Significance
Resistance evolution caused by CRISPR/Cas9 gene-drive systems has a major impact on both the future scientific design of such gene-drive systems and on the politics of regulating experimentation and use of such systems. In our study, we show that in-frame drive-resistant alleles can be produced readily and inherently in a suppression gene-drive system. The rate at which such alleles emerge will determine the maximum size of the population that could be targeted for collapse and elimination. Here, we provide a potential target site and the modeling framework for implementation and optimization of a suppression gene-drive strategy to control Mediterranean fruit fly populations.
Keywords: homing endonuclease, integrated pest management, molecular entomology, sex reversal, Tephritid fruit flies
Abstract
The use of a site-specific homing-based gene drive for insect pest control has long been discussed, but the easy design of such systems has become possible only with the recent establishment of CRISPR/Cas9 technology. In this respect, novel targets for insect pest management are provided by new discoveries regarding sex determination. Here, we present a model for a suppression gene drive designed to cause an all-male population collapse in an agricultural pest insect. To evaluate the molecular details of such a sex conversion-based suppression gene drive experimentally, we implemented this strategy in Drosophila melanogaster to serve as a safe model organism. We generated a Cas9-based homing gene-drive element targeting the transformer gene and showed its high efficiency for sex conversion from females to males. However, nonhomologous end joining increased the rate of mutagenesis at the target site, which resulted in the emergence of drive-resistant alleles and therefore curbed the gene drive. This confirms previous studies that simple homing CRISPR/Cas9 gene-drive designs will be ineffective. Nevertheless, by performing population dynamics simulations using the parameters we obtained in D. melanogaster and by adjusting the model for the agricultural pest Ceratitis capitata, we were able to identify adequate modifications that could be successfully applied for the management of wild Mediterranean fruit fly populations using our proposed sex conversion-based suppression gene-drive strategy.
The use of CRISPR-Cas9 systems as a homing-based gene-drive tool to alter the genotype of insect populations has theoretically (1–5) and practically (6–8) been shown to be feasible. These systems can potentially allow the spread of any desired trait in a wild population of target species even if the desired phenotype imposes a fitness cost (2, 4, 5, 8). Therefore, the spread of lethality or sterility traits that could result in suppression and eventually collapse of the target population should be possible. This has recently attracted special attention in pest and disease vector control (1, 3, 6–8). However, the effort had focused mainly on disease-vector mosquitoes such as Anopheles (7, 8). In homing CRISPR/Cas9 gene-drive (HCGD) systems, a CRISPR/Cas9 homing element (CHE) composed of at least the Cas9 endonuclease-coding sequence and a guide RNA (gRNA) is integrated in the host genome at the gRNA target site. In the heterozygous state, Cas9 introduces an RNA-guided double-strand break in the wild-type allele (similar to homing endonucleases) which then will be repaired either by homology-directed repair (HDR) or error-prone mechanisms such as nonhomologous end joining (NHEJ). In the former case, the CHE allele serves as the repair template and is copied into the homologous chromosome. Directing this process to the germline will result in super-Mendelian inheritance driving the CHE and any accompanying genes into the population. Therefore, the highly customizable nature of CRISPR/Cas9 allows simple design of HCGDs to drive any desired trait, even those resulting in sterility, into wild populations as long as the cost of this phenotype does not surpass a certain threshold (1, 4).
In a recent study, Hammond et al. (8) identified a set of genes whose knockout resulted in female-specific sterility in Anopheles. However, they found that only one of these genes could be used as a target for HCGDs to achieve an efficient drive of female-specific sterility into the population. The remaining sterility genes imposed a very strong cost on the carriers that eventually resulted in the elimination of the drive allele from the population. As predicted by mathematical population genetics models, the spread of female-specific sterility traits in a population using HCGDs should eventually result in a population collapse and local or global elimination of the target species (1, 8). Another proposed strategy to achieve this goal is to design drive elements that alter the population’s sex ratio toward males. Surprisingly, such gene-drive elements have naturally been observed in some organisms. In Aedes aegypti, for example, a type of drive element known as a “Killer-Y chromosome” is able to shatter the X chromosome during spermatogenesis, and therefore all offspring of mosquitoes carrying such a chromosome will be male. To replicate this phenomenon, Galizi et al. (9) employed a specific homing endonuclease, I-Ppol, to specifically shatter the X chromosome during spermatogenesis of Anopheles gambiae. By generating transgenic males carrying an engineered version of such a homing endonuclease gene (HEG) on somatic chromosomes, they have shown that at high initial load frequencies these flies will result in population collapse in cage experiments. They proposed that integration of such a HEG on the Y chromosome could be an effective gene-drive strategy for population control of An. gambiae. The distortion of the sex ratio using an X chromosome-specific CRISPR/Cas9 system has also been shown to be successful in An. gambiae (10).
