Significance
Oxidative stress is a critical contributor to aging-associated diseases, including neurodegeneration, cancer, and cardiovascular disease. Here, we demonstrate that the p53 family transcription factor TAp73 contributes to the oxidative stress response by participating in the control of protein synthesis. Regulation of mRNA translation ensures a prompt and efficient method to overcome stress, and TAp73 depletion results in aberrant ribosomal biogenesis and impaired protein synthesis. In particular, TAp73 is important for maintaining active translation of mitochondrial transcripts in response to oxidative stress, thus promoting mitochondrial activity that contributes to adaptation to stress conditions. Our data therefore reveal an unexpected role for TAp73 in regulating protein synthesis responsible for its homeostatic ability.
Keywords: p53 family, TAp73, translation, ROS, mitochondria
Abstract
TAp73 is a transcription factor that plays key roles in brain development, aging, and cancer. At the cellular level, TAp73 is a critical homeostasis-maintaining factor, particularly following oxidative stress. Although major studies focused on TAp73 transcriptional activities have indicated a contribution of TAp73 to cellular metabolism, the mechanisms underlying its role in redox homeostasis have not been completely elucidated. Here we show that TAp73 contributes to the oxidative stress response by participating in the control of protein synthesis. Regulation of mRNA translation occupies a central position in cellular homeostasis during the stress response, often by reducing global rates of protein synthesis and promoting translation of specific mRNAs. TAp73 depletion results in aberrant ribosomal RNA (rRNA) processing and impaired protein synthesis. In particular, polysomal profiles show that TAp73 promotes the integration of mRNAs that encode rRNA-processing factors in polysomes, supporting their translation. Concurrently, TAp73 depletion causes increased sensitivity to oxidative stress that correlates with reduced ATP levels, hyperactivation of AMPK, and translational defects. TAp73 is important for maintaining active translation of mitochondrial transcripts in response to oxidative stress, thus promoting mitochondrial activity. Our results indicate that TAp73 contributes to redox homeostasis by affecting the translational machinery, facilitating the translation of specific mitochondrial transcripts. This study identifies a mechanism by which TAp73 contributes to the oxidative stress response and describes a completely unexpected role for TAp73 in regulating protein synthesis.
TAp73 is a member of the p53 family of transcription factors that has important functions in development (1), reproduction (2, 3), and cancer (4, 5). TAp73 plays a key role in aging, as TAp73−/− mice exhibit accelerated aging, oxidative stress, and mitochondrial defects that are attributed to insufficient expression of the mitochondrial protein cytochrome-C–oxidase–subunit-4 (COX4i1), a direct TAp73 target gene (6). Furthermore, TAp73 promotes the expression of glucose-6–phosphate dehydrogenase (G6PD) and glutaminase-2 (GLS-2), thus stimulating antioxidant and biosynthetic pathways (3, 7, 8). In cancer, TAp73 functions as a tumor suppressor, acting both in the maintenance of genomic stability (3) and in opposing angiogenesis by promoting HIF-1α (hypoxia inducible factor 1α) degradation (5, 9). This evidence implicates TAp73 as an important factor for maintaining cellular homeostasis and reinforces its role as a tumor suppressor.
Regulation of protein synthesis allows cells to exert tight control on transcripts translated under defined stress conditions (10). A master regulator of these processes is the mammalian target of rapamycin (mTOR) kinase, which coordinates the cellular responses to a range of cell stresses including amino acid deprivation and oxidative stress (11). Importantly, mTOR regulates both the global and selective translation of transcripts (12). Translation is also regulated at the elongation stage by the eukaryotic elongation factor 2 kinase (eEF2K), which directly phosphorylates its substrate, eukaryotic elongation factor 2 (eEF2), thereby inhibiting translation elongation (13). Like mTOR, eEF2K is also known to promote the translation of specific transcripts (14). The eEF2K/eEF2 axis is involved in reducing translation elongation rates following ribosomal stress and dysfunctional ribosome biogenesis (15, 16); eEF2K is indeed down-regulated as a compensatory mechanism in cells that exhibit defective ribosomal RNA (rRNA) processing (17).
Although cross-talk between mTOR and TAp73 has been recently reported (18) to regulate autophagy, a direct role for TAp73 in translational regulation has never been addressed. Here, we unravel a mechanism involving mRNA translation by which TAp73 contributes to the oxidative stress response. TAp73 depletion results in aberrant rRNA processing, reduced translation elongation, and impaired protein synthesis. Concurrently, TAp73 knockdown increases the cellular sensitivity to oxidative stress that correlates with reduced ATP levels, hyperactivation of 5′ adenosine monophosphate-activated protein kinase (AMPK), and translational defects. TAp73 maintains mitochondrial function by acting as a regulator of the translation of selected mRNAs encoding mitochondrial proteins. Therefore, the transcription factor TAp73 plays an important role in regulating mRNA translation, thus contributing to the oxidative stress response.
Results
TAp73 Depletion Results in Reduced Tolerance to Oxidative Stress and Impaired Protein Synthesis.
