competition binding curves of nifrofam with [3H]cytisine binding in rat brain regions.
Based on the crystal structure of the α4β2* receptor,22 the high-affinity binding sites sit at the interface between the α4 and β2 subunits in the receptor complex. Binding of the pyrrolidine ring nitrogen is likely at the α4 subunit side and since this part of the molecule remains the same in the case of nifrolidine, nifrodansyl and nifrofam, high affinity for the α4β2* receptor sites is maintained. Variations in the binding affinity between nifrodansyl and nifrofam may be due to the larger structural requirements and the constraints at the β2 subunit side of the site.
The binding affinity studies were carried out using [3H]cytisine which has been reported to have a high affinity for α2β2 (1.07 nM), α4β2 (1.51 nM), α2β4 (5.41 nM) and α4β4 (2.10 nM) nAChR subtypes.23 Rat brain mRNA regional distribution suggests the predominant receptor subtype is the α4β2 nAChR in the brain regions reported here.24 However significant differences in binding affinities between nifrodansyl and nifrofam in the frontal cortex and striatum were observed, compared to other brain regions. This may be due to binding to other receptor subtypes in these brain regions. Further studies are required to characterize subtype selectivity of nifrodansyl and nifrofam.
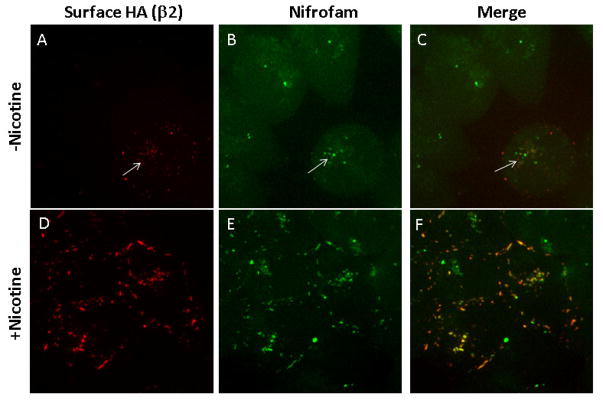
To examine the cellular distribution of α4β2* nAChRs with high-affinity binding sites, we labeled live HEK cells stably expressing α4β2 receptors with nifrofam (Fig 7). We compared nifrofam labeling in cells with and without exposure to nicotine for 17 hours to test whether nicotine upregulation of the receptors altered the labeling consistent with the upregulation. Nifrofam labeling was increased after nicotine exposure consistent with the 4–5-fold increases of 125I-epibatidine binding observed with nicotine-mediated upregulation.25, 26 Together with the nifrofam labeling, the α4β2 receptors on the cell surface were labeled with antibodies that recognized the β2 subunits tagged with the hemagglutinin (HA) epitope (Fig 7A, D; labeled surface anti-HA), which also increased with upregulation caused by a change in receptor conformation increasing binding sites numbers per receptor and increases in receptor numbers.25, 26 The anti-HA antibodies labeled the α4β2* receptors on the cell surface. In addition, it appears that a small amount of the labeled α4β2* receptors were internalized over the 30 minute and the labeling had the appearance of an endosomal pool. During the assembly and maturation of α4β2* nAChRs, high-affinity binding sites can form on the receptors and the number of high-affinity binding sites is increased or “upregulated” with long-term nicotine exposure.
Figure 7. α4β2HA stable cells - live labeling and live cell imaging.
HEK cells stably expressing α4β2HA nicotinic receptor subtypes were untreated (top panel, A–C) or treated with nicotine (10 μM for 17 hr; bottom panel, D–F). Nicotine was washed off prior to labeling the live cells with antibody or ligand. Cell surface receptors were labeled with anti-HA (β2 subunit has an HA tag at the extracellular C-terminus tail) followed by Nifrofam (100 nM) and Alexa Fluor 568 anti-rabbit secondary antibody. Images were taken on Marianas Yokogawa type spinning disk confocal microscope using 100X objective. Images are z-projects of 25 slices taken with 0.2 μm step size. Nifrofam (B, E) labeling showed high degree of co-localization with surface expressed β2 subunits (A, D). Labeling with anti-HA antibodies and nifrofam was performed for 30 minutes at 37°C.
