Significance
Phosphoenolpyruvate carboxykinase (PEPCK) has a unique position in cell metabolism by allowing the flexible use of nutrients for anabolic biosynthetic reactions. PEPCK activity has just recently been described in cancer cells. It has been shown to promote cancer cell survival under nutrient deprivation, a typical feature in solid cancers, as well as cancer growth. To decipher the metabolic downstream pathways of PEPCK is critical for a better understanding of cancer cell adaptive responses under low nutrient stress. Here, we show that mitochondrial PEPCK (PEPCK-M) mediates the synthesis of glycerol phosphate from noncarbohydrate precursors, and that PEPCK-M is needed to maintain levels of glycerophospholipids, major constituents of biomembranes, in starved lung cancer cells.
Keywords: cancer, metabolism, glyceroneogenesis, PEPCK, PCK2
Abstract
Cancer cells are reprogrammed to consume large amounts of glucose to support anabolic biosynthetic pathways. However, blood perfusion and consequently the supply with glucose are frequently inadequate in solid cancers. PEPCK-M (PCK2), the mitochondrial isoform of phosphoenolpyruvate carboxykinase (PEPCK), has been shown by us and others to be functionally expressed and to mediate gluconeogenesis, the reverse pathway of glycolysis, in different cancer cells. Serine and ribose synthesis have been identified as downstream pathways fed by PEPCK in cancer cells. Here, we report that PEPCK-M–dependent glycerol phosphate formation from noncarbohydrate precursors (glyceroneogenesis) occurs in starved lung cancer cells and supports de novo glycerophospholipid synthesis. Using stable isotope-labeled glutamine and lactate, we show that PEPCK-M generates phosphoenolpyruvate and 3-phosphoglycerate, which are at least partially converted to glycerol phosphate and incorporated into glycerophospholipids (GPL) under glucose and serum starvation. This pathway is required to maintain levels of GPL, especially phosphatidylethanolamine (PE), as shown by stable shRNA-mediated silencing of PEPCK-M in H23 lung cancer cells. PEPCK-M shRNA led to reduced colony formation after starvation, and the effect was partially reversed by the addition of dioleyl-PE. Furthermore, PEPCK-M silencing abrogated cancer growth in a lung cancer cell xenograft model. In conclusion, glycerol phosphate formation for de novo GPL synthesis via glyceroneogenesis is a newly characterized anabolic pathway in cancer cells mediated by PEPCK-M under conditions of severe nutrient deprivation.
In cancer cells, metabolic pathways are rewired to allow the rapid synthesis of cellular building blocks. Despite the formation of a new blood vessel network (angiogenesis), the demand for nutrients often exceeds supply. Thus, cancer cells constantly have to adapt to fluctuations in nutrient and oxygen supply (1–3). Phosphoenolpyruvate carboxykinase (PEPCK) has recently been shown by us and others to play an unexpected role in the metabolic flexibility of cancer cells (4–7). As the only enzyme catalyzing the conversion of oxaloacetate (OAA) to the glycolytic/gluconeogenic intermediate phosphoenolpyruvate (PEP), PEPCK is required for the initial step of gluconeogenesis in the liver and other organs (8, 9). In addition, PEPCK allows the utilization of noncarbohydrate carbon sources to feed important biosynthetic pathways branching from gluconeogenesis/glycolysis (10). Two isoforms of PEPCK exist: a cytoplasmic isoform (PEPCK-C, encoded by PCK1) and a mitochondrial isoform (PEPCK-M, encoded by PCK2) (8, 9). In our previous work, we showed that PEPCK-M is expressed in lung cancer cells and in human non-small cell lung cancer (NSCLC) samples and mediates the conversion of stable isotope-labeled lactate to PEP in the direction of gluconeogenesis (4). Furthermore, we found that PEPCK-M enhances lung cancer cell survival under glucose restriction (4). A proproliferative, prosurvival role of PEPCK-M in MCF-7 breast cancer cells has been described (5). Recently, PEPCK-M silencing has been reported to reduce proliferation of lung cancer cells under glucose-limiting conditions and to profoundly inhibit lung cancer cell xenograft growth in vivo (6). PEPCK-C has been found to be expressed and to have a protumorigenic function in colon cancer cells in vivo and to enhance metastasis formation by tumor-repopulating melanoma cells in vivo (7, 11). Interestingly, in the latter model, PEPCK-M overexpression reduced metastasis formation (12). In contrast, cancers originating from gluconeogenic tissues with high basal PEPCK activity, like liver cancer or renal cell carcinoma, showed a down-regulation of PEPCK-C (13) or a reduction of the gluconeogenesis gene fructose-1,6-bisphosphatase 1 (14).
A better knowledge of metabolic downstream pathways fed by PEPCK is a key to understanding cancer cell adaptive responses under low nutrient stress. Four major potential PEPCK downstream pathways exist: the synthesis of serine and its further conversion to glycine, glycerol phosphate synthesis, the synthesis of ribose phosphate, and glycogen synthesis. The conversion of glutamine to serine and glycine via PEPCK-M has recently been found in lung cancer cells in the absence of glucose (6). However, PEPCK-C has recently been shown to mediate ribose phosphate synthesis from glutamine in colon carcinoma cells in medium containing moderately low levels of glucose (2.5 mM), while less conversion of glutamine to ribose was found at high glucose (25 mM) (7). Although glyceroneogenesis, the synthesis of glycerol phosphate via PEPCK, was described in the liver and in adipocytes 50 y ago (15), it has not been studied in cancer cells, to the best of our knowledge. Both PEPCK isoforms have been shown to mediate glyceroneogenesis (15, 16). It is classically believed to occur during lipolysis to allow the reesterification of excess fatty acids (15). Glycerol phosphate not only forms the backbone of triglycerides but is also needed for the biosynthesis of glycerophospholipids (GPL), the key constituents of biomembranes (17). According to present concepts, glycerol phosphate derived from glucose is utilized for GPL biosynthesis in cancer cells (18, 19). In the present study, we show that in cancer cells expressing PEPCK-M, noncarbohydrate precursors like glutamine or lactate are used for the synthesis of the GPL backbone under glucose-restricted conditions.