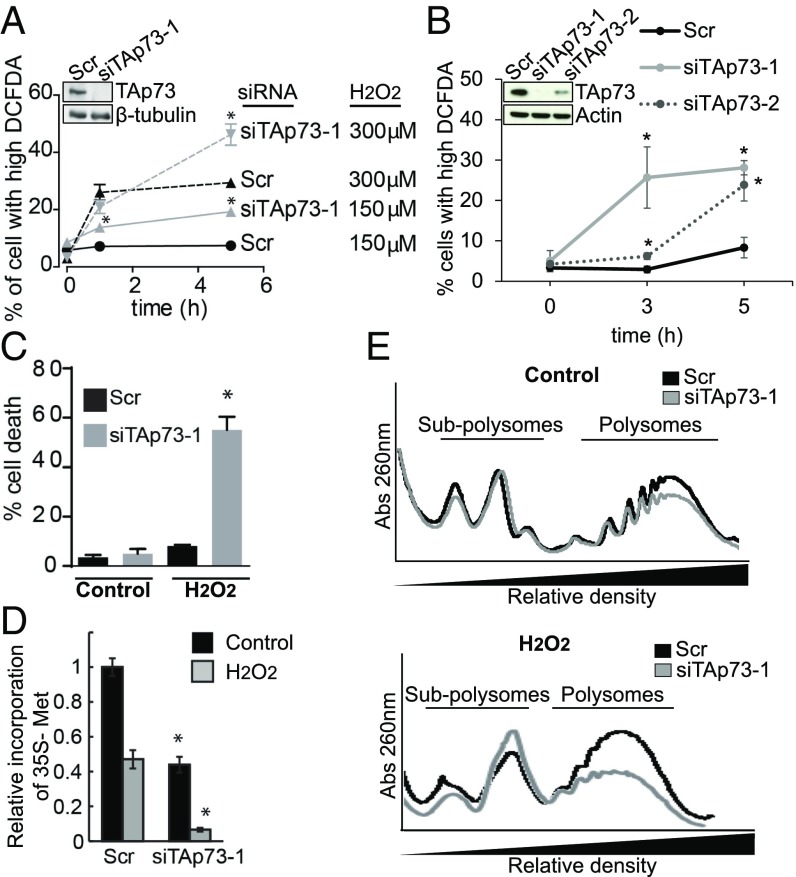
We used two different siRNAs specific for the TA (transactivation domain) isoform to knockdown (KD) TAp73 expression in HEK293T cells to investigate the role of this transcription factor in the oxidative stress response (Fig. 1 A and B). This cell line expresses high levels of TAp73, with barely detectable levels of the ΔN isoform (SI Appendix, Fig. S1A) and, equally importantly, has high expression of the Simian Vacuolating Virus 40 Tag protein which inactivates p53 (19). TAp73 KD cells accumulated more reactive oxygen species (ROS) than control cells following treatment with different concentrations of H2O2 (150 and 300 μM) or a sublethal dose of the superoxide-inducer Doxorubicin (Fig. 1 A and B and SI Appendix, Figs. S1B and S2 B–D) and displayed increased cell death following exposure to H2O2 (Fig. 1C and SI Appendix, Fig. S2A). During stress, such as an oxidative imbalance, translation regulation occupies a central position in restoring cellular homeostasis (10, 11), reducing overall protein synthesis and promoting the translation of a specific subset of mRNAs (20). We pulsed TAp73 KD and control cells with 35S-labeled methionine and cysteine and measured their incorporation in de novo synthesized proteins to explore a possible effect of TAp73 on protein synthesis. TAp73-depleted cells exhibited reduced protein synthesis (Fig. 1D and SI Appendix, Fig. S3A), which was particularly prominent under oxidative stress conditions (150 μM H2O2 for 3 h). As expected, after H2O2 treatment, a decrease in protein synthesis rate was also observed in control cells (10). We previously reported (6) that TAp73 knockout MEFs exhibit increased intracellular basal ROS. However, ROS-scavenger N-acetyl cysteine (NAC) treatment did not restore protein synthesis in TAp73 KD cells (SI Appendix, Fig. S3B), thus ruling out the possibility that the protein synthesis reduction was caused by increased basal ROS. We then examined the distribution of RNAs using sucrose-density gradient ultracentrifugation to separate translating ribosomes (polysomes) from nontranslating fractions that contain the dissociated ribosomal subunits (subpolysomes). Consistent with the decrease in protein synthesis rates, we observed a relatively small reduction in the number of polysomes in TAp73 KD cells under resting conditions (Fig. 1E, Top, and SI Appendix, Fig. S3C). The H2O2 treatment reduced the polysomal/subpolysomal ratio in control cells (SI Appendix, Fig. S3D) and induced a decrease in the number of polysomes in TAp73 KD cells (Fig. 1E, Bottom), indicating a substantial translational block when cells were challenged with prooxidant agents in the absence of TAp73. Together, our data reveal an unexpected role for TAp73 in regulating protein synthesis and suggest that this role is important in the oxidative stress response.
Fig. 1.
TAp73 KD affects the oxidative stress response and protein synthesis. (A and B) HEK293T cells were transfected with siRNAs targeting TAp73 or scrambled control for 72 h and then were challenged with 300 μM or 150 μM H2O2. Cells were washed and incubated in the presence of CM-DCFDA for 30 min and then analyzed by flow cytometry. Percentage of cells with high DCFDA is shown (mean ± SD, n = 3; *P < 0.05 vs. Scr). KD efficiency was tested (Top). (C) Cell death was determined using PI (propidium iodide) staining and flow cytometry (300 μM H2O2 for 8 h) (mean ± SD, n = 3; *P < 0.05 vs. Scr). (D) [35S]-methionine and cysteine incorporation in the proteome was measured under control conditions and after 150 μM H2O2 treatment for 3 h (mean ± SD, n = 3; *P < 0.05 vs. Scr). (E) Representative polysome profiles obtained by sucrose-density ultracentrifugation of HEK293T cell lysates transfected as in A. (Top) Control conditions. (Bottom) Cells were treated with 150 μM H2O2 for 3 h after silencing.
