Significance
Retrotransposons replicate through RNA intermediates that are reverse transcribed and inserted at new genomic locations. LINE-1 (L1) elements constitute ∼17% of the human genome, making them the most successful retrotransposons in the human genome by mass. The activity of L1s was shown first in the germline or during early embryogenesis. More recent studies demonstrate a wider prevalence of L1 expression in somatic cells including neurons, aging cells, and different types of cancer. In this study, we developed the MapRRCon pipeline and performed a comprehensive computational analysis of L1 transcriptional regulators using ENCODE ChIP-seq datasets. We revealed the binding of various transcription factors, including Myc and CTCF, to the 5′ UTR promoter of the youngest human L1 family (L1HS) and their potential functional impact on L1HS expression.
Keywords: LINE-1, Myc, CTCF, ChIP-seq, ENCODE
Abstract
Transposable elements (TEs) represent a substantial fraction of many eukaryotic genomes, and transcriptional regulation of these factors is important to determine TE activities in human cells. However, due to the repetitive nature of TEs, identifying transcription factor (TF)-binding sites from ChIP-sequencing (ChIP-seq) datasets is challenging. Current algorithms are focused on subtle differences between TE copies and thus bias the analysis to relatively old and inactive TEs. Here we describe an approach termed “MapRRCon” (mapping repeat reads to a consensus) which allows us to identify proteins binding to TE DNA sequences by mapping ChIP-seq reads to the TE consensus sequence after whole-genome alignment. Although this method does not assign binding sites to individual insertions in the genome, it provides a landscape of interacting TFs by capturing factors that bind to TEs under various conditions. We applied this method to screen TFs’ interaction with L1 in human cells/tissues using ENCODE ChIP-seq datasets and identified 178 of the 512 TFs tested as bound to L1 in at least one biological condition with most of them (138) localized to the promoter. Among these L1-binding factors, we focused on Myc and CTCF, as they play important roles in cancer progression and 3D chromatin structure formation. Furthermore, we explored the transcriptomes of The Cancer Genome Atlas breast and ovarian tumor samples in which a consistent anti-/correlation between L1 and Myc/CTCF expression was observed, suggesting that these two factors may play roles in regulating L1 transcription during the development of such tumors.
Much of the human genome is derived from retrotransposons, self-propagating sequences resident within our genome. Moreover, retrotransposons continually engage in complex host–parasite relationships during evolution (1–5). In the current human genome assembly, about 45% of our total DNA has clear-cut homology to consensus sequences of retroelements (6–8), whereas other studies suggest the proportion of the human genome derived from repeats is over 75% (7). Three families of retrotransposons are still highly active today in the human genome: LINE-1 (L1), Alu, and SVA. All these elements require a combination of host factors and ORF1 and ORF2 proteins encoded by the L1 element to retrotranspose into the genome (5); thus, L1 elements represent the only autonomous retroelement in the human genome. Because L1 encodes enzymatic proteins essential for the formation of new insertions, studying the cellular regulation of autonomous L1 is critical to better understand the transposons’ impact on the human genome and transcriptome.
A full-length L1 element is about 6 kb long and consists of a 5′ UTR/promoter, two ORFs (ORF1 and ORF2), and a 3′ UTR containing a poly(A) tail. Following transcription by RNA polymerase II, translation produces ORF1 and ORF2 proteins (ORF1p and ORF2p). ORF1p is required for L1 retrotransposition and functions as a chaperone protein or an ssRNA-binding protein (reviewed in ref. 9). ORF2p has two recognized enzymatic domains, an endonuclease domain (10) and a reverse transcriptase domain (11), and both domains play important roles during the actual insertion step called “target-primed reverse transcription” (TPRT) (12, 13). Because TPRT happens directly by cleavage and primer extension of genomic DNA targets, TPRT initiates from the 3′ end of the L1 RNA and often fails to reach the 5′ end. As a result most existing L1 insertions are 5′-truncated and therefore lack a promoter and are transcriptionally inactive.
Importantly, the 5′ UTR promoter of L1 has unique features. In addition to serving as the 5′ UTR, this “downstream” sequence contains the L1 promoter in its entirety. That is, the L1 promoter is unique in that all promoter elements are downstream of the transcription start site (14–16). Adding further complexity, the promoter actively promotes transcription of both the sense and antisense strands and thus produces a series of mRNAs that read into adjacent host DNA (17) and even produces an antisense strand-encoded “ORF0” protein (17, 18). Because of the unique architecture of its 5′ UTR promoter, L1 brings along its own package of regulatory sequences when it retrotransposes into a new genomic location. Thus, we expect that all L1 transcriptional regulators will bind 5′ regulatory/transcribed sequences. This allows the identification of L1-interacting factors by screening for key binders of the 5′ UTR promoter without mapping to individual L1 copies.
Despite the importance of studying the transcriptional regulation of L1 and predicting binding sites for transcription factors (TFs) (19), only a few previous studies have identified critical TFs binding to the L1 5′ UTR promoter. These include YY1 (20, 21), RUNX3 (22), p53 (23, 24), SRY (25), MeCP2, Oct4, Sox2, Nanog, and KLF4 (26–28); all have been demonstrated to regulate L1 transcription in specific human cells types, such as Sp1 and Sp3 in rats (29 and reviewed in ref. 30) and Sin3A in mouse ES cells (31). Evolutional analysis reveals conservation of TF-binding sites among human-specific L1s, although the mutation rate of the L1 5′ UTR promoter is higher than that of the L1 ORFs (32), probably due, at least in part, to the enrichment of CpG dinucleotides. This suggests that possible core regulatory network/features may exist that control L1 activity in diverse cell types.
Because of L1’s ability to induce genome instability and mutagenic outcomes, its activity is generally suppressed somatically (33, 34), probably via extensive cytosine methylation at CpGs (35). However, L1 expression is highly up-regulated in cancer cells, in line with the common observation of global hypomethylation in tumors (36, 37). Despite the correlation of L1 activity with cancer progression observed in most tumor types (37), L1’s role in cancer initiation and progression remains unclear. In addition, because of the variability of different cancer cell lines, it is difficult to determine which common factors/pathways drive L1 activity. Thus, a comprehensive picture of the control of L1 expression is needed to define common or unique regulators in different cell types. In addition to cancer cells, human ES cells (hESCs) exhibit a permissive environment for L1 retrotransposition (38–42). Comparison of the L1 regulatory network in hESCs and cancer cells might help further identify essential regulators, since cancer cells, which also tend to be somewhat “dedifferentiated,” may exploit similar pathways to activate L1 expression.
The ENCODE project (encodeproject.org/ENCODE/) (43) has produced numerous ChIP-sequencing (ChIP-seq) datasets that map the genomic locations of TF binding and histone modifications in various types of tissues and cell lines. In the standard ChIP-seq pipeline, transposable element (TE)-associated reads are discarded when aligning at multiple locations and thus cannot be unambiguously assigned. For example, previous studies of TF-binding profiles for human endogenous retroviruses excluded multiple aligned reads (44). However, those reads are extremely valuable in understanding retrotransposon-interacting TFs, and a TE-savvy method is required to analyze the “junk” repetitive (low mappability) reads from deep-sequencing datasets.