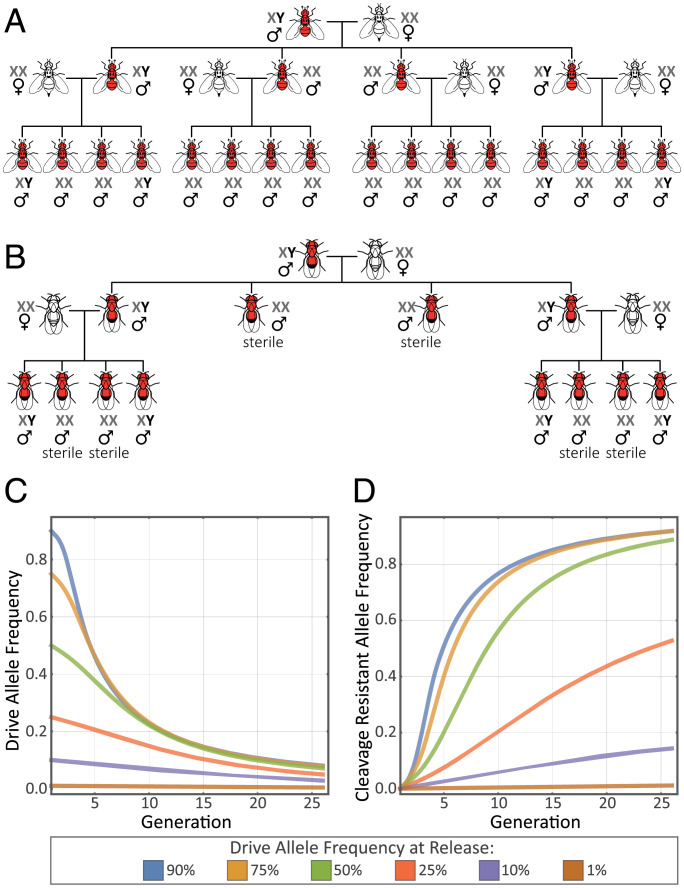
Here, we propose an independent approach that converts female individuals into fertile males by disturbing the developmental sex-determination pathways, which distorts the sex ratio without adverse effects on the reproductive success of carrier males. A prime target gene to achieve this goal is transformer (tra). tra plays a pivotal role in female sex determination in different insect orders, including Diptera (11). In a devastating agricultural fruit pest, the Mediterranean fruit fly, Ceratitis capitata (“medfly”), tra-knockdown XY males develop normally, while XX individuals develop as fertile males (12). Therefore, C. capitata XX males carrying a CHE-targeted tra locus could further spread the CHE to all their progeny (Fig. 1A), resulting in an effective gene drive without any direct effect on the fecundity of individuals carrying the drive element. This in theory could lead to an all-male population collapse that can be used for controlling the wild population of this aggressive pest.
Fig. 1.
Insect suppression gene drive based on forced all-male offspring. (A) In C. capitata, a tra-targeting CHE with both germline and somatic expression causes super-Mendelian inheritance of the red fluorescence-marked CHE-null allele but also results in the transformation of XX individuals into males and in theory leads to a subsequent collapse of the population. (B) In D. melanogaster, homing into tra in somatic cells transforms XX individuals into sterile pseudomales, which halts the spread of the selfish element but allowed us to safely study the dynamics and molecular consequences of using CHEs in a suppression gene-drive system. (C) Predicted transience of a Cas9-based homing construct targeting the tra locus in D. melanogaster. Predictions are based on the introduction of tranCHE/+ males at frequencies of 1–90% into a population of otherwise wild-type males and females in equal proportion. We assume a Cas9-mediated cleavage efficiency of 100%, a probability of accurate HDR following cleavage of 90%, one-third of drive-resistant alleles (NHEJ products) being in-frame indels, and no fitness cost associated with in-frame drive-resistant alleles. A construct having these parameter values and released in the form of tranCHE/+; XY males is expected to be reduced to an allele frequency of less than 10% within ∼25 generations with a trajectory tending toward elimination regardless of the introduction frequency. (D) At high release frequencies, the presence of the drive allele results in the generation and establishment of cleavage-resistant alleles in the population. At low release frequencies, which may occur because of accidental escapes, the drive allele will be eliminated early, and the cleavage-resistance alleles will appear at only negligible frequencies. This indicates that D. melanogaster is a safe model organism for the evaluation of a tra-based suppression gene drive causing sex conversion.
Because of the strict guidelines on gene-drive experiments and to adhere to recommendations of scientific communities (13–15), we decided to test this gene-drive strategy first using Drosophila melanogaster as a model organism. In D. melanogaster, tra-mutant XX individuals develop into infertile pseudomales (16), not giving rise to further progeny (Fig. 1B). Since the cost of this infertility is significantly higher than the threshold tolerated by gene-drive systems (1, 4), a CHE targeting the tra locus in D. melanogaster, despite its ability to show super-Mendelian inheritance in individual crosses, is not able to drive into a population (Fig. 1 C and D). This biological confinement allows us to employ D. melanogaster as a safe model organism for studying the limitations of our suggested suppression gene-drive systems at the molecular level in the laboratory and thereby experimentally identify parameters that might need to be adjusted to achieve an efficient suppression gene-drive system in C. capitata.
In our study, we found that targeting tra works as an efficient means of sex conversion in D. melanogaster. However, the early onset of the formation of in-frame drive-resistant alleles compromises drive efficiency. Based on our observations, we simulated the use of a tra-based suppression gene-drive system for control of C. capitata populations and showed that HCGD systems employing multiple gRNAs that target the tra locus can serve as an effective pest-control strategy for C. capitata.
Results
Design of a tra-Based Sex Conversion-Suppression Gene-Drive System.
