Significance
SHORTROOT (SHR) and SCARECROW (SCR) orchestrate a transcriptional program that is essential for ground tissue patterning. The regulation of the transcriptional output of SHR/SCR remains unclear. We show that MED31, a subunit of the plant Mediator coactivator complex, bridges the communication between SHR/SCR and the RNA polymerase II general transcriptional machinery. MED31 and SHR bind to the same protein domain of SCR, and the relative abundance of MED31 and SHR determines the dynamic formation of a MED31/SCR/SHR ternary complex. We studied the role of the MED31/SCR/SHR ternary complex in controlling the spatiotemporal expression of CYCLIND6;1 during ground tissue patterning. This study provides insight into the mechanisms by which master transcriptional regulators control organ patterning.
Keywords: Mediator, root development, transcription regulation, SCARECROW, SHORTROOT
Abstract
Stem cell specification in multicellular organisms relies on the precise spatiotemporal control of RNA polymerase II (Pol II)-dependent gene transcription, in which the evolutionarily conserved Mediator coactivator complex plays an essential role. In Arabidopsis thaliana, SHORTROOT (SHR) and SCARECROW (SCR) orchestrate a transcriptional program that determines the fate and asymmetrical divisions of stem cells generating the root ground tissue. The mechanism by which SHR/SCR relays context-specific regulatory signals to the Pol II general transcription machinery is unknown. Here, we report the role of Mediator in controlling the spatiotemporal transcriptional output of SHR/SCR during asymmetrical division of stem cells and ground tissue patterning. The Mediator subunit MED31 interacted with SCR but not SHR. Reduction of MED31 disrupted the spatiotemporal activation of CYCLIND6;1 (CYCD6;1), leading to defective asymmetrical division of stem cells generating ground tissue. MED31 was recruited to the promoter of CYCD6;1 in an SCR-dependent manner. MED31 was involved in the formation of a dynamic MED31/SCR/SHR ternary complex through the interface protein SCR. We demonstrate that the relative protein abundance of MED31 and SHR in different cell types regulates the dynamic formation of the ternary complex, which provides a tunable switch to strictly control the spatiotemporal transcriptional output. This study provides valuable clues to understand the mechanism by which master transcriptional regulators control organ patterning.
In the reference plant Arabidopsis thaliana, the primary root is composed of single-cell layers of epidermis, cortex, endodermis, and pericycle surrounding the vascular tissues (1). This regular pattern is established and maintained by different sets of stem cells residing in the root meristem. These stem cells surround a group of mitotically less active quiescent center (QC) cells (2). QC cells and the actively dividing stem cells form a specialized cellular microenvironment called the stem cell niche (3–7). Within the stem cell niche, the cortex/endodermis initials (CEI) and their immediate daughter cells (CEID) undergo formative asymmetrical cell division (ACD) to generate the separate endodermis and cortex cell layers, which collectively constitute the ground tissue (3–7).
As in animal systems, the gene expression program that determines stem cell fate and organ patterning in plants is controlled by a small number of master transcription factors (6, 7). SHORTROOT (SHR) and SCARECROW (SCR) play critical roles in ground tissue patterning by controlling the proper specification, maintenance, and ACD of the CEI and/or CEID (8–12). Mutations of SHR and SCR lead to related defects in ground tissue patterning (8–12). SHR is transcriptionally expressed in the stele, and the encoded protein moves to the outer adjacent cell layer, where SCR sequesters SHR to the nucleus by forming an SHR/SCR complex (13). This complex orchestrates a transcriptional program that controls the spatiotemporal ACD of the CEI and/or CEID for ground tissue patterning (14–17).
Direct transcriptional targets of the SHR/SCR complex have been identified. A group of plant-specific INDETERMINATE DOMAIN C2H2 zinc finger transcription factors (BIRD) are transcriptional targets of the SHR/SCR complex during ground tissue specification and patterning (18–21). Several members of the BIRD family, including JACKDAW (JKD) and MAGPIE (MGP), physically interact with SHR and/or SCR, suggesting that these proteins are important players in the SHR/SCR complex (19–23).
CYCLIND6;1 (CYCD6;1), whose temporal expression coincides with the onset of specific ACDs of the CEID that pattern the ground tissue, is a direct transcriptional target of the SHR/SCR complex (24). The spatiotemporal activation of CYCD6;1 is controlled by a bistable switch involving SHR, SCR, and the cell differentiation factor RETINOBLASTOMA-RELATED (RBR) (25).
Despite these research advances, the precise mechanism underlying SHR/SCR-mediated transcriptional regulation remains enigmatic. In particular, the process by which SHR/SCR relays regulatory signals to the RNA polymerase II (Pol II) general transcription machinery to transcribe specific genes in a cell type- and developmental stage-specific manner remains unclear.
Mediator is an evolutionarily conserved, multisubunit coactivator complex that is essential for Pol II-dependent gene transcription (26–31). Since its identification in yeast (32, 33) and animals (34), Mediator has been extensively investigated for its ability to orchestrate the transcription factor-dependent assembly of the Pol II preinitiation complex (PIC) via discrete interactions with signal-dependent transcription factors and Pol II (26–29). Novel functions are continuously ascribed to the yeast and animal Mediator in controlling almost every stage of Pol II-dependent gene transcription, including epigenetic regulation, transcriptional elongation and termination, mRNA processing, noncoding RNA activation, and chromatin loop formation (28, 30, 31, 35–37). These functions suggest that Mediator acts as a pivotal coordinator of transcriptional regulation.
The plant Mediator complex was not biochemically isolated until 2007 (38). Despite the identification of a number of plant Mediator subunits that are implicated in the regulation of plant development and adaptive responses (39–47), mechanistic studies on the functions of plant Mediator are still in their infancy.
Considering the essential role of Mediator in bridging the cross-talk between gene-specific transcription factors and the general transcription machinery during gene transcription, we hypothesized that a specific Mediator subunit might directly interact with SHR and/or SCR. Notably, we found that the plant Mediator subunit MED31 directly interacts with SCR but not with SHR. Here, we report that MED31 plays an essential role in SHR/SCR-directed spatiotemporal activation of CYCD6;1 and ground tissue patterning. Genetic and biochemical analyses indicated that MED31 is involved in the formation of a ternary MED31/SCR/SHR transcription complex, and this ternary complex is dynamic because MED31 and SHR compete for binding to SCR. The dynamic interaction of MED31 with SCR determines the transcriptional output of the SHR/SCR transcription complex, which spatiotemporally coincides with stem cell ACD during ground tissue patterning.
Results
MED31 Directly Interacts with SCR but Not SHR.