Both nifrodansyl and nifrofam were used in preliminary binding studies in mouse brain slices.27 Although dansyl group as a fluorophore has been used in the development of fluorescent probes for pancreatic islet cells and other targets,28–30 detection of binding to α4β2* nAChR sites with nifrodansyl proved to be difficult, primarily due to the inability to achieve sufficient excitation. The dansyl group has been shown to have an excitation wavelength of approx. 330–350 nm and an emission wavelength of approx. 500–520 nm. Two-photon fluorescence microscopy may be useful in future experiments using nifrodansyl.29
Studies with fluorescein (FAM) labeled molecules and macromolecules have been actively pursued for both in vitro and in vivo applications.31 In vitro binding of nifrofam to mouse brain sections were carried out in order to evaluate α4β2* nAChRs located in the auditory cortex. Preliminary findings shown in Fig-8 indicates localization of nifrofam to α4β2* nAChRs located in the auditory cortex regions. This is consistent with the presence of α4β2* nAChRs in this brain region and also correlates to binding of PET agents such as [18F]nifene in mouse brain slices.10 Currently experiments are underway in order to optimize the specific binding of nifrofam while reducing the extent of nonspecific binding and evaluate nicotine pretreatment effects.
Figure 8. Mouse brain slice fluorescence images of Nifrofam.
(A). Binding of nifrofam (0.5 μM) to distinct cell bodies (shown by arrows; 20x) in mouse brain slices auditory cortex region. (B). Adjacent slices treated with ACSF alone (in the absence of nifrofam) did not reveal any distinct fluorescent cell bodies.
The higher excitation wavelength of 465 nm is more amenable to fluorescence imaging devices as demonstrated in our previously reported albumin-FAM derivatives.32 Our preliminary findings of nifrofam binding to the α4β2* nAChR expressed on HEK cells and in mouse brain sections shows promise in further evaluation of nifrofam cellular and subcellular binding. Further studies are also underway to establish selectivity of nifrofam binding to nAChR subtypes.
In conclusion, we have successfully developed fluorescent probes for the α4β2* nAChRs by modifying our reported PET probes. Our findings suggest that the replacing the fluorine atom with larger fluorophores moieties is tolerable at the receptor binding site with minimal loss in affinity. This feature potentially opens up the possibility of incorporation of various boron-dipyrromethene (BODIPY) fluorophores in order to red-shift fluorescence imaging of α4β2* nAChRs which can be potentially useful in deep tissue as well as in vivo imaging.33 The potential to further increase the affinity also exists by using higher affinity analogs to nifrolidine.8 Our preliminary studies both in the stably expressed α4β2* nAChR on HEK cells and mouse brain slices show promise in further understanding the physiological role of this receptor system.
Acknowledgments
This research was financially supported by a grant from NIH/NIA AG029479 (JM), NIH/NIDA DA044760 (WG and JM) and NIH/NIDCD DC13200 (RM).