The proposed CHE is composed of a spCas9-coding sequence under the control of a suitable promoter, as explained below, a gRNA targeting the first exon of tra under the control of a Pol III promoter, and a fluorescent marker to identify the genomic integration (SI Appendix, Fig. S1C). The activity of this CHE unit will be similar to that of homing endonucleases and would be able to perform homing into the wild-type tra allele. For our tra-targeting CHE to drive in a population, it is essential that Cas9 is expressed in the germ cells to promote homing into the wild-type tra allele by HDR. To achieve sex conversion, however, tra needs to be inactivated in the somatic cells of XX individuals. Thus two scenarios in XX individuals heterozygous for the drive allele are plausible: (i) Cas9 protein is expressed only in a fraction of the cells, and its activity results in the development of mosaic intersex individuals or (ii) Cas9 is expressed in all somatic cells and uniformly destroys the wild-type tra allele, resulting in the development of XX males. In C. capitata, the latter will result in development of fertile XX males (12), which can further spread the drive allele into the population (Fig. 1A). It is important to note that it is irrelevant whether the mutation of the wild-type tra allele in the somatic cells is based on HDR or NHEJ as long as the mutation disrupts the function of tra and thereby causes sex conversion.
Therefore, the combination of germline homing at the tra locus (which results in the spread of the drive allele) and somatic targeting of the wild-type tra allele (which results in sex conversion) is needed to enable our proposed suppression gene-drive strategy to be effective. To achieve this, different types of promoters or combinations thereof could be used. (i) A germline-specific promoter could be combined with an early zygotic promoter from a cellularization gene for high and ubiquitous blastoderm expression (17). Such cellularization promoters have already been successfully applied for transgenic approaches in C. capitata (18). It is important to note that these early cellularization genes are not expressed in the primordial germ cell (PGC) nuclei (19, 20), which are therefore not exposed to NHEJ-based mutation in the early embryo (21, 22). In D. melanogaster, one of these cellularization genes, Sry-α, is in fact expressed both in a somatically limited way in the blastoderm and in the PGCs at later developmental stages (23, 24), and its promoter therefore might be sufficient for both germline homing and somatic sex conversion. (ii) Since Pol II-dependent transcription is actively suppressed in the PGCs (25), a ubiquitous cell cycle-specific promoter, such as the DNApol-α180 promoter (26), could result in uniform targeting of all cells during development except early-stage PGCs. The paternal-only transmission of our proposed gene-drive strategy is likely to help overcome the problem of DNA cleavage at early embryonic stages when HDR is unlikely to occur (21, 22) and therefore is expected to result in both uniform sex conversion and germline homing. (iii) Since the target gene tra is expressed in the somatic cells at very early embryonic stages, the genomic context might mediate suitable amounts of expression independently of the introduced promoter. Thus, the introduction of a germline-specific promoter, such as the Rcd-1r promoter, which had previously been shown to result in efficient homing-based gene drive in D. melanogaster (27), might by itself be sufficient to drive Cas9 expression for both purposes.
D. melanogaster as a Safe Model System for Evaluation of a tra-Based Suppression Gene Drive.
In our experiments, we followed the recommended physical containment procedures (15) (SI Appendix, SI Materials and Methods). Moreover, since in D. melanogaster XX males are always sterile, the somatic sex conversion imposes a strong fitness cost on the XX individuals carrying the drive allele, which impedes the spread of the drive allele in the population (1, 4), rendering D. melanogaster a safe model system to study this suppression gene-drive strategy at the molecular level (Fig. 1B). Nevertheless, to ensure that the use of a CHE against the tra locus in D. melanogaster is indeed biologically confined in case of an unlikely accidental escape, a deterministic model for an ideal scenario (homing efficiency of 90% and assuming that one-third of NHEJ events result in the formation of in-frame indels) based on predicted phenotypic outcomes of the drive in D. melanogaster was used. The modeling graphs demonstrate that, because of its high fitness cost, even at 90% initial frequency a CHE targeting the tra locus not only is unable to drive into a population but also is actively eliminated from the population (Fig. 1C). In this example, the presence of the drive allele at high frequencies may result in the generation of cleavage-resistant alleles, which theoretically could alter the genetic makeup of the population at the targeted locus (28). However, our results indicate that at the low release frequencies (<1%) that are expected in case of an accidental release, the drive allele becomes eliminated from the population at very early stages without any significant effect on the wild population (Fig. 1D). Therefore, it is safe to assume that such a drive system is biologically confined in D. melanogaster and thus meets the recommendations for gene-drive experiments (13–15, 28).
Implementation of the tra-Based Suppression Gene-Drive System in D. melanogaster.
Since our sex conversion-based gene-drive system requires both somatic and germline Cas9 activity, we tested three different promoters (SI Appendix, Fig. S1): (i) the Sry-α promoter, (ii) the DNApol-α180 promoter, and (iii) the Rcd-1r promoter. We also included the 3′ UTR of the β2 Tubulin (βTub85D) gene at the 3′ end of the Cas9 transcript, as it had been shown to increase the homing efficiency in D. melanogaster (27). Moreover, the first intron of αTub84B was inserted upstream of the Cas9 coding sequence to further enhance Cas9 expression (29).