TAp73 Regulates Translation Elongation.
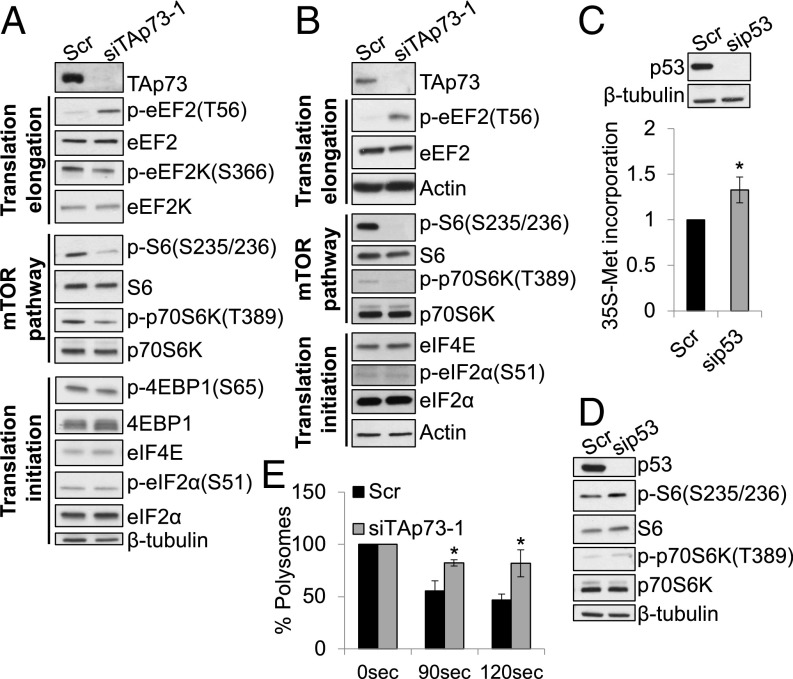
We next asked how TAp73 affects protein synthesis. Despite the large decrease in the global translation rate, we only observed a marginal reduction in the amount of polysomes in TAp73 KD cells (Fig. 1E, Top). These profiles appeared to be greatly different from cells in which translation initiation is impaired [e.g., during apoptosis or genotoxic stress (21, 22)]. Indeed, in this case, the polysomal fraction is considerably reduced and is associated with a concomitant large increase in the amount of subpolysomes. Based on these observations, we noticed that mRNAs were retained in the polysomal fraction of TAp73 KD cells, implying a possible translation elongation defect. Thus, we assessed whether TAp73 preferentially affected translation elongation or initiation by evaluating the expression and phosphorylation of eEF2 and its kinase eEF2K, in addition to the initiation factors 4EBP1 (4E binding protein 1) and eIF4E (eukaryotic initiation factor 4E). Consistent with the results shown in Fig. 1E, we observed an increase in the inhibitory phosphorylation of eEF2 in TAp73 KD cells compared with control cells, thus strongly suggesting an elongation blockade (Fig. 2A and SI Appendix, Fig. S4 A and B). In agreement with this evidence, inhibitory phosphorylation of eEF2K was reduced following TAp73 depletion (Fig. 2A). We also confirmed these data in the H1299 p53-null lung cancer cell line (Fig. 2B). Moreover, we observed a reduction in the activity of the mTOR pathway in TAp73 KD cells, as demonstrated by reduced p70S6K and S6 phosphorylation (Fig. 2A and SI Appendix, Fig. S4A). Consistent with the unaffected 4EBP1 phosphorylation and the unaltered eIF4E expression (Fig. 2 A and B and SI Appendix, Fig. S4A), the m7-GTP pull down of the eIF4F cap-binding complex (23) did not reveal any disruption in the assembly after TAp73 KD (SI Appendix, Fig. S5A). Furthermore, TAp73 depletion did not lead to any changes in eIF2α (eukariotic initiation factor 2α) phosphorylation, suggesting that the endoplasmic reticulum (ER) stress response is not influenced by TAp73 (Fig. 2 A and B). Lastly, KD of the best characterized member of the family, p53, resulted in increased protein synthesis and mTOR activity (Fig. 2 C and D), indicating the specificity of the TAp73 effects. Taken together, these data strongly imply an involvement of translation elongation blockade in reducing global protein synthesis after TAp73 KD. We measured the relative rate of translation elongation in TAp73 KD and control cells using the harringtonine runoff assay to validate this hypothesis. Harringtonine blocks translation initiation, allowing only ribosomes committed to elongation to terminate their mRNAs while preventing reinitiation; cycloheximide (CHX), which blocks elongation, is then added at different time points, and the polysome abundance is calculated following sucrose gradient ultracentrifugation of lysates. An increased amount of polysomes remaining at 90 and 120 s before CHX addition was observed in TAp73 KD cells compared with control cells, confirming a reduction in the rate of elongation (Fig. 2E and SI Appendix, Fig. S5B). Overall, our results reveal an important role for TAp73 in promoting translation elongation.
Fig. 2.