Footnotes
Presented in part at Annual Society of Nuclear Medicine and Molecular Imaging Meeting, Denver, Colorado, June 10–14, 2017
Declaration of Conflict of Interest
The authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kutlu MG, Gould TJ. Physiol Behav. 2016;155:162–171. doi: 10.1016/j.physbeh.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posadas I, Lopez-Hernandez B, Cena V. Curr Neuropharmacol. 2013;11:298–314. doi: 10.2174/1570159X11311030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kant R, Constantinescu CC, Parekh P, Pandey SK, Pan ML, Easwaramoorthy B, Mukherjee J. EJNMMI Res. 2011;1:6. doi: 10.1186/2191-219X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pichika R, Easwaramoorthy B, Collins D, Christian BT, Shi B, Narayanan TK, Potkin SG, Mukherjee J. Nucl Med Biol. 2006;33:295–304. doi: 10.1016/j.nucmedbio.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Hillmer A, Wooten WD, Slesarev MS, Ahlers EO, Barnhart TE, Schneider ML, Mukherjee J, Christian BT. J Cereb Blood Flow Metab. 2013;33:1806–1814. doi: 10.1038/jcbfm.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee J, Lao P, Betthauser T, Samra GK, Pan M-L, Patel IH, Liang C, Metherate R, Christian BT. J Comp Neurol. 2018;526:80–95. doi: 10.1002/cne.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay S, Xue B, Pichika R, Collins D, Bagnera R, Leslie FM, Christian BT, Shi B, Narayanan TK, Potkin SG, Mukherjee J. J Nucl Med. 2005;46:130–140. [PubMed] [Google Scholar]
- 8.Pichika R, Easwaramoorthy B, Christian BT, Shi B, Narayanan TK, Collins D, Mukherjee J. Nucl Med Biol. 2011;38:1183–1192. doi: 10.1016/j.nucmedbio.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pichika R, Kuruvilla SA, Patel N, Vu K, Sinha S, Easwaramoorthy B, Narayanan TK, Shi B, Christian B, Mukherjee J. Nucl Med Biol. 2013;40:117–125. doi: 10.1016/j.nucmedbio.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieszczad KM, Kant R, Constantinescu CC, Pandey SK, Kawai HD, Metherate R, Weinberger NM, Mukherjee J. Synapse. 2012;66:418–434. doi: 10.1002/syn.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandl J, Sakr E, Kotzyba-Hibert F, Krieger F, Bertrand S, Bertrand D, Vogel H, Goeldner M, Hovius R. Angew Chem Int Ed. 2007;46:3505–3508. doi: 10.1002/anie.200604807. [DOI] [PubMed] [Google Scholar]
- 12.Hone AJ, Whiteaker P, Christensen S, Xiao Y, Meyer EL, McIntosh JM. J Neurochem. 2009;111:80–89. doi: 10.1111/j.1471-4159.2009.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renda A, Nashmi R. J Vis Exp. 2012;60 doi: 10.3791/3516. pii: 3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drenan RM, Nashmi R, Imoukhuede P, Just H, McKinney S, Lester HA. Mol Pharm. 2008;73:27–41. doi: 10.1124/mol.107.039180. [DOI] [PubMed] [Google Scholar]
- 15.Girod R, Jareb M, Moss J, Role L. J Neurosci Meth. 2003;122:109–122. doi: 10.1016/s0165-0270(02)00232-7. [DOI] [PubMed] [Google Scholar]
- 16.Lin NH, Li Y, He Y, Holladay MW, Kuntzweiler T, Anderson DJ, Campbell JE, Arneric SP. Bioorg Med Chem Lett. 2001;11:631–633. doi: 10.1016/s0960-894x(01)00030-0. [DOI] [PubMed] [Google Scholar]