To allow the simple generation of various strains in an isogenic background for these promoters, we used a transgenesis approach similar to that demonstrated in Anopheles (SI Appendix, Fig. S2) (8). First, a tranDOCK strain was established by site-specific integration of a recombinase-mediated cassette exchange (RMCE) docking site into the first exon of the tra gene using an efficient gRNA (SI Appendix, Fig. S1 A and B). Second, to generate the homing strains for each of the promoters, RMCE was performed in tranDOCK embryos using ϕC31 integrase. All individuals that carried the CHE allele (tranCHE) were found either to be males or to show a mosaic intersex phenotype, indicating that targeting the tra locus is indeed an efficient sex-conversion strategy in D. melanogaster (SI Appendix, Fig. S3A).
To assess the efficiency of each promoter in performing gene drive as well as inducing somatic sex conversion, 10 males from each of the tranCHE strains and the tranDOCK strain were individually crossed with w− virgins, and the ratio of females in the F1 generation from each single cross was determined (SI Appendix, Fig. S1D). The results show that all three promoters can block female development of heterozygous (tranCHE/+) XX individuals (somatic sex conversion) and drive into the next generation (germline activity). However, since HDR in the germline is of key importance for the molecular study of gene drive, we continued our experiments with the Rcd-1r strain.
To evaluate the drive efficiency of this CHE and the rate at which the tra locus is targeted to cause sex conversion, 12 heterozygous (tranCHE/+) males were crossed individually with virgin w− flies. Screening the F1 progeny revealed that up to 92% of the individuals carried the DsRed eye marker (on average 78%, corresponding to a homing efficiency of 56%), and up to 96% (on average 89%) were males/intersexes (SI Appendix, Fig. S3 B and C). These results further confirmed that our proposed suppression gene-drive strategy is indeed able to perform super-Mendelian inheritance, similar to findings in another recent study in D. melanogaster (21).
Evolution of Cleavage Drive-Resistant tra Alleles.
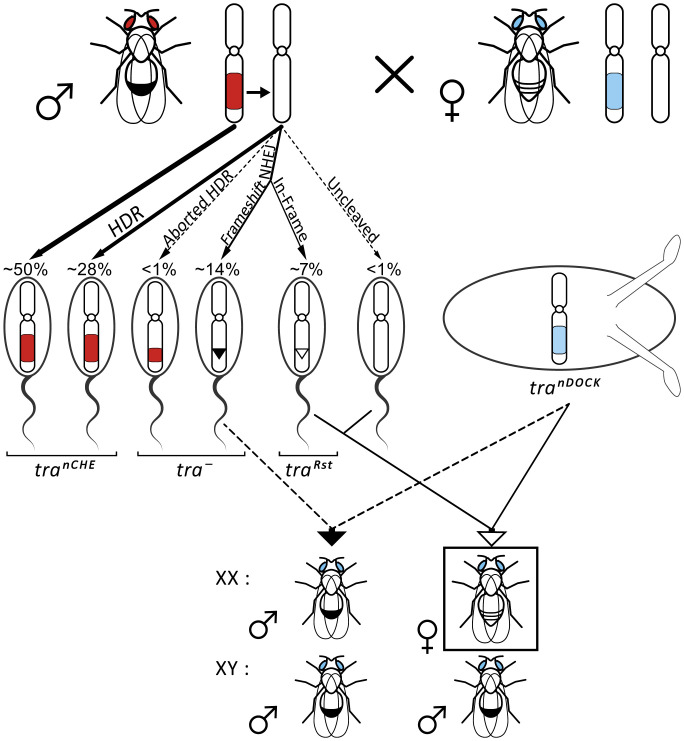
While we found our system to be highly efficient for sex conversion in D. melanogaster, we noticed during routine screening of the stocks the appearance of female flies with the DsRed eye marker phenotype. This was contrary to our previous observation and expectations that all heterozygous (tranCHE/+) XX flies should develop at least an intersex phenotype. Two scenarios could explain the presence of females with the DsRed eye marker: (i) an aborted or imperfect HDR, during which the DsRed eye marker is copied faithfully while an essential part of the drive element was lost or mutated, which would result in a dead CHE allele (tranD), or (ii) the presence of an in-frame mutation in the tra allele, which abolishes the recognition site of the gRNA without affecting the function of the tra-encoded protein (traRst). Such mutations are likely to emerge from in-frame indel mutations as a result of NHEJ events induced by the CHE itself.
To check these hypotheses, virgin females with the DsRed eye marker were isolated and individually crossed with w− males. One of these crosses did not show any signs of an active drive system, with about 50% of the offspring showing the DsRed eye marker. Molecular analysis of the mother and some female offspring from this cross revealed a large deletion in the CHE as the result of an aborted HDR event (SI Appendix, Fig. S4A). The other crosses, however, showed an efficient super-Mendelian inheritance, indicating the presence of an active CHE in the mother, which could be a sign of the presence of a traRst allele in the mother. Sequencing the tra locus of these mothers confirmed the presence of in-frame indel mutations in the recognition site of the gRNA (similar to sequences in SI Appendix, Fig. S4B). By crossing such females carrying an active CHE with tranCHE males, we were able to obtain homozygous tranCHE/tranCHE males. When these homozygous males were crossed with w− virgins, all offspring were either male or intersex (SI Appendix, Fig. S3A), which further confirmed the high sex-conversion efficiency of this tra-targeting CHE.
