Significance
The mitochondrial electron transport chain (ETC) of Plasmodium falciparum malaria parasites contains targets for antimalarial drug development including cytochrome B (PfCytB) and NADH dehydrogenase 2 (PfNDH2). Atovaquone (ATQ), a widely used antimalarial drug, is a hydroxynaphthoquinone inhibitor of PfCytB; substituted quinolone compounds (CK-2-68 and RYL-552) have also been developed as putative inhibitors of PfNDH2. Unexpectedly, our experiments yielded mutations of PfCytB, not PfNDH2, in parasites selected with these quinolones. ATQ selections yielded different PfCytB mutants that showed little change or slightly increased sensitivity to CK-2-68 or RYL-552. Differential patterns of drug response in various PfCytB mutants suggest that combinations of ETC inhibitors may help counter drug-resistant malaria.
Keywords: malaria, drug resistance, NADH dehydrogenase 2, mitochondrial electron transport chain
Abstract
Malaria control is threatened by a limited pipeline of effective pharmaceuticals against drug-resistant strains of Plasmodium falciparum. Components of the mitochondrial electron transport chain (ETC) are attractive targets for drug development, owing to exploitable differences between the parasite and human ETC. Disruption of ETC function interferes with metabolic processes including de novo pyrimidine synthesis, essential for nucleic acid replication. We investigated the effects of ETC inhibitor selection on two distinct P. falciparum clones, Dd2 and 106/1. Compounds CK-2-68 and RYL-552, substituted quinolones reported to block P. falciparum NADH dehydrogenase 2 (PfNDH2; a type II NADH:quinone oxidoreductase), unexpectedly selected mutations at the quinol oxidation (Qo) pocket of P. falciparum cytochrome B (PfCytB). Selection experiments with atovaquone (ATQ) on 106/1 parasites yielded highly resistant PfCytB Y268S mutants seen in clinical infections that fail ATQ-proguanil treatment. In contrast, ATQ pressure on Dd2 yielded moderately resistant parasites carrying a PfCytB M133I or K272R mutation. Strikingly, all ATQ-selected mutants demonstrated little change or slight increase of sensitivity to CK-2-68 or RYL-552. Molecular docking studies demonstrated binding of all three ETC inhibitors to the Qo pocket of PfCytB, where Y268 forms strong van der Waals interactions with the hydroxynaphthoquinone ring of ATQ but not the quinolone ring of CK-2-68 or RYL-552. Our results suggest that combinations of suitable ETC inhibitors may be able to subvert or delay the development of P. falciparum drug resistance.
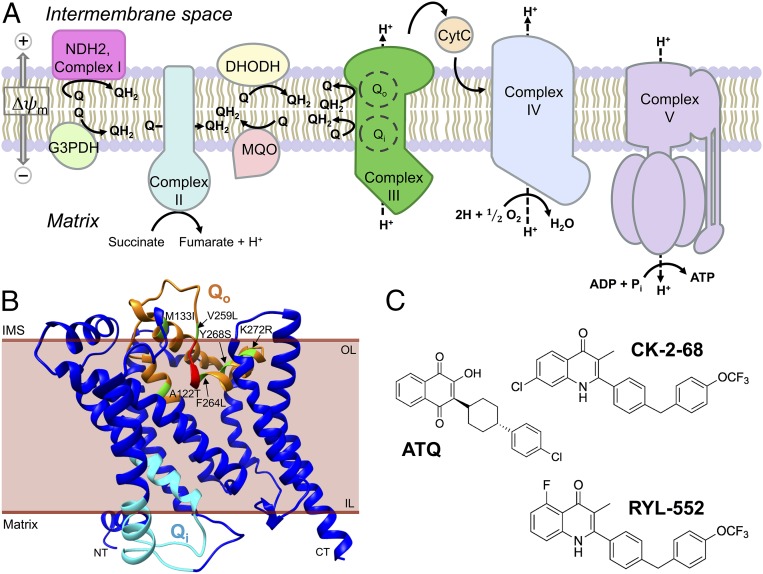
Essential life processes of eukaryotic cells depend upon the electron transport chain (ETC) lodged in the inner membrane of the double membrane-bound mitochondrion, where an evolutionarily conserved cascade of oxidation-reduction activities creates a membrane potential and proton-motive energy for ATP production (1). Structural and functional properties of the ETC components, and of the assembled multisubunit respiratory complexes, provide drug targets in their differences between eukaryotic pathogens and mammalian cells (2). The ETCs of the Plasmodium falciparum malaria parasite (PfETC) and human (hETC) offer a case in point: Both include five canonical respiratory complexes (Fig. 1A), but with notable differences including that some of the PfETC complexes have fewer subunits than the hETC (3). Malate-quinone oxidoreductase is found in the PfETC instead of a malate dehydrogenase as in the hETC (4), and unique structural aspects of P. falciparum dihydroorotate dehydrogenase (DHODH) are being exploited for new antimalarial drug candidates (5, 6). Additional important PfETC targets for antimalarial drug discovery include P. falciparum cytochrome B (PfCytB, in complex III) and the P. falciparum NADH dehydrogenase 2 (PfNDH2, alternative complex I) (7, 8).
Fig. 1.
Features of the PfETC, predicted structure of PfCytB, and chemical structures of inhibitors used for selection. (A) Components of the PfETC. CytC, soluble cytochrome C; G3PDH, glycerol-3-phosphate dehydrogenase; MQO, malate-quinone oxidoreductase; Q, ubiquinone; QH2, ubiquinol; Δψm, mitochondrial membrane potential. (B) Three-dimensional ribbon model of PfCytB (mal_mito_3) predicted by iterative threading using known crystal structures deposited in the PDB archive (62–64). Qo residues are highlighted in orange, PEWY motif in red, Qi residues in cyan, and mutations selected by drug pressure in green. CT, carboxy terminus; IL, inner leaflet; IMS, inner membrane space; NT, amino terminus; OL, outer leaflet. (C) ATQ is a hydroxynaphthoquinone; CK-2-68 and RYL-552 are both substituted quinolones.