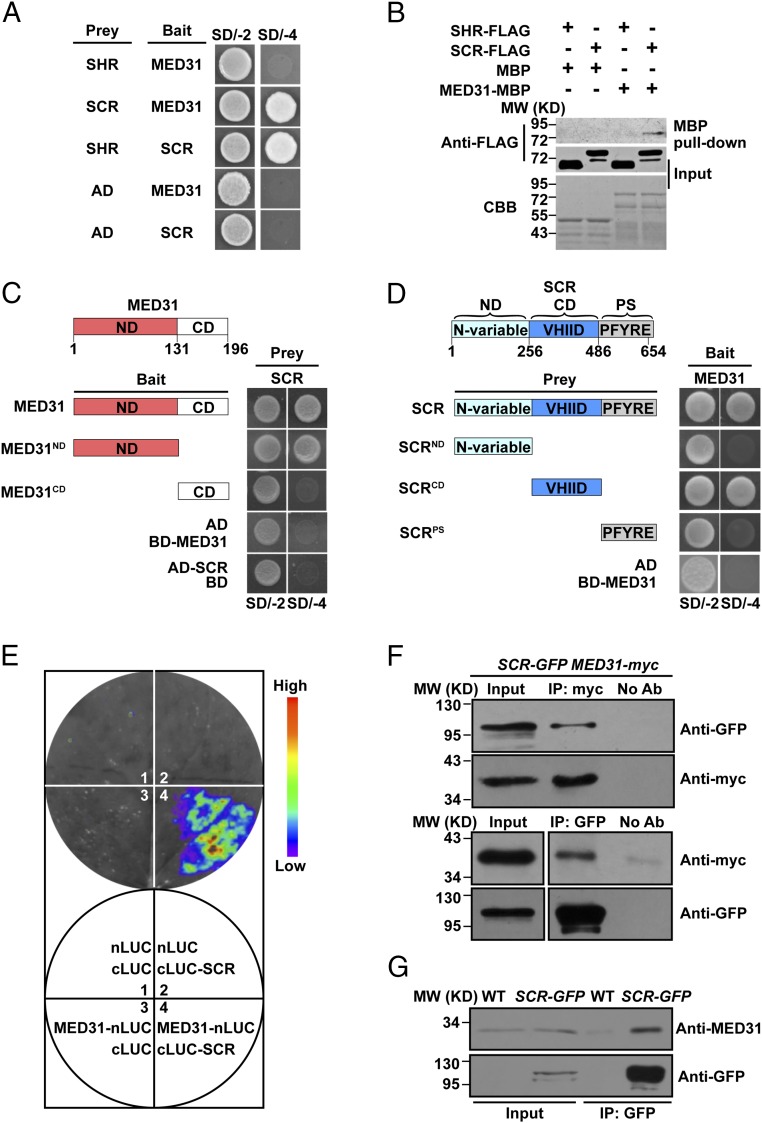
The direct linkage between the SHR/SCR complex and the Pol II transcription machinery was examined by designing a yeast two-hybrid assay to identify Mediator subunits interacting with SHR and/or SCR. For this purpose, SHR or SCR was fused to the GAL4 activation domain as bait to screen a library of Arabidopsis Mediator subunits. The results showed that MED31 interacted with SCR but not SHR (Fig. 1A and SI Appendix, Fig. S1 A and B). To confirm these observations, in vitro pull-down experiments were performed using purified maltose-binding protein (MBP)–tagged MED31 (MED31-MBP) and FLAG epitope-tagged SHR (SHR-FLAG) or SCR (SCR-FLAG). MED31-MBP pulled down SCR-FLAG but not SHR-FLAG (Fig. 1B), indicating that MED31 interacted with SCR in vitro. Domain mapping indicated that the highly conserved N-terminal domain of MED31 (SI Appendix, Fig. S1 C and D) and the central domain (CD) of SCR were involved in the MED31/SCR interaction (Fig. 1 C and D). Considering that the CD of SCR is also responsible for its interaction with SHR (16), these results suggested that MED31 and SHR may bind to the same domain of SCR.
Fig. 1.
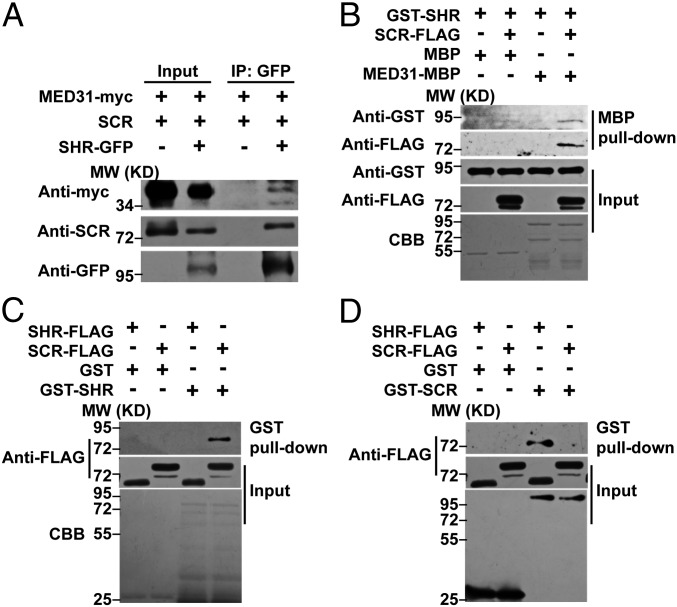
MED31 directly interacts with SCR but not SHR. (A) Yeast two-hybrid (Y2H) assays showing that MED31 interacts with SCR but not SHR. The yeast transformants were dropped onto SD/-Trp/-Leu (SD/-2) and SD/-4 media to assess protein–protein interactions. AD, GAL4 activation domain. (B) In vitro pull-down assays showing that MED31 directly interacts with SCR but not SHR. SCR-FLAG was pulled down by MED31-MBP immobilized on amylose resin. Protein bound to amylose resin was eluted and analyzed by anti-FLAG antibody. CBB, Coomassie brilliant blue staining; MW, molecular weight (mass). (C) Domains of MED31 involved in the MED31/SCR interaction were mapped using Y2H assays. Yeast cells cotransformed with pGBKT7-MED31 or pGBKT7-MED31 derivatives (baits) and pGADT7-SCR (prey) were dropped onto SD/-2 and SD/-4 media to assess interactions with SCR. CD, C-terminal domain; ND, N-terminal domain. (D) Domains of SCR involved in the MED31/SCR interaction were mapped using the Y2H assay. Yeast cells cotransformed with pGADT7-SCR or pGADT7-SCR derivatives (baits) and pGBKT7-MED31 (prey) were dropped onto SD/-2 and SD/-4 media to assess interactions with MED31. CD, central domain; ND, N-variable domain; PS, PFYRE/SAW domain. (E, Top) LUC complementation imaging assays showing that MED31 interacts with SCR. (E, Bottom) N. benthamiana leaves coinfiltrated with the different combinations are shown. The pseudocolor bar shows the range of luminescence intensity. (F) Co-IP assays of MED31 with SCR in N. benthamiana. MED31-myc and SCR-GFP were coinfiltrated into N. benthamiana leaves. Protein samples were immunoprecipitated with anti-myc antibody and immunoblotted with anti-GFP antibody (Top) or immunoprecipitated with anti-GFP antibodies and immunoblotted with anti-myc antibodies (Bottom). (G) Co-IP assays of MED31 with SCR in Arabidopsis. Protein extracts from 6 dags after germination (DAG) WT and SCR-GFP seedlings were immunoprecipitated with anti-GFP antibodies. For F and G, samples before (input) and after immunoprecipitation (IP) were immunoblotted with the indicated antibodies. All experiments in A–G were repeated at least three times with similar results.
To determine whether MED31 interacts with SCR in planta, we conducted firefly luciferase (LUC) complementation imaging (48) assays in Nicotiana benthamiana leaves. MED31 and SCR were fused to the N- and C-terminal halves of LUC, respectively. N. benthamiana cells coexpressing MED31-nLUC and cLUC-SCR displayed strong fluorescence signals, whereas those coexpressing nLUC and cLUC-SCR or MED31-nLUC and cLUC displayed no signal (Fig. 1E), confirming that the MED31/SCR interaction occurred in vivo.
Next, we performed reciprocal coimmunoprecipitation (co-IP) experiments in N. benthamiana leaves, which showed that green fluorescent protein (GFP)-tagged SCR (SCR-GFP) was immunoprecipitated by MED31-myc and vice versa (Fig. 1F). In co-IP assays using SCR-GFP plants and anti-MED31 antibodies, native MED31 was pulled down by SCR-GFP, confirming the interaction between MED31 and SCR in planta (Fig. 1G). Taken together, these results indicated that MED31 directly interacts with SCR but not SHR; therefore, we next investigated the potential role of MED31 in SHR/SCR-directed gene transcription and ground tissue patterning.
Knockdown of MED31 Impairs Ground Tissue Patterning.
Phylogenetic analyses indicated that MED31 is conserved among plant species as well as in yeast and animals (SI Appendix, Fig. S1 C and D). The sequence conservation of MED31 resides within an ∼69-aa N-terminal motif that contains a remarkable number of absolutely conserved residues (49) (SI Appendix, Fig. S1C). Notably, it has been shown that MED31 is essential for mammalian development by controlling cell fate establishment and cell proliferation during early embryogenesis (50, 51).