- 17.All chemicals and solvents were of analytical or HPLC grade from Aldrich Chemical Co. and Fisher Scientific. Electrospray mass spectra were obtained on a Model 7250 mass spectrometer (Micromass LCT). Proton NMR spectra were recorded on a Bruker OMEGA 500 MHz spectrometer. Analytical thin layer chromatography (TLC) was carried out on silica coated plates (Baker-Flex, Phillipsburg, NJ). Chromatographic separations were carried out on preparative TLC (silica gel GF 20×20 cm 2000 micron thick; Alltech Assoc. Inc., Deerfield, IL) or silica gel flash column or semi-preparative reverse-phase columns using the Gilson high performance liquid chromatography (HPLC) systems. 5-(3′-azidopropyl)-3-(1-BOC-2-(S)-pyrrolidinylmethoxy)pyridine 7: 3-(1-tert-Butoxycarbonyl-2-(S)-pyrrolidinylmethoxy)-5-(3′(4-methylbenzenesulfonyloxy)-propyl)pyridine 6 (34.2 mg, 70 μmol; prepared previously, Chattopadhyay et al. 2005) was reacted with sodium azide (100 mg, 1.54 mmol) in dimethylformamide (1 mL containing 0.2 mL water). The reaction was heated for 24 hours and extracted with dichloromethane. The dichloromethane extract was purified on preparative silica gel TLC using 9:1 dichloromethane-methanol to provide pure 7 as an oil in 70% yield. NMR (CDCl3, 500 MHz) δ ppm: Mass spectra (m/z, %), 362 ([M + H]+, 70%), 384 ([M + Na]+, 53%). 1H NMR 8.20 (s, 1H), 8.11 (s, 1H), 7.45 (s, 1H), 4.11 (m, 2H), 3.95 (m, 1H), 3.65 (m, 2H), 3.39 (t, 2H, J=6Hz, CH2N3), 2.78 (m, 2H), 2.00 (m, 6H), 1.47 (s, 9H). 5-(3′-aminopropyl)-3-(1-BOC-2-(S)-pyrrolidinylmethoxy)pyridine 8: The substituted azide 7 (20 mg; 55 μmol) was taken in anhydrous THF (1 mL) into which 0.1 mL of 2M lithium borohydride was added. The reaction was stirred at room temperature for 24 hours. The reaction was quenched with water and extracted with dichloromethane. The organic extract was purified on preparative silica gel TLC using 9:1 dichloromethane-methanol to provide pure 8 as a yellow oil in 60% yield. Mass spectra (m/z, %), 336 ([M + H]+, 20%). NMR (CDCl3, 500 MHz) δ ppm: 1H NMR δ 8.20 (s, 1H), 8.11 (s, 1H), 7.45 (s, 1H), 4.11(m, 2H), 3.95 (m, 1H), 3.65 (m, 2H), 2.90 (t, 2H, CH2NH2), 2.79 (m, 2H), 1.99 (m, 6H), 1.47 (s, 9H). 5-(3′-propyldansylamide)-3-(1-BOC-2-(S)-pyrrolidinylmethoxy)pyridine 9: The amine 8 (15 mg; 45 μmol) was dissolved in dichloromethane (1 mL). To this solution, dansyl chloride (24 mg; 89 μmol) was added. The solution turned bright yellow and was stirred at room temperature for 24 hours. The reaction was then washed with saturated sodium bicarbonate and extracted with dichloromethane. The organic extract was purified on preparative silica gel TLC using 9:1 dichloromethane-methanol to provide pure 9 as an oil in 80% yield. Mass spectra (m/z, %): 569 ([M+H]+, 100%). NMR (CDCl3, 500 MHz) δ ppm: 1H NMR δ 8.43 (d, 1H, J=8Hz), 8.30 (d, 1H, J=8Hz), 8.20 (s, 1H), 8.11 (s, 1H), 8.10 (d, 1H), 7.60 (m, 2H), 7.45 (s, 1H), 7.20 (d, 1H), 4.05 (d, 2H), 4.11(m, 2H), 3.95 (m, 1H), 3.65 (m, 2H), 2.90 (t, 2H), 2.81 (s, 6H), 2.79 (m, 2H), 1.99 (m, 6H), 1.47 (s, 9H). 5-(3′-propyldansylamide)-3-(2-(S)-pyrrolidinylmethoxy)pyridine (Nifrodansyl) 4: The dansylamide 9 (5 mg; 9 μmol) was taken in dichloromethane (2 mL) into which 0.1 mL of trifluoroacetic acid was added. The reaction was stirred at room temperature for 24 hours. The reaction was quenched with saturated sodium bicarbonate and extracted with dichloromethane. The organic extract was purified on preparative silica gel TLC using 9:1 dichloromethane-methanol to provide >95% pure 4 in 60% yield as a brown oil. Mass spectra (m/z, %): 469 (45%, [M + H]+). NMR (CDCl3, 500 MHz) δ ppm: 1H NMR δ 8.40 (d, 1H, J=8.9Hz), 8.29 (d, 1H, J=8Hz), 8.20 (s, 1H), 8.11 (s, 1H), 8.10 (d, 1H), 7.60 (m, 2H), 7.45 (s, 1H), 7.20 (d, 1H, J=6Hz), 4.03 (d, 2H, J=7Hz), 4.10 (m, 2H), 3.95 (m, 1H), 3.65 (m, 2H), 2.90 (m, 2H), 2.81 (s, 6H), 2.79 (m, 2H), 1.99 (m, 6H).