PfCytB occurs as part of the cytochrome bc1 assembly with the Rieske iron sulfur protein in complex III. The quinone reduction site (Qi) reduces ubiquinone at the mitochondrial matrix side of PfCytB, whereas the quinol oxidation site (Qo) catalyzes electron removal from ubiquinol at the intermembrane space (IMS; Fig. 1B). Enzymatic activities of the Qi and Qo sites are coupled to the translocation of hydrogen atoms into the IMS and are essential to mitochondrial membrane potential and parasite viability (2). Atovaquone (ATQ), a substituted hydroxynaphthoquinone compound (2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxy-1,4-naphthalenedione; Fig. 1C), competitively displaces ubiquinol from the PfCytB Qo site, producing mitochondrial membrane depolarization, abrogation of pyrimidine biosynthesis, and parasite death (9–13). Other Qo site inhibitors are available although none are currently approved for use in human malaria treatment (7). Among Qi site antagonists, antimycin A (AMA) has low micromolar activity against P. falciparum; more effective Qi inhibitors with efficacies in the nanomolar range have recently been described (14–16). One Qi inhibitor, ELQ300, has entered preclinical studies (17, 18).
PfNDH2 is a bacterial-like type II NADH:quinone oxidoreductase that differs markedly from the human multisubunit type I enzyme. Unlike mammalian complex I, PfNDH2 is rotenone-insensitive, is not protonmotive, and lacks a transmembrane domain (19). PfNDH2 reduces ubiquinone to ubiquinol for downstream use by the respiratory complexes, thereby contributing to cellular ATP production (20). Based on the ubiquinone analog 1-hydroxy-2-dodecyl-4(1H)quinolone, CK-2-68, a bisaryl quinolone compound (Fig. 1C), was developed with inhibitory action in an end-point PfNDH2 bench assay; CK-2-68 exhibited submicromolar half-maximal effective concentration (EC50) levels against P. falciparum parasite cultures (21). In CK-2-68 selection experiments on the P. falciparum K1 clone, a parasite line was obtained with a threefold decrease in drug sensitivity and a reported PfNDH2 V203I substitution (21), which occurs in the NADH catalytic region apart from the quinone binding domain. A structurally similar compound, RYL-552 (Fig. 1C), was also found to have strong activity against malaria parasites and was shown to bind PfNDH2 in surface plasmon resonance studies and in cocrystallization experiments (22).
Use of ATQ as monotherapy rapidly fails from PfCytB mutations (23, 24). However, resistance arises less readily in the presence of proguanil, which acts synergistically with ATQ against the parasite mitochondrial function and is converted to cycloguanil, an active metabolite that inhibits dihydrofolate reductase in the parasite’s essential folate pathway (25). To explore comparative features of resistance that can develop to ATQ, CK-2-68, and RYL-552, we performed systematic studies with single-step selection pressure on two distinct P. falciparum clones, 106/1 and Dd2. Our results with these inhibitors show various PfCytB mutations but no PfNDH2 mutations, indicating important activity of CK-2-68 and RYL-552 on the PfCytB target. Further, differential patterns of response by these various mutants suggest that combinations of ETC inhibitors may provide a strategy to subvert or delay the development of drug resistance.
Results
Selection of ETC Inhibitor-Resistant Mutants of P. falciparum.
To study responses and resistance obtained by exposure to ATQ, CK-2-68, and RYL-552, we subjected the P. falciparum Dd2 and 106/1 clones to continuous concentrations (3–100× EC50) of these compounds for periods up to 60 d (Table 1). Resistant populations were selected from both Dd2 and 106/1. Sequencing showed no change of the pfndh2 (PF3D7_0915000) coding region in any of these populations, despite the previous report of a PfNDH2 V203I substitution following CK-2-68 pressure (21) (PfCytB sequence not reported). In contrast to our finding of no PfNDH2 mutations, nonsynonymous codon changes occurred in the pfcytB (mal_mito_3) gene of all resistant mutants. These mutations differed between the Dd2 and 106/1 selections, although the experiments were performed simultaneously and employed the same culture conditions.
Table 1.
Electron transport chain inhibitor selection of Plasmodium falciparum cytochrome B mutations
| Line | Compound | Selection concentration | Inoculum | No. of positive/no. of flasks | Day* (generation no.)† | PfCytB mutation | Codon change | PfNDH2 mutation |
| Dd2 | ATQ | 5 nM (10× EC50) | 3.0 × 108 | 4/4 | 16 (8) | M133I | ATG → ATT | None |
| 3.0 × 108 | 16 (8) | K272R | ATG → AGA | None | ||||
| 1.9 × 108 | 18 (9) | M133I | ATG → ATA | None | ||||
| 2.5 × 108 | 18 (9) | M133I | ATG → ATT | None | ||||
| RYL-552 | 150 nM (8× EC50) | 1.8 × 108 | 1/4 | 22 (11) | F264L | TTT → TTA | None | |
| 106/1 | ATQ | 100 nM (100× EC50) | 1.8 × 108 | 1/4 | 25 (12) | Y268S | TAT → TCT | None |
| CK-2-68 | 300 nM (4.3× EC50) | 1.7 × 108 | 1/4 | 59 (29) | A122T | GCT → ACT | None | |
| RYL-552 | 150 nM (3× EC50) | 1.8 × 108 | 4/4 | 28 (14) | V259L | GTA → TTA | None | |
| 1.3 × 108 | 42 (21) | A122T | GCT → ACT | None | ||||
| 1.8 × 108 | 45 (22) | V259L | GTA → TTA | None | ||||
| 1.8 × 108 | 56 (28) | V259L | GTA → TTA | None |
Day parasites were first observed by microscopy of thin blood smears.