Fusion of promoter and GUS (β-glucuronidase) indicated that MED31 is richly expressed in the root meristem (SI Appendix, Fig. S2 A and B) and that a MED31-GFP fusion protein localized to the nucleus (SI Appendix, Fig. S2C). Interestingly, we found that treatment with the protein synthesis inhibitor cycloheximide led to reduced MED31 protein abundance, whereas treatment with the proteasome inhibitor MG132 led to increased MED31 protein abundance (SI Appendix, Fig. S2 D–G), suggesting that MED31 is subject to proteasome-dependent degradation. The physical interaction of MED31 with SCR prompted us to investigate the potential role of MED31 in root radial patterning using a genetic approach. An efficient CRISPR/Cas9 gene editing system (52) was used to generate the knockout mutant med31-c (SI Appendix, Fig. S3 A–E). The med31-c mosaic mutants showed defective root growth (SI Appendix, Fig. S3 F–J). Homozygous med31-c mutants showed an embryonic lethal phenotype, which could be complemented by a MED31-GFP fusion driven by the MED31 promoter (pMED31::MED31:GFP) (SI Appendix, Fig. S3 B–D′). These results indicated that Arabidopsis MED31 plays an essential role in early embryo development.
RNA interference (RNAi) was used to generate a collection of MED31-RNAi lines (the RNAi strategy is described in SI Appendix). We obtained multiple lines with similar short root phenotypes, coinciding with their reductions of MED31 transcript (SI Appendix, Fig. S4). The MED31-RNAi line L28, in which MED31 expression was substantially reduced at both the transcript and protein levels (SI Appendix, Fig. S4 A and B), was designated as MED31-RNAi and used for further analyses.
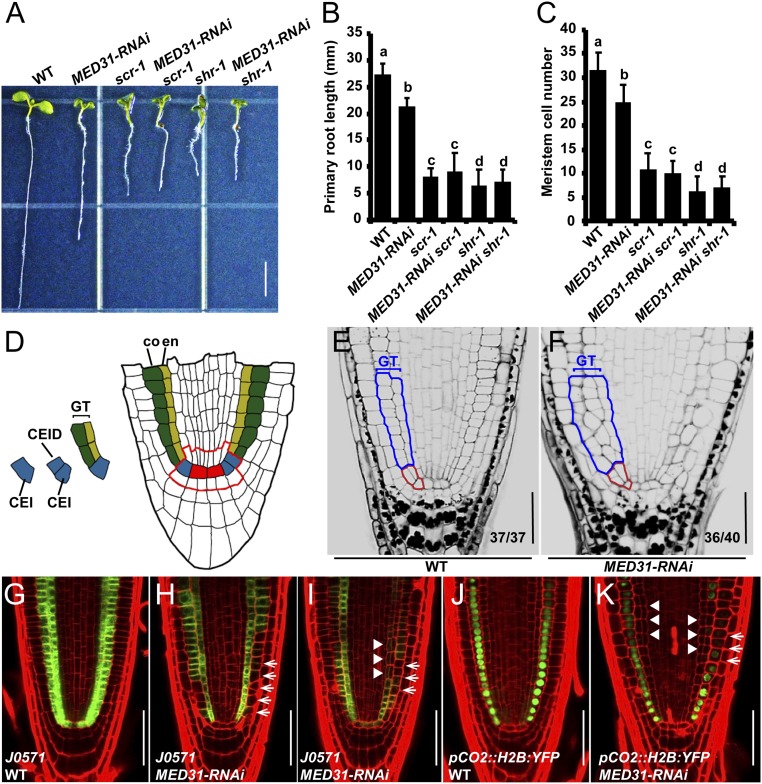
Similar to the shr-1 and scr-1 mutants (8, 9, 11, 12), MED31-RNAi plants showed stunted root growth and reduced root meristem size (Fig. 2 A–C). In the stem cell niche of wild-type (WT) roots, the CEI divides anticlinally and generates the CEID (Fig. 2 D and E). The CEID then undergoes periclinal ACD and generates an inner layer of endodermis and an outer layer of cortex, constituting the well-organized ground tissue (Fig. 2 D and E). Considering that SCR plays a critical role in stem cell ACD and ground tissue patterning, we investigated whether MED31-RNAi roots showed similar defects. In the presumptive stem cell region of MED31-RNAi roots, the regular ACD pattern of the CEI and CEID was disrupted, despite the fact that no obvious delayed ACD of the CEI was observed (Fig. 2 E and F). In the upper ground tissue region of MED31-RNAi roots, the regular cellular organization was also disrupted (Fig. 2 E and F). Notably, the aberrant ground tissue patterning in MED31-RNAi roots correlated with nonstereotyped cell shapes as well as extra cell layers (Fig. 2 E and F).
Fig. 2.
Reduction of MED31 impairs ground tissue patterning. (A) Phenotypes of the indicated genotypes at 5 DAG. (Scale bar, 5 mm.) (B) Primary root length of the indicated genotypes at 5 DAG. (C) Quantification of the meristem cell number of the indicated genotypes at 5 DAG. Data shown are the average and SD (n = 20) in B and C. Samples with different letters (a–d) are significantly different at P < 0.01. (D) Median longitudinal view of root stem cell niche organization and asymmetrical divisions defining the ground tissue lineage. Red, QC; blue, CEI and CEID; green, cortex (co); yellow, endodermis (en); GT, ground tissue. The red line outlines the root stem cell niche. Modified pseudo-Schiff propidium iodide (mPS-PI) staining of WT (E) and MED31-RNAi (F) root tips at 6 DAG is shown. Red lines outline the CEI and CEID. Blue lines outline the ground tissue. (Scale bars, 50 μm.) J0571 expression in WT (G) and MED31-RNAi (H and I) at 6 DAG is shown. White arrows indicate the abnormal expression in ground tissue of MED31-RNAi, and arrowheads indicate the irregular cell divisions in MED31-RNAi. (Scale bars, 50 μm.) The expression pattern of pCO2::H2B:YFP in the WT (J) and MED31-RNAi (K) at 6 DAG is shown. White arrows indicate the interrupted CO2 expression in ground tissue of MED31-RNAi, and arrowheads indicate the supernumerary cells in MED31-RNAi. (Scale bars, 50 μm.)
Closer observation revealed an obvious delayed ACD of the CEI in med31-c mosaic lines compared with the WT (SI Appendix, Fig. S3 G and H, arrowheads). We also observed severe patterning defects in the upper ground tissue region of med31-c mosaic roots (SI Appendix, Fig. S3 I and J, arrowheads). Together, these results revealed that knockdown of MED31 leads to aberrant ground tissue patterning.
To better visualize these patterning defects, several cell type-specific markers were introduced into the MED31-RNAi background. Expression of J0571 (24, 53), which is specifically expressed in the endodermal and cortical layers of WT roots (Fig. 2G), was dramatically reduced in MED31-RNAi roots (Fig. 2 H and I, arrows). Misshaped cortex cells with a disorganized cellular arrangement were observed in MED31-RNAi roots, and the expression of J0571 in these irregular cells was reduced or undetectable (Fig. 2I, arrowheads). Similarly, the general expression levels of pCO2::H2B:YFP (15), which is specifically expressed in the cortical layer of WT roots (Fig. 2J), were also markedly reduced in MED31-RNAi roots (Fig. 2K, arrows). Ectopic cells were observed between the endodermis and cortex layers in MED31-RNAi roots, and the expression of pCO2::H2B:YFP was almost undetectable in these supernumerary cells (Fig. 2K, arrowheads). Taken together, these observations indicated that MED31 plays a critical role in regulating cell-type specification and ground tissue patterning.
MED31 Acts in the SHR/SCR Pathway to Regulate Ground Tissue Patterning.