- 18.5-(3′-propylfluoresceincarboxamide)-3-(2-(S)-pyrrolidinylmethoxy)pyridine (nifrofam) 5: The amine 8 (3 mg; 9 μmol mmol) was dissolved in DMSO (0.2 mL). To this solution, fluorescein NHS ester (8 mg; 17 μmol) was added along with 0.1 mL water. The solution turned bright orange and was stirred at room temperature for 24 hours. The reaction was extracted with dichloromethane and purified on preparative silica gel TLC using 9:1 dichloromethane-methanol to provide pure 5-(3′-propylfluoresceincarboxamide)-3-(1-BOC-2-(S)-pyrrolidinylmethoxy)pyridine. Mass spectra (m/z, %): 694 ([M+H]+, 100%). 5-(3′-propylfluoresceincarboxamide)-3-(1-BOC-2-(S)-pyrrolidinylmethoxy)pyridine was taken in dichloromethane (2 mL) into which 0.1 mL of trifluoroacetic acid was added. The reaction was stirred at room temperature for 24 hours. The reaction was quenched with saturated sodium bicarbonate and extracted with dichloromethane. The organic extract was purified on preparative silica gel TLC using 1:1 dichloromethane-methanol to provide >90% pure nifrofam 5 as a orange oil in 50% yield. Mass spectra (m/z, %): 594 ([M+H]+, 100%). NMR (CD3OD, 500 MHz) δ 8.20 (s, 1H), 8.11 (s, 1H), 7.75 (d, 1H, J=8Hz), 7.5 0 (d, 1H, J=8Hz), 7.45 (s, 1H), 6.90 (d, 2H, J=8.5Hz), 6.60 (d, 1H, J=8.5Hz), 6.50 (d, 2H), 6.45 (s, 2H), 4.10(m, 2H), 3.95 (m, 1H), 3.65 (m, 2H), 2.85 (2H), 2.79 (m, 2H), 1.99 (m, 6H), 1.49 (s, 9H).
- 19.All animal studies were approved by the Institutional Animal Care and Use Committee of University of California-Irvine. Ex vivo rat brain slices were prepared at 10 μm thick using a Leica 1850 cryotome. Autoradiographic studies using [3H]cytisine and drug (nifrodansyl and nifrofam) concentrations were carried out by exposing tissue radiolabeled brain sections on storage phosphor screens (Perkin Elmer Multisensitive, Medium MS). The apposed phosphor screens were read and analyzed by OptiQuant acquisition and analysis program of the Cyclone Storage Phosphor System (Packard Instruments Co., Boston, MA). Region-of-interest of same size were drawn and analyzed on brain regions using OptiQuant software and binding of [3H]cytisine measured in Digital Light Units/mm2 (DLU/mm2). Data was analyzed using following procedure: (a) the non-specific binding of 3H-cytisine was subtracted for all samples; (b) the specific binding was normalized to 100% (no competitive ligand) and (c) the binding isotherms were fit to the Hill equation (KELL BioSoft software (v 6), Cambridge, U.K.).