Here, we generated an L1-interacting TF/histone mark landscape by analyzing the entire human ENCODE ChIP-seq database. We developed a method, “MapRRCon” (mapping repeat reads to a consensus), to specifically identify TFs binding to L1 sequences and in particular to the L1 5′ UTR promoter. We identified a remarkably long and diverse list of TFs, possibly reflecting a general opening of L1 chromatin in certain cell types and, consequently, promiscuous and nonphysiologic binding of many factors to the 5′ UTR promoter (45–47). However, we also identified a set of TFs that are activated in many cell types in which L1 is transcribed and which are known to interact in other contexts. Among the list of identified binding proteins, the oncoprotein Myc was a major binder in various cell types, and, importantly, Myc RNA levels were significantly anticorrelated with those of L1 in breast and ovarian tumors. Additionally, we observed that CTCF binds to the 5′ UTR promoter and 3′ UTR of L1s, colocalizing with Myc and RNA polymerase II. siRNA-knockdown experiments further supported the involvement of both Myc and CTCF in regulating L1 transcription. This landscape provides a comprehensive resource of L1 regulators in various cell types and identifies key components of the remodeled L1 regulatory network in cancer cells.
Results
Exploring Binding Factors to L1HS Using MapRRCon.
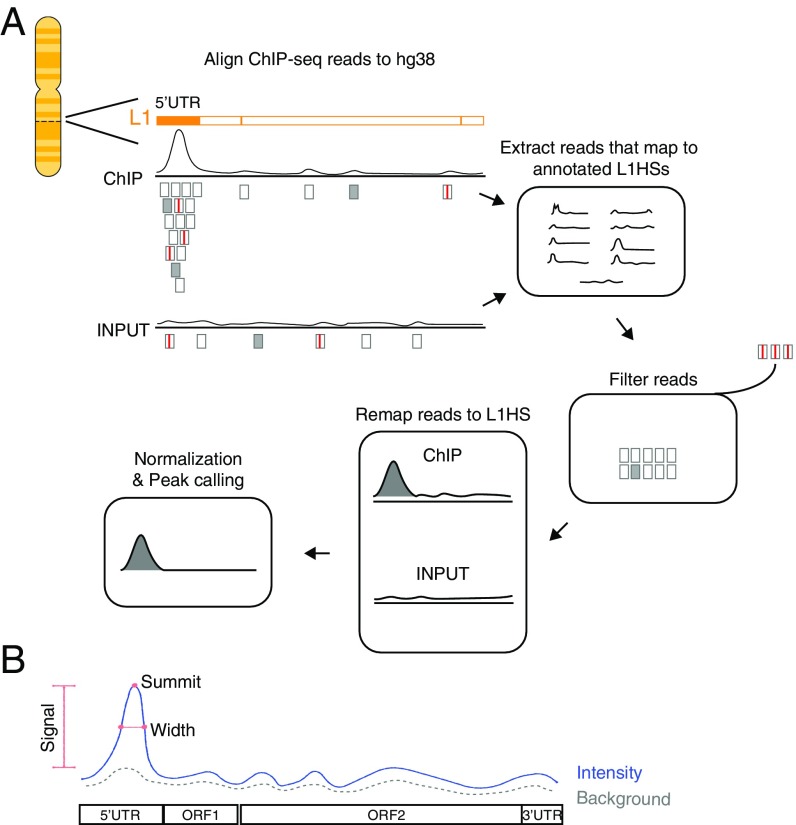
To optimize alignment of ChIP-seq reads to L1HS consensus sequence, we developed MapRRCon, which aligns ChIP-seq datasets to a predetermined unmasked reference genome and extracts information of the target binding at repetitive elements. In this study we specifically focused on factors binding to human L1 sequences. We first aligned ChIP-seq data to the human reference sequence hg38 containing annotated L1HS locations. This step not only assigns uniquely mapped reads to their genomic locations but also randomly distributes reads with multiple genomic hits. In the case of the youngest human LINE-1 subfamily (L1HS) there are 1,620 annotated sites in the reference genome, and most of the reads, being repetitive, are not uniquely mapped. The reads mapping to these 1,620 sites are extracted based on their genomic locations (unique or randomly assigned by the alignment algorithm) and filtered to eliminate possible contamination by other L1 subfamilies or similar sequences. We removed reads with the following features: more than three mismatches, any indels, or partial alignments (soft clipping) (boxes with red lines in Fig. 1A). After extracting and filtering, these L1 reads were subsequently mapped to the L1HS consensus sequence to generate a coverage profile. For each TF, we generate two coverage profiles from both the Input and ChIP datasets and performed median-based normalization (Fig. 1A). ChIP-seq peaks within the L1HS consensus sequence are called using a signal-processing algorithm developed in-house, and a true peak is defined as being present in the normalized data (Fig. 1B). This peak-finding method was benchmarked against manual peak-picking and was highly accurate and robust in detecting peaks from short-sequence datasets (SI Appendix, Fig. S1).
Fig. 1.
The pipeline of MapRRCon. (A) In this pipeline, ChIP-seq data are first aligned to the human reference sequence hg38. Both unique reads (filled gray boxes) and multiply aligned reads (hollow gray boxes) are then extracted and mapped to the 1,620 annotated L1HS sites based on their genome coordinates. We exclude reads with partial alignment (soft clipping), more than three mismatches, or any indels (boxes with red lines). Filtered reads are subsequently mapped to the L1HS consensus sequence to obtain compiled reads. Finally, we generate coverage profiles for both ChIP and Input data and then perform median-based normalization (Methods). The normalized data are used for peak calling. (B) We developed a peak-calling algorithm that is suitable for short sequences such as L1s. Peaks are detected by applying a smoothing filter and finding positions where the smoothed signal has maxima. The peaks are filtered using two thresholds on the original signal: signal intensity (blue line) minus background intensity (dotted gray line) larger than 1 and an rmsd ratio between signal and background larger than 1.3. The width of the peak (red line) is defined by the location where the signal drops to 25% of its maximum.
In Silico Screening of TF Factors That Interact with LINE-1 Using MapRRCon.
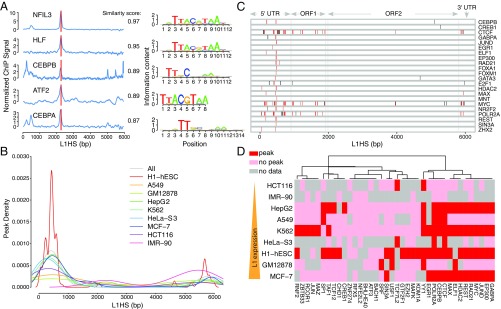
Using MapRRCon, we screened the entire human ENCODE database for factors that interact with L1 and identified dozens of TFs and chromatin marks that associate with the L1 sequences (Fig. 2). Naturally, the binding patterns obtained reflect binding to an unknown subset of element copies, since the sequences are present at multiple genomic locations. Thus, an important limitation of our study is that we are unable to state exactly which genomic L1 copies are bound. Nevertheless, this analysis provides a wealth of other types of information. We screened 512 TFs in 118 biosamples spanning the entire human ENCODE ChIP-seq database for TFs as of November 2017. Remarkably, 165 of these TFs (32% of the TFs tested) showed clear evidence of sequence-specific binding of L1. This should be considered a minimum, as many antibodies were tested in a limited number of biosamples, and expansion to other cell lines, tissues, or tumors will likely reveal additional binders. Although the coverage of our analysis is biased toward certain factors that have been more extensively evaluated using ChIP under various conditions—while others have limited datasets and some are absent altogether because no antibody against them exists—these data provide interesting maps of TF and chromatin mark landscapes across the L1 population. The majority of these binding profiles map to the 5′ UTR promoter of L1, as is consistent with two possible interpretations: (i) these might represent specific transcriptional regulators of L1 or (ii), since L1 is highly expressed in stem and cancer cell samples, some and perhaps most of these may represent opportunistic but not necessarily biologically relevant bindings reflected in ChIP-seq studies (45–47). We also identified 26 TFs that do not bind to the 5′ UTR promoter of L1 but, interestingly, showed highly specific peaks in the coding regions (ORF1 and ORF2) of the L1 sequence. The identification of these peaks and factors, which would not have been identified in classical reporter assays developed to measure L1 promoter activities (i.e., the L1 5′ UTR promoter driving the expression of a reporter gene), indicates a strength of MapRRCon compared with these approaches: its agnosticism toward effects on expression. Among the internal binders, we found five basic leucine zipper (bZIP) TFs that bind to the same location on the L1 sequence—the beginning of the ORF2 gene. Interestingly, two of these bZIPs also bound to the 5′ UTR promoter, suggesting a possible distinct role for internal binding (Fig. 2A). The internal colocalization can be explained by bZIP-binding motifs sharing high similarity with each other and by these TFs often forming homo- or heterodimers (48) when associated with DNA. Therefore, we cannot rule out the possibility that some of them are indirectly recruited to the DNA via protein–protein interactions. Although it is difficult to interpret the function of the binding of bZIPs to L1s, our discovery of binding factors from the same family showing a set of overlapping peaks suggests that our screen is robust and comprehensive.