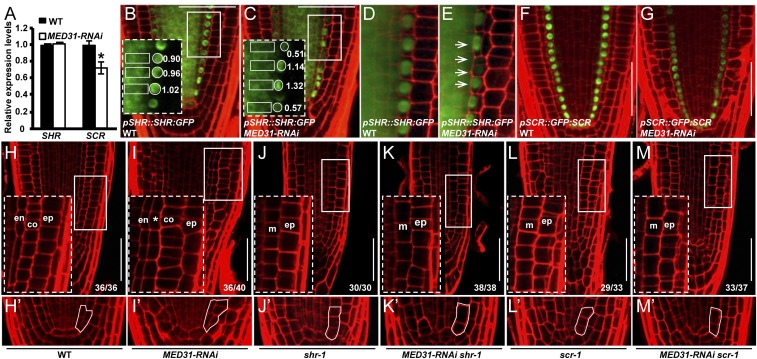
Considering the established role of SHR and SCR in ground tissue patterning, we compared the expression of SHR and SCR between MED31-RNAi and WT. Reverse transcription-quantitative PCR (RT-qPCR) assays indicated that the SHR transcript levels did not show a significant change in MED31-RNAi roots (Fig. 3A). We then transferred the pSHR::SHR:GFP protein fusion (14) into MED31-RNAi and measured SHR-GFP fluorescence in the stele, which is the source of the mobile SHR protein. The levels of SHR-GFP fluorescence displayed negligible alteration in the stele of MED31-RNAi roots (Fig. 3 B–E). To ask whether MED31 plays a role in SHR movement into the endodermal cells, we measured the SHR-GFP fluorescence ratio between the endodermis and the stele as previously described (54). In 5-d-old WT roots, the endodermal-to-stele ratio of the SHR-GFP fluorescence was stable (∼1.0); in MED31-RNAi roots, on the other hand, the endodermal-to-stele ratio of the SHR-GFP fluorescence showed sporadic variations throughout the ground tissue (0.13–1.43) (Fig. 3 B–E and SI Appendix, Fig. S5 A–D), revealing an important function of MED31 in maintaining the uniform distribution of the SHR protein in individual endodermal cells. Significantly, we found that endodermal cells showing altered SHR protein accumulation correlate with irregular periclinal division, which leads to the formation of extra cell layers in MED31-RNAi roots (Fig. 3 C and E). Closer observation revealed that in yet-to-divide or dividing cells (before nuclear breakdown), the endodermal-to-stele ratio of SHR-GFP was low, whereas in recently divided cells, the endodermal-to-stele ratio of SHR-GFP was high (Fig. 3 C and E).
Fig. 3.
MED31 acts in the SHR/SCR pathway to regulate ground tissue patterning. (A) RT-qPCR analysis showing the relative expression levels of SHR and SCR in WT and MED31-RNAi roots. Total RNA was extracted from 5-mm root tip sections of 5 DAG seedlings. Error bars represent SD from three independent experiments (Student’s t test, *P < 0.05). pSHR::SHR:GFP expression in WT (B and D) and MED31-RNAi (C and E) at 6 DAG is shown. (B and C, Insets) SHR-GFP levels in endodermal cells are shown in areas surrounded by the white dashed rectangles. Numbers indicate the endodermal-to-stele ratios of SHR-GFP fluorescence. Arrows in E show variations of SHR-GFP levels in endodermal cells of MED31-RNAi. (Scale bars, 50 μm.) pSCR::GFP:SCR expression in WT (F) and MED31-RNAi (G) at 6 DAG is shown. (Scale bars, 50 μm.) Root apical meristem phenotypes of WT (H), MED31-RNAi (I), scr-1 (J), MED31-RNAi scr-1 (K), shr-1 (L), and MED31-RNAi shr-1 (M) at 6 DAG are shown. (H′–M′) Magnifications of stem cell niche areas in H–M. (H–M, Insets) Root radial patterning is shown in areas surrounded by the white rectangles. Abbreviations: co, cortex; en, endodermis; ep, epidermis; m, mutant cell layer in shr-1 and scr-1; *, irregular cell layer in MED31-RNAi. (Scale bars, 50 μm.)
We then examined whether MED31-RNAi affects SCR expression. The expression of pSCR::GFP:SCR and pSCR::GFP was considerably lower in the endodermis of MED31-RNAi than in the WT (Fig. 3 F and G and SI Appendix, Fig. S5 E and F). Consistently, RT-qPCR assays showed that the transcript levels of SCR were significantly lower in the root tips of MED31-RNAi than in those of the WT (Fig. 3A), indicating that knockdown of MED31 affects the expression of SCR. As SCR is a direct target of SHR/SCR (16), the reduced transcript level of SCR, in the MED31-RNAi genotype, could be due to the compromised transcriptional efficiency of the SHR/SCR transcription complex. RT-qPCR assays showed that the expression of several known SHR/SCR targets, including SCL3, RLK, JKD, MGP, and NUC (21, 24), was also significantly reduced in MED31-RNAi plants (SI Appendix, Fig. S5 G–I).
To explore the genetic relationship between MED31 and the SHR/SCR pathway, we generated MED31-RNAi scr-1 and MED31-RNAi shr-1 double-mutant lines. At 6 days after germination (DAG), the root growth of MED31-RNAi scr-1 was comparable to that of scr-1 and the root growth of MED31-RNAi shr-1 was comparable to that of shr-1 (Fig. 2 A and B), indicating that MED31-RNAi had no additive effects with scr-1 or shr-1 in regulating root growth. These results support that MED31 acts in the SHR/SCR pathway to regulate root growth.
Next, we compared the ground tissue phenotypes of the MED31-RNAi scr-1 and MED31-RNAi shr-1 double-mutant lines with those of their parental lines (Fig. 3 H–M′). In shr-1 and scr-1 mutants, the CEI/CEID did not divide correctly and generated a single cell layer (Fig. 3 H, H′, J, J′, L, and L′). The MED31-RNAi shr-1 double mutant shared a similar phenotype of a single ground tissue layer with shr-1 (Fig. 3 K and K′), and the MED31-RNAi scr-1 double mutant shared a similar phenotype of a single ground tissue layer with scr-1 as well (Fig. 3 M and M′). This supported that MED31 acts in the SHR/SCR pathway to regulate ground tissue patterning.
MED31 Differentially Regulates the Expression of CYCD6;1 in the CEI and CEID or the Upper Ground Tissue.