Maximum generation number was calculated as the first day parasites were observed, divided by 2. This calculation assumes the mutant was in the population at the start of selection.
In the ATQ pressure experiments, four independent populations were obtained from Dd2: a PfCytB K272R mutant and three M133I mutants with an ATG → ATT or ATG → ATA codon change (Table 1). However, ATQ pressure on 106/1 yielded one population carrying PfCytB Y268S, a mutation frequently detected in clinical failures of atovaquone-proguanil (26, 27) and recently obtained from human isolates by in vitro selection (28).
In pressure experiments with the substituted quinolones, CK-2-68 selection was unsuccessful with Dd2 parasites but yielded a PfCytB A122T mutant from 106/1 (Table 1). Selection with RYL-552, however, returned resistant populations from both Dd2 and 106/1. These included a PfCytB F264L mutant from Dd2, and V259L as well as A122T mutants from 106/1. We note that V259L has also been reported from selections of Dd2 parasites with the 4(1H) pyridone, ELQ400 (29), and with a chemotype I acridone, T13 (30).
ETC Inhibitor Response Profiles.
Clones were isolated from all selected lines by limiting dilution and verified by microsatellite typing (SI Appendix, Table S1). EC50 concentrations of antimalarial compounds against these clones were determined by SYBR Green assays and compared with the responses of Dd2 and 106/1 as well as standard P. falciparum 3D7 and HB3 clones (Table 2). Results showed similar nanomolar EC50 sensitivities of the Dd2, 106/1, 3D7, and HB3 control parasites to ATQ, RYL-552, and CK-2-68. Activities of the Qi antagonist AMA, chloroquine (CQ), and the PfDHODH inhibitor DSM1 were also in the expected ranges for these four controls (31–35). The clones from the ETC inhibitor-selected lines retained the same CQ responses as those of the original Dd2 (CQ-resistant) and 106/1 (CQ-sensitive) clones (Table 2).
Table 2.
EC50 of Plasmodium falciparum clones
| EC50, nM | ||||||||
| Clone | Selection agent | PfCytB mutation | ATQ | CK-2-68 | RYL-552 | AMA | DSM1 | CQ |
| Controls | ||||||||
| HB3 | 0.4 ± 0.1 | 44 ± 3 | 17 ± 0.1 | 381 ± 78 | 69 ± 6 | 3.1 ± 0.4 | ||
| 3D7 | 0.7 ± 0.1 | 34 ± 4 | 22 ± 3 | 250 ± 15 | 155 ± 8 | 12 ± 2 | ||
| Dd2 lineage | ||||||||
| Dd2 | 0.4 ± 0.02 | 57 ± 3 | 18 ± 2 | 221 ± 58 | 134 ± 22 | 214 ± 31 | ||
| DA-3H6M133I | ATQ | M133I | 10 ± 2 | 61 ± 9 | 3.3 ± 1 | 243 ± 25 | 207 ± 21 | 244 ± 40 |
| DA-4K272R | ATQ | K272R | 55 ± 7 | 12 ± 4 | 11 ± 3 | 45 ± 5 | 155 ± 31 | 209 ± 37 |
| DR-4H5F264L | RYL-552 | F264L | 1.1 ± 0.2 | 71 ± 12 | 91 ± 7 | 63 ± 15 | 137 ± 16 | 201 ± 21 |
| 106/1 lineage | ||||||||
| 106/1 | 1.1 ± 0.1 | 69 ± 7 | 50 ± 5 | 417 ± 58 | 189 ± 13 | 28 ± 0.2 | ||
| 6A-4F12Y268S | ATQ | Y268S | 6,760 ± 1,600 | 38 ± 2 | 75 ± 10 | 108 ± 16 | 93 ± 27 | 39 ± 3 |
| 6C-2A7A122T | CK-2-68 | A122T | 0.7 ± 0.1 | 225 ± 16 | 50 ± 4 | 208 ± 12 | 221 ± 57 | 20 ± 2 |
| 6R-3H8V259L | RYL-552 | V259L | 8.2 ± 0.5 | 209 ± 7 | 202 ± 17 | 137 ± 22 | 98 ± 48 | 32 ± 2 |
| 6R-4E5A122T | RYL-552 | A122T | 0.7 ± 0.1 | 220 ± 6 | 158 ± 2 | 293 ± 36 | 107 ± 23 | 30 ± 3 |
Mean ± SD; n = 3–7.
The EC50 values of clones from the mutant populations were elevated, as expected, when tested with the same ETC inhibitors used for their selection: 25–6,150× with ATQ against DA-3H6M133I, DA-4K272R, or 6A-4F12Y268S; 3.3× with CK-2-68 against 6C-2A7A122T; and 3.2–5.1× with RYL-552 against DR-4H5F264L, 6R-3H8V259L, or 6R-4E5A122T (Table 2). In comparisons for cross-resistance, clones from ATQ-selected populations showed no decreased susceptibility to CK-2-68 or RYL-552; instead, 1.6–5.5× increases of susceptibility were observed with one or both of these compounds (clones DA-3H6M133I; DA-4K272R; 6A-4F12Y268S). Regarding the ATQ EC50 values of the clones from CK-2-68– or RYL-552–selected lines, these were 1.6× reduced (6R-4E5A122T; 6C-2A7A122T) or 2.8–7.5× increased (DR-4H5F264L; 6R-3H8V259L) relative to EC50 values of the original Dd2 or 106/1 parasites.
AMA responses showed reductions in all PfCytB mutant clones except DA-3H6M133I: These included 1.4–4.9× EC50 reductions in the DA-4K272R, DR-4H5F264L, 6A-4F12Y268S, 6C-2A7A122T, 6R-3H8V259L, and 6R-4E5A122T clones (Table 2). DSM1 EC50 values of all clones showed little or no change from original Dd2 or 106/1 parasites and also were in the range of the HB3 and 3D7 responses. Drug responses of all clones remained stable in repeat assays over several months’ time, including prolonged cultivation periods interrupted by cycles of cryopreservation and thawing.
pfcoxI, pfcoxIII, pfcytB, and Pfdhod Copy Number Determinations.