Fig. 2.
Landscape of L1-interacting TFs. (A) We identified a group of TFs (bZIPs) enriched at the beginning of ORF2. Blue lines show the ChIP profiles of each TF in HepG2 cells, and the vertical red bar indicates the location of known motifs; logos are shown on the Right. (B) Peak enrichment of each cell line is made by combining all the TF peaks called from each dataset of that cell line, and the distribution is generated using the kernel function. (C) All the binding TFs in MCF-7 cells are plotted along the L1HS DNA sequence. Red bars indicate a peak location that has been identified repeatedly for multiple TFs in our analysis; black bars indicate unique peak locations for a specific TF. (D) Heatmap of selected TFs in nine commonly used ENCODE cell lines. The cell lines are sorted based on their relative L1 expression level from RNA-seq, and the color indicates whether each TF has binding peaks in the L1 promoter. The TFs are ordered by calculating the Euclidean distance and are hierarchically clustered using Ward’s method. It is clear that TF binding to the 5′ UTR promoter is not highly correlated with the L1 expression level.
In general, we found that more TFs bind L1 sequences in hESCs than in tissues and primary cells. Also, cancer lines in general showed high levels of TF binding to L1, consistent with recent reports showing very widespread expression of ORF1p in cancer cells and tumors (37, 49). However, we did observe considerable variation in TF binding among the cancer lines screened, suggesting substantial binding heterogeneity (SI Appendix, Fig. S2A and Dataset S1).
We found that 83.6% (138 TFs; 27% of the screened TFs) of the TFs that bind L1 sequences (165 TFs; 32.2% of the screened TFs) bind the L1 5′ UTR promoter in at least one biosample. We next examined the distribution of all TFs in each cell line and found that the binding was highly clustered within a promoter subregion around position 450 in the 5′ UTR promoter of the L1HS consensus sequence; this enrichment was even stronger in H1 hESCs than in other cell lines (Fig. 2B). The clustering of TFs at the L1 5′ UTR promoter could also be observed when aligning binding peaks of individual TFs; for example, in MCF-7 cells the peaks around position 450 could be seen for nearly all the TFs that bound L1, although peaks for those TFs also existed in other regions (Fig. 2C). This finding is consistent with previous research showing that nucleotide positions 390–526 of the L1 5′ UTR promoter are critical for effective L1 transcription (16).
Among the commonly used ENCODE cell lines, a few (MCF-7, K562, HepG2, and H1-hESCs) showed TF enrichment at the L1 5′ UTR promoter compared with the other cell lines (Fig. 2D). We first hypothesized that this may reflect the open chromatin state at the L1 5′ UTR promoter in these cell lines that allowed higher accessibility for TFs. To test this hypothesis, we quantified L1 expression levels in a few of the ENCODE cell lines that had the highest amount of ChIP-seq data, using ENCODE RNA-sequencing (RNA-seq) datasets and ranking them based on L1 expression (Fig. 2D). L1 expression did not correlate with the number of TFs bound to its 5′ UTR promoter; for instance, MCF-7 cells showed the highest L1 expression among these cell lines but had a low number of TF peaks compared with H1-hESCs, K562, and HepG2 cells; GM12878 cells also exhibited less TF binding although L1 expression was high (Fig. 2D and SI Appendix, Fig. S3A). By comparing the binding profiles with the individual TF genes’ expression, we excluded the possibility that the TF binding on L1 was driven by the expression level of the TF under consideration (SI Appendix, Fig. S3B).
Motif Analysis of TF Binding to L1.
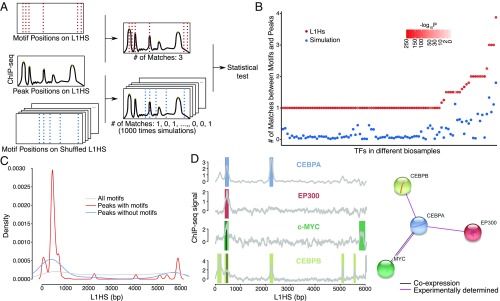
To uniquely identify TF-binding sites, we searched for DNA motifs. We therefore analyzed the overlaps between TF peaks and the appropriate TF DNA motifs. We used a motif database which combines a few of the known databases, including Jaspar and TRANSFAC(R), as well as motifs newly discovered from a subset of ENCODE ChIP-seq datasets (50). This database analyzed the same sets of ChIP-seq experiments used for MapRRCon but uses a different set of reads [unique reads for the Kheradpour and Kellis database (50); multiply aligned reads for MapRRCon]. Use of the Kheradpour and Kellis database provided extra power for our motif analysis (Discussion). We asked whether the colocalization of a TF peak and its DNA motif was significantly different from random. Randomized sequences were obtained by shuffling L1HS 1,000 times and calculating the number of matches between the TF signal profile and motifs found in each simulated sequence (Fig. 3A, blue dotted lines). After comparing the number of true matches (Fig. 3A, red dotted lines) with simulated matches, we found that simulated values were significantly lower in all samples (Fig. 3B). This simulation supports the ability of our pipeline to robustly identify TF peaks that align perfectly with expected DNA motifs on L1HS sequence.
Fig. 3.
Motifs underlying TF-binding peaks. (A) To test whether the match of motif and TF peaks from ChIP-seq is significantly different from random, we performed simulation by shuffling the L1HS sequence 1,000 times and looked for motifs in those shuffled sequences. The number of matches between motif and peak location was counted and compared with the number of true matches (number of matches between the unshuffled L1HS sequence and ChIP-seq peaks). P values were calculated using the Wilcoxon one-sample signed rank test. (B) The number of true matches (red dots) and simulated matches (blue dots) are plotted for each TF–cell line pair. The simulated matches are averaged, and P values are indicated by the intensity of redness. (C) The distribution of motifs and peaks is plotted along the L1HS sequence. The ChIP-seq peaks are categorized into two groups based on whether the specific TF motifs can be found at the peak locations. (D, Left) A small network of four physically interacting TFs colocalizes at the L1 promoter. Two motifs, c-Myc and CEBPB (black lines), are found under peaks. Color bars indicate the peak locations for each TF, and gray lines show the ChIP profiles. (Right) The scheme was generated by STRING database v10.5.