Results
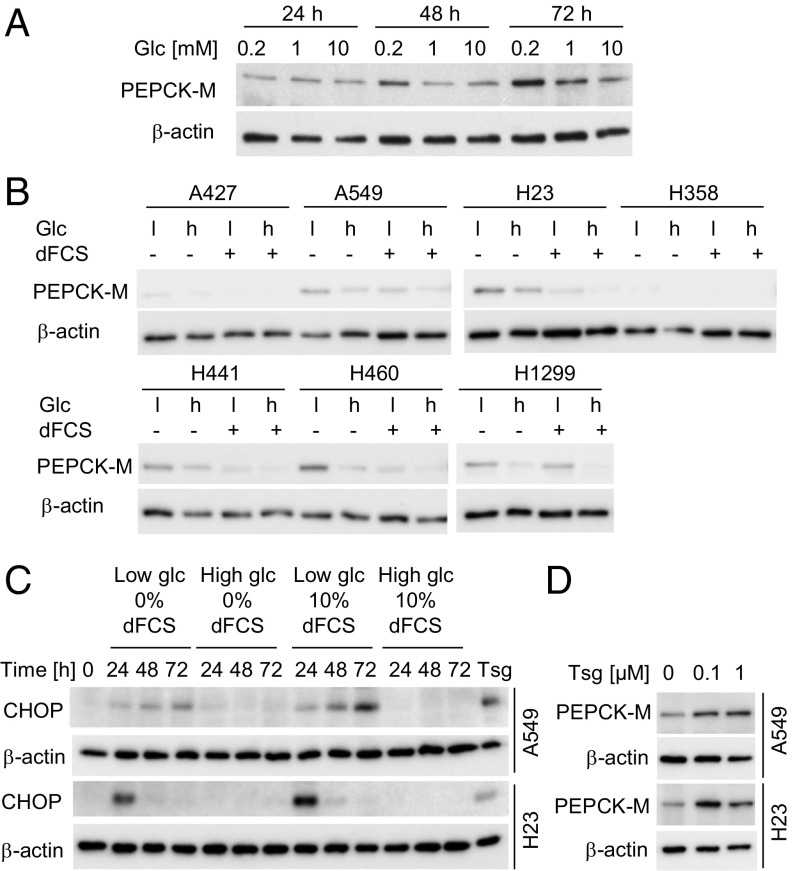
To study the role of PEPCK-M in adaptive mechanisms of lung cancer cells to nutrient deprivation, we analyzed the expression of PEPCK-M under different concentrations of glucose in serum-containing or serum-free medium. PEPCK-M expression peaked at 0.2 mM glucose in A549 lung cancer cells in serum-free medium, albeit it was still present at 10 mM glucose (Fig. 1A). When we analyzed PEPCK-M in a panel of NSCLC cell lines, we found that PEPCK-M protein was up-regulated in the majority of cell lines under combined glucose and serum starvation (Fig. 1B and SI Appendix, Fig. S1A). Likewise, PEPCK-M mRNA was up-regulated under glucose and serum deprivation, suggesting at least a partial regulation of PEPCK-M on the transcriptional level (SI Appendix, Fig. S1A). However, mRNA and protein expression did not correlate in all conditions (SI Appendix, Fig. S1A), thus also posttranslational regulation may occur, which has been described for the cytoplasmic isoform of PEPCK (20). PEPCK-M has been shown to be up-regulated by endoplasmic reticulum (ER) stress in different cancer cell lines (5). We found that glucose deprivation led to a time-dependent up-regulation of C/EBP-homologous protein (CHOP), a marker of ER stress and the resulting unfolded protein response (21), in A549 and H23 cells (Fig. 1C). Furthermore, thapsigargin, an ER stress inducer, led to an increase of PEPCK-M (Fig. 1D), suggesting that ER stress may at least contribute to PEPCK-M up-regulation under starvation.
Fig. 1.
PEPCK-M expression is up-regulated in lung cancer cells under glucose and serum starvation and is associated with ER stress. (A) PEPCK-M expression in A549 cells cultured in serum-free medium containing different concentrations of glucose (Glc). (B) PEPCK-M expression in seven NSCLC cell lines after treatment with high (10 mM) or low (0.2 mM) Glc in the presence or absence of dialyzed FCS (dFCS) for 48 h. (C) Time course of CHOP expression in A549 and H23 cells under different conditions. Cells treated with 1 µM thapsigargin (Tsg) for 5 h served as a positive control. (D) Effect of thapsigargin (ER stress inducer) on PEPCK-M expression after 48 h of treatment in normal growth medium.