TAp73 KD impairs translation elongation. (A) Immunoblot analysis of lysates obtained from HEK293T cells transfected with the indicated siRNAs for 72 h. (B) Immunoblot analysis of lysates obtained from H1299 cells transfected with the indicated siRNAs for 48 h. (C) [35S]-methionine and cysteine incorporation in A549 (p53wt) cells transfected with siRNA targeting p53 or scrambled control for 72 h (mean ± SD, n = 3; *P < 0.05 vs. Scr). KD efficiency was tested (Top). (D) Immunoblot analysis of lysates obtained from A549 cells transfected as in C. (E) Ribosome runoff assay performed in HEK293T cells transfected as in A (mean ± SD, n = 3; *P < 0.05 vs. Scr 90 s and vs. Scr 120 s, respectively).
TAp73 Supports the Translation of Ribosomal Biogenesis Factors.
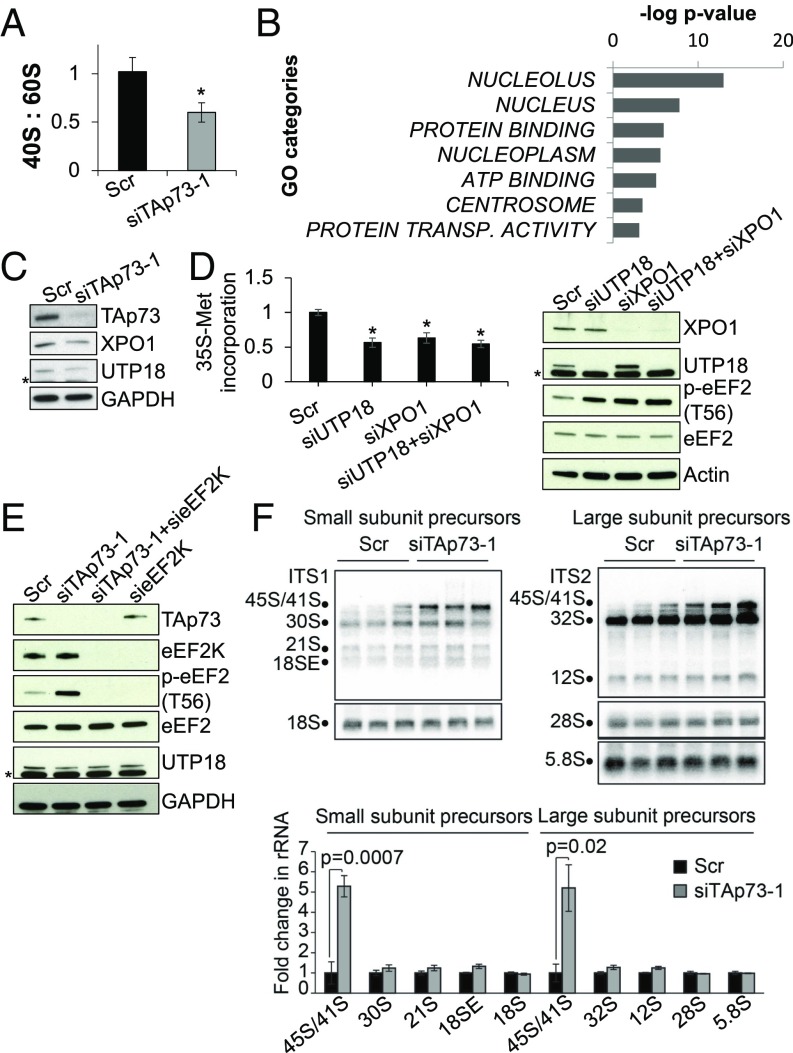
Both defective rRNA processing and ribosomal stress inhibit translation elongation. We asked whether TAp73 is involved in ribosome biogenesis to further explore the mechanisms underlying the TAp73-dependent translation elongation defects. Therefore, we examined the ribosome profiles, which showed a significant reduction in the 40S:60S ratio following TAp73 KD, indicative of a defect in ribosome assembly (Fig. 3A). We performed a microarray analysis of total RNA from TAp73 KD to assess the involvement of TAp73 in this process. As expected, TAp73 KD resulted in a massive change in the cellular transcriptome (SI Appendix, Fig. S8A and Dataset S1). However, the gene ontology (GO) analysis did not reveal changes that explained our observations, suggesting that the observed effect did not directly involve transcriptional modifications. We therefore asked whether changes in the cellular translatome might account for the ribosome assembly defect. Thus, we used RNAs collected from sub- and polysomal fractions of control and TAp73 KD cells and a microarray approach. TAp73-dependent changes in the translatome were observed following TAp73 KD (SI Appendix, Fig. S8 B and C and Dataset S2). GO analysis revealed several categories that were enriched in the list of transcripts that exhibited a reduction in polysomal distribution, of which the top category was “nucleolus” (Fig. 3B). One of the major functions of the nucleolus is rRNA processing; therefore, we hypothesized that TAp73 may play a role in this process. Several transcripts whose proteins are known to play a role in rRNA processing and biogenesis, including UTP18, XPO1, DKC1, and NOL9, exhibited reduced expression in polysomes, indicating a reduction in their translation (SI Appendix, Fig. S6A and Dataset S2). We focused on UTP18 and XPO1, both critical factors in ribosome biogenesis. UTP18 is an important component of the small subunit processome involved in the nucleolar processing of the pre-18S rRNA, while XPO1 is the nuclear pre-40S exportin. We observed decreased expression both of UTP18 and XPO1 in TAp73-depleted cells (Fig. 3C). The KD of these factors was able to recapitulate the reduction in global translation rate (Fig. 3D) and was associated with eEF2 phosphorylation. Furthermore, the simultaneous depletion of eEF2K and TAp73 did not restore the expression of UTP18, thus demonstrating that the elongation block is a consequence and not a cause of the defect in rRNA processing and ribosome assembly (Fig. 3E and SI Appendix, Fig. S6B).