To further evaluate the drive-resistant allele hypothesis and to estimate the rate at which resistant alleles may emerge from NHEJ events, we crossed heterozygous virgins carrying the docking-null allele (tranDOCK/+) with heterozygous (tranCHE/+) driver males (Fig. 2). By looking at the progeny that carry the tranDOCK allele (marked by ECFP fluorescence) but lack a tranCHE allele (DsRed fluorescence), we confined our analysis to situations of non-HDR at the paternal wild-type tra allele. Sequencing the tra allele in non-DsRed, ECFP females of the first generation resulted in the discovery of various independent in-frame indel mutations (SI Appendix, Fig. S5A). This suggests that drive-resistant alleles, traRst, are readily created as a result of NHEJ in heterozygous males that carry the CHE allele. To determine the frequency at which these traRst alleles are generated, we crossed four heterozygous (tranCHE/+) males individually to tranDOCK/+ virgins and sequenced all progeny that showed only an ECFP fluorescence. We identified in-frame indels (traRst) in up to 10% of all progeny, representing about one-third of all NHEJ events. The relative high emergence rate of such traRst alleles in the F1 progeny demonstrates the rapid evolution of resistance as a direct consequence of an active homing CHE (SI Appendix, Fig. S5B) and confirms similar results from other groups (21, 30).
Fig. 2.
CHE targeting of the homologous gene locus. By analyzing the ECFP/non-DsRed progeny of tranCHE/+ males and tranDOCK/+ virgins, we focused on non-HDR targeting events at a single tra locus. Estimation of each genotype frequency based on the observed efficiency values is indicated above sperm illustrations. Molecular analysis of the CHE target site in F1 female progeny (boxed) identified independent NHEJ-derived in-frame indels that resulted in drive-resistant functional alleles (SI Appendix, Fig. S5A).
Resistant Allele Dynamics and Spread.
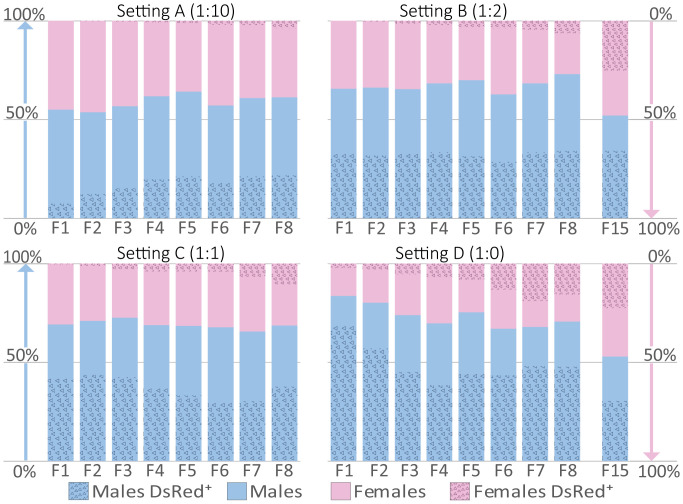
To estimate the dynamics of resistance allele emergence and spread in a population, we crossed (in five replicates each) w− virgins with four different ratios of heterozygous tranCHE/+ males to w− males and followed the progeny for up to 15 generations. Thereby we documented the sex ratios as well as the spread of the DsRed-marked tranCHE allele, whose presence in females indicates potential drive-resistant traRst alleles (Fig. 3 and SI Appendix, Figs. S6–S8). The ratio of such DsRed-positive females increased progressively over the generations, corresponding to the expected selective increase of resistance allele frequency. To characterize the molecular basis of the resistance to HCGD, we sequenced the tra locus from DsRed-fluorescent females from all the experimental settings at generation F6 and observed a diverse set of in-frame mutations representing drive-resistant traRst alleles (SI Appendix, Fig. S4B). We also selected one setting for a molecular time-course analysis (setting D, replicate 4, at generations F1, F2, F6, and F13) and found that such mutations were heritable (SI Appendix, Fig. S5C). The diversity of these in-frame indels across experimental settings and generations shows that these traRst alleles are constantly created, independently of each other, at the site of cleavage. Interestingly, we already had observed DsRed-fluorescent females at the F1 generation of this replicate. These possessed a wild-type tra allele (SI Appendix, Fig. S5C) but contained a large deletion in the Cas9 gene of the tranCHE allele, which was likely the result of a rare, aborted HDR event (similar to that in SI Appendix, Fig. S4A). Following the populations to generation F15, we found an almost regular 1:1 ratio of males to females in all the replicates (Fig. 3 and SI Appendix, Fig. S8), independent of the original frequency of tranCHE allele inoculation (settings B and D).
Fig. 3.
Dynamics of sex ratio and indicated resistance allele spread in population experiments. w− virgins were crossed with various ratios of CHE (tranCHE/+) and wild-type (w−) males (settings A–D). For each setting five replicates were carried out (SI Appendix, Fig. S6). Progeny were screened for sex and the presence of the DsRed eye marker for up to 15 generations. In setting D, where only tranCHE males were used, a sex ratio of over 80% males was achieved within one generation, indicating the collapse potential of this forced male offspring-only system. In XX embryos, tranCHE attacks the wild-type tra locus in somatic cells, resulting in intersex individuals (SI Appendix, Fig. S2). Thus, only females carrying nonfunctional defective tranCHE or drive-resistant functional traRst alleles can show the DsRed marker. Therefore, the DsRed marker serves as an indicator for the presence of the traRst allele in females, and the rise in the percentage of DsRed females indicates the spread of resistance in the population. Screening the F15 progeny in settings B and D showed that the populations adapted to the presence of the tranCHE homing allele with the female sex ratio returning to about 50% (SI Appendix, Fig. S6).