We then classified peaks into two groups: (i) TFs with peaks that colocalize with their own motifs and (ii) TFs with peaks that do not colocalize with their own DNA motifs and therefore cannot be explained simply by direct binding. For this latter class, there might be indirect binding. Alternatively, the specific TFs may lack known DNA motifs (Fig. 3C). We plotted the distribution of peaks of these two groups (group 1 in red and group 2 in blue in Fig. 3C) against the distribution of all TF motifs found in L1HS (gray line in Fig. 3C). The TFs in group 1 were highly promoter-enriched, as expected from our previous analysis (Fig. 2). The TFs in group 2 also clustered mainly in the promoter, suggesting that these TFs were recruited to the L1 promoter by features other than the presence of their motifs. The distribution of all motifs identified on L1HS further validated this hypothesis, as we did not observe any motif enrichment at the same region (Fig. 3C, gray line). Based on this analysis, we could not rule out the possibility that some of the binding motifs may not exist in the database or that other nonmotif features might contribute to the binding of TFs to L1. We analyzed the TF protein interactome to identify possible corecruitment not explained by the presence of DNA motifs in the L1 sequence. An example of this corecruitment was a small interacting cluster centered on CEBPA protein known to interact with CEBPB, EP300, and Myc; remarkably, these four TFs occupied the same L1 promoter subregion in HepG2 cells (Fig. 3D). As we found motifs for Myc and CEBPB but not for CEBPA or EP300 at the peak location, the recruitment of CEBPA and EP300 could potentially be explained by physical interactions with Myc or CEBPB. Furthermore, there is evidence that CEBPA and CEBPB can form heterodimers (51), supporting the idea of indirect binding of the identified L1 TFs to DNA.
Myc Represses L1 Transcriptional Activity.
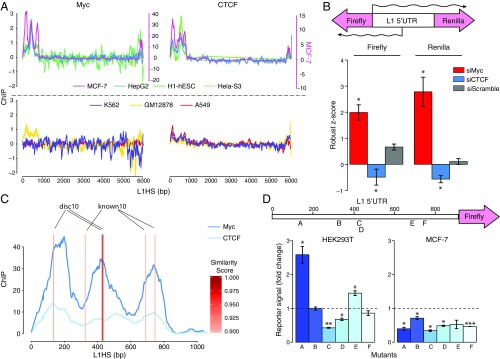
Among the factors bound to the 5′ UTR promoter of L1s, we focused on Myc oncoprotein, as it has been shown to be involved in various cellular processes including growth control, differentiation, and apoptosis and its overexpression is often observed in tumors (52–55). We found that Myc preferentially occupied the L1 promoter in several cell lines. However, Myc binding was cell-type specific, as Myc was expressed but did not bind L1 promoters in certain cell lines. Although a few cell lines showed Myc binding at position 450, in MCF-7 cells, a breast cancer cell line, we identified two additional strong binding peaks at positions 150 and 700 (Fig. 4A). To test whether Myc binding has any functional impact on L1 transcription, we used siRNA to knock down Myc in HEK293 cells containing an integrated luciferase reporter. The reporter was designed to have the L1 promoter driving Renilla luciferase and firefly luciferase from its sense and antisense promoters, respectively (Fig. 4B). As the knockdown experiments were performed in a high-throughput manner, we used the robust z-score of a plate-wide median as a measurement, and the normalization was done within each plate (Methods). We found that knocking down Myc in HEK293 cells increased the promoter activity of the L1 5′ UTR promoter in both orientations (Fig. 4B). This result suggests that Myc acts as a transcriptional repressor at L1 promoters in these cells.
Fig. 4.
c-Myc and CTCF colocalize at the L1 5′ UTR to regulate L1 promoter activity. (A) The coverage of c-Myc and CTCF ChIP-seq signals is plotted along L1HS in seven cell lines. The four cell lines in the upper plots show binding peaks for both TFs, whereas the three cell lines in the lower plots do not. (B, Upper) The diagram illustrates the reporter construct, which is a L1 promoter sequence attached to a luciferase reporter at both ends. Wavy lines indicate sense and antisense transcripts. Renilla luciferase measures the forward promoter activity, and firefly luciferase measures the reverse promoter activity. (Lower) The luciferase signal is normalized to the plate median (Methods) for each knockdown. *P > 0.01, **P > 0.001, ***P ≤ 0.001. (C) The ChIP-seq signal of c-Myc and CTCF is plotted on the L1HS along with the locations of sequences that are highly similar to their identified motifs. The similarity is indicated by color gradient; the sequences that are similar to c-Myc discovery motif 10 (disc10) and known motif 10 (known10) are marked (see text). (D) L1 5′ UTR promoter activities in different c-Myc motif mutants measured by reporter assay. (Upper) The reporter construct in which a L1 5′ UTR promoter drives firefly luciferase is shown. The letters A–F indicate the six motifs we identified in the L1 5′ UTR promoter and the locations of their matched sequences. (Lower) The bar graph shows the fold change of reporter signals normalized to wild type (dotted line). *P < 0.05, **P ≤ 0.05, ***P ≤ 0.001.
We asked whether the effect of Myc binding on the L1 promoters was due to the presence of its DNA motifs at the promoters. After scanning the L1HS consensus sequence, we found six putative Myc-binding motifs located within the 5′ UTR promoter region, corresponding to two known Myc motifs present in the Kheradpour and Kellis database (“discovery 10” and “known 10”) (Discussion). Interestingly, five of these six motifs are present in very close proximity to the Myc-binding peak summits we defined from MapRRCon analysis in MCF-7 cells (Fig. 4C). To interrogate the contribution of these six motifs to regulation of L1 transcription, we generated mutations that had low similarity scores to the identified Myc motifs and did not create new binding motifs in the surrounding sequences. All six mutated sequences were cloned into the L1 5′ UTR promoter upstream of the firefly luciferase reporter gene. Renilla luciferase driven by a constitutive promoter was also transfected, and the Renilla signals were used to normalize transfection efficiency. We measured the firefly signals after 48 h of transfection and normalized those signals to the Renilla signals. We observed various effects resulting from different mutations of the Myc motifs. Although no evidence of complete disruption of Myc binding is seen, consistent with the knockdown results, we observed elevated promoter activity in motif A and E mutants; contrarily, motif C and D mutants showed decreased signals, whereas mutations in motifs B and F did not alter L1 promoter activity (Fig. 4D). This result suggests that the multiple Myc-binding sites in the L1 5′ UTR promoter might cooperate to form a complex regulatory network. In addition, because of the overall enrichment of TFs we observed at the promoter (Fig. 2 B and C), mutating Myc-binding motifs may also disrupt or create sites for other TFs, complicating interpretation (Discussion). To add another level of complexity, we conducted similar experiments in the MCF-7 cell line. Strikingly, all the mutants showed a decreased level of L1 5′ UTR promoter-mediated transcription, suggesting cell-type–specific regulation of L1 expression. Considering global expression-level differences between the different cell lines, it is possible that mutating the same sequences in the L1 5′ UTR promoter will have opposite effects in the two cell lines due to the binding of different TFs and TF interactors.