The mitochondria of Plasmodium spp. contain an estimated 20–150 copies of a 6-kb chromosome in a composite of linear branched, lariat, and circular repeats (36, 37). Of the ETC proteins in P. falciparum, three, cytochrome B (PfCytB), cytochrome oxidase I (PfCoxI), and cytochrome oxidase III (PfCoxIII), are encoded by the 6-kb mitochondrial genome, whereas other members of the ETC are encoded by nuclear genes and imported from the cytosol. To check for gene copy number increases as a possible mechanism of drug resistance, we used quantitative PCR (qPCR) to assess copy number variation (CNV) in the pfcoxI (mal_mito_1), pfcoxIII (mal_mito_2), and pfcytB mitochondrial genes, as well as pfdhod (PF3D7_0915000) of the nuclear genome. Results relative to the reference single copy seryl tRNA ligase gene (pfserRS; PF3D7_1216000) are presented in SI Appendix, Table S2.
Our overall copy number average for pfcytB, 38 ± 8, is somewhat higher than the previous estimate (37). A pfcytB copy number of 48 ± 10 was obtained for the ATQ-selected DA-3H6M133I clone, but this number was not significantly higher than in Dd2 (34 ± 6) by χ2 test (P = 0.87). Results from pfcoxI and pfcoxIII qPCR likewise showed little or no evidence for copy number increases after selection, with overall average copy numbers of 26 ± 6 and 33 ± 7, respectively.
The nuclear pfdhod gene is normally single-copy in P. falciparum but amplification has been found with increases in ATQ EC50 levels upon DSM1 selection (28, 35, 38). We tested for pfdhod CNV in the clones listed in Table 2. No clones showed more than a single copy of this gene (SI Appendix, Table S2), providing no evidence for a contribution of pfdhod amplification in the clones’ resistance to ATQ, CK-2-68, or RYL-552.
Docking Analysis of ATQ, CK-2-68, and RYL-522 Bound to PfCytB.
The mutations obtained with ATQ, CK-2-68, and RYL-552 selection map in or near the Qo pocket of PfCytB (Fig. 1B). Mutations V259L and F264L occur to either side of the catalytic PEWY motif (residues 260–263, P. falciparum numbering; SI Appendix, Fig. S1), which is essential for hydrogen transfer by complex III (39).
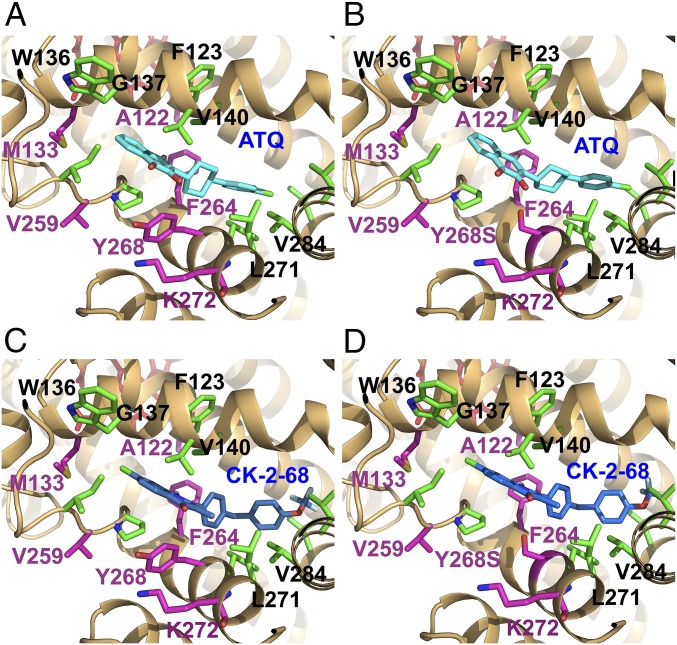
We used AutoDock 4.0 software to model predicted binding of ATQ, CK-2-68, and RYL-552 to the crystal structures of Saccharomyces cerevisiae CytB determined with bound ATQ [ATQ-ScCytB; Protein Data Bank (PDB) ID code 4PD4] or alone (ScCytB; PDB ID code 1KB9) (40, 41). ATQ docking into ATQ-ScCytB, computationally stripped of the drug, was blocked by a shifted position of the F123 residue in the Qo pocket. In contrast, AutoDock positioned ATQ in unbound ScCytB (PDB ID code 1KB9) in nearly the same position as reported from crystallized ATQ-ScCytB (SI Appendix, Fig. S2A) (41). Next, we remodeled the ScCytB Qo pocket to better resemble PfCytB by making amino acid replacements ScCytB L275F, M295V, F296L, and I299L (corresponding to the residues at PfCytB positions 264, 284, 285, and 288) in the unbound PDB ID code 1KB9 structure. ATQ readily inserted into this unbound, predicted PfCytB (Fig. 2A), with an estimated binding free energy of −11.3 kcal/mol, and a predicted inhibition constant (Ki) of 4.8 nM (Table 3). Much higher predicted Ki or complete loss of ATQ binding were computed for the Y268S, Y268C, and Y268N forms of the PfCytB Qo pocket, consistent with a strong binding contribution of Y268 contact with the naphthoquinone rings and with the high levels of ATQ resistance observed for P. falciparum parasites that carry PfCytB position 268 mutations (42, 43) (Fig. 2B and Table 3).
Fig. 2.