A tra-Based Suppression Gene-Drive System for C. capitata.
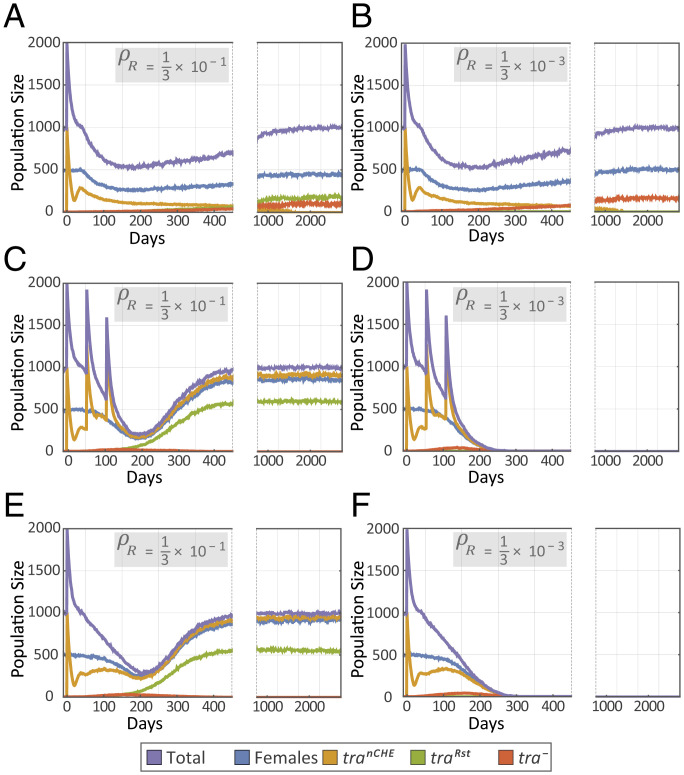
Having shown the capability of our proposed CHE in inducing sex conversion in D. melanogaster and after identifying potential weaknesses of the system due to resistance evolution, we simulated the outcome of using our proposed method as a pest-control strategy in C. capitata (Fig. 4). Our population dynamics simulation results indicate that the evolution of in-frame drive-resistant alleles at rates that we observed in D. melanogaster would indeed impede a population collapse in C. capitata (Fig. 4A), even if multiple releases were implemented in quick succession (Fig. 4C). To tackle the issue of in-frame drive-resistant alleles (these are problematic because they prevent homing while still allowing tra expression and hence are not removed due to a selective advantage), we considered the use of multiple gRNAs to target the tra gene to reduce the proportion of resistant alleles that are in-frame. Using multiple gRNAs may not have a drastic effect on the overall NHEJ rate but will reduce the in-frame resistant allele formation rate exponentially with each additional gRNA, as each new target site would have to obtain an in-frame mutation that does not affect the function of the protein (22, 31). Our simulation study predicts that by using multiple gRNAs, and thereby reducing the generation rate of in-frame resistance alleles by at least two orders of magnitude, the effectiveness of the system is greatly improved. A single release is still not sufficient to achieve a population collapse in C. capitata (Fig. 4B); however, three releases in quick succession are sufficient (Fig. 4D).
Fig. 4.
Predicted dynamics of a Cas9-based homing system targeting the tra locus in C. capitata. Predictions are based on the population genetics model depicted in SI Appendix, Fig. S10 combined with the population dynamics model depicted in SI Appendix, Fig. S11 in which the life cycle of C. capitata is divided into four stages, egg, larva, pupa, and adult, with density-dependent mortality occurring at the larval stage (SI Appendix, Table S1). Homing occurs only in tranCHE/+ heterozygotes, where “+” represents the wild-type allele and “tranCHE” represents the intact drive allele. We assume a Cas9-mediated cleavage efficiency of 100% and a probability of accurate HDR following cleavage of 90% [NHEJ rate (δ) = 0.1]. By default, in-frame drive-resistant alleles (traRst) account for one-third of generated resistant alleles, although this proportion may be reduced through gRNA multiplexing. The remaining cleavage-resistant alleles are out-of-frame or other mutations that result in a tra−-null allele. The equilibrium population size of C. capitata is 1,000. Releases consist of 1,000 tranCHE; XY males at a single time or at intervals. In A–D, the scenario in which tranCHE; XX individuals are infertile intersexes is considered. (A) For a homing efficiency of 90% and an in-frame resistant allele generation rate (ρR = δθ, where δ is the NHEJ rate and θ is the fraction of NHEJs that produce in-frame indels) of one-third of 10%, a single release of 1,000 tranCHE; XY males results in temporary population suppression, halving the adult population size, with the population rebounding over a period of several years. (B) Decreasing the in-frame drive-resistant allele generation rate, ρR, by two orders of magnitude to 1/300 of 10%, and hence increasing the out-of-frame resistant allele generation rate, ρB = δ(1 − θ), to ∼10%, the population suppression is still only moderate and transient. (C) If three releases of 1,000 tranCHE; XY males are carried out in succession, the extent of population suppression is much greater (>75% suppression); however at a ρR of one-third of 10%, the population still rebounds over a period of several years with an increase in the frequency of traRst alleles. (D) If three consecutive releases are carried out for a construct with the decreased in-frame drive-resistant allele generation rate, population elimination can be achieved within ∼1 year after the last release. (E and F) The scenario in which tranCHE; XX individuals are fertile males is considered. For a homing efficiency of 90% and an in-frame resistant-allele generation rate, ρR, of one-third of 10%, a single release of 1,000 tranCHE; XY males results in temporary population suppression, as in-frame drive-resistant alleles become prevalent, preventing population elimination (E). However, if the in-frame resistant-allele generation rate, ρR, is reduced by two orders of magnitude, to 1/300 of 10%, the emergence of in-frame drive-resistant alleles is unlikely, and the population can be eliminated following a single release of 1,000 tranCHE; XY males (F).