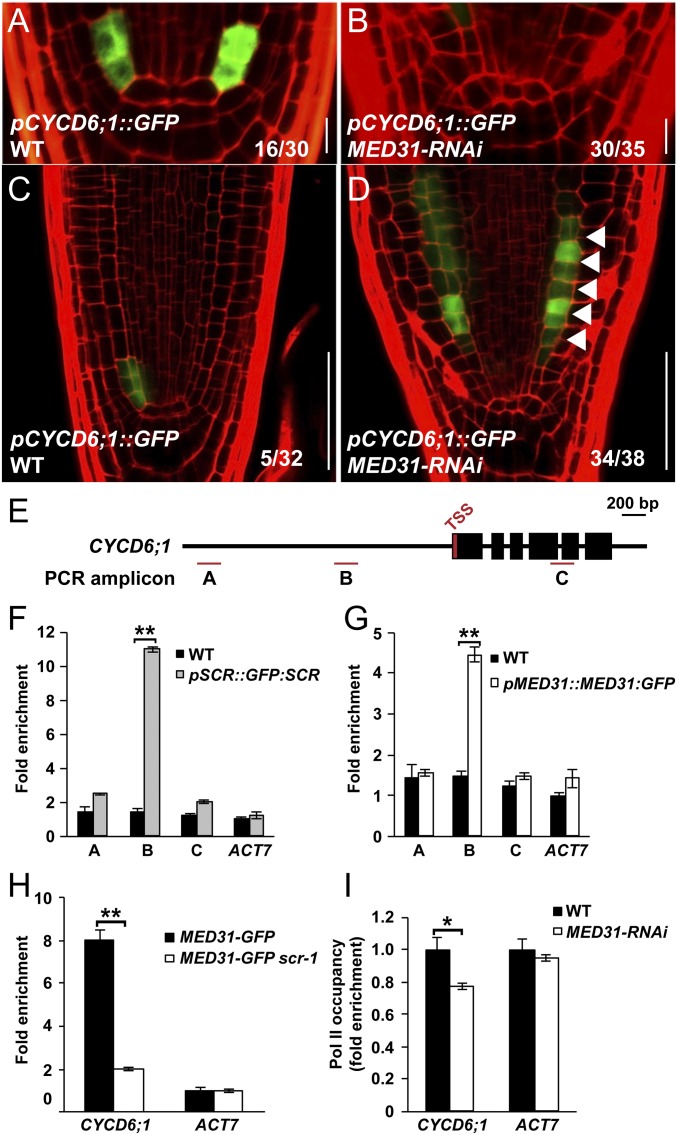
CYCD6;1 is a direct transcriptional target of SHR/SCR, and this cell cycle gene plays an important role in stem cell ACD and ground tissue patterning (24). In line with the irregular ACDs of the CEI and CEID (Fig. 2 E and F), the strict spatiotemporal expression pattern of pCYCD6;1::GFP in the CEI and CEID was largely abolished in MED31-RNAi roots (Fig. 4 A and B, 16 of 30 in WT and 30 of 35 in MED31-RNAi), indicating a critical role of MED31 in SHR/SCR-regulated spatiotemporal activation of CYCD6;1 in the CEI and CEID.
Fig. 4.
MED31 is involved in SHR/SCR-regulated spatiotemporal activation of CYCD6;1. pCYCD6;1::GFP expression in the root stem cell niche of WT (A) and MED31-RNAi (B) at 4 DAG is shown. (Scale bars, 10 μm.) Expression of pCYCD6;1::GFP in the root meristem of WT (C) and MED31-RNAi (D) at 6 DAG is shown. White arrowheads indicate extra cell divisions with pCYCD6;1::GFP expression. (Scale bars, 50 μm.) (E) Schematic diagram of the CYCD6;1 and PCR amplicons (indicated as letters A–C) used for ChIP-qPCR. TSS, transcription start site. ChIP-qPCR results show the enrichment of SCR (F) and MED31 (G) on the chromatin of CYCD6;1. Sonicated chromatins from 5 DAG seedlings were precipitated with anti-GFP antibodies (Abcam). The precipitated DNA was used as a template for qPCR analysis, with primers targeting different regions of the CYCD6;1 as shown in E. (H) ChIP-qPCR results showing that SCR mutation impairs the recruitment of MED31 to the promoter regions of SCR targets. Chromatins were extracted from MED31-GFP and MED31-GFP/scr-1 seedlings at 5 DAG and precipitated with anti-GFP antibodies, respectively. (I) ChIP-qPCR results showing that reduction of MED31 impairs Pol II-directed transcription of CYCD6;1. Chromatin was extracted from WT and MED31-RNAi seedlings at 5 DAG and precipitated with anti-CTD antibodies (Abcam), respectively. In F–I, the ChIP signal was quantified as the percentage of total input DNA by qPCR. In F–H, the ChIP signal was normalized to ACT7 in the indicated genotypes, respectively. In I, the ChIP signal was normalized to WT. Error bars represent SD from three independent experiments. Asterisks indicate significant differences, according to Student’s t test (*P < 0.05; **P < 0.01).
In contrast to the abolished pCYCD6;1::GFP expression in the CEI and CEID, ectopic pCYCD6;1::GFP expression occurred in the upper ground tissue in MED31-RNAi roots (Fig. 4 C and D, five of 32 in WT and 34 of 38 in MED31-RNAi). Notably, as with the altered accumulation of the SHR protein (Fig. 3 C and E), the ectopic expression of pCYCD6;1::GFP in the upper endodermis always coincided with extra periclinal divisions of these cells (Fig. 4D, arrowheads). These results indicated that MED31 negatively regulates the expression of CYCD6;1 in the upper ground tissue. Given that the overall expression levels of CYCD6;1 in whole seedlings of MED31-RNAi were comparable to those in the WT (SI Appendix, Fig. S6A), our results suggest that the altered expressions of CYCD6;1 are specifically restricted to the root tip.
To determine the genetic relationship of MED31 and CYCD6;1 and its role in regulating ground tissue patterning, we generated a MED31-RNAi cycd6;1 double-mutant line. The extra periclinal divisions in the ground tissue of MED31-RNAi were largely rescued by cycd6;1 (SI Appendix, Fig. S6 B–F), suggesting that MED31 functions in ground tissue patterning by regulating the spatiotemporal expression of CYCD6;1. Taken together, the above results indicated that MED31 plays an important role in the SHR/SCR-mediated spatiotemporal regulation of CYCD6;1 expression in the CEI and CEID as well as in the upper ground tissue.
MED31 Is Recruited to the CYCD6;1 Promoter in an SCR-Dependent Manner.
Our findings that MED31 physically interacts with SCR and plays a critical role in the SCR-regulated expression of CYCD6;1 prompted us to investigate whether MED31 was recruited to the CYCD6;1 promoter region bound by SCR (24). Consistent with a previous report (24), chromatin immunoprecipitation (ChIP)-qPCR assays using pSCR::GFP:SCR and anti-GFP antibodies showed an enrichment peak of SCR at ∼1,000 bp upstream of the CYCD6;1 promoter (Fig. 4 E, fragment B and F). Parallel ChIP-qPCR assays using pMED31::MED31:GFP plants revealed that the enrichment pattern of MED31 on the CYCD6;1 promoter overlapped with that of SCR (Fig. 4 E, fragment B and G), indicating that MED31 and SCR were recruited to the same region of the CYCD6;1 promoter. The MED31 enrichment on the CYCD6;1 promoter was markedly reduced in scr-1 null mutant compared with WT, indicating that the recruitment of MED31 to the CYCD6;1 promoter is dependent on the function of SCR (Fig. 4H). Consistently, ChIP-qPCR assays revealed that MED31 was also recruited to the promoters of other known SHR/SCR targets in an SCR-dependent manner (SI Appendix, Fig. S7 A–C).
Considering the established role of Mediator in bridging the interaction of DNA-bound transcription factors and Pol II for PIC formation (31, 55), we hypothesized that knockdown of MED31 could impair the recruitment of Pol II to the CYCD6;1 promoter. ChIP-qPCR assays showed that the recruitment of the C-terminal domain of Pol II (42, 56) to the CYCD6;1 promoter was more decreased in MED31-RNAi than in WT (Fig. 4I). This identified an important role of MED31 in recruiting Pol II to the CYCD6;1 promoter during the SHR/SCR-regulated transcription of this cell cycle gene.
Collectively, our data support a mechanism by which the physical interaction of MED31 with SCR occurs on SHR/SCR target promoters, and this interaction is critical for SHR/SCR-regulated gene transcription.
MED31 Forms a Ternary Complex with SHR and SCR in an SCR-Dependent Manner.
Results showing that MED31 physically interacts with SCR (Fig. 1 A and B), together with the long-standing observation that SHR and SCR act as a heterodimer (16), suggested the possibility that MED31 forms a ternary complex with SHR and SCR in vivo. To test this, we performed co-IP assays by coexpressing MED31-myc, SCR, and SHR-GFP in N. benthamiana leaves. As shown in Fig. 5A, both MED31-myc and SCR coimmunoprecipitated with SHR-GFP, indicating that MED31 indeed exists in the same complex with SHR and SCR in vivo.
Fig. 5.