Fig. 3.
TAp73 depletion results in aberrant rRNA processing. (A) 40S:60S ratio in polysome profiles of HEK293T cells transfected with the indicated siRNAs for 72 h (mean ± SD, n = 4; *P < 0.05). (B) GO analysis of transcripts that exhibit reduction on polysomes following TAp73 KD (Dataset S2). (C) Immunoblot analysis of UTP18 and XPO1 in lysates from HEK293T cells transfected as in A (star indicates nonspecific band). (D) [35S]-methionine and cysteine incorporation in HEK293T cells transfected with siRNAs targeting ribosome biogenesis factors for 72 h (mean ± SD, n = 3; *P < 0.05 vs. Scr) (star in blot analysis indicates nonspecific band). (E) Immunoblot analysis of double-knockdown experiment in lysates obtained from HEK293T transfected with the indicated siRNAs (star indicates nonspecific band). (F) Total RNAs of HEK293T cells transfected as in A were size-resolved and used for Northern blotting. Radio-labeled oligonucleotide probes revealed the abundance of small (ITS1) and large (ITS2) ribosomal subunit rRNAs precursors and the mature 28S, 18S, and 5.8S rRNAs (Top). Each lane is a biological replicate. Note that the 45S/41S species is a precursor of both small and large subunits. The abundance of each rRNA species was quantified by densitometry (Bottom) (mean ± SD, n = 3; P values are shown in the figure).
We then analyzed rRNA precursors in TAp73-depleted and control cells by Northern blotting to examine the defect in rRNA processing. Consistent with our hypothesis, we observed impaired rRNA processing in TAp73 KD cells, as evidenced by a fivefold increase in the earliest rRNA precursors (45S/41S), which were detected using two independent probes (Fig. 3F). An increase was also observed for other pre-rRNA species upstream of 18S, 5.8S, and 28S RNA (30S, 21S, 18SE, 32S, and 12S), although the difference was not significant. Taken together, these data reveal an unknown function of TAp73 in promoting translation by facilitating the association of nucleolar protein-encoding transcripts with polysomes.
TAp73 Supports Mitochondrial Activity Under Oxidative Stress Conditions.
We previously showed that TAp73-depleted cells exhibit increased sensitivity to oxidative stress (Fig. 1). A greater accumulation of ROS following H2O2 or Doxorubicin treatments was also detectable in mitochondria of TAp73 KD cells, where increased levels of superoxide were observed (SI Appendix, Figs. S2C and S7A). Since defects in oxidative phosphorylation elicit oxidative stress in vitro (6, 24), we therefore asked whether the reduced tolerance to oxidative stress is associated to defects in mitochondrial activity. TAp73 depletion did not affect oxygen consumption under resting conditions; however, a reduction was observed in TAp73-depleted cells under oxidative stress (SI Appendix, Fig. S7B). Importantly, following rotenone–succinate stimulation, no differences were observed in oxygen consumption between TAp73 KD and control cells, suggesting a defect in mitochondrial complex I (SI Appendix, Fig. S7B).
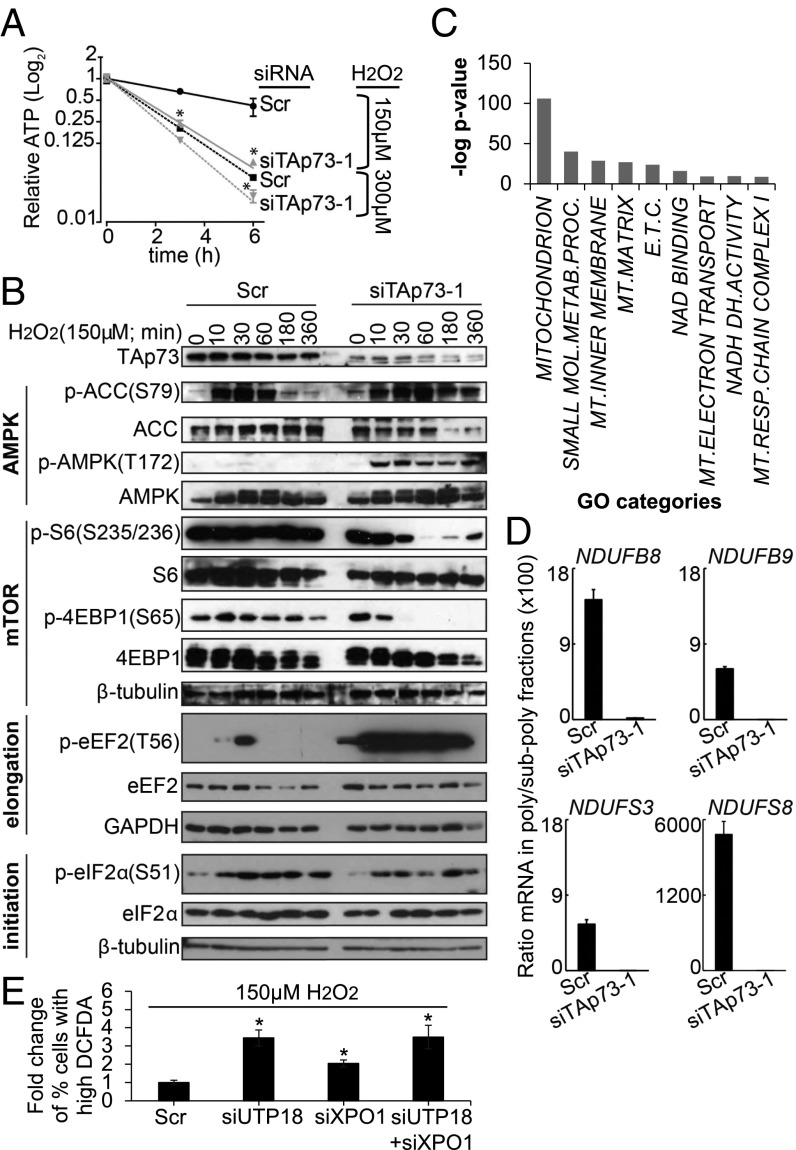
Consistent with the oxygen consumption analysis, we observed a greater decrease in ATP levels in TAp73 KD cells than in control cells following the H2O2 treatment (Fig. 4A). Based on these data, we conclude that TAp73 supports mitochondrial activity and energy production under oxidative stress conditions.