Docking models of ATQ and CK-2-68 in the PfCytB Qo pocket. Compounds were docked to the predicted PfCytB structure using remodeled ScCytB (PDB ID code 1KB9) and AutoDock 4.0 software. (A) ATQ (cyan) docks in the predicted wild-type PfCytB Qo pocket. (B) ATQ in the Y268S mutant pocket is shifted and turned slightly from its position in the Y268 structure. (C and D) CK-2-68 (blue) docks similarly in the predicted wild-type and mutant Y268S structures of predicted PfCytB. In all images, residues that have known mutations are highlighted in magenta and ATQ binding residues in green.
Table 3.
Docking results generated by AutoDock 4.0
| Protein | Ligand | ΔGbind* | Ki,† nM | EC50 (Ref.) |
| ScCytB | ATQ | −10.3 | 28 | 50 nM (12) |
| PfCytB | ATQ | −11.3 | 4.8 | 0.5–13 nM (24) |
| PfCytBY268S | ATQ | −9.4 | 134 | 4–29 μM (24) |
| PfCytBY268C | ATQ | DND‡ | ||
| PfCytBY268N | ATQ | DND‡ | ||
| PfCytB | CK-2-68 | −10.3 | 30 | 34–69 nM (12) |
| PfCytBY268S | CK-2-68 | −9.8 | 69 | 38 nM |
| PfCytB | RYL-552 | −10.0 | 49 | 17–50 nM |
| PfCytBY268S | RYL-552 | −9.6 | 101 | 75 nM |
ΔGbind, Predicted free energy of binding (kcal/mol).
Ki, Predicted inhibition constant.
DND, does not dock.
For CK-2-68, the best fit calculation oriented the quinolone ring similarly to that of ATQ’s naphthoquinone in wild-type predicted PfCytB, but with a relative shift to the Y268 phenolic ring of 1.7 Å while maintaining a 5.2 Å distance between the ring centers (Fig. 2C). Combined effects of this shift with the volume and electron withdrawing effect of the chloro substituent may reduce the effect of Y268 interaction. CK-2-68 contacts with other Qo pocket residues support binding with an estimated free energy of −10.3 kcal/mol and a predicted Ki of 29.6 nM (Table 3). In the PfCytB Y268S calculation, the best fit orientation of CK-2-68 remained similar, although with a somewhat less favorable free energy of binding (−9.8 kcal/mol) and predicted Ki (69 nM) relative to CK-2-68 in wild-type PfCytB (Fig. 2D and Table 3).
Docking calculations oriented the fluoro-substituted RYL-552 quinolone in a position similar to that of the ATQ naphthoquinone with the same ring-ring distance from Y268 (5.5 Å; SI Appendix, Fig. S2B). As with CK-2-68 binding, the RYL-552 interaction with Y268 was less than in ATQ binding (Table 3). Slightly less favorable free energy of binding and predicted Ki was calculated for RYL-552 binding to the PfCytB Y268S structure relative to wild type (−9.6 vs. −10.0 kcal/mol; 101 vs. 49 nM, Table 3 and SI Appendix, Fig. S2C).
In these calculations, van der Waals forces predominated in the estimated free energies of binding (SI Appendix, Table S3). In silico subtraction of 7-chloro and 5-fluoro substituents of CK-2-68 and RYL-552 showed marked reduction of binding to both Y268 and Y268S forms of PfCytB; however, there was little benefit from the in silico placement of either a 7-chloro or 5-fluoro substituent on the ATQ naphthoquinone ring (SI Appendix, Fig. S3).
Discussion
Our selection experiments with ETC inhibitors on two distinct P. falciparum clones, Dd2 and 106/1, identified a variety of drug-resistant mutants at or near the Qo site of PfCytB in complex III. PfCytB mutations after pressure with the two substituted quinolones, CK-2-68 and RYL-552, were unexpected, as both compounds have been developed and described as inhibitors of PfNDH2 (21, 22). PfCytB A122T was obtained by CK-2-68 selection, and PfCytB F264L, V259L, and A122T were obtained by RYL-552 selection, but no changes in PfNDH2 were detected in any of our experiments. These findings indicate that the Qo site of PfCytB is an important in vivo target of CK-2-68 and RYL-552, although our results do not exclude an activity of these quinolones on PfNDH2.
Pressure with ATQ, a substituted hydroxynaphthoquinone, yielded various PfCytB mutants with K272R, M133I (from two different codon changes), or Y268S. Different EC50 increases were observed with these mutations: 25× (M133I), 138× (K272R), and 6,150× (Y268S), consistent with the association of the latter mutation to highly resistant atovaquone-proguanil failures (24, 42, 44, 45). Assays with the quinolone compounds, however, showed heightened sensitivity of the M133I mutant to RYL-552 and of the Y268S mutant to CK-2-68, whereas the K272R mutant had heightened sensitivity to both drugs. Conversely, the A122T resistance mutants selected by CK-2-68 or by RYL-552 showed increased sensitivity to ATQ.
In this work, no evidence for amplification of pfcoxI, pfcoxIII, pfcytB, or pfdhod was found in ETC inhibitor-resistant mutants. Mitochondrial heteroplasmy and mutations that arise with replication of mtDNA during mitosis may cause clinical failures after ETC inhibitor treatment (28, 46). Reduced susceptibility of the V259L mutants to ATQ, CK-2-68, and RYL-552 suggests a general effect of this mutation on the binding ability of the PfCytB Qo site. PfCytB mutations may cause fitness reductions, e.g., by compromised phenotypes of respiration that could impair transmission through mosquitoes or otherwise affect stage developments in the parasite life cycle. It was reported that PfCytB M133I and V259L mutants, as well as P. berghei CytB (PbCytB) M133I, Y268C, and Y268N mutants, failed to transmit through Anopheles stephensi mosquitoes (47). However, more recent studies have demonstrated that PbCytB M133I parasites, resistant to both menoctone and atovaquone, transmit through mosquitos to mice and back again (48). Additionally, P. chabaudi CytB Y268N mutants are reported to transmit through A. stephensi to mice (49).