Under the conditions showing peak PEPCK-M expression, 0.2 mM glucose and serum starvation, cell numbers did not increase or only slightly increased over time (SI Appendix, Fig. S1B), suggesting that proliferation was inhibited or greatly reduced. Nucleotide biosynthesis and the demand for ribose phosphate is usually high in proliferating but low in nonproliferating cells (22), thus we concentrated on glycerol phosphate synthesis as another putative PEPCK downstream intermediate. We hypothesized that the synthesis of glycerol phosphate is maintained under glucose and serum starvation from noncarbohydrate precursors by the action of PEPCK-M (glyceroneogenesis).
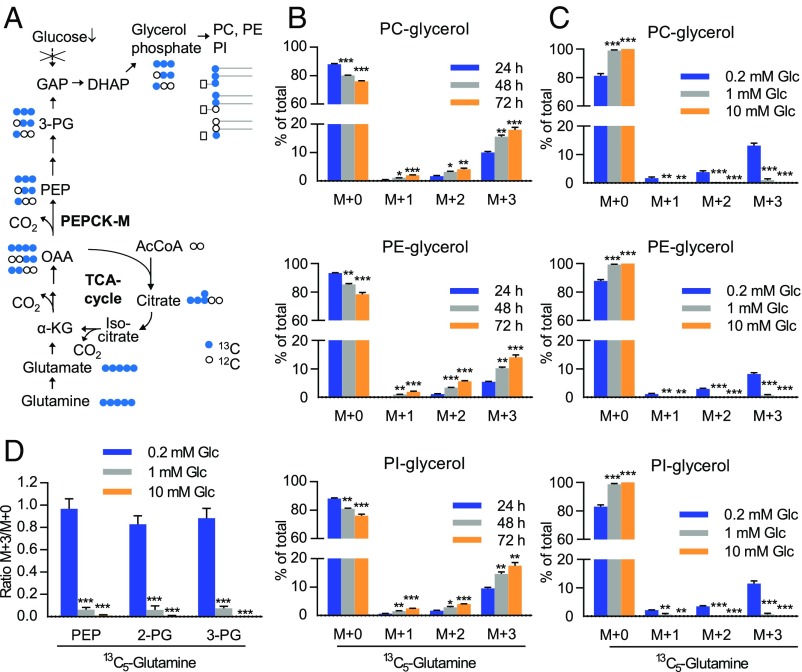
To prove that glyceroneogenesis is activated, we utilized fully 13C-labeled glutamine in A549 and H23 lung cancer cells under serum-free, low glucose conditions. We analyzed isotopologue abundance of GPL-bound glycerol using mass spectrometry (MS). The metabolic pathway for the conversion of glutamine to glycerol phosphate is shown in Fig. 2A. Labeled carbons were transferred to the glycerol backbone of the major classes of GPL, phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), in serum-free, low glucose (0.2 mM) medium in a time-dependent manner (Fig. 2B). Fully labeled glycerol (M+3) was the most abundant isotopologue. Additionally, glycerol backbone containing one or two 13C (M+1 and M+2, respectively) was detected, resulting from label dilution after additional turns of the tricarboxylic acid (TCA) cycle (Fig. 2B). Glyceroneogenesis occurred preferentially at very low glucose levels (0.2 mM), while it was absent at 10 mM (Fig. 2C). Overall levels of PC, PE, or PI were not significantly altered under low compared with high glucose conditions in serum-free medium (SI Appendix, Fig. S2A). We also evaluated 13C label enrichment from glutamine at the level of the glycerol phosphate precursors PEP, 2-phosphoglycerate (2-PG), and 3-phosphoglycerate (3-PG). All glyceroneogenic intermediates were fully labeled by 13C5-glutamine at a high rate under glucose starvation, but label enrichment was low under 10 mM glucose (Fig. 2D). These data indicate that the contribution of gluconeogenesis/glyceroneogenesis to these pools is lower under high glucose. This might be attributable, at least partially, to the lower expression of PEPCK-M under high glucose conditions. Glutamine was converted to glutamate, the first step in the pathway, under all glucose levels tested, albeit the ratio of fully labeled to unlabeled glutamate was highest under starvation (SI Appendix, Fig. S2B). When we examined the fatty acid moieties of PE, we found 13C-labeling in 13C5-glutamine–treated cells under low as well as normal glucose levels in serum-free medium, indicating the conversion of glutamine to fatty acids that are utilized for GPL synthesis (SI Appendix, Fig. S2C). This agrees with previous data from the literature on the use of glutamine carbons for fatty acid synthesis in cancer cells, which either requires the conversion of α-ketoglutarate to citrate along the TCA cycle in the conventional direction (23) or involves reductive carboxylation (24, 25). PE head group fragment ions lacking glycerol showed weak 13C labeling at one or two carbon positions (SI Appendix, Fig. S3A). This indicates a low rate incorporation of carbons from glutamine into the ethanolamine moiety. In all classical cell culture systems, including our experimental model, glucose levels decline over time due to rapid consumption (SI Appendix, Fig. S3C). Therefore, medium replacement was performed every 24 h in all cell culture experiments.
Fig. 2.