Fig. 4.
TAp73 depletion results in loss of ATP, activation of the AMPK pathway, and inhibition of mTOR under oxidative stress. (A) HEK293T cells were transfected with siRNA targeting TAp73 or scrambled control for 72 h, and ATP levels were measured after treatment with H2O2 as indicated (mean ± SD, n = 3; *P < 0.05 vs. Scr). (B) Lysates obtained from HEK293T cells transfected with the indicated siRNAs and treated with 150 μm H2O2 for the indicated time points were analyzed by immunoblot using the indicated antibodies. (C) GO analysis of transcripts that exhibit reduction on polysomes following TAp73 KD and treatment with 150 μM H2O2 for 3 h (Dataset S2). (D) HEK293T cells were transfected with the indicated siRNAs for 72 h. Sucrose-density ultracentrifugation was performed following treatment with 150 μM H2O2 for 3 h, and subpolysomal and polysomal fractions were collected. mRNAs levels were measured in each fraction by RT-qPCR (mean ± SD, n = 3). (E) ROS analysis in HEK293T cells transfected with siRNAs targeting ribosome biogenesis factors for 72 h and then treated with 150 μM H2O2 for 3 h (mean ± SD, n = 3; *P < 0.05).
TAp73 Influences the mTOR Pathway and eEF2-Mediated Protein Synthesis Under Oxidative Stress Conditions.
One important arm of the ATP sensor, AMPK, is the mTOR pathway, of which AMPK is a negative regulator. Because TAp73 depletion resulted in accelerated reduction of ATP levels in cells treated with H2O2, this change might be reflected in a more robust activation of AMPK and phosphorylation of its downstream targets. Indeed, following H2O2 treatment, TAp73 KD cells exhibited a more robust activation of AMPK than control cells, as reflected by AMPK phosphorylation and ACC (acetyl-CoA carboxylase) phosphorylation, which lasted for a longer time (Fig. 4B and SI Appendix, Fig. S4D). Moreover, a strong reduction in the activity of the mTOR pathway was observed in the TAp73 KD cells, as reflected in the phosphorylation status of two major effectors of the mTOR pathway, S6 and 4EBP1 (Fig. 4B and SI Appendix, Fig. S4 C and D). Thus, TAp73 is important for the ability of the cell to sustain the activation of the mTOR pathway and reduce the activation of AMPK under oxidative stress conditions. AMPK is known to negatively regulate translation elongation by activating eEF2K (13, 14). Following the H2O2 treatment, eEF2 phosphorylation was increased in TAp73-depleted cells compared with control cells, as the increase was much more substantial than the level observed under basal conditions (Fig. 4B; 300 min in SI Appendix, Fig. S4D). These results showed the ability of TAp73 to control translation elongation via AMPK, even under oxidative stress conditions. Energetic stress also triggers the ER stress response and eIF2α phosphorylation (25), resulting in the inhibition of translation initiation. We observed a rapid increase in eIF2α phosphorylation in both TAp73-depleted and control cells following H2O2 exposure, and this signal persisted for the duration of the experiment (Fig. 4B). However, there were no observable differences in the eIF2α response. Thus, we concluded that this arm of the stress response is not sensitive to TAp73 depletion and that translation initiation downstream of eIF2α is blocked by H2O2 independent of TAp73. Together, mTOR inhibition, eIF2α phosphorylation, and eEF2 hyperphosphorylation led to a massive block of translation and a reduction in the global protein synthesis rate (Fig. 1).
TAp73 Regulates the Translatome Under Oxidative Stress Conditions.
According to our results, TAp73 influences the cellular response to prooxidant insults. In particular, under oxidative stress conditions, TAp73 expression supports mitochondrial activity, ATP synthesis, and the activation of the mTOR pathway. Analysis of the total RNA from cells ±TAp73 and ±H2O2 (3 h, 150 mM; sublethal dose) revealed that TAp73 KD resulted in a significant change of cellular transcriptome under resting conditions; however, H2O2 treatment did not cause any significant transcriptional changes (SI Appendix, Fig. S8A). This suggests that the cellular response to H2O2 under these conditions does not involve transcriptional changes, and, perhaps, TAp73 contribution to redox response is transcriptional-independent.