MED31 is an important component of the SHR/SCR complex. (A) Co-IP assay showing that SHR associates with SCR and MED31 in N. benthamiana. MED31-myc, SCR, and SHR-GFP were coinfiltrated into N. benthamiana leaves. Protein samples were immunoprecipitated with anti-GFP antibody and immunoblotted with anti-myc and anti-SCR antibodies. MED31-myc– and SCR-coinfiltrated N. benthamiana leaves were used as negative controls. (B) In vitro pull-down assays showing that MED31 forms a ternary complex with SHR and SCR. GST-SHR and SCR-FLAG were pulled down by MED31-MBP immobilized on amylose resin. Proteins bound to amylose resin were eluted and analyzed by anti-GST and anti-FLAG antibodies, respectively. (C) In vitro pull-down assays showing that SHR directly interacts with SCR but not itself. SCR-FLAG was pulled down by GST-SHR immobilized on a GST-Bind resin. Protein bound to the GST-Bind resin was eluted and analyzed using an anti-FLAG antibody. (D) In vitro pull-down assay showing that SCR directly interacts with SHR but not itself. SHR-FLAG was pulled down by GST-SCR immobilized on a GST-Bind resin. Protein bound to the GST-Bind resin was eluted and analyzed using an anti-FLAG antibody. CBB, Coomassie brilliant blue staining; MW, molecular weight (mass).
To exclude the possibility that MED31 forms a ternary complex with SHR/SCR through a linkage protein other than SCR, we performed in vitro pull-down assays using purified MED31-MBP, GST-SHR, and SCR-FLAG. The results indicated that MED31-MBP could pull down GST-SHR in the presence of SCR-FLAG but failed to pull down GST-SHR in the absence of SCR-FLAG (Fig. 5B). These results substantiated that the formation of the MED31/SCR/SHR ternary complex strictly depends on the interface protein SCR.
The finding that MED31 exists in the same complex with SHR and SCR in vivo prompted us to investigate the precise details of their interactions. For this purpose, we first performed in vitro pull-down assays to examine the interactions of SHR and SCR with themselves or with each other. The results showed that GST-SHR could pull down SCR-FLAG, whereas it failed to pull down SHR-FLAG, indicating that SHR interacts with SCR but not with itself (Fig. 5C). In parallel experiments, GST-SCR pulled down SHR-FLAG, whereas it failed to pull down SCR-FLAG (Fig. 5D), indicating that SCR interacts with SHR but not with itself. These results are consistent with the notion that SHR and SCR form a heterodimer involved in the regulation of stem cell ACD and ground tissue patterning (16). Taken together, these data support that MED31 forms a ternary complex with the SHR/SCR heterodimer in vivo, and the SCR interface serves as a bridge between MED31 and SHR in the MED31/SCR/SHR ternary complex.
MED31 and SHR Compete with Each Other for Binding to SCR.
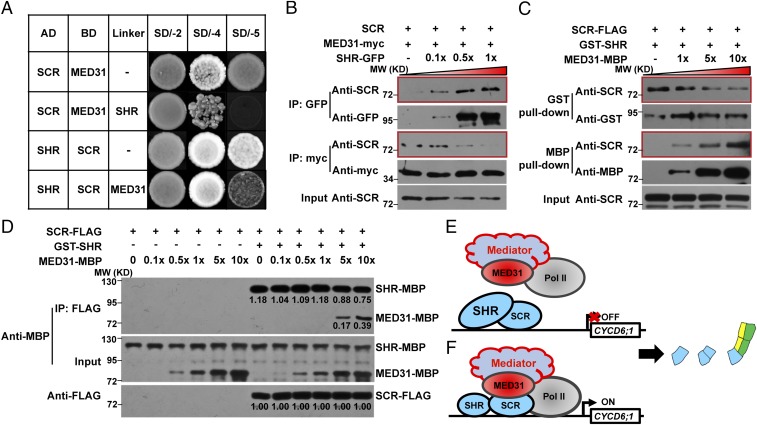
Since MED31 (Fig. 1D) and SHR (16) bind the same protein domain of SCR, we tested whether MED31 and SHR affect each other in binding SCR. Yeast three-hybrid assays detected the SCR and MED31 interaction on the selection synthetically defined (SD) medium SD/-Ade/-His/-Trp/-Leu (SD/-4) (Fig. 6A). Induction of SHR expression on the selection medium SD/-Ade/-His/-Trp/-Leu/-Met (SD/-5) abolished the MED31/SCR interaction (Fig. 6A), suggesting that SHR competitively inhibits the MED31/SCR interaction in yeast cells. In parallel experiments, the SHR/SCR interaction was detected on the selection medium SD/-4 (Fig. 6A). Induction of MED31 expression on the selection medium SD/-5 substantially reduced the SHR/SCR interaction (Fig. 6A), suggesting that MED31 also interferes with the SHR/SCR interaction in yeast cells. These results suggest that SHR and MED31 affect each other in the interaction with SCR in yeast cells.
Fig. 6.
MED31 and SHR compete with each other for binding to SCR. (A) Yeast three-hybrid (Y3H) assays showing that addition of SHR dramatically reduced the MED31/SCR interaction (Top two panels) and addition of MED31 reduced the SHR/SCR interaction (Bottom two panels). (Top two panels) Yeast cells cotransformed with pGADT7-SCR and pBridge-MED31-SHR were dropped onto SD/-Trp/-Leu (SD/-2) and SD/-4 media to assess the MED31/SCR interaction. The cotransformed yeast cells were dropped onto SD/-5 medium to induce SHR. (Bottom two panels) Yeast cells cotransformed with pGADT7-SHR and pBridge-SCR-MED31 were dropped onto SD/-2 and SD/-4 media to assess the SHR/SCR interaction. The cotransformed yeast cells were dropped onto SD/-5 medium to induce MED31. AD, activation domain fusion; BD, binding domain fusion. (B) Co-IP assay showing that the gradually increasing addition of SHR disrupted the MED31/SCR interaction in N. benthamiana. MED31-myc and SCR were coexpressed in N. benthamiana leaves. SHR-GFP was added to SCR-MED31-myc protein extracts according to the gradient as shown. Protein samples were immunoprecipitated with anti-GFP antibodies or anti-myc antibodies and immunoblotted with anti-SCR antibodies to detect the SHR/SCR interaction and MED31/SCR interaction, respectively. MW, molecular weight (mass). (C) In vitro pull-down assays showing that high concentrations of MED31 affected SHR/SCR interaction. For each sample, the amounts of SCR-FLAG and GST-SHR were equal, and MED31-MBP was added according to the indicated gradient. SCR-FLAG was pulled down by GST-SHR immobilized on a GST-Bind resin or MED31-MBP immobilized on an amylose resin. Proteins were eluted and analyzed using an anti-FLAG antibody. (D) In vitro quantitative co-IP assays showing that the SHR/SCR interaction was stronger than the MED31/SCR interaction. For each sample, the amounts of SCR-FLAG and SHR-MBP were equal, and MED31-MBP was added according to the indicated gradient. SCR-FLAG was immunoprecipitated with anti-FLAG antibodies and immunoblotted with anti-MBP antibodies to analyze MED31-MBP and SHR-MBP. Bands were quantified using ImageJ software (NIH). (E and F) Proposed mechanism by which MED31 regulates SHR/SCR-mediated dynamic activation of CYCD6;1 in the CEI and CEID. (E) When the relative protein abundance between SHR and MED31 is high enough to prevent MED31/SCR interaction, CYCD6;1 expression is turned off. (F) When the relative protein abundance between SHR and MED31 is low enough to enable MED31/SCR interaction, CYCD6;1 expression is turned on.
To test whether SHR and MED31 compete with each other for interacting with SCR in vivo, we performed co-IP assays in N. benthamiana leaves. For these experiments, protein extracts from N. benthamiana leaves coexpressing MED31-myc and SCR were incubated with gradients of the separately expressed SHR/GFP fusion protein and immunoprecipitated with anti-GFP or anti-myc antibodies. Results indicated that the interaction between MED31-myc and SCR was weakened by increasing amounts of SHR-GFP (Fig. 6B). In parallel experiments, the interaction between SHR-GFP and SCR was only mildly attenuated by increasing amounts of MED31-myc (SI Appendix, Fig. S8). These results confirmed that SHR potently impairs the binding of MED31 to SCR in vivo, and the binding affinity between SHR and SCR is stronger than that between MED31 and SCR.