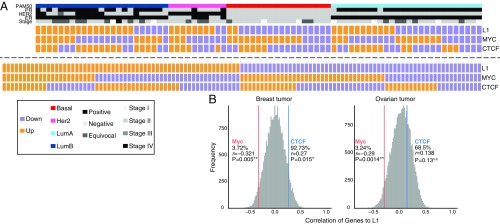
Based on the findings described above, we hypothesized that the binding of Myc at the L1 5′ UTR promoter overall represses L1 expression. To test whether this repression was also observed in patient samples, we examined The Cancer Genome Atlas (TCGA) dataset for the relationship between the expression of Myc and L1 in tumors. We selected 77 breast and 127 ovarian tumor samples from TCGA and analyzed L1 transcription from RNA-seq data. We applied the principle of MapRRCon analysis (alignment to the genome followed by alignment to L1HS consensus) to RNA-seq reads to filter out DNA contaminations. We first aligned RNA-seq reads to hg38 using STAR (56) and extracted reads aligned to 1,620 annotated L1Hs locations. These reads were then realigned to the L1HS consensus sequence using stringent alignment criteria (Methods). We were able to detect L1 transcripts in almost all breast and ovarian tumors. We excluded possible genomic contamination from truncated L1s (the majority of the L1 sequences present in the genome) and observed no 3′ bias along L1HS reads (SI Appendix, Fig. S4). Consistent with the knockdown data of Myc, we found that MYC expression level was significantly anticorrelated with L1 expression in both breast and ovarian tumors (Fig. 5). In addition, distinct L1 regulation machinery may exist in different breast cancer subtypes, as more L1 transcripts were detected in the Her2 subtype and few were detected in the basal subtype (Fig. 5A). This result suggests that Myc may be a major regulator of L1 transcription in cancer development.
Fig. 5.
The MYC and L1 expression levels are anticorrelated among different breast tumors. (A) We analyzed the RNA-seq data of 77 breast tumors (Upper) and 127 ovarian tumors (Lower) and compared the expression of L1 with that of MYC or CTCF. The heatmap is shown as binary colors, which indicate whether the expression level is above or below the mean of all samples within the same cancer type. The heatmap of breast tumors is clustered by four subtypes; their diagnostic markers and stages are also shown. (B) Spearman’s correlation coefficient (r) is calculated between each TF and L1 (gray bars) to form a background distribution; red and blue lines indicate the correlation of MYC and CTCF, respectively, compared with the overall distribution. P values are calculated by a correlation test.
CTCF Colocalizes with Myc on the L1 Promoter and 3′ UTR.
We found that CTCF protein colocalizes with Myc in multiple cell lines including MCF-7, HepG2, H1-hESC, and HeLa-S3 cells. MCF-7 cells, in particular, showed three distinct binding peaks for both Myc and CTCF on the L1 5′ UTR promoter (Fig. 4A). Contrary to the effect of Myc knockdown, depletion of CTCF by siRNA treatment reduced L1 promoter activity (Fig. 4B). The same observation was obtained when analyzing the expression of CTCF and L1 proteins in TCGA patient data (CTCF exhibited a positive correlation with L1, although the correlation was not as significant as the Myc and L1 anticorrelation) (Fig. 5). This positive effect on expression was in line with studies showing that CTCF sites have been identified on the promoter of the MYC gene and that CTCF acts as a repressor for MYC expression (57, 58). Thus, knocking down CTCF may be a secondary consequence of lower MYC expression, which in turn increases L1 transcription (Fig. 4B). However, transcriptional control of MYC by CTCF would not explain the observed colocalization at L1 promoters. As the well-known function of CTCF is to act as an insulator protein that blocks the interaction of enhancers and promoters (59–62), it is possible that the binding to L1 also creates complex DNA structures, either inter- or intramolecularly. Indeed, a previous study analyzing topologically associated domains (TADs) showed that MCF-7 TAD boundaries are enriched for several oncoproteins, including Myc (63). This is consistent with our observations about the specific recruitment of CTCF and Myc on L1 and suggests a possible functional interaction between Myc and CTCF. In addition, Cohesin subunit Rad21, a protein subunit that works together with CTCF (64, 65) to assist long-range interactions, was also found at the L1 promoter with CTCF in multiple cell lines (SI Appendix, Fig. S5A), and Cohesin subunit Rad21 overexpression in tumor cells has previously been shown to be associated with increased L1 expression (66).
CTCF bound not only to the 5′ UTR of L1 but also to the L1 3′ UTR (the same recruitment profile was observed for Myc). This observation made us hypothesize that CTCF may mediate intermolecular (anchoring two L1 copies by binding their 5′ UTR promoters) or intramolecular (anchoring the 5′ UTR promoter and 3′ UTR of the same L1) interactions. A previous study (67) showed that gene loops enhanced transcriptional directionality at bidirectional promoters by physically bringing together promoter and terminator and allowing RNA polymerase to reload onto promoters efficiently after finishing the previous round of transcription. Although the current resolution of Hi-C data is insufficient to detect L1 intramolecular interactions, the formation of gene loops of L1 was consistent with the two following observations: (i) RNA polymerase colocalized with CTCF at both the L1 promoter and the 3′ UTR (SI Appendix, Fig. S5B), and (ii) knocking down Ssu72, a factor critical for gene-loop formation (67), decreases the level of L1 transcription to a similar degree as knockdown of CTCF (SI Appendix, Fig. S5C). This finding may suggest a mechanism of L1 transcription in cancer cells, but more studies are necessary to address it better.
MapRRCon Analysis of Histone Marks of L1HS.
We also tested MapRRCon in analyzing ChIP-seq datasets of histone marks. After analyzing 14 histone marks in 115 biosamples, we noticed that a few active transcription/open chromatin histone marks (H3K9ac, H3K27ac, H3K4me2, H3K4me3, and H3K4me1) were more represented among our sample cohort, with a slight enrichment in immortalized cells and stem cells, which had higher L1 expression than tissue and primary cell samples. However, we observed a high degree of variability in L1 histone marks among cancer cell samples, as is consistent with a diversity of regulatory networks in different cells. It was also notable that three hESC lines showed very consistent patterns across all histone marks, as did neural progenitor cells, which also reportedly exhibit high L1 expression (68–70). Repressive markers such as H3K9me3 were generally absent from L1s. We also identified an enrichment of H2A.Z (encoded by the H2AFZ gene) on L1HS specifically in hESCs and in a few other high L1-expressing cell lines, including breast cancer and prostate cancer cell lines (i.e., MCF-7 and PC-3, respectively) (SI Appendix, Figs. S2B and S6 and Dataset S2). This mark is also reportedly associated with mouse L1 promoters and exhibited forward feedback regulation with Myc in breast cancer cells (71, 72).
Discussion
Our study provides a valuable resource of potential L1 regulators and their binding regions at L1 sequences in a large number of cell types. This database can be further expanded as additional ChIP-seq datasets are generated. We developed the MapRRCon pipeline to screen factors interacting with L1 sequences and also used it to analyze RNA-seq data to lower or eliminate DNA contamination from the analysis. This method not only identified known binders such as YY1 (20, 21), Nanog (26, 27), Sin3A (31), RUNX3 (22), and SP1 (29) (SI Appendix, Fig. S7) but also reported 175 additional TFs that bind within the L1HS sequence, the great majority of which had not previously been identified as L1 regulators. We further validated the effect of two factors, Myc and CTCF, on L1 transcription using siRNA-knockdown reporter assays. We also explored the correlation of their expression levels with L1 expression in tumors and thus provided support for the hypothesis they are important regulators controlling L1 expression in cancer cells. This resource may provide insight into and knowledge about L1 regulatory networks in various cancer types.