- 20.Kuruvilla SA, Hillmer A, Wooten DW, Patel A, Christian BT, Mukherjee J. Amer J Nucl Med Mol Imaging. 2014;4:354–364. [PMC free article] [PubMed] [Google Scholar]
- 21.Pabreza LA, Dhawan S, Kellar KJ. Mol Pharmacol. 1991;39:9–12. [PubMed] [Google Scholar]
- 22.Morales-Perez CL, Noviello CM, Hibbs RE. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chellappan SK, Xiao Y, Tueckmantel W, Kellar KJ, Kozikowski AP. J Med Chem. 2006;49:2673–2676. doi: 10.1021/jm051196m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wada E, Wada K, Boulter J, Deneris E, Heineman S, Patrick J, Swanson LW. J Comp Neurol. 1989;284:314–335. doi: 10.1002/cne.902840212. [DOI] [PubMed] [Google Scholar]
- 25.Govind AP, Vallejo YF, Stolz JR, Yan JZ, Swanson GT, Green WN. eLife. 2017;6:e25651. doi: 10.7554/eLife.25651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallejo YF, Buisson B, Bertrand D, Green WN. J Neurosci. 2005;25:5563–5572. doi: 10.1523/JNEUROSCI.5240-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Following decapitation under halothane anesthesia, brains were removed and placed in ice cold artificial cerebrospinal fluid (ACSF; in mM: 125 NaCl, 2.5 KCl, 1.25 KH2PO4, 25 NaHCO3, 1.2 MgSO4, 2 CaCl2 and 10 dextrose) bubbled with 95% O2-5% CO2. While submerged in ACSF, the brain was blocked by removing the anterior 25% and the cerebellum with coronal cuts. The blocked brain was removed from ACSF and glued to the stage of a vibrating tissue slicer (Leica VT1000S) with the anterior cut surface facing down. The brain was re-immersed in cold ACSF, and slices initially were cut in 400–550 μm-thick coronal sections. As the auditory forebrain became visible (evidenced by the appearance of the medial geniculate body), a sagittal cut was made to separate left and right hemispheres, and sectioning continued. Coronal, 250–300 μm-thick sections containing auditory cortex and thalamus were retained and placed in a holding chamber filled with 0.5 μM nifrofam in ACSF, or ACSF only. Slices were incubated 60 min at room temperature, continuously bubbled with 95% O2-5% CO2. After incubation, sections were placed on microscope slides for imaging (Zeiss Axioskop with Zeiss Axiocam digital camera and fluorescent light source; X-Cite; 120 Q Series, EXFO Photonic Solution; filter set 38 HE for eGFP EX BP 470/40, BS FT 495, EM BP 525/50). Software (Zeiss AxoVision) was used to acquire and store images.
- 28.Martino L, Masini M, Novelli M, Beffy P, Bugliani M, Marselli L, Masiello P, Marchetti P, De Tata V. PLoS ONE. 2012;7:e36188. doi: 10.1371/journal.pone.0036188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang HJ, Hsu CL, Yang JY, Yang WY. PLoS ONE. 2012;7:e32693. doi: 10.1371/journal.pone.0032693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenmogue LC, Maltais R, Poirier D. Bioorg Med Chem Lett. 2016;26:2179–2183. doi: 10.1016/j.bmcl.2016.03.069. [DOI] [PubMed] [Google Scholar]
- 31.Belykh E, Martirosyan NL, Yagmurlu K, Miller EJ, Eschbacher JM, Izadyazzdanabadi M, Bardonova LA, Byvaltsev VM, Nakaji P, Preul MC. Front Surg. 2016;3:55. doi: 10.3389/fsurg.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey SK, Kaur J, Easwaramoorthy B, Shah A, Coleman RA, Mukherjee J. Mol Imag. 2014;13:1–7. [PubMed] [Google Scholar]
- 33.Deliolanis NC, Kasmieh R, Wurdinger T, Tannous BA, Shah K, Ntziachristos V. J Biomed Opt. 2008;13:044008. doi: 10.1117/1.2967184. [DOI] [PMC free article] [PubMed] [Google Scholar]