Utilization of glutamine as a carbon source for GPL glycerol backbone biosynthesis in starved lung cancer cells. (A) Metabolic pathway for the biosynthesis of glycerol phosphate from glutamine (glyceroneogenesis) via PEPCK-M. (B) Incorporation of carbons from 13C5-glutamine into the glycerol backbone of different GPL classes in A549 cells grown in low glucose (0.2 mM) medium. (C) Modulation by exogenous glucose. (B and C) Data are mean ± SEM from three independent experiments. (D) A549 cells were incubated with 13C5-glutamine for 12 h in medium containing different concentrations of glucose after preincubation without label for 60 h. Label enrichment is shown as mean ± SEM from four experiments. *P < 0.05, **P < 0.01, ***P < 0.001 on one-way ANOVA with Dunnett post hoc analysis versus 24 h (B) or 0.2 mM Glc (C and D).
Under glucose starvation (0.2 mM glucose), exogenous lactate showed a small but steady decline over 4 d, if medium change was omitted, compared with a rapid increase of lactate in cell culture supernatants in medium containing 10 mM glucose (SI Appendix, Fig. S3D). To test whether lactate is a precursor for glyceroneogenesis, we used uniformly labeled 13C-lactate as a tracer (metabolic pathway shown in SI Appendix, Fig. S4A, green circles). We found that, under low glucose serum-free conditions, lactate also was utilized for glycerol-backbone biosynthesis, albeit at lower levels than glutamine (SI Appendix, Fig. S4 B and C). The addition of lactate did not significantly affect glyceroneogenesis from glutamine (SI Appendix, Fig. S4B). Summarizing the contributions of glutamine and lactate to the glycerol backbone of phospholipids, we found that in lung cancer cells under starvation, a large proportion of GPL (up to 25–30%) contained carbons derived either from glutamine or lactate (SI Appendix, Fig. S4C). The additional contribution of lactate to glycerol M+1 or M+2 (4–13%, gray bars in SI Appendix, Fig. S4C) may either represent a net contribution via pyruvate carboxylation or a contribution via acetyl-CoA formation and consecutive labeling of TCA intermediates, since these pathways are not distinguishable. In the latter case, no net contribution of carbons occurs since the carbons are lost as CO2 (SI Appendix, Fig. S4A). Glycerol phosphate from glutamine or lactate was not only incorporated into PE, but also into plasmalogen PE, characterized by the presence of a vinyl ether linkage instead of an ester linkage at the sn-1 position (SI Appendix, Fig. S5) (26). In summary, glutamine and lactate contributed to a considerable proportion of glycerol backbones of different GPL classes.
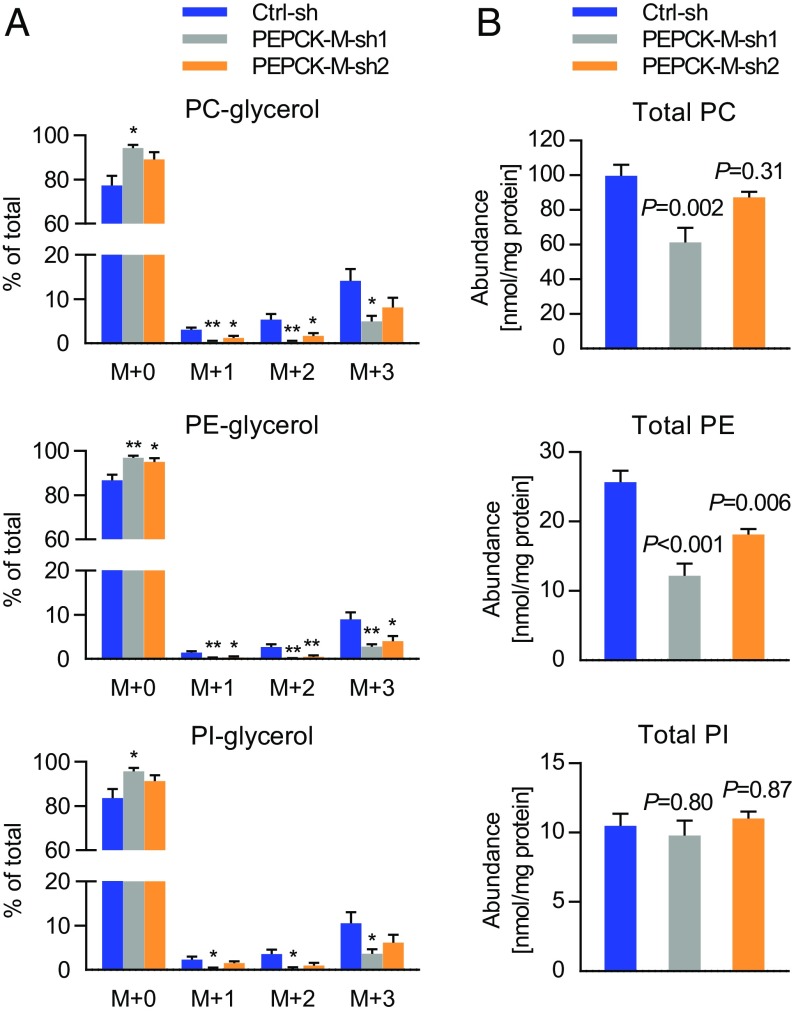
PEPCK-M silencing inhibited the incorporation of labeled glutamine into the glycerol backbone of PE, and partially also PC and PI (Fig. 3A). Importantly, when absolute phospholipid levels were measured, we found a 30–50% reduction of PE levels by PEPCK-M silencing under deprivation of serum and glucose (Fig. 3B and SI Appendix, Fig. S6A). Other phospholipid classes were less severely affected (Fig. 3B and SI Appendix, Fig. S6A). To prove the specificity of the effect of shRNA-mediated silencing on PE levels, we transfected shRNA-expressing cells with either a control vector or a shRNA-resistant PEPCK-M allele before applying a starvation period of 96 h. PEPCK-M shRNA led to reduced cellular PE, while no significant difference was found if the shRNA-resistant PEPCK-M allele was coexpressed with PEPCK-M shRNA (SI Appendix, Fig. S6 B and C). Total plasmalogen levels were not consistently reduced by both PEPCK-M shRNA constructs (SI Appendix, Fig. S6D). PEPCK-M provides glycerol phosphate needed for fatty acid esterification. Elevated levels of free fatty acids are toxic to cells (27). However, levels of free fatty acids were not significantly altered by PEPCK-M silencing (SI Appendix, Fig. S6E).
