Compounds derived from the oxidative metabolism of polyunsaturated fatty acids are synthesized in response to external stimuli and serve important roles in controlling diverse processes in both plants and animals. Members of the eicosanoid family of lipid mediators have been studied extensively with respect to their biosynthesis from C20 fatty acids and their function in the regulation of cell differentiation, immune responses, and homeostasis in animal systems (1). In plants, oxygenated derivatives of C18 and C16 fatty acids participate in the regulation of many defense-related and developmental processes. Research on fatty acid-based signaling systems in plants has focused mainly on the hormonally active compound, jasmonic acid (JA). A rapidly growing body of literature indicates that plant defense responses against insect herbivores (Fig. 1) and some microbial pathogens are orchestrated by signaling pathways involving the biosynthesis and subsequent action of JA. In this issue of PNAS, Stintzi et al. (2) used an elegant biochemical genetic approach to determine whether the cyclopentenone precursor of JA, 12-oxo-phytodienoic acid (OPDA), is also a physiological signal for defense. Their demonstration that OPDA confers broad-spectrum resistance in the absence of JA marks a major advance in our understanding of jasmonate-signaled responses.
Figure 1.
Jasmonic acid and related compounds regulate plant defense responses against hostile invaders. This photograph shows a tobacco hornworm (Manduca sexta) attacking a tomato plant.
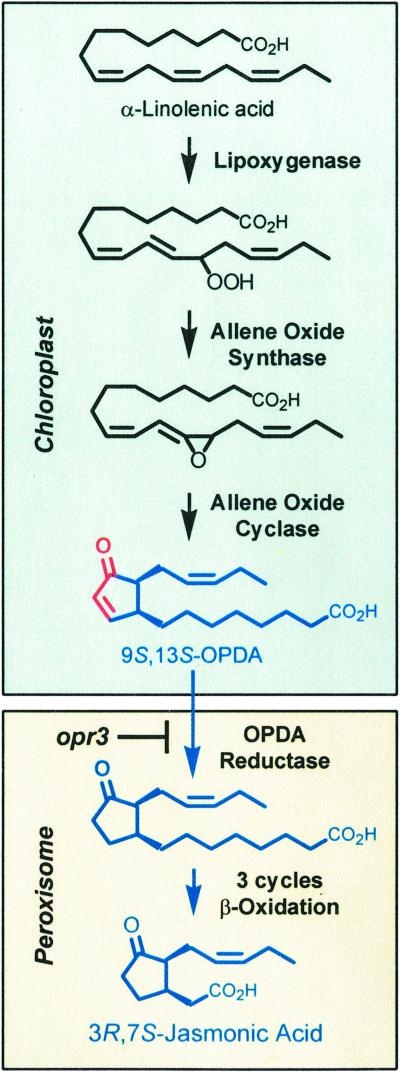
JA is a terminal product of the octadecanoid pathway (ref. 3; Fig. 2). This series of reactions is initiated by lipoxygenase, which adds molecular oxygen to linolenic acid. The resulting 13-hydroperoxide is converted in the chloroplast to a specific stereoisomer of OPDA (9S,13S-OPDA) by the sequential action of allene oxide synthase and allene oxide cyclase. The next step in the pathway involves reduction of the cyclopentenone ring of OPDA by OPDA reductase (OPR). OPR likely catalyzes this reaction in peroxisomes (4), where β-oxidation enzymes complete the synthesis of JA. The jasmonate family of compounds includes biologically active cyclopentenones (e.g., OPDA) and cyclopentanones (e.g., JA) of related structure and biosynthetic origin.
Figure 2.
The octadecanoid pathway for biosynthesis of jasmonic acid. Members of the jasmonate family of oxylipins are shown in blue. The reactive α,β-unsaturated carbonyl group in the cyclopentenone ring of OPDA is highlighted in red. The biosynthetic step blocked by the opr3 mutation is also indicated.
Several lines of evidence support an essential role for the octadecanoid pathway in various aspects of defense. First, treatment of plants with exogenous JA (or methyl-JA) results in major reprogramming of gene expression, including defense-related genes that are activated by wounding and pest attack (5–8). Second, endogenous levels of JA increase rapidly in response to wounding and other biotic stress (3). Finally, mutants defective in either the biosynthesis or perception of JA are dramatically compromised in resistance to numerous plant invaders (9–14). These findings have led to the general assumption that JA is the physiological signal for several wound- and pathogen-induced responses. A few studies, however, indicate that OPDA is also active as a signal without prior metabolism to JA. Some of the best evidence for this comes from studies of the tendril coiling response of Bryonia to mechanical stimulation. Dose-response studies with intermediates of the octadecanoid pathway provide strong evidence that OPDA is the physiologically relevant signal for this response (15, 16).