The above-mentioned simulations are for the scenario in which tranCHE/+; XX individuals are infertile intersexes; however, we also explored the case in which these individuals are fertile males (Fig. 4 E and F). In D. melanogaster, we observed that heterozygous (tranCHE/+) XX individuals develop into mosaic intersex individuals (SI Appendix, Fig. S3A). This is likely because the ectopic expression of Cas9 under the control of the Rcd-1r promoter in only a proportion of the cells results in a mosaic phenotype. Since the intersex-based infertility of tranCHE XX individuals places a fitness load on the system and reduces the drive (by preventing its occurrence in XX individuals), we propose the use of an early embryonic promoter, such as Sry-α, for the expression of Cas9. Expression from Sry-α in germ cells will allow gene drive to occur, and the early blastoderm expression guarantees a uniform destruction of the wild-type tra allele in all cells of the embryo at a very early stage; therefore heterozygous (tranCHE/+) XX individuals could develop into fertile medfly males. This in turn reduces the fitness load associated with the drive allele and increases the drive (by allowing super-Mendelian inheritance of the drive allele to occur in tranCHE/+; XX individuals). Our simulation shows that enabling the fertility of heterozygous (tranCHE/+) XX individuals does enhance the effectiveness of the system in collapsing a C. capitata population following a single release, provided that the in-frame drive-resistant allele generation rate is reduced by using multiple gRNAs (Fig. 4 E and F). The tolerable generation rate of in-frame resistant alleles depends on the size of the targeted medfly population. Our simulations predict the extent by which this rate must be reduced to achieve a population collapse as a function of population size (SI Appendix, Fig. S9).
Discussion
Our mathematical modeling has shown that two main factors—the formation rate of the in-frame resistance allele and the fitness of heterozygous (tranCHE/+) sex-converted XX individuals—can have a significant effect on the expected outcome of a release in the wild. When heterozygous XX individuals are infertile, our model predicts that a population collapse can be achieved only if multiple inundative releases of the driver males are performed (Fig. 4D). While this limitation could potentially be overcome by using an early embryonic stage promoter such as Sry-α (Fig. 4F), this may not be desirable to ensure the local treatment of an insect pest population without the potential concern about the elimination of an entire species. Similar considerations have also been brought forward by Prowse et al. (32) with respect to fighting invasive vertebrate species.
In addition, we have shown that in our system it is the generation rate of in-frame drive-resistant alleles, rather than the overall NHEJ rate, that has a significant impact on the outcome of release scenarios. This is of significant importance for species such as D. melanogaster (and possibly for related pest species such as Drosophila suzuki) that might seem semirefractory toward homing-based gene-drive strategies (27), as it indicates that future designs may not necessarily require an extremely high homing rate but that only lowering the formation rate of the in-frame resistance allele and thus employing multiple gRNAs (22, 31) might be sufficient for an efficient suppression gene-drive strategy in such species.
Our results support the idea that using a CHE to target genes that are essential for female-specific development in insects, such as tra, can effectively result in a gender-biased population, finally resulting in a population collapse. This provides a basis for further development of similar suppression gene-drive strategies to introduce a gender bias in wild populations of insect pests such as the medfly or disease vectors. If such a gender bias can be sustained long enough, species-specific elimination of the target species can be achieved. If the HCGDs explored here were applied to efficient pest-control management, the strategy in which tranCHE/+; XX individuals are infertile intersexes is safer, because it requires multiple releases to achieve population collapse and hence will cause a population collapse only where these releases are carried out.
Overall, we provide here an example, an implementation strategy, and the mathematical modeling required for the design and optimization of a homing-based sex conversion-suppression gene-drive approach for local or global species-specific elimination of insect pest or disease vector species. Moreover, we show that only lowering the formation rate of in-frame drive-resistant alleles by employing multiple gRNAs may be sufficient to achieve an effective suppression gene-drive outcome, which has important implications for the design of such systems in species that exhibit a low homing rate in their germ cells.
Materials and Methods
Detailed methods on cloning, transgenesis, screening, molecular analysis, stock keeping of D. melanogaster strains, population modeling, and simulations can be found in SI Appendix, SI Materials and Methods.