Effects from the mutations and results of our molecular docking studies provide insights into the differential activities of the substituted hydroxynaphthoquinone (ATQ) versus the halogenated quinolone compounds (CK-2-68, RYL-552) on mutants of PfCytB. The Y268S substitution greatly reduced inhibition of PfCytB by ATQ but not by CK-2-68 or RYL-552. π–π binding interactions between the aromatic rings of ATQ and Y268 may be disrupted by this mutation, in addition to effects of the Y → S change on pocket volume and hydrophobicity. Nevertheless, the computed Ki increase from the model (from 4.8 nM to 134 nM) does not reach the magnitude of the experimentally reported EC50 increases (from 0.5–13 nM to 4–29 µM; Table 3). This suggests additional factors involved in drug response, e.g., limits on compound access and/or uptake of high drug concentrations to the PfCytB target in parasitized erythrocytes.
Our observed EC50 and calculated Ki results using wild-type and Y268S parasites indicate that CK-2-68 and RYL-552 binding to PfCytB is much less dependent on van der Waals interactions with the Y268 ring than is ATQ. This finding may help to explain the lack of cross-resistance of these compounds with ATQ in the Y268S mutant. Of other mutations in the Qo pocket, ATQ was 138-fold less potent against our K272R mutant; arginine’s longer sidechain may contribute a steric displacement or an alteration of charge influence on the binding inhibition. M133I was 25-fold less susceptible to ATQ, a finding potentially attributable to a change in pocket volume. Both mutants are more susceptible to RYL-552, consistent with opposing effects on the binding of ATQ and RYL-552 by the substitutions of K272R or M133I.
Combination chemotherapy strategy is often based on drugs against different molecular targets; however, combination of different drugs against the same target has also been proposed as a strategy to counter resistance that might arise against either drug alone. For example, mutations of high-level pyrimethamine-resistant Plasmodium vivax strains are reported to confer enhanced sensitivity to WR99210, suggesting that pairings of alternative dihydrofolate reductase inhibitors may slow the spread of the resistant parasites (50). Differential resistance of P. falciparum dihydrofolate reductase– thymidylate synthase (PfDHFR–TS) mutants to pyrimethamine and cycloguanil has been described (51), and new searches for drug combinations have been boosted by knowledge of the conflicting mutations required to resist pyrimethamine or WR99210 (52). Moreover, certain compounds are known to have increased efficacy against drug-resistant parasites carrying mutations in the PfCRT chloroquine resistance transporter or in PfDHODH (53, 54). Taken together, these lines of evidence and the results from our present study suggest that mutations in drug binding sites frequently give rise to differential resistance and sensitivity. Effective treatments against drug-resistant malaria may therefore be possible with combinations of alternative ETC inhibitors that have distinct chemical structures. In the case of PfCytB mutants, successful development of these inhibitor combinations could help protect ATQ as one of the most important compounds in the antimalarial pharmacopeia.
Methods
P. falciparum Cultivation.
106/1 and Dd2 parasites were cultivated by standard methods (55, 56) with modifications. Cultures were maintained at 5% hematocrit with human erythrocytes (Virginia Blood Services) in complete medium composed of RPMI 1640 (Sigma-Aldrich), 1% Albumax II wt/vol (Life Technologies), 0.02 mg/mL−1 gentamycin solution (KD Medical), and 0.21% sodium bicarbonate. Incubation of the cultures was at 37 °C under a 90% nitrogen/5% carbon dioxide/5% oxygen atmosphere.
Selection of Resistant Parasites and Cloning by Limiting Dilution.
ATQ was purchased at Sigma Aldrich, CK-2-68 was synthesized by Asis Chem, and RYL-552 (22) was a kind gift from Yu Rao (School of Pharmaceutical Sciences, Tsinghua University, Beijing). The 10 mM stocks of these compounds were held at −20 °C in dimethyl sulfoxide (DMSO; Sigma-Aldrich). Sixty-day selections of resistant parasites were performed essentially as described (57, 58); for further details, see the SI Appendix. Expansion of the recovered parasites for cloning, cryopreservation, and DNA extraction was performed in selection medium. Cloning by limiting dilution (59) was performed without antimalarial pressure.
Dose–Response Assays.
DSM1 (5-Methyl-N-(2-naphthyl)[1,2,4] triazolo [1,5-a] pyrimidin-7-amine) (33) was obtained from the Malaria Research and Reference Reagent Resource Center (BEI Resource); CQ and AMA were purchased from Sigma-Aldrich. Stock solutions of the compounds were prepared at 10 mM in DMSO and held at −20 °C. Drug response profiles were assessed in vitro using the previously described 72-h malaria SYBR Green I assay (60, 61) (SI Appendix). Half-maximal effective concentrations were determined using the variable slope sigmoidal function feature from Prism version 7 (GraphPad Software Inc.). EC50 data were obtained from three to seven independent experiments.
PfCytB Modeling and Autodocking Calculations.
Structure modeling was performed by iterative threaded assembly refinement using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) (62–64). The structure was annotated and recolored using Chimera software (UCSF, www.rbvi.ucsf.edu/chimera) (65). Predicted transmembrane domains from the primary sequence were used for placement of the model relative to the inner mitochondrial membrane; Qo and Qi sites were identified as previously described (66). Automated docking simulations were conducted using the AutoDock 4.0 program suite (67). The yeast bc1 crystal structures, PDB ID codes 1KB9 and 4PD4, were used for ligand docking studies after replacement of four Saccharomyces cerevisiae with P. falciparum residues to model the PfCytB Qo pocket. Docking of compounds was performed using the Lamarckian genetic algorithm to minimize both steric clashes and docking energies, then to redock the molecules (further details provided in the SI Appendix). A ligand binding to the protein with the lowest root-mean-square deviation (rmsd) to the reference was considered to be docked.
Microsatellite Typing.