We hypothesized that further increased doses of MED31 may display a competitive effect on the SHR/SCR interaction. To test this, we performed an in vitro pull-down assay in which the amounts of the SHR and SCR proteins were kept constant in each sample, whereas the concentration of MED31 was increased in a gradient. In this assay, recombinant GST-SHR or MED31-MBP was used to pull down SCR-FLAG synthesized by in vitro transcription/translation reactions. Results showed that increasing concentrations (i.e., five- and 10-fold the amount of GST-SHR) of MED31-MBP reduced the interaction between SCR-FLAG and GST-SHR (Fig. 6C), validating our hypothesis.
To further substantiate the above observations, we performed a quantitative in vitro co-IP assay in which synthetic SCR-FLAG was used to pull down recombinant SHR-MBP and MED31-MBP (Fig. 6D). Low concentrations of MED31-MBP (i.e., 0-, 0.1-, 0.5-, or 1.0-fold the amount of SHR-MBP) had a minor impact on the interaction between SHR-MBP and SCR-FLAG, whereas higher doses of MED31-MBP (i.e., five- and 10-fold the amount of SHR-MBP) reduced the interaction between SHR-MBP and SCR-FLAG to 0.75- or 0.64-fold compared with control, respectively (fivefold, 0.88/1.18; 10-fold, 0.75/1.18) (Fig. 6D). These results confirmed that the SHR/SCR interaction was stronger than the MED31/SCR interaction. The differences in the strength of the interaction of MED31 or SHR with the scaffold protein SCR may affect the dynamic formation of the MED31/SCR/SHR ternary complex.
Discussion
MED31 Shows Differential Effects on CYCD6;1 Expression Between Stem Cells and the Upper Ground Tissue.
Our results reveal that MED31 can act as an activator for CYCD6;1 in the CEI and CEID, while acting as a repressor in the upper ground tissue, and that the change in MED31 function likely correlates with different levels of SHR. We propose a working model to explain the differential effects of MED31 in regulating CYCD6;1 expression in different cell types. In the CEI and CEID, relatively high SHR protein levels weaken the interaction of MED31 with SCR, and therefore impair the ability of MED31 to “bridge” the communication between this “gene-specific” transcriptional activator and the Pol II general transcription machinery. At this stage, CYCD6;1 expression is shut off (Fig. 6E). In response to developmental cues, SHR protein levels are relatively low, enabling MED31 to interact with SCR (impaired in MED31-RNAi) and thereby activate CYCD6;1 expression (Fig. 6F). This model highlights that the dynamic movement of SHR into the CEI and CEID could change the local SHR-to-MED31 protein ratio, which determines the transcriptional output of the MED31/SCR/SHR ternary complex.
Moreover, our results support that the above scenario also operates in the upper ground tissue. In the endodermis layer of WT roots, the SHR protein abundance is relatively high and the SHR-to-MED31 protein ratio in individual cells is uniformly controlled to a threshold ratio, which keep the CYCD6;1 expression in check (SI Appendix, Fig. S9A). However, in the presumptive endodermis layer of MED31-RNAi roots, the SHR-to-MED31 protein ratio is impaired, and thereby leads to sporadic activation of CYCD6;1 expression (SI Appendix, Fig. S9B).
MED31 Is a Conserved Stem Cell Regulator Between Plants and Animals.
Gene transcription regulated by Pol II plays a crucial role in stem cell ACD and organ patterning. While much effort is devoted to gene-specific transcription factors and their transcriptional targets, we know surprisingly little about the function of protein complexes that interact with Pol II during gene transcription, such as the Mediator complex. Here, we provide biochemical and genetic evidence that the MED31 subunit of the plant Mediator complex is a critical component of the extensively investigated SHR/SCR transcriptional complex involved in regulating ACD of the CEI/CEID and ground tissue patterning. We further reveal that MED31 is required for the accurate spatiotemporal activation of CYCD6;1 expression and is recruited to SHR/SCR target promoters in an SCR-dependent manner. These data support the notion that the MED31 interface of Mediator directly links the context-specific transcriptional activators with the Poll II general transcription machinery during SHR/SCR-regulated gene transcription.
Although MED31 shows differential effects on CYCD6;1 expression in stem cells and the upper ground tissue (Fig. 4 C and D), our RT-qPCR results indicate that this Mediator subunit generally acts as a positive regulator of SHR/SCR transcriptional targets, including SCR, SCL3, RLK, JKD, MGP, and NUC (Fig. 3 A, F, and G and SI Appendix, Fig. S5 E–I). In this regard, the mode of action of MED31 is in contrast to that of the RBR protein, which also physically binds SCR but negatively regulates SHR/SCR transcriptional targets (25, 57). Therefore, adding to the knowledge that RBR acts as a repressor of SHR/SCR activity, the present study demonstrated that MED31 acts as a coactivator of SHR/SCR activity. Considering that both MED31 (Fig. 1D) and RBR (25) bind to the CD of SCR, the relationship between MED31 and RBR with respect to their binding to SCR needs to be elucidated in future studies to understand the underlying mechanism and physiological significance.
Similar to animal MED31, which plays an essential role in embryonic development (49, 50), we showed that knockout of MED31 leads to embryonic lethality in Arabidopsis (SI Appendix, Fig. S3 B–D′). Although the mechanisms underlying the involvement of animal MED31 in stem cell regulation are not fully understood, the data suggest conserved functions of MED31 in early embryogenesis between animals and plants. In line with this, recent structural studies in yeast showed that the highly conserved MED31 is involved in the formation of a unique structure (i.e., MED7/31 submodule) that directly binds the Pol II CTD in the PIC during transcription initiation (55, 58). Considering that the MED7 subunit is also conserved between yeast and plants, it is necessary and important in future studies to explore whether MED31 functions alone or together with other Mediator subunits, such as MED7, to regulate root and embryo development.
Considering that the master stem cell regulators SHR and SCR are plant-specific proteins (6) and the functions of MED31 and RBR can be conserved in plants and animals (57), the present results support a scenario in which the regulators SHR and SCR that specify stem cells are recruited from plant-specific protein families, whereas their coactivator (i.e., MED31) or repressor (i.e., RBR) is conserved between the plant and animal kingdoms.
The Dynamic Formation of the MED31/SCR/SHR Ternary Complex Controls the Spatiotemporal Transcriptional Output of SHR/SCR.
Although it is generally believed that SHR and SCR function as transcriptional regulators, the precise details by which these GIBBERELLIC-ACID INSENSITIVE (GAI), REPRESSOR of GAI (RGA), and SCR (GRAS) family proteins activate target gene transcription remain largely unknown. Consistently, a recent structural study suggested that SHR and SCR function as transcriptional cofactors that indirectly bind to their target promoters via their interacting BIRD transcription factors (22).
We explored the mechanism underlying the regulation of the transcriptional output of SHR/SCR by linking SHR/SCR with the Mediator complex. We provided evidence that MED31 is involved in the dynamic formation of a MED31/SCR/SHR ternary complex. We used the well-established SHR/SCR/CYCD6;1 transcriptional module to demonstrate how the dynamic ternary complex strictly controls the spatiotemporal transcriptional output of SHR/SCR (Fig. 6 E and F). The fact that MED31 directly binds the Pol II CTD in the PIC during transcription initiation (55) suggests that the transcriptional output of SHR/SCR is only turned on when MED31 physically interacts with SCR (presumably the abundance of MED31 is high enough at this stage to compete with SHR for binding to SCR). This stage is the only one at which MED31 can recruit the Pol II general transcriptional machinery to SHR/SCR target promoters for PIC formation. The transcriptional output of SHR/SCR is turned off when MED31 is unable to physically interact with SCR (presumably the SHR abundance is high enough at this stage to compete with MED31 for binding to SCR). At this stage, the Pol II general transcriptional machinery cannot be recruited to SHR/SCR target promoters.