Hence, we asked whether the sensitivity of the TAp73-depleted cells could be explained by their altered translational program in response to the H2O2 treatment. Using polysome profiling and a microarray analysis of H2O2-treated cells, we determined that a significant number of transcripts related to mitochondria were lost from the polysomal fraction in the TAp73 KD cells under oxidative stress conditions (Fig. 4C, SI Appendix, Fig. S8 B and C, and Dataset S2). In agreement with the reduced mitochondrial activity (SI Appendix, Fig. S7B), we observed that mRNAs for the mitochondrial complex I subunits NDUFB8, NDUFB9, NDUFS3, and NDUFS8 lost their polysomal distribution in TAp73 KD cells following the H2O2 treatment (Fig. 4D). Thus, TAp73 may play an important role in protecting mitochondria from oxidative stress by facilitating the translation of mitochondrial proteins under these conditions. To examine further the link between defective translation and reduced tolerance to oxidative stress, we measured ROS in cells depleted for rRNA biogenesis factors, UTP18 and XPO1, following H2O2 treatment. Our data show that KD of either or both was associated with increased accumulation of ROS compared with control, thus suggesting that the impaired translational machinery resulting from TAp73 depletion underpinned the defective response to oxidative stress (Fig. 4E). Based on these data, we conclude that TAp73 plays an important role in the translation of mitochondrial proteins during oxidative stress, thus promoting mitochondrial activity and coupling the latter to cellular pathways regulating translation. Overall, TAp73 promotes protein translation under oxidative stress conditions, sustaining mitochondrial activity and thus enhancing the capability of the cells to cope with redox insults (SI Appendix, Fig. S9).
Discussion
The transcription factor TAp73 is a p53 family member known to function as an important homeostasis-maintaining component in the context of cellular metabolism and oxidative stress (6, 8) by acting as a transcriptional regulator of several genes. Here we demonstrate that TAp73 contributes to cellular homeostasis and supports the cell’s ability to cope with oxidative stress by impacting protein synthesis.
Using ribosome profiling, we found that TAp73 expression is necessary to retain nucleolus-specific transcripts in the polysomal fraction. Thus, TAp73 depletion resulted in aberrant synthesis of nucleolar proteins and defective rRNA processing. The mechanism by which TAp73 affects this process is unknown, and we did not identify any direct involvement of transcription. This implies that further investigation will be necessary to fully understand the role of TAp73 in controlling the cellular translatome.
How does TAp73 promote protein synthesis? As aberrant rRNA processing leads to defective ribosomal assembly and reduced translation, TAp73 plausibly promotes protein synthesis by facilitating the translation of nucleolar proteins. In agreement with recent studies (15, 16, 26), we report that defective rRNA processing resulted in increased eEF2 phosphorylation and reduced translation elongation. Furthermore, depletion of two TAp73 target transcripts (UTP18, XPO1) recapitulated TAp73-dependent effects on the translation elongation pathway and global protein synthesis. Thus, reduced expression of ribosome biogenesis factors, such as UTP18, appears to be a primary defect upstream of elongation block, supported by our data obtained following eEF2K knockdown. In this regard, the suppression of translational elongation is a key part of the cellular response to ribosomal stress. We ruled out an involvement of basal ROS accumulation produced by TAp73 depletion on the rate of mRNA translation: treatment with the antioxidant NAC did not revert the decrease in protein synthesis rates.
Notably, the effects observed for TAp73 on protein synthesis and translation regulation pathways are specific for this member of the p53 family and are different from those of p53. The latter has been described as a repressor of global protein synthesis (27), showing inhibitory effects on translation initiation and ribosome biogenesis. In this regard, our data consistently indicate that p53 depletion results in a slight increase in protein synthesis rates.
The loss of TAp73 leads to defective ribosomes and inefficient protein synthesis that could alter the cellular capability to promptly respond to stress, such as ROS. Indeed, analysis of the translatome during oxidative stress reveals a significant shift in mitochondrial protein-encoding transcripts to the subpolysomal fraction, thus impairing mitochondrial activity in TAp73 KD cells. As functional mitochondria are necessary for the cellular response to oxidative damage (6, 24), it is reasonable to conclude that corrupted mitochondrial metabolism may be responsible for the reduced tolerance to redox imbalance in TAp73-depleted cells. Importantly, our data show that the loss of rRNA biogenesis factors (UTP18, XPO1) sensitizes cells to oxidative stress, as demonstrated by the accumulation of more ROS under H2O2 treatment. Although this approach does not fully recapitulate the complexity of TAp73 knockdown in terms of regulated genes and cell response, these data highlight the fact that fully functional ribosomes are required to maintain oxidative balance. At this stage, it is difficult to speculate how defective ribosomes, downstream of TAp73 loss, impair the translation of a specific subset of transcripts. In this regard, it is very tempting to propose a mechanism involving both the specificity in 5′-UTR sequences (28) and mechanisms of surveillance of mRNA translation (29, 30), with the latter able to recognize and dissociate defective/stalled ribosomes along the mRNA with consequent redistribution of transcripts.
TAp73 depletion reduces ATP levels under oxidative stress, possibly explaining the activation of AMPK. The robust up-regulation of the AMPK pathway in TAp73-depleted cells is associated with a sustained down-regulation of its downstream target mTOR, a reduction that correlates with strong repression of the protein synthesis rate. Moreover, the 4EBP1 arm of the mTOR pathway exhibited the greatest alterations in TAp73 KD cells under oxidative stress conditions. Interestingly, signaling through this branch of the mTOR pathway is also responsible for selective mRNA translation promoted by mTOR (12).