Supplementary Material
Acknowledgments
We thank Anna K. Crater, Jinfeng Shao, and Jeremy Sims for consultations on aromatic ring chemistry; Christine Figan for microsatellite typing advice; and Yu Rao, Maojun Yang, and Lubin Jiang for RYL-552 and discussions. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Disease, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804492115/-/DCSupplemental.
References
- 1.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 2.Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- 3.Fisher N, Antoine T, Ward SA, Biagini GA. Mitochondrial electron transport chain of Plasmodium falciparum. In: Hommel M, Kremsner PG, editors. Encyclopedia of Malaria. Springer; New York: 2014. pp. 1–14. [Google Scholar]
- 4.Mogi T, Kita K. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol Int. 2010;59:305–312. doi: 10.1016/j.parint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Coteron JM, et al. Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem. 2011;54:5540–5561. doi: 10.1021/jm200592f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavadai E, et al. Identification of new human malaria parasite Plasmodium falciparum dihydroorotate dehydrogenase inhibitors by pharmacophore and structure-based virtual screening. J Chem Inf Model. 2016;56:548–562. doi: 10.1021/acs.jcim.5b00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nixon GL, et al. Targeting the mitochondrial electron transport chain of Plasmodium falciparum: New strategies towards the development of improved antimalarials for the elimination era. Future Med Chem. 2013;5:1573–1591. doi: 10.4155/fmc.13.121. [DOI] [PubMed] [Google Scholar]
- 8.Stocks PA, et al. Novel inhibitors of the Plasmodium falciparum electron transport chain. Parasitology. 2014;141:50–65. doi: 10.1017/S0031182013001571. [DOI] [PubMed] [Google Scholar]
- 9.Fry M, Pudney M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4′-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80) Biochem Pharmacol. 1992;43:1545–1553. doi: 10.1016/0006-2952(92)90213-3. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava IK, Rottenberg H, Vaidya AB. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J Biol Chem. 1997;272:3961–3966. doi: 10.1074/jbc.272.7.3961. [DOI] [PubMed] [Google Scholar]
- 11.Gutteridge WE, Dave D, Richards WH. Conversion of dihydroorotate to orotate in parasitic protozoa. Biochim Biophys Acta. 1979;582:390–401. doi: 10.1016/0304-4165(79)90131-4. [DOI] [PubMed] [Google Scholar]
- 12.Kessl JJ, et al. Molecular basis for atovaquone binding to the cytochrome bc1 complex. J Biol Chem. 2003;278:31312–31318. doi: 10.1074/jbc.M304042200. [DOI] [PubMed] [Google Scholar]
- 13.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 14.Vallières C, et al. HDQ, a potent inhibitor of Plasmodium falciparum proliferation, binds to the quinone reduction site of the cytochrome bc1 complex. Antimicrob Agents Chemother. 2012;56:3739–3747. doi: 10.1128/AAC.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capper MJ, et al. Antimalarial 4(1H)-pyridones bind to the Qi site of cytochrome bc1. Proc Natl Acad Sci USA. 2015;112:755–760. doi: 10.1073/pnas.1416611112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukens AK, et al. Diversity-oriented synthesis probe targets Plasmodium falciparum cytochrome b ubiquinone reduction site and synergizes with oxidation site inhibitors. J Infect Dis. 2015;211:1097–1103. doi: 10.1093/infdis/jiu565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miley GP, et al. ELQ-300 prodrugs for enhanced delivery and single-dose cure of malaria. Antimicrob Agents Chemother. 2015;59:5555–5560. doi: 10.1128/AAC.01183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stickles AM, et al. Atovaquone and ELQ-300 combination therapy as a novel dual-site cytochrome bc1 inhibition strategy for malaria. Antimicrob Agents Chemother. 2016;60:4853–4859. doi: 10.1128/AAC.00791-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biagini GA, Viriyavejakul P, O’neill PM, Bray PG, Ward SA. Functional characterization and target validation of alternative complex I of Plasmodium falciparum mitochondria. Antimicrob Agents Chemother. 2006;50:1841–1851. doi: 10.1128/AAC.50.5.1841-1851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boysen KE, Matuschewski K. Arrested oocyst maturation in Plasmodium parasites lacking type II NADH:ubiquinone dehydrogenase. J Biol Chem. 2011;286:32661–32671. doi: 10.1074/jbc.M111.269399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biagini GA, et al. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc Natl Acad Sci USA. 2012;109:8298–8303. doi: 10.1073/pnas.1205651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, et al. Target elucidation by cocrystal structures of NADH-ubiquinone oxidoreductase of Plasmodium falciparum (PfNDH2) with small molecule to eliminate drug-resistant malaria. J Med Chem. 2017;60:1994–2005. doi: 10.1021/acs.jmedchem.6b01733. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999;33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 24.Korsinczky M, et al. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000;44:2100–2108. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava IK, Vaidya AB. A mechanism for the synergistic antimalarial action of atovaquone and proguanil. Antimicrob Agents Chemother. 1999;43:1334–1339. doi: 10.1128/aac.43.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plucinski MM, et al. Novel mutation in cytochrome B of Plasmodium falciparum in one of two atovaquone-proguanil treatment failures in travelers returning from same site in Nigeria. Open Forum Infect Dis. 2014;1:ofu059. doi: 10.1093/ofid/ofu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingasia LA, et al. Molecular characterization of the cytochrome b gene and in vitro atovaquone susceptibility of Plasmodium falciparum isolates from Kenya. Antimicrob Agents Chemother. 2015;59:1818–1821. doi: 10.1128/AAC.03956-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel SV, et al. 2017. Mitochondrial heteroplasmy is responsible for atovaquone drug resistance in Plasmodium falciparum. bioRxiv:10.1101/232033.