Fig. 3.
Glyceroneogenesis and phosphatidylethanolamine (PE) levels are reduced by PEPCK-M silencing. (A) 13C5-Glutamine was used as a tracer in H23 cells stably expressing nonsilencing shRNA (control shRNA, Ctrl-sh) or two different constructs of PEPCK-M shRNA (PEPCK-M-sh1 and PEPCK-M-sh2) in serum-free, low glucose (0.2 mM) medium. Enrichment of the label in the pool of GPL-bound glycerol is shown as mean ± SEM from four independent experiments. *P < 0.05, **P < 0.01. (B) Total levels of phospholipids in H23 cells stably expressing nonsilencing shRNA (Ctrl-sh) or two different constructs of PEPCK-M shRNA (PEPCK-M-sh1 and PEPCK-M-sh2) grown in 0.2 mM glucose for 96 h with daily medium replacement. Data are mean ± SEM from five independent experiments. (A and B) Group comparisons were performed with one-way ANOVA and Dunnett post hoc analysis versus Ctrl-sh.
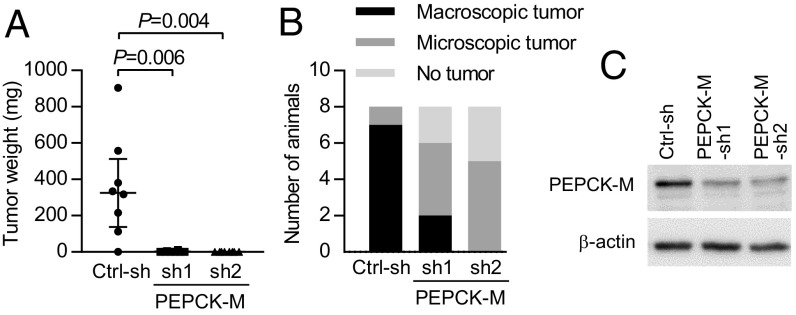
Recently it has been shown by Vincent et al. (6) that PEPCK-M silencing inhibits s.c. growth of A549 and NCI-H1299 lung cancer cell xenografts in vivo. We examined whether the glyceroneogenic or other metabolic functions of PEPCK-M might play a role in tumor growth in vivo using H23 cells expressing two different PEPCK-M shRNA constructs. PEPCK-M shRNAs prevented H23 lung cancer cells from forming macroscopic tumors after s.c. injection into nude mice (Fig. 4 and SI Appendix, Fig. S7). The mean tumor weight was greatly reduced by PEPCK-M silencing (Fig. 4A). In 2 of 8 mice injected with cells expressing PEPCK-M-sh1 and in 0 of 8 mice injected with PEPCK-M-sh2, macroscopically visible tumors were found. In contrast, visible and palpable tumors were found in 7 of 8 mice injected with control shRNA-expressing cells (Fig. 4B). In approximately half of the animals injected with PEPCK-M–silenced cells, no tumor was detectable at the site of injection while tumor growth did not exceed a microscopic size in the remaining animals (Fig. 4B and SI Appendix, Fig. S7 A and C). Interrogating the role of PEPCK-M in lung tumor growth, we modified the classical colony formation assay by applying a period of glucose and serum starvation after plating of the cells, followed by a recovery period in normal growth medium. Glucose and serum starvation led to a reduced colony formation capability (Fig. 5A). Colony formation was further reduced by stable expression of two different constructs of PEPCK-M shRNA, compared with control shRNA, after 7 d of treatment with low glucose media with reduced or absent serum (Fig. 5A). No significant effect of PEPCK-M shRNA was observed in normal growth medium (Fig. 5A). The effect of PEPCK-M shRNA under starvation was robust and also occurred when starvation (0.2 mM glucose in serum-free medium) was shortened to 3 d (Fig. 5B). The glutaminase inhibitor CB-839 significantly reduced colony formation. The effect was more pronounced under starvation, compared with nonstarvation conditions (SI Appendix, Fig. S8A).
Fig. 4.
PEPCK-M silencing substantially inhibits s.c. tumor growth. (A and B) Growth of s.c. tumors in nude mice (n = 8 animals per group) injected with H23 cells stably expressing control shRNA, or two different constructs of PEPCK-M shRNA (PEPCK-M-sh1 and PEPCK-M-sh2). (A) Tumor weight at the end of the experiment (median and interquartile range). P values were calculated by Mann–Whitney U test with Bonferroni correction for multiple comparisons. (B) Number of mice with macroscopic tumors, microscopic tumors found on histological examination, or without tumor. (C) Efficiency of PEPCK-M silencing in cells in vitro.
Fig. 5.