The inability to maintain an appropriate redox homeostasis in cells depleted of TAp73 results in cell death when they are exposed to high doses of hydrogen peroxide and for long periods. Toxic levels of ROS activate a caspase-independent cell death, defined as “oxeiptosis” (31). Although we did not investigate the mechanism that drives cell death, it is possible that analogous molecular events may lead to death also in our model.
ROS can mediate “nonspecific” damage to DNA with consequent mutations and higher probability of oncogenic events. In this context, our findings indicate a mechanism mediated by mRNA translation that might be critical for the well-known function of TAp73 as a tumor suppressor (3).
In summary, our work reveals an unexplored role for the transcription factor TAp73 in regulating key pathways linked to mRNA translation. We present evidence that TAp73 promotes protein synthesis, possibly by facilitating the translation of transcripts involved in rRNA processing. Moreover, under oxidative stress conditions, TAp73 supports a specific translational program required to maintain mitochondrial activity, helping cells to cope with redox stress. In addition to the well-established transcriptional role of TAp73 in maintaining the oxidative balance, our work describes a mechanism for TAp73-dependent response to oxidative stress involving regulation of protein synthesis (SI Appendix, Fig. S9).
Materials and Methods
Cell Culture.
HEK293T and A549 cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Sigma-Aldrich) and penicillin/streptomycin (1 U/mL) (Gibco). H1299 cells were maintained in RPMI medium 1640 (Gibco) supplemented with 10% FBS and penicillin/streptomycin (1 U/mL).
Determination of Protein Synthesis Rates.
Cells in methionine–cysteine-free medium (Sigma-Aldrich) were pulsed with 1.11 MBq/mL [35S]-labeled methionine–cysteine (Hartman Analytical) for 30 min, washed with PBS, and lysed in PLB buffer (Passive Lysis Buffer, Promega), and then trichloroacetic acid (TCA) was added. Insoluble proteins were captured on glass microfiber filter papers (Whatmann), and radioactivity was determined by scintillation counting (National Diagnostics) and normalized to the total protein content.
Mitochondrial Activity.
Cells were transfected with siRNAs for 72 h and then treated with 150 μM H2O2 for 3 h. Cell pellets were collected and respiration was measured using Oroboros, as previously described (6).
Full material and methods are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by grants from the Medical Research Council, United Kingdom, and Associazione Italiana per la Ricerca contro il Cancro (AIRC): AIRC 2014 IG15653 (to G.M.), AIRC 5xmille MCO9979 (to G.M.), and AIRC 2011 IG11955 (to G.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718531115/-/DCSupplemental.
References
- 1.Agostini M, et al. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13–17. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoue S, et al. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Natl Acad Sci USA. 2014;111:1843–1848. doi: 10.1073/pnas.1323416111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Amelio I, et al. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1α degradation. Proc Natl Acad Sci USA. 2015;112:226–231. doi: 10.1073/pnas.1410609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rufini A, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26:2009–2014. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du W, et al. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol. 2013;15:991–1000. doi: 10.1038/ncb2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amelio I, et al. p73 regulates serine biosynthesis in cancer. Oncogene. 2014;33:5039–5046. doi: 10.1038/onc.2013.456. [DOI] [PubMed] [Google Scholar]
- 9.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): Determinants of cancer progression. Trends Biochem Sci. 2015;40:425–434. doi: 10.1016/j.tibs.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 11.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proud CG. Signalling to translation: How signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- 14.Leprivier G, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight JR, et al. Cooling-induced SUMOylation of EXOSC10 down-regulates ribosome biogenesis. RNA. 2016;22:623–635. doi: 10.1261/rna.054411.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight JR, et al. Eukaryotic elongation factor 2 kinase regulates the cold stress response by slowing translation elongation. Biochem J. 2015;465:227–238. doi: 10.1042/BJ20141014. [DOI] [PubMed] [Google Scholar]
- 17.Liu R, et al. Impairing the production of ribosomal RNA activates mammalian target of rapamycin complex 1 signalling and downstream translation factors. Nucleic Acids Res. 2014;42:5083–5096. doi: 10.1093/nar/gku130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenbluth JM, Mays DJ, Jiang A, Shyr Y, Pietenpol JA. Differential regulation of the p73 cistrome by mammalian target of rapamycin reveals transcriptional programs of mesenchymal differentiation and tumorigenesis. Proc Natl Acad Sci USA. 2011;108:2076–2081. doi: 10.1073/pnas.1011936108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci USA. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushell M, et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Powley IR, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Roux PP, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung HJ, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;12:1–5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez E, Powell ML, Bevington A, Herbert TP. A decrease in cellular energy status stimulates PERK-dependent eIF2alpha phosphorylation and regulates protein synthesis in pancreatic beta-cells. Biochem J. 2008;410:485–493. doi: 10.1042/BJ20071367. [DOI] [PubMed] [Google Scholar]
- 26.Gismondi A, et al. Ribosomal stress activates eEF2K-eEF2 pathway causing translation elongation inhibition and recruitment of terminal oligopyrimidine (TOP) mRNAs on polysomes. Nucleic Acids Res. 2014;42:12668–12680. doi: 10.1093/nar/gku996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcel V, Catez F, Diaz JJ. p53, a translational regulator: Contribution to its tumour-suppressor activity. Oncogene. 2015;34:5513–5523. doi: 10.1038/onc.2015.25. [DOI] [PubMed] [Google Scholar]
- 28.Sinvani H, et al. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015;21:479–492. doi: 10.1016/j.cmet.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soudet J, Gélugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holze C, et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat Immunol. 2018;19:130–140. doi: 10.1038/s41590-017-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.