MED31 and SHR affect each other for binding to SCR, and the interaction between SHR and SCR is stronger than that between MED31 and SCR (Fig. 6). Therefore, our model predicts that the relative protein abundance of SHR and MED31 in the CEI and CEID may serve as an important parameter that affects the dynamic formation of the MED31/SCR/SHR ternary complex, which eventually determines the transcriptional output of SHR/SCR. In line with this prediction, it is well-recognized that SHR acts as a mobile, dose-dependent signal, and its protein can move from the stele to the CEI and CEID (16). In addition, SHR protein can be degraded during cell division (25), and our data indicated that MED31 is subject to proteasome-dependent degradation (SI Appendix, Fig. S2 E–G). In this context, we speculate that, in response to developmental cues, the relative protein abundance of SHR and MED31 in the CEI and CEID changes dynamically, switching the transcriptional output of SHR/SCR on or off. It will be very interesting in future studies to elucidate how the relative protein abundance of SHR and MED31 is regulated in response to developmental cues.
Furthermore, we revealed a critical function of MED31 in maintaining the uniform distribution of the SHR protein in each endodermis cell (Fig. 3 C and E). Notably, in the presumptive endodermis layer of MED31-RNAi roots, variations of SHR protein abundance always correlate with ectopic activation of CYCD6;1 expression (Fig. 4D), as well as with irregular periclinal division of the corresponding cells (Fig. 3 C and E), suggesting that the MED31/SCR/SHR ternary complex also operates in regulating the patterning of the upper ground tissue. In line with this scenario, it has been elegantly shown that SHR acts in a dose-dependent manner to regulate CYCD6;1 expression and promotes periclinal divisions to form middle cortex along the endodermis (54). Considering that SCR plays a pivotal role in regulating SHR movement and nuclear residence (16) and that MED31 physically and functionally interacts with SCR (this study), it is reasonable to speculate that the function of MED31 in maintaining the uniform endodermal distribution of SHR is achieved through collaboration with its interacting partner SCR. It is of great importance in future studies to explore the underlying mechanisms.
Materials and Methods
All seedlings were grown on half-strength Murashige and Skoog medium with 1% sucrose under a 16-h light/8-h dark cycle at 22 °C. Plant materials, plasmid construction methods, cytological techniques, biochemical methods, and all other experimental procedures are described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Philip Benfey for sharing research materials. This work was supported by the National Basic Research Program of China (Grant 2015CB942900), the National Natural Science Foundation of China (Grant 31320103910), and the Tai-Shan Scholar Program from the Shandong Provincial Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800592115/-/DCSupplemental.
References
- 1.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Scheres B, et al. Embryonic origin of the Arabidopsis primary root and root-meristem initials. Development. 1994;120:2475–2487. [Google Scholar]
- 3.van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- 4.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 5.Benfey PN, Scheres B. Root development. Curr Biol. 2000;10:R813–R815. doi: 10.1016/s0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- 6.Scheres B. Stem-cell niches: Nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- 7.Dinneny JR, Benfey PN. Plant stem cell niches: Standing the test of time. Cell. 2008;132:553–557. doi: 10.1016/j.cell.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Benfey PN, et al. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- 9.Scheres B, et al. Mutations affecting the radial organization of the Arabidopsis root display specific defects throughout the embryonic axis. Development. 1995;121:53–62. [Google Scholar]
- 10.Scheres B, Benfey PN. Asymmetric cell division in plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:505–537. doi: 10.1146/annurev.arplant.50.1.505. [DOI] [PubMed] [Google Scholar]
- 11.Di Laurenzio L, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 12.Helariutta Y, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 15.Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui H, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 17.Yu Q, et al. Cell-fate specification in Arabidopsis roots requires coordinative action of lineage instruction and positional reprogramming. Plant Physiol. 2017;175:816–827. doi: 10.1104/pp.17.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levesque MP, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch D, et al. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long Y, et al. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell. 2015;27:1185–1199. doi: 10.1105/tpc.114.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Risueno MA, et al. Transcriptional control of tissue formation throughout root development. Science. 2015;350:426–430. doi: 10.1126/science.aad1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano Y, et al. Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nat Plants. 2017;3:17010. doi: 10.1038/nplants.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long Y, et al. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature. 2017;548:97–102. doi: 10.1038/nature23317. [DOI] [PubMed] [Google Scholar]
- 24.Sozzani R, et al. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Ramírez A, et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell. 2012;150:1002–1015. doi: 10.1016/j.cell.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 30.Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen BL, Taatjes DJ. The Mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelleher RJ, 3rd, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 33.Flanagan PM, Kelleher RJ, 3rd, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 34.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlsten JO, Zhu X, Gustafsson CM. The multitalented Mediator complex. Trends Biochem Sci. 2013;38:531–537. doi: 10.1016/j.tibs.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta. 2013;1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin JW, Wang G. The Mediator complex: A master coordinator of transcription and cell lineage development. Development. 2014;141:977–987. doi: 10.1242/dev.098392. [DOI] [PubMed] [Google Scholar]
- 38.Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K. Diverse roles of the Mediator complex in plants. Semin Cell Dev Biol. 2011;22:741–748. doi: 10.1016/j.semcdb.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 40.Samanta S, Thakur JK. Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front Plant Sci. 2015;6:757. doi: 10.3389/fpls.2015.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Li L, Qu LJ. Plant Mediator complex and its critical functions in transcription regulation. J Integr Plant Biol. 2016;58:106–118. doi: 10.1111/jipb.12377. [DOI] [PubMed] [Google Scholar]
- 42.Chen R, et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 2012;24:2898–2916. doi: 10.1105/tpc.112.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An C, et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc Natl Acad Sci USA. 2017;114:E8930–E8939. doi: 10.1073/pnas.1710885114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Z, Guan H, Leal F, Grey PH, Oppenheimer DG. Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLoS One. 2013;8:e53924. doi: 10.1371/journal.pone.0053924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhawan R, et al. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell. 2009;21:1000–1019. doi: 10.1105/tpc.108.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillmor CS, et al. The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development. 2010;137:113–122. doi: 10.1242/dev.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai Z, et al. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat Commun. 2014;5:3064. doi: 10.1038/ncomms4064. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linder T, Gustafsson CM. The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator. J Biol Chem. 2004;279:49455–49459. doi: 10.1074/jbc.M409046200. [DOI] [PubMed] [Google Scholar]
- 50.Bosveld F, van Hoek S, Sibon OC. Establishment of cell fate during early Drosophila embryogenesis requires transcriptional Mediator subunit dMED31. Dev Biol. 2008;313:802–813. doi: 10.1016/j.ydbio.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Risley MD, Clowes C, Yu M, Mitchell K, Hentges KE. The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Dev Biol. 2010;342:146–156. doi: 10.1016/j.ydbio.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 52.Yan L, et al. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant. 2015;8:1820–1823. doi: 10.1016/j.molp.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Haseloff J. GFP variants for multispectral imaging of living cells. Methods Cell Biol. 1999;58:139–151. doi: 10.1016/s0091-679x(08)61953-6. [DOI] [PubMed] [Google Scholar]
- 54.Koizumi K, Hayashi T, Wu S, Gallagher KL. The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc Natl Acad Sci USA. 2012;109:13010–13015. doi: 10.1073/pnas.1205579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nozawa K, Schneider TR, Cramer P. Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature. 2017;545:248–251. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- 56.Li P, Tao Z, Dean C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 2015;29:696–701. doi: 10.1101/gad.258814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wildwater M, et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 58.Koschubs T, et al. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J. 2009;28:69–80. doi: 10.1038/emboj.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.