Exogenous PE partially rescues colony formation under starvation in H23 cells expressing PEPCK-M shRNA. (A) H23 cells stably expressing Ctrl-sh or PEPCK-M shRNA (PEPCK-M-sh1 and PEPCK-M-sh2) were plated for colony formation and treated with different glucose and serum concentrations for 7 d followed by recovery in normal growth medium. The number of seeded cells per well was adjusted to avoid coalescence of colonies. (B) A starvation period of 72 h in serum-free medium containing 0.2 mM glucose was followed by 2-wk recovery in normal growth medium. Scans of cell culture plates are shown at Right. (A and B) Groups were compared using one-way ANOVA with Dunnett post hoc analysis versus Ctrl-sh. (C) Partial rescue by exogenous DOPE (PE 36:2) added during starvation. Group comparisons were made using two-sided, unpaired Student’s t tests. (A–C) Data are mean ± SEM from four independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., nonsignificant. (D) Proposed model for the synthesis of the GPL backbone from glyceroneogenesis via PEPCK-M as an alternative pathway under starvation.
To explore a potential role of perturbed phospholipid homeostasis for reduced colony forming capability of PEPCK-M–silenced cells, we added 1,2-dioleoyl-glycero-3-phosphoethanolamine (DOPE, PE 36:2), the most abundant PE phospholipid species, exogenously during the colony formation assay. Supplementation with DOPE during starvation partially rescued colony formation in PEPCK-M–silenced H23 cells (Fig. 5C and SI Appendix, Fig. S8B). The maximum rescue was observed at 2.5 µg/mL DOPE (Fig. 5C and SI Appendix, Fig. S8B), while a higher concentration was not effective, indicating that the cells are dependent on optimal levels of DOPE during adaptation to low glucose and colony formation. Glycerol is a potential noncarbohydrate precursor for glycerol phosphate that is transported across cell membranes. However, the phosphorylation of glycerol requires glycerol kinase activity. Glycerol kinase, an enzyme predominantly expressed in the liver, was expressed at very low levels in H23 cells compared with HepG2 liver adenoma cells and accordingly exogenous glycerol did not rescue colony formation (SI Appendix, Fig. S8 C and D). Glycerol phosphate required for de novo phospholipid backbone synthesis is generally believed to originate from glycolysis in cancer cells. Based on the data in this study, we propose a model in which cancer cells utilize glyceroneogenesis from noncarbohydrate precursors via PEPCK-M as an alternative pathway to generate glycerol phosphate required for GPL de novo synthesis under starvation (Fig. 5D).
Discussion
In the present study, we show that despite glucose deprivation stress, lung cancer cells continue to synthesize GPL de novo via PEPCK-M using glutamine or lactate instead of glucose as carbon sources for the glycerol backbone. In this study, we did not assess the use of other potential glyceroneogenic precursors, e.g., other amino acids. Multiple metabolic pathways converge at the level of PEPCK, which acts as the sole enzyme converting the TCA cycle intermediate OAA to the glycolytic/gluconeogenic intermediate PEP. The precursor of OAA may depend on the available carbon source and on flux through upstream reactions (e.g., glutaminolysis and TCA cycle). The use of lactate, however, requires the oxidation of lactate to pyruvate, followed by carboxylation to OAA by pyruvate carboxylase or to malate by malic enzyme. Both enzymes have been described to be active in lung cancer (28, 29). Although PEPCK-C expression is low or absent in lung cancer cells (4, 6), a contribution of PEPCK-C to the pathway cannot be ruled out. However, PEPCK-M silencing significantly inhibited the incorporation of labeled glutamine into the glycerol backbone of PE and partially also PC and PI.
Cancer cells have been shown to possess high turnover rates for GPL (30). Different tumors have been found to contain high levels of the PC biosynthetic intermediate phosphocholine in vivo (31), and inhibitors of choline metabolism have been shown to inhibit tumor growth in preclinical models (31). The results of this study suggest that GPL de novo synthesis might be critical for cancer cells under low nutrition stress despite temporarily reduced proliferation. Membrane biogenesis and phospholipid turnover are required in different stages of proliferating cells, but also in resting cells. During cell division, biomembranes have to expand dramatically by de novo synthesis. However, also in the G1 phase of the cell cycle, GPL are continuously synthetized de novo, while their synthesis is balanced by concomitant degradation (32, 33). Interestingly, it has been shown in yeast that GPL biosynthesis enzymes are spatially reorganized but remain active upon starvation (34). The composition of membrane phospholipids and the concentration of the phospholipid precursor phosphatidic acid are continuously monitored in cells (35, 36). A proper GPL composition is important for stress responses like the unfolded protein response during ER stress (37). Additionally, de novo GPL synthesis protects cancer cells from oxygen radical-mediated death, since newly synthesized GPL are more saturated and less prone to peroxidation (38).