To investigate the potential role of OPDA in defense, Stintzi et al. took advantage of the Arabidopsis opr3 mutant (also known as dde1), which fails to metabolize OPDA to JA. The opr3/dde1 mutant (hereafter referred to as opr3) was identified on the basis of a defect in male gametophyte development (4, 17). A T-DNA insertion in the mutant disrupts the OPR3 locus encoding one of three known OPR isozymes. Biochemical and genetic studies have established that OPR3 catalyzes the reduction of 9S,13S-OPDA, the physiologically relevant precursor of JA (4, 17, 18). The possibility that other OPR isozymes can substitute for OPR3 in the biosynthesis of JA was ruled out by two observations. First, recombinant forms of OPR1 and OPR2 do not use 9S,13S-OPDA as a substrate (18). Second, wounded leaves of opr3 plants accumulate 9S,13S-OPDA but lack detectable levels of 3R,7S-JA (2). Although further metabolism (e.g., β-oxidation) of OPDA in the absence of OPR3 remains a formal possibility, these findings demonstrate that opr3 plants are completely blocked in the conversion of OPDA to JA (Fig. 2). Accordingly, this mutant provides a useful genetic tool to disentangle the physiological effects of endogenous OPDA from those of JA.
Mutants of Arabidopsis have been instrumental in establishing a role for JA in defense against a broad spectrum of pests. Of particular importance have been the fad3 fad7 fad8 “triple” mutant, which fails to synthesize the linolenate precursor of JA (19), and the coi1 mutant, which is deficient in the JA signaling cascade (20). Both mutants are highly compromised in resistance to the dipteran insect Bradysia impatiens and the nectrotrophic fungus Alternaria brassicicola. Based on these results and the JA-deficient phenotype of opr3, one might have predicted that opr3 plants would also be compromised in resistance. When this was put to the test, however, opr3 plants exhibited a wild-type level of resistance to both Bradysia and Alternaria. This striking result indicates that resistance in opr3 plants is mediated by a signal other than JA, the most likely candidate being OPDA.
The result indicates that resistance in opr3 plants is mediated by a signal other than JA, the most likely candidate being OPDA.
Two independent signaling pathways have been shown to regulate the expression of wound-responsive genes in Arabidopsis (6, 21). One pathway relies on wound-induced JA biosynthesis and subsequent signaling through COI1. A second pathway regulates the expression of a distinct set of target genes independently of JA and COI1. To further investigate the molecular basis of resistance in opr3 plants, cDNA microarray analysis was used to examine the expression of numerous defense-related genes in response to wounding and applied OPDA. Results of these experiments point to two important conclusions. First, the COI1 pathway can receive input from both JA and OPDA. Second, full activation of wound responsive genes involves the combined action of JA and OPDA.
Although both JA and OPDA work through COI1 to control expression of many of the same genes, these two signals also effect different responses. For example, not all COI1-dependent genes are expressed in opr3 plants in response to wounding or exogenous OPDA. The most striking example of this phenomenon is the VSP gene, which is activated by JA but not OPDA. Prior insight into the differential action of JA and OPDA has come from studies showing that the strict requirement for JA in anther and pollen maturation cannot be fulfilled by OPDA (4), and that JA and OPDA induce different patterns of volatiles in leaves of lima beans (22). These and other studies (15, 16) point to the existence of cellular mechanisms that can distinguish JA from OPDA.
Interestingly, exogenous OPDA and wounding activate several COI1-independent genes whose expression is not affected by JA. Stintzi and coworkers suggest that this effect is mediated by the defining structural feature of cyclopentenones, namely the α,β-unsaturated carbonyl group located in the cyclopentenone ring (Fig. 2). Of relevance to this hypothesis is the fact that many biological actions of cyclopentenone prostaglandins, including effects on gene expression, require the α,β-unsaturated carbonyl moiety (23). The electrophilic properties of this reactive center render cyclopentenones susceptible to conjugate addition reactions (Michael addition) with various intracellular targets. Structure-activity studies will be necessary to determine the extent to which such a mechanism accounts for the unique biological activity of OPDA, or other cyclopentenones produced from related biosynthetic pathways (24, 25).
The function of JA, OPDA, and other oxylipins (26, 27) as signals for defense supports the concept that host responses to hostile invaders are fine-tuned by a complex mix of signals, termed the oxylipin signature (24). A recent study by Hause et al. (28) highlights the fact that oxylipin profiles are shaped not only by external stimuli, but also by developmental cues that exert control on the octadecanoid pathway in specific tissues and cell types. Future advances in our understanding of the jasmonate signaling pathway will likely come from research aimed at filling several important knowledge gaps. First, what specific gene products and metabolites account for jasmonate-mediated resistance to various pests? The JA-independent resistance phenotype of opr3 plants may provide a useful starting point for such studies. Second, what are the control points that govern the synthesis and accumulation of jasmonates? Of potential relevance to this question is the recent observation that the major fraction of OPDA occurs esterified in chloroplast glycerolipids (29). Finally, what are the mechanisms by which jasmonate signals are perceived and transduced into diverse functional responses? Answers to these questions promise to shed new light on the intriguing similarities and evolutionary origins of fatty acid-based signaling processes that are essential for the well being of both plants and animals.