- 29.Stickles AM, et al. Inhibition of cytochrome bc1 as a strategy for single-dose, multi-stage antimalarial therapy. Am J Trop Med Hyg. 2015;92:1195–1201. doi: 10.4269/ajtmh.14-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huezo S. 2015. Investigation of dual stage acridones as a potent malaria treatment. MS thesis (Dominican University of California, San Rafael, CA)
- 31.Wellems TE, et al. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 32.Johnson DJ, et al. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol Cell. 2004;15:867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips MA, et al. Triazolopyrimidine-based dihydroorotate dehydrogenase inhibitors with potent and selective activity against the malaria parasite Plasmodium falciparum. J Med Chem. 2008;51:3649–3653. doi: 10.1021/jm8001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher N, et al. Cytochrome b mutation Y268S conferring atovaquone resistance phenotype in malaria parasite results in reduced parasite bc1 catalytic turnover and protein expression. J Biol Chem. 2012;287:9731–9741. doi: 10.1074/jbc.M111.324319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guler JL, White J, 3rd, Phillips MA, Rathod PK. Atovaquone tolerance in Plasmodium falciparum parasites selected for high-level resistance to a dihydroorotate dehydrogenase inhibitor. Antimicrob Agents Chemother. 2015;59:686–689. doi: 10.1128/AAC.02347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaidya AB, Arasu P. Tandemly arranged gene clusters of malarial parasites that are highly conserved and transcribed. Mol Biochem Parasitol. 1987;22:249–257. doi: 10.1016/0166-6851(87)90056-9. [DOI] [PubMed] [Google Scholar]
- 37.Preiser PR, et al. Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 1996;15:684–693. [PMC free article] [PubMed] [Google Scholar]
- 38.Ross LS, et al. In vitro resistance selections for Plasmodium falciparum dihydroorotate dehydrogenase inhibitors give mutants with multiple point mutations in the drug-binding site and altered growth. J Biol Chem. 2014;289:17980–17995. doi: 10.1074/jbc.M114.558353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mather MW, Henry KW, Vaidya AB. Mitochondrial drug targets in apicomplexan parasites. Curr Drug Targets. 2007;8:49–60. doi: 10.2174/138945007779315632. [DOI] [PubMed] [Google Scholar]
- 40.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 41.Birth D, Kao WC, Hunte C. Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action. Nat Commun. 2014;5:4029. doi: 10.1038/ncomms5029. [DOI] [PubMed] [Google Scholar]
- 42.Fivelman QL, Butcher GA, Adagu IS, Warhurst DC, Pasvol G. Malarone treatment failure and in vitro confirmation of resistance of Plasmodium falciparum isolate from Lagos, Nigeria. Malar J. 2002;1:1. doi: 10.1186/1475-2875-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musset L, Bouchaud O, Matheron S, Massias L, Le Bras J. Clinical atovaquone-proguanil resistance of Plasmodium falciparum associated with cytochrome b codon 268 mutations. Microbes Infect. 2006;8:2599–2604. doi: 10.1016/j.micinf.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Looareesuwan S, et al. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am J Trop Med Hyg. 1996;54:62–66. doi: 10.4269/ajtmh.1996.54.62. [DOI] [PubMed] [Google Scholar]
- 45.Musset L, et al. Apparent absence of atovaquone/proguanil resistance in 477 Plasmodium falciparum isolates from untreated French travellers. J Antimicrob Chemother. 2006;57:110–115. doi: 10.1093/jac/dki420. [DOI] [PubMed] [Google Scholar]
- 46.Cottrell G, Musset L, Hubert V, Le Bras J, Clain J. Atovaquone-Proguanil Treatment Failure Study Group Emergence of resistance to atovaquone-proguanil in malaria parasites: Insights from computational modeling and clinical case reports. Antimicrob Agents Chemother. 2014;58:4504–4514. doi: 10.1128/AAC.02550-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodman CD, et al. Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science. 2016;352:349–353. doi: 10.1126/science.aad9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blake LD, et al. Menoctone resistance in malaria parasites is conferred by M133I mutations in cytochrome b that are transmissible through mosquitoes. Antimicrob Agents Chemother. 2017;61:e00689-17. doi: 10.1128/AAC.00689-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonso A, et al. Plasmodium chabaudi chabaudi malaria parasites can develop stable resistance to atovaquone with a mutation in the cytochrome b gene. Malar J. 2010;9:135. doi: 10.1186/1475-2875-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hastings MD, Sibley CH. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc Natl Acad Sci USA. 2002;99:13137–13141. doi: 10.1073/pnas.182295999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Japrung D, Leartsakulpanich U, Chusacultanachai S, Yuthavong Y. Conflicting requirements of Plasmodium falciparum dihydrofolate reductase mutations conferring resistance to pyrimethamine-WR99210 combination. Antimicrob Agents Chemother. 2007;51:4356–4360. doi: 10.1128/AAC.00577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lukens AK, et al. Harnessing evolutionary fitness in Plasmodium falciparum for drug discovery and suppressing resistance. Proc Natl Acad Sci USA. 2014;111:799–804. doi: 10.1073/pnas.1320886110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 56.Haynes JD, Diggs CL, Hines FA, Desjardins RE. Culture of human malaria parasites Plasmodium falciparum. Nature. 1976;263:767–769. doi: 10.1038/263767a0. [DOI] [PubMed] [Google Scholar]
- 57.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cooper RA, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 59.Rosario V. Cloning of naturally occurring mixed infections of malaria parasites. Science. 1981;212:1037–1038. doi: 10.1126/science.7015505. [DOI] [PubMed] [Google Scholar]
- 60.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooper RA, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol. 2007;63:270–282. doi: 10.1111/j.1365-2958.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, et al. The I-TASSER suite: Protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Vaidya AB, Lashgari MS, Pologe LG, Morrisey J. Structural features of Plasmodium cytochrome b that may underlie susceptibility to 8-aminoquinolines and hydroxynaphthoquinones. Mol Biochem Parasitol. 1993;58:33–42. doi: 10.1016/0166-6851(93)90088-f. [DOI] [PubMed] [Google Scholar]
- 67.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.