Our analysis of GPL abundance in serum- and glucose-starved cells revealed that individual GPL classes are differentially affected by PEPCK-M silencing. PE levels showed the most severe decline by PEPCK-M silencing under starvation. PE comprises ∼25% of mammalian phospholipids and is enriched in mitochondrial inner membranes (39, 40). Furthermore, the cone-shaped phospholipid has been found to be important for membrane fusion and remodeling (39, 40). PE itself or PE-derived ethanolamine is covalently linked to diverse proteins, including signaling proteins, and lipidation of ubiquitin-like protein LC3 by PE is a prerequisite for autophagosome formation (39, 40). Recently, PE synthesized in mitochondria has been found to be important for mitochondrial function (41). The cause for the preferential reduction of PE by PEPCK-M silencing in vitro is not known. Different turnover rates and tight regulation of the phospholipid composition under conditions of starvation and/or PEPCK-M silencing might be the underlying cause. The decrease was observed with two different constructs of PEPCK-M shRNA and affected all three most abundant PE species, PE 36:2 (dioleyl-PE), PE 35:1, and PE 34:1, as well as total PE levels. The inability of PEPCK-M–silenced H23 cancer cells to form macroscopic tumors precluded the analysis of the GPL levels and composition after PEPCK-M silencing in vivo. This limitation should be circumvented in future studies using inducible silencing strategies. As another limitation of the study, the phenotype in PEPCK-M shRNA-expressing cells observed in vivo and in colony formation experiments in vitro could potentially be attributed to off-target effects, although we obtained similar results with two different shRNA constructs. Reexpression of PEPCK-M in PEPCK-M–silenced cells in a glucose-starvation dependent manner, e.g., by stable integration of appropriate cDNA expression constructs, could be utilized in future studies to validate the specificity of the effects of PEPCK-M silencing on the phenotype.
A direct comparison of PEPCK downstream pathways, the glyceroneogenesis pathway, the ribose or serine synthesis pathways, and other possible downstream pathways, like glycogen synthesis, was not performed in this study. We focused on a detailed analysis of the phospholipid synthesis pathway, which has not been extensively studied in cancer cells. The severely compromised tumor growth in PEPCK-M–silenced cells indicates that the microenvironmental pressures enhancing PEPCK-M expression and activity might be effective also in lung cancers in vivo. However, the availability of different nutrients and their range of concentrations in the dynamic metabolic microenvironment of human cancers is still a matter of debate. Interestingly, recent detailed studies on nutrient use in human lung cancers revealed a high consumption of noncarbohydrate nutrients and fuels including lactate (42, 43). Further studies are warranted to clarify in which types of human cancer PEPCK-M or PEPCK-C are activated, and what is their metabolic function in vivo. The present study adds an important piece to the puzzle by showing that glyceroneogenesis mediated by PEPCK-M followed by GPL backbone synthesis is part of a metabolic adaptation program in lung cancer cells.
Materials and Methods
Stable Isotopic Labeling and Mass Spectrometry.
All procedures for stable isotopic labeling and liquid chromatography/MS are described in SI Appendix, Supplementary Materials and Methods. Lipid extraction was performed as described (44), GPL chromatography was modified after ref. 45. Reversed phase liquid chromatographic/MS was performed for quantitative analysis of GPL (46). Data were processed using Lipid Data Analyzer (47, 48) and annotated according to the official LIPID MAPS shorthand nomenclature (49). Mass isotopologue abundance was corrected for the natural abundance of 13C using IsoCor software (50).
Quantitative Real-Time PCR (qPCR).
Details for qPCR can be found in SI Appendix, Supplementary Materials and Methods.
Western Blot.
Details for Western blot are provided in SI Appendix, Supplementary Materials and Methods.
shRNA-Mediated Silencing.
For stable expression of PEPCK-M shRNA or nonsilencing shRNA, H23 cells were transfected with different commercially available shRNA plasmids targeting PEPCK-M or a nonsilencing control shRNA followed by antibiotics selection (Qiagen). In a set of experiments, phenotypic rescue by transient transfection of shRNA-expressing cells with either an empty vector or a plasmid containing shRNA-resistant PEPCK-M cDNA, was studied. Details for plasmids, site-directed mutagenesis, transfection, selection and maintenance are found in SI Appendix, Supplementary Materials and Methods.
Subcutaneous Growth of Xenotransplants in Nude Mice.
All animal experiments were approved by the Austrian Federal Ministry of Science, Research, and Economy. Male athymic nude mice (Hsd:Athymic Nude – Foxn1nu) 8 wk of age and specific pathogen-free were obtained from Envigo. Mice were injected s.c. with H23 cells expressing PEPCK-M shRNA (two different constructs) or control shRNA. For details see SI Appendix, Supplementary Materials and Methods.
Proliferation and Colony Formation Assays.
Procedures for the assessment of cell proliferation (cell numbers) and colony formation (51) are described in SI Appendix, Supplementary Materials and Methods.
Statistics.