Acknowledgments
We thank Simon Bullock, Marc F. Schetelig, and Marita Büscher for kindly providing plasmids and D. melanogaster strains; Sean Wu and Jared Bennett for helpful contributions to the modeling framework used in this work; Gerd Vorbrüggen and one reviewer for very insightful comments that helped improve our manuscript; and Herbert Jäckle at the Department of Molecular Developmental Biology at the Max Planck Institute for Biophysical Chemistry for support. H.M.M.A. was supported by the German Academic Exchange Service, and E.M.C. received a Stipend of the Excellence Foundation for the Promotion of the Max Planck Society (IMPRS Molecular Biology). J.M.M. was supported by a grant from the Innovative Genomics Institute, University of California, Berkeley, San Francisco and Defense Advanced Research Projects Agency Safe Genes Program Grant HR0011-17-2-0047.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713825115/-/DCSupplemental.
Change History
January 7, 2022: The author line has been updated.
References
- 1.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unckless RL, Clark AG, Messer PW. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics. 2017;205:827–841. doi: 10.1534/genetics.116.197285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17:146–159. doi: 10.1038/nrg.2015.34. [DOI] [PubMed] [Google Scholar]
- 4.Deredec A, Burt A, Godfray HCJ. The population genetics of using homing endonuclease genes in vector and pest management. Genetics. 2008;179:2013–2026. doi: 10.1534/genetics.108.089037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unckless RL, Messer PW, Connallon T, Clark AG. Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics. 2015;201:425–431. doi: 10.1534/genetics.115.177592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gantz VM, Bier E. Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112:E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond A, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galizi R, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun. 2014;5:3977. doi: 10.1038/ncomms4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galizi R, et al. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci Rep. 2016;6:31139. doi: 10.1038/srep31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geuverink E, Beukeboom LW. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex Dev. 2014;8:38–49. doi: 10.1159/000357056. [DOI] [PubMed] [Google Scholar]
- 12.Pane A, Salvemini M, Delli Bovi P, Polito C, Saccone G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development. 2002;129:3715–3725. doi: 10.1242/dev.129.15.3715. [DOI] [PubMed] [Google Scholar]
- 13.Oye KA, et al. Regulating gene drives. Science. 2014;345:626–628. doi: 10.1126/science.1254287. [DOI] [PubMed] [Google Scholar]
- 14.Lunshof J. Regulate gene editing in wild animals. Nature. 2015;521:127. doi: 10.1038/521127a. [DOI] [PubMed] [Google Scholar]
- 15.Akbari OS, et al. Safeguarding gene drive experiments in the laboratory. Science. 2015;349:927–929. doi: 10.1126/science.aac7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown EH, King RC. Studies on the expression of the transformer gene of Drosophila melanogaster. Genetics. 1961;46:143–156. doi: 10.1093/genetics/46.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn C, Wimmer EA. A transgene-based, embryo-specific lethality system for insect pest management. Nat Biotechnol. 2003;21:64–70. doi: 10.1038/nbt769. [DOI] [PubMed] [Google Scholar]
- 18.Schetelig MF, Caceres C, Zacharopoulou A, Franz G, Wimmer EA. Conditional embryonic lethality to improve the sterile insect technique in Ceratitis capitata (Diptera: Tephritidae) BMC Biol. 2009;7:4. doi: 10.1186/1741-7007-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 20.Lécuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Champer J, et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 2017;13:e1006796. doi: 10.1371/journal.pgen.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Champer J, et al. Reducing resistance allele formation in CRISPR gene drives. Proc Natl Acad Sci USA. 2018;115:5522–5527. doi: 10.1073/pnas.1720354115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweisguth F, Yanicostas C, Payre F, Lepesant J-A, Vincent A. cis-regulatory elements of the Drosophila blastoderm-specific serendipity α gene: Ectopic activation in the embryonic PNS promoted by the deletion of an upstream region. Dev Biol. 1989;136:181–193. doi: 10.1016/0012-1606(89)90140-1. [DOI] [PubMed] [Google Scholar]
- 24.Fisher B, et al. 2012. BDGP insitu homepage. Available at insitu.fruitfly.org/cgi-bin/ex/insitu.pl. Accessed April 19, 2018.
- 25.Timinszky G, Bortfeld M, Ladurner AG. Repression of RNA polymerase II transcription by a Drosophila oligopeptide. PLoS One. 2008;3:e2506. doi: 10.1371/journal.pone.0002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- 27.Chan YS, Huen DS, Glauert R, Whiteway E, Russell S. Optimising homing endonuclease gene drive performance in a semi-refractory species: The Drosophila melanogaster experience. PLoS One. 2013;8:e54130. doi: 10.1371/journal.pone.0054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble C, Adlam B, Church GM, Esvelt KM, Nowak MA. 2017. Current CRISPR gene drive systems are likely to be highly invasive in wild populations. bioRxiv, 10.1101/219022. [DOI] [PMC free article] [PubMed]
- 29.Duncker BP, Davies PL, Walker VK. Introns boost transgene expression in Drosophila melanogaster. Mol Gen Genet. 1997;254:291–296. doi: 10.1007/s004380050418. [DOI] [PubMed] [Google Scholar]
- 30.Hammond AM, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 2017;13:e1007039. doi: 10.1371/journal.pgen.1007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble C, Olejarz J, Esvelt K, Church G, Nowak M. Evolutionary dynamics of CRISPR gene drives. Sci Adv. 2017;3:e1601964. doi: 10.1126/sciadv.1601964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prowse TAA, et al. Dodging silver bullets: Good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proc Biol Sci. 2017;284:20170799. doi: 10.1098/rspb.2017.0799. [DOI] [PMC free article] [PubMed] [Google Scholar]