MapRRCon analysis is not restricted to L1HS sequences but can be easily adapted both to other host species and to other types of repetitive sequences such as Alu. Although the term “MapRRCon” is designed to emphasize the mapping to consensus sequences, the genome-wide prealignment is an important step for reducing background. Due to the rapid genomic evolution of retrotransposons and the historical proliferation of related but extinct subfamilies of elements (1, 73–75), it is critical to map the reads to the unmasked human reference genome first, to avoid overly aggressive mapping of reads caused by direct alignment to L1HS. We have compared the reads mapped to the consensus sequences with or without prealigning for both ChIP-seq and RNA-seq datasets and found that prealignment greatly increased the signal-to-noise ratio. The elimination of this high noise was not essential for strong binders (e.g., Myc), as we could still observe their enrichment even when we mapped reads directly to the consensus; however, the peak-to-noise ratio was significantly reduced. Limitations of the method include the inability to map to specific copies. Previous studies have shown that the local chromatin environment and upstream flanking sequences can influence L1 transcription in a cell-type–specific manner (49, 76), indicating that additional flanking-region binders might indirectly regulate L1 transcription; such TFs will be missed by MapRRCon.
Although this study aims to reveal factors that bind retrotransposition-competent L1s, most of which belong to the L1HS family, we cannot exclude the possibility that a subset of reads that map to L1HS may come from other closely related L1 families. For instance, the L1PA1 consensus sequence is very closely related to L1HS (∼1 bp difference per 600 bp), making them virtually indistinguishable during read alignment. Comparing the consensus sequences of the L1PA1–L1PA7 families with L1HS, we observed that TF-binding sites are clustered in relatively conserved regions in the L1HS 5′ UTR promoter (SI Appendix, Fig. S8A), suggesting evolutionary conservation in recruiting TFs to L1s.
MapRRCon analysis depends largely on L1HS annotations made by RepeatMasker (www.repeatmasker.org/). To determine whether these 1,620 L1HS sequences specifically belong to the L1HS family, we performed phylogenetic analysis on the 332 RepeatMasker-annotated full-length L1HS sequences along with consensus sequences identified by RepeatMasker as belonging to a family within the range L1HS to L1PA7. The phylogenetic tree showed that most of these elements cluster with the L1HS, L1PA1, or L1PA2 5′ UTR promoter sequences; however, 41 elements clustered with older L1 families (L1PA3–LAPA12) (SI Appendix, Fig. S8B). This indicates that MapRRCon is likely unable to definitively distinguish whether TFs are bound to L1HS versus L1PA1/L1PA2; however, many fewer “contaminating reads” belonging to families more ancient than L1PA3 are included using our standard parameter settings.
To study the evolutionary conservation of TF binding to L1s, we applied MapRRCon to a subset of datasets from hESCs using the L1PA2–L1PA7 consensus sequences. We observed a significant decrease in the number of bound TFs on L1PA4–L1PA7 compared with L1HS, L1PA2, and L1PA3, among which L1PA6 and L1PA7 are largely free of peaks in their 5′ UTR promoter regions (SI Appendix, Fig. S9A and Dataset S3). Even among the more closely related families, L1HS clearly stands out with the highest peak values. Comparing the binding profiles, we found that most L1HS peaks are also seen in L1PA2 but with decreased signal. For instance, Myc binding is strongest at L1HS and disappears as evolutionary distance increases (SI Appendix, Fig. S9B), further confirming that most of the TFs found bind younger/active L1 family members.
The motif database (50) exploited here used 427 ENCODE ChIP-seq datasets to perform de novo motif discovery, based on a subset of the ChIP-seq data analyzed here. Importantly, motif discovery was restricted to uniquely mapped reads, while most of our analyzed reads were not evaluated. It is possible that some DNA motifs assigned to specific TFs were identified because the considered TF colocalized with other proteins responsible for direct DNA binding. In our case, we found that one Myc motif (“discovery motif 10”) was very similar to known CTCF motifs but was distinct from other Myc motifs. In our analysis, this motif resides at the strongest Myc/CTCF peak summit. We therefore conclude that Myc discovery motif 10 actually represents a CTCF motif. This observation may partly explain the effect of mutating individual Myc motifs (Fig. 4D) and supports the idea that small sequence changes in the L1 5′ UTR promoter can alter the binding of multiple factors, as we probably also mutated CTCF-binding sites in addition to Myc-binding sites.
One of the major concerns when quantifying L1 expression from RNA-seq datasets is genomic DNA contamination, which can produce a nontrivial background, as L1 insertions exist in great abundance compared with single-copy genes. By applying MapRRCon (extracting reads from the RepeatMasker-annotated L1HS insertions first and then aligning to the consensus sequence) to the RNA-seq analysis, we found that genomic contamination is largely reduced, as seen by decreased 3′ coverage bias on the L1HS consensus (SI Appendix, Fig. S4). Furthermore, because (i) the hg38 reference is itself a consensus sequence and (ii) many individual L1 insertions are still nonfixed due to their ongoing retrotransposition activity, allowing four mismatches in each read helps account for polymorphisms of L1HS insertions. However, there are still obstacles that both MapRRCon and traditional methods are unable to overcome, such as the inability to distinguish reads that arise from readthrough transcripts.
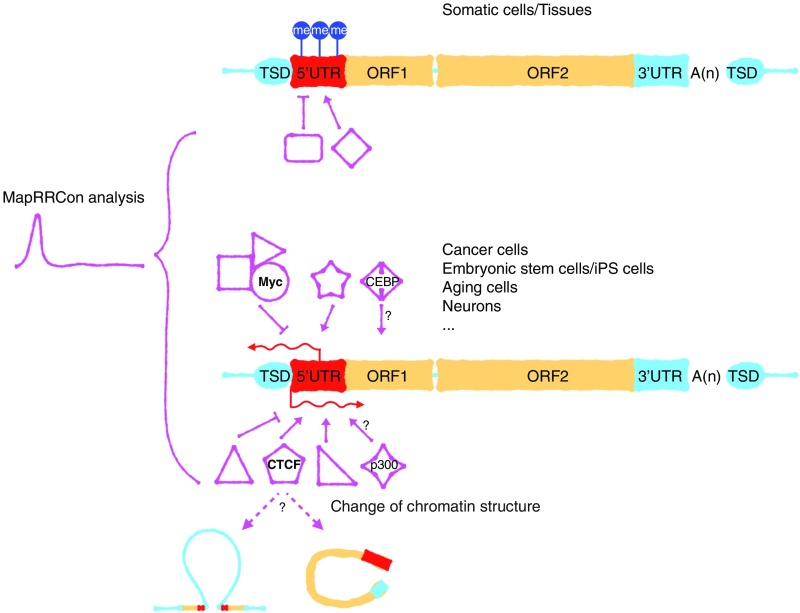
In summary, this resource provides a wealth of information on TFs that bind to a highly repetitive human sequence, L1. In terms of cell types in which such binding is observed, it is striking that little binding is seen in normal tissues and primary differentiated cells, as is consistent with the lack of evidence for L1 mRNA expression. Conversely, ES cells, cancer cells, and tumors show extensive evidence of both expression and binding of multiple TFs. The fact that roughly one-third of all TFs seem to bind the L1 sequence specifically, as judged by ChIP-seq peaks, is exciting but mystifying. A subset of these likely represents some type of spurious binding. However, we present here an initial analysis of some key factors, including the CEBP proteins, p300 acetyltransferase, Myc, and CTCF, that strongly supports the direct involvement of some or all of these factors in the control of L1 expression. We anticipate that many new findings on the relationship between host TFs and the control of TE activity will be revealed by expanding these studies to other TFs, host species, and transposable element types (Fig. 6). Finally, the remarkable repertoire of TFs that bind these elements in cancer cells raises an interesting question: Could the binding of large quantities of TFs by open TE chromatin, made accessible by global changes in DNA and histone modifications, shape tumorigenic transcriptional states?