Acknowledgments
I thank Kurt Stepnitz for assistance with the photograph in Fig. 1. This work was supported by a grant from the National Institutes of Health (RO1GM57795).
Footnotes
See companion article on page 12837.
References
- 1.Smith W L, DeWitt D L, Garavito R M. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 2.Stintzi A, Weber H, Reymond P, Browse J, Farmer E E. Proc Natl Acad Sci USA. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. . (First Published October 9, 2001; 10.1073/pnas.211311098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaller F. J Exp Bot. 2001;52:11–23. [PubMed] [Google Scholar]
- 4.Stintzi A, Browse J. Proc Natl Acad Sci USA. 2000;97:10625–10630. doi: 10.1073/pnas.190264497. . (First Published September 5, 2000; 10.1073/pnas.190264497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farmer E E, Ryan C A. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reymond P, Weber H, Damond M, Farmer E E. Plant Cell. 2000;12:707–720. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenk P M, Kazan K, Wilson I, Anderson J P, Richmond T, Somerville S C, Manners J M. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermsmeier D, Schittko U, Baldwin I T. Plant Physiol. 2001;125:683–700. doi: 10.1104/pp.125.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe G A, Lightner J, Browse J, Ryan C A. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McConn M, Creelman R A, Bell E, Mullet J E, Browse J. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penninckx I A M A, Eggermont K, Terras F R G, Thomma B P H J, Samblanx G W D, Buchala A, Metraux J-P, Manners J M, Broekaert W F. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijayan P, Shockey J, Lévesque C A, Cook R J, Browse J. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pieterse C M, van Wees S C, van Pelt J A, Knoester M, Laan R, Gerrits H, Weisbeek P J, van Loon L C. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Royo J, León J, Vancanneyt G, Albar J P, Rosahl S, Ortego F, Castañera P, Sánchez-Serrano J J. Proc Natl Acad Sci USA. 1999;96:1146–1151. doi: 10.1073/pnas.96.3.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiler E W, Albrecht T, Groth B, Xia Z-Q, Luxem M, Liss H, Andert L, Spengler P. Phytochemistry. 1993;32:591–600. [Google Scholar]
- 16.Blechert S, Bockelmann C, Füβlein M, Schrader T v, Stelmach S B, Niesel U, Weiler E W. Planta. 1999;207:470–479. [Google Scholar]
- 17.Sanders P M, Lee P Y, Biesgen C, Boone J D, Beals T P, Weiler E W, Goldberg R B. Plant Cell. 2000;12:1041–1061. doi: 10.1105/tpc.12.7.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaller F, Biesgen C, Mussig C, Altmann T, Weiler E W. Planta. 2000;210:979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- 19.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titarenko E, Rojo E, León J, Sánchez-Serrano J J. Plant Physiol. 1997;11:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch T, Krumm T, Jung V, Engelberth J, Boland W. Plant Physiol. 1999;121:153–162. doi: 10.1104/pp.121.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straus D S, Glass C K. Med Res Rev. 2001;21:185–210. doi: 10.1002/med.1006. [DOI] [PubMed] [Google Scholar]
- 24.Weber H, Vick B A, Farmer E E. Proc Natl Acad Sci USA. 1997;94:10473–10478. doi: 10.1073/pnas.94.19.10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamberg M. Lipids. 2000;35:353–363. doi: 10.1007/s11745-000-532-z. [DOI] [PubMed] [Google Scholar]
- 26.Bate N J, Rothstein S J. Plant J. 1998;16:561–569. doi: 10.1046/j.1365-313x.1998.00324.x. [DOI] [PubMed] [Google Scholar]
- 27.Krumm T, Bandemer K, Boland W. FEBS Lett. 1995;377:523–529. doi: 10.1016/0014-5793(95)01398-9. [DOI] [PubMed] [Google Scholar]
- 28.Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C. Plant J. 2000;24:113–126. doi: 10.1046/j.1365-313x.2000.00861.x. [DOI] [PubMed] [Google Scholar]
- 29.Stelmach B A, Muller A, Hennig P, Gebhardt S, Schubert-Zsilavecz M, Weiler E W. J Biol Chem. 2001;276:12832–12838. doi: 10.1074/jbc.M010743200. [DOI] [PubMed] [Google Scholar]