Data were compiled and analyzed with the software package SPSS, version 23.0. Group differences were calculated using two-sided, unpaired Student’s t test, one-group Student’s t test, one-way ANOVA with post hoc analysis or Mann–Whitney u test, as appropriate. P values smaller than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank P. Vesely and G. Höfler (Medical University of Graz) and R. Zechner (University of Graz) for their advice and discussions, B. Schwemberger (Medical University of Graz) for technical support, and I. Vidakovic, H. Lindermuth, and J. Lesko (Medical University of Graz) for their help. The study was supported by the Austrian Science Fund Grant P 28692-B31 (to K.L.) and by a project grant from MEFOgraz (to K.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719871115/-/DCSupplemental.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 3.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leithner K, et al. PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene. 2015;34:1044–1050. doi: 10.1038/onc.2014.47. [DOI] [PubMed] [Google Scholar]
- 5.Méndez-Lucas A, Hyroššová P, Novellasdemunt L, Viñals F, Perales JC. Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) is a pro-survival, endoplasmic reticulum (ER) stress response gene involved in tumor cell adaptation to nutrient availability. J Biol Chem. 2014;289:22090–22102. doi: 10.1074/jbc.M114.566927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincent EE, et al. Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol Cell. 2015;60:195–207. doi: 10.1016/j.molcel.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Montal ED, et al. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol Cell. 2015;60:571–583. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Kalhan SC, Hanson RW. What is the metabolic role of phosphoenolpyruvate carboxykinase? J Biol Chem. 2009;284:27025–27029. doi: 10.1074/jbc.R109.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark R, Kibbey RG. The mitochondrial isoform of phosphoenolpyruvate carboxykinase (PEPCK-M) and glucose homeostasis: Has it been overlooked? Biochim Biophys Acta. 2014;1840:1313–1330. doi: 10.1016/j.bbagen.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balsa-Martinez E, Puigserver P. Cancer cells hijack gluconeogenic enzymes to fuel cell growth. Mol Cell. 2015;60:509–511. doi: 10.1016/j.molcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, et al. Upregulation of cytosolic phosphoenolpyruvate carboxykinase is a critical metabolic event in melanoma cells that repopulate tumors. Cancer Res. 2015;75:1191–1196. doi: 10.1158/0008-5472.CAN-14-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo S, et al. Downregulation of PCK2 remodels tricarboxylic acid cycle in tumor-repopulating cells of melanoma. Oncogene. 2017;36:3609–3617. doi: 10.1038/onc.2016.520. [DOI] [PubMed] [Google Scholar]
- 13.Shi H, et al. The oncoprotein HBXIP suppresses gluconeogenesis through modulating PCK1 to enhance the growth of hepatoma cells. Cancer Lett. 2016;382:147–156. doi: 10.1016/j.canlet.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Li B, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson RW, Reshef L. Glyceroneogenesis revisited. Biochimie. 2003;85:1199–1205. doi: 10.1016/j.biochi.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Escós M, et al. Kinetic and functional properties of human mitochondrial phosphoenolpyruvate carboxykinase. Biochem Biophys Rep. 2016;7:124–129. doi: 10.1016/j.bbrep.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg JM, Tymoczko JL, Stryer L. Biochemistry. W.H. Freeman; Houndmills, UK: 2011. The biosynthesis of membrane lipids and steroids; pp. 787–820. [Google Scholar]
- 18.Natter K, Kohlwein SD. Yeast and cancer cells–Common principles in lipid metabolism. Biochim Biophys Acta. 2013;1831:314–326. doi: 10.1016/j.bbalip.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menon D, et al. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J Biol Chem. 2017;292:6303–6311. doi: 10.1074/jbc.M116.772988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XZ, et al. Identification of novel stress-induced genes downstream of chop. EMBO J. 1998;17:3619–3630. doi: 10.1093/emboj/17.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemons JM, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodhi IJ, Semenkovich CF. Peroxisomes: A nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zechner R, et al. FAT SIGNALS–Lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sellers K, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren JG, et al. Knockdown of malic enzyme 2 suppresses lung tumor growth, induces differentiation and impacts PI3K/AKT signaling. Sci Rep. 2014;4:5414. doi: 10.1038/srep05414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane AN, Fan TW, Xie Z, Moseley HN, Higashi RM. Isotopomer analysis of lipid biosynthesis by high resolution mass spectrometry and NMR. Anal Chim Acta. 2009;651:201–208. doi: 10.1016/j.aca.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng M, Bhujwalla ZM, Glunde K. Targeting phospholipid metabolism in cancer. Front Oncol. 2016;6:266. doi: 10.3389/fonc.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackowski S. Coordination of membrane phospholipid synthesis with the cell cycle. J Biol Chem. 1994;269:3858–3867. [PubMed] [Google Scholar]
- 33.Zhang XH, et al. Disruption of G1-phase phospholipid turnover by inhibition of Ca2+-independent phospholipase A2 induces a p53-dependent cell-cycle arrest in G1 phase. J Cell Sci. 2006;119:1005–1015. doi: 10.1242/jcs.02821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suresh HG, et al. Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae. Mol Biol Cell. 2015;26:1601–1615. doi: 10.1091/mbc.E14-11-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 36.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 37.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rysman E, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–8126. doi: 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- 39.Bohdanowicz M, Grinstein S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol Rev. 2013;93:69–106. doi: 10.1152/physrev.00002.2012. [DOI] [PubMed] [Google Scholar]
- 40.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831:543–554. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Tasseva G, et al. Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hensley CT, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faubert B, et al. Lactate metabolism in human lung tumors. Cell. 2017;171:358–371.e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triebl A, et al. Quantitation of phosphatidic acid and lysophosphatidic acid molecular species using hydrophilic interaction liquid chromatography coupled to electrospray ionization high resolution mass spectrometry. J Chromatogr A. 2014;1347:104–110. doi: 10.1016/j.chroma.2014.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triebl A, Trötzmüller M, Hartler J, Stojakovic T, Köfeler HC. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;1053:72–80. doi: 10.1016/j.jchromb.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartler J, et al. Lipid data analyzer: Unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27:572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- 48.Hartler J, et al. Deciphering lipid structures based on platform-independent decision rules. Nat Methods. 2017;14:1171–1174. doi: 10.1038/nmeth.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liebisch G, et al. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millard P, Letisse F, Sokol S, Portais JC. IsoCor: Correcting MS data in isotope labeling experiments. Bioinformatics. 2012;28:1294–1296. doi: 10.1093/bioinformatics/bts127. [DOI] [PubMed] [Google Scholar]
- 51.Guzmán C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9:e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.