Fig. 6.
A TF landscape provides information on L1 regulation. The structure of L1 is shown in boxes containing the 5′ UTR (red), ORF1 and ORF2 (orange), and the 3′ UTR (blue). L1 expression is often suppressed in somatic cells and tissues by DNA methylation or other cellular factors. Upon tumorigenesis or induction of pluripotency, L1 sometimes becomes expressed due to the change in the genetic environment. This reactivation of L1 depends on multiple TFs and chromatin remodelers and is cell-type specific. The L1 activity will further influence gene expression, chromatin structure, and genome instability. me, DNA methylation; TSD, target site duplication.
Methods
MapRRCon Pipeline.
The MapRRCon analysis pipeline comprises six major steps: (i) optional: preparation of the sequencing reads; (ii) sequence alignment to the reference genome; (iii) read extraction based on annotated L1 genomic locations; (iv) sequence alignment to the L1Hs consensus sequence; (v) two-step normalization and quality control; and (vi) peak calling. To process ENCODE ChIP-seq datasets, we downloaded ENCODE ChIP-seq data from the ENCODE website (https://www.encodeproject.org) using the following search filters: Assay = ChIP-seq; Project = ENCODE; Organism = Homo sapiens; Available data = fastq/bam. For any experiment, if unfiltered BAM files (aligned reads) were available, we used them directly in step 3; otherwise, we used FASTQ files (raw reads) and started from step 1. We matched ChIP and Input datasets based on experiment accession numbers and removed ChIP data that did not have associated Input data. After mapping the extracted reads to L1HS, we excluded datasets that had no L1 reads and also removed locations that had coverages less than 10 for each generated profile.
-
i)
Low-quality base pairs and adaptor sequences of single-end or paired-end reads are trimmed using Trimmomatic (77). This step is optional in the pipeline, as quality controls are also performed in steps 3 and 5.
-
ii)
The processed reads are aligned to the hg38/GRCh38 (December 2013) human reference genome assembly using BWA-MEM (78); the alignment options followed the ENCODE standard ChIP-seq analysis pipeline.
-
iii)
Reads aligned to 1,620 RepeatMasker-annotated L1HS sites are extracted using an in-house–developed Java script. Meanwhile, reads containing the following features are filtered out: (i) more than three mismatches if the read length is shorter than 50 bp or more than four mismatches if the read length is longer than 50 bp; (ii) reads that contain insertions or deletions; and (iii) reads that contain soft clipping (partial match). Importantly, this step is not limited to annotated L1HS locations; it is also able to take any genomic intervals (in UCSC.bed file format) and extract reads that belong to the input regions. In addition, we also provide an option to specify the number of allowed mismatches to fit more diverse purposes.
-
iv)
Extracted reads are aligned to the L1 consensus sequence using BWA-MEM with default parameter setting. The coverage distribution of the aligned reads at each position of the L1 consensus sequence is generated using BEDTools (79) genomecov with option −d.
-
v)
Two-step normalization is performed using an in-house–developed R script. The read coverage at each position was first normalized by the number of reads mappable to the reference genome (this number can be obtained in the output file of step iii) and then is normalized against the measured background (Input DNA). The median coverage of all base positions on the L1 consensus sequence in the ChIP sample is calculated. The same division is also performed for the Input sample, followed by subtraction of the Input sample from the ChIP sample at each position. We also excluded positions that have coverage less than 10 in the ChIP or Input distribution.
-
vi)
ChIP-seq peaks within the L1 consensus sequence are called using an in-house–developed signal-processing algorithm (provided as a Python script), and a true peak is defined as present in the normalized data. The signal background was estimated by calculating the rmsd of the signal in a sliding window (width = ±80 bp) across L1Hs, and the rmsd of the background was estimated from the distribution of rmsds by finding the rmsd where the distribution dips to 20% of its maximum (i.e., disregarding large rmsd values that correspond to peaks for the background estimation). To find peaks, the signal is smoothed using a smoothing filter (Hanning filter with width = ±80 bp) and differentiated to find local maxima, i.e., where the derivative of the smoothed signal is zero and the second derivative is negative. The peaks are filtered using two thresholds on the original unsmoothed signal: signal minus background intensity larger than 1 and an rmsd ratio between signal and background larger than 1.3. The algorithm is insensitive to the choice of width of smoothing filter and the width of window for rmsd calculations within a range of 40–120 bp. Information about the peaks, including peak location, height above background, width (defined as where the signal drops to 25% of its maximum), and signal-to-noise ratio, is extracted from the original signal ratio.
RNA-Seq Analysis.
BAM files of 77 breast tumor and 127 ovarian tumor samples were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/), which had been aligned to hg38 using a TCGA-harmonized pipeline. Reads mapped to 1,620 RepeatMasker-annotated L1HS regions were extracted from the BAM files. Reads containing the following features were filtered out: secondary alignments, clipping alignments, indels, or more than four mismatches. BAM files containing the L1 reads subset were converted to FASTQ files using SAMtools (80). Converted FASTQ files were subsequently aligned to the L1HS consensus sequence using STAR (56). We used the same parameter settings for this realignment step as for the TCGA-harmonized pipeline. Read counts of L1HS were then generated using HTSeq-count. We calculated fragments per kilobase of transcript per million mapped reads (FPKM) values and performed samplewise normalization. For the cell line RNA-seq data, FASTQ files from ENCODE were first aligned to hg38 with the TCGA-harmonized pipeline, and then the procedure described above was followed.
Motif Analysis.
The Position Weight Matrix (PWM) of each TF was downloaded from Motif Browser (compbio.mit.edu/encode-motifs/) (50). The motif similarity score was calculated by the following steps: (i) the weights of each position in the target sequence were extracted from the PWM; (ii) we summed up the extracted weights at each position and divided the value by the maximum of summed weights. We considered sequences that have a similarity score ≥0.9 as true DNA motifs in our analysis.
Cell Culture and Stable Cell Lines.
The luciferase 293T-REx reporter cell line and HEK293T and MCF-7 cells were maintained in DMEM supplemented with 10% FBS and 4 mM l-glutamine. The luciferase 293T-REx reporter cell line stably expresses the 5′ UTR of L1rp (L1 element in retinitis pigmentosa) flanked by Renilla luciferase in the forward orientation and firefly luciferase in the antisense orientation. The construction of the luciferase 293T-REx reporter cell line was previously described in ref. 81, and the MCF-7 cell line was a generous gift from Benjamin G. Neel at NYU Langone Health, New York. The HEK293T cell line used was reported previously (82). The luciferase 293T-REx reporter cell line was used in the siRNA experiments; HEK293T and MCF-7 cells were used in the motif-mutant reporter assay.
Motif Mutant Constructs.
We identified six putative Myc-binding regions in the L1 5′ UTR promoter sequence according to their high similarity score (>0.9) to Myc motifs. To disrupt the motifs, we shuffled each of the six sequences together with its 5-bp flanking regions to maintain the same nucleotide composition. Among the mutated sequences generated for each region, we selected the one that had the lowest similarity score and proceeded with experimental validation. The wild-type reporter was generated by inserting a DNA fragment containing the L1rp 5′ UTR promoter driving firefly luciferase in the forward direction. The mutant L1 5′ UTR promoter sequences (plus 40-bp homology arms) were synthesized using the BioXp 3200 System (SGI-DNA) and ligated into the pcDNA5/FRT (Thermo Fisher Scientific) between the KpnI and BstXI restriction sites using Gibson Assembly master mix (New England Biolabs). The Renilla construct (transfection control) was generated by cloning the Renilla sequence into the pCEP4 mammalian expression vector (Thermo Fisher Scientific) under the CMV promoter at the HindIII site.
Luciferase Reporter Assay.
HEK293T cells (0.075 million) and MCF-7 cells (0.004 million) were plated in each well of a 96-well plate. The next day these cells were cotransfected with the Renilla construct and with each of the reporter constructs using FuGENE HD transfection reagent (Promega) according to the manufacturer’s recommendations. Replicates were done within the same plate. Forty-eight hours after transfection, we lysed the cells and measured the luciferase activity with the Dual-Glo system (Promega). Firefly signal was first normalized to the Renilla signal within a well and then to the wild-type well.
Knockdown Experiments.
The luciferase 293T-REx reporter cell line was used for the knockdown experiments. We plated 2,500 cells in each well of a 384-well plate; at the same time, cells were transfected with siRNA control or siRNA against specific proteins (Life Technologies). DharmaFECT transfection reagent (0.1 μL per well) (Dharmacon) was used for siRNA transfection. Forty-eight hours after knockdown, the firefly and Renilla luciferase activities were measured with the Dual-Glo system. This experiment was designed in a format ideal for a whole-genome siRNA knockdown screen, and the robust z-score of the values in each well was calculated.
Data Access.
All ChIP-seq and cell line RNA-seq datasets are available on the ENCODE website (https://www.encodeproject.org) (43). All the RNA-seq datasets of breast and ovarian tumors are available on the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). The MapRRCon analyzed results on L1HS and closely related elements are freely available to access and visualize via a web tool (maprrcon.org). The lists of TF/histone mark peaks were uploaded as Datasets S1–S3. In-house scripts related to MapRRCon pipeline step 3, 5, 6 can be downloaded from the MapRRCon website.
Supplementary Material
Acknowledgments
We thank the High-Performance Computing Facility and the cluster at the Institute for Systems Genetics at NYU Langone Health for bioinformatics support, Matt Maurano for advice on high-performance computing, Carmine Fedele and Benjamin Neel for sharing the MCF7 cell line and advice on cell culture, and Molly Hammell and Jason D. Fernandes for helpful discussions on the challenges of mapping repetitive DNA reads. This work was supported by NIH Grants P50GM107632 and P01AG051449 (to J.D.B. and D.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722565115/-/DCSupplemental.
References
- 1.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 3.St Laurent G, 3rd, Hammell N, McCaffrey TA. A LINE-1 component to human aging: Do LINE elements exact a longevity cost for evolutionary advantage? Mech Ageing Dev. 2010;131:299–305. doi: 10.1016/j.mad.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang CRL, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2003;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 6.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 7.de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- 9.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7:706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 11.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 12.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 13.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minakami R, et al. Identification of an internal cis-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res. 1992;20:3139–3145. doi: 10.1093/nar/20.12.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrova EA, et al. Sense transcripts originated from an internal part of the human retrotransposon LINE-1 5′ UTR. Gene. 2012;511:46–53. doi: 10.1016/j.gene.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denli AM, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163:583–593. doi: 10.1016/j.cell.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Thornburg BG, Gotea V, Makałowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006;365:104–110. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 20.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 22.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wylie A, et al. p53 genes function to restrain mobile elements. Genes Dev. 2016;30:64–77. doi: 10.1101/gad.266098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris CR, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchénio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 28.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 29.Fedorov AV, Lukyanov DV, Podgornaya OI. Identification of the proteins specifically binding to the rat LINE1 promoter. Biochem Biophys Res Commun. 2006;340:553–559. doi: 10.1016/j.bbrc.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 30.Mita P, Boeke JD. How retrotransposons shape genome regulation. Curr Opin Genet Dev. 2016;37:90–100. doi: 10.1016/j.gde.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Rica L, et al. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol. 2016;17:234. doi: 10.1186/s13059-016-1096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Mun S, Meyer TJ, Han K. High levels of sequence diversity in the 5′ UTRs of human-specific L1 elements. Comp Funct Genomics. 2012;2012:129416. doi: 10.1155/2012/129416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. The distribution of a germline methylation marker suggests a regional mechanism of LINE-1 silencing by the piRNA-PIWI system. BMC Genet. 2012;13:31. doi: 10.1186/1471-2156-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinomoto M, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 36.Ashktorab H, et al. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics. 2014;9:503–512. doi: 10.4161/epi.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodić N, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184:1280–1286. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Hurk JAJM, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 40.Brouha B, et al. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito J, et al. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 2017;13:e1006883. doi: 10.1371/journal.pgen.1006883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain D, Baldi S, Zabel A, Straub T, Becker PB. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 2015;43:6959–6968. doi: 10.1093/nar/gkv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park D, Lee Y, Bhupindersingh G, Iyer VR. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One. 2013;8:e83506. doi: 10.1371/journal.pone.0083506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acharya A, Rishi V, Moll J, Vinson C. Experimental identification of homodimerizing B-ZIP families in Homo sapiens. J Struct Biol. 2006;155:130–139. doi: 10.1016/j.jsb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 49.Philippe C, et al. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. eLife. 2016;5:166. doi: 10.7554/eLife.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jakobsen JS, et al. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 2013;23:592–603. doi: 10.1101/gr.146399.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4:a014241. doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 54.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filippova GN, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klenova EM, et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 60.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S, Yu N-K, Kaang B-K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47:e166. doi: 10.1038/emm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barutcu AR, et al. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 2015;16:214. doi: 10.1186/s13059-015-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 65.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 66.Xu H, et al. Cohesin Rad21 mediates loss of heterozygosity and is upregulated via Wnt promoting transcriptional dysregulation in gastrointestinal tumors. Cell Rep. 2014;9:1781–1797. doi: 10.1016/j.celrep.2014.10.059. [DOI] [PubMed] [Google Scholar]
- 67.Tan-Wong SM, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macia A, et al. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 2017;27:335–348. doi: 10.1101/gr.206805.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rangasamy D. Histone variant H2A.Z can serve as a new target for breast cancer therapy. Curr Med Chem. 2010;17:3155–3161. doi: 10.2174/092986710792231941. [DOI] [PubMed] [Google Scholar]
- 72.Rangasamy D. Distinctive patterns of epigenetic marks are associated with promoter regions of mouse LINE-1 and LTR retrotransposons. Mob DNA. 2013;4:27. doi: 10.1186/1759-8753-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 75.Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- 76.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: Variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 2004;14:2253–2260. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mita P, et al. URI regulates KAP1 phosphorylation and transcriptional repression via PP2A phosphatase in prostate cancer cells. J Biol Chem. 2016;291:25516–25528. doi: 10.1074/jbc.M116.741660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dai L, Taylor MS, O’Donnell KA, Boeke JD. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol Cell Biol. 2012;32:4323–4336. doi: 10.1128/MCB.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.