Significance
Bacterial operons must solve a fundamental problem: how to produce discordant amounts of proteins from cotranscribed genes. Here, we used a genome-wide approach to map operon mRNA isoforms with nucleotide resolution in Bacillus subtilis and to quantify their abundances in different genetic backgrounds. These results revealed RNA endonucleolytic cleavages located in between cotranscribed genes that lead to differential transcript stability and abundance. The RNA endonuclease’s activity toward operon mRNA maturation requires a three-protein Y-complex. Interestingly, the Y-complex has few effects on other targets of the RNA endonuclease. The dichotomy of targets suggests that the Y-complex acts as a specificity factor for the general endonuclease, a role that could be broadly conserved in other Firmicutes, including the human pathogen Staphylococcus aureus.
Keywords: Bacillus subtilis, RNase Y, mRNA processing, mRNA degradation, Y-complex
Abstract
Endonucleolytic cleavage within polycistronic mRNAs can lead to differential stability, and thus discordant abundance, among cotranscribed genes. RNase Y, the major endonuclease for mRNA decay in Bacillus subtilis, was originally identified for its cleavage activity toward the cggR-gapA operon, an event that differentiates the synthesis of a glycolytic enzyme from its transcriptional regulator. A three-protein Y-complex (YlbF, YmcA, and YaaT) was recently identified as also being required for this cleavage in vivo, raising the possibility that it is an accessory factor acting to regulate RNase Y. However, whether the Y-complex is broadly required for RNase Y activity is unknown. Here, we used end-enrichment RNA sequencing (Rend-seq) to globally identify operon mRNAs that undergo maturation posttranscriptionally by RNase Y and the Y-complex. We found that the Y-complex is required for the majority of RNase Y-mediated mRNA maturation events and also affects riboswitch abundance in B. subtilis. In contrast, noncoding RNA maturation by RNase Y often does not require the Y-complex. Furthermore, deletion of RNase Y has more pleiotropic effects on the transcriptome and cell growth than deletions of the Y-complex. We propose that the Y-complex is a specificity factor for RNase Y, with evidence that its role is conserved in Staphylococcus aureus.
Bacterial RNA endonucleases, such as RNase Y in Bacillus subtilis and RNase E in Escherichia coli, are well understood for their role in global mRNA turnover and stable RNA processing (1–8). In addition to these canonical functions, the maturation of a small number of mRNA species is also known to involve RNA endonucleases (9–15). For example, endonucleolytic cleavages in the 5′ untranslated regions (UTRs) of genes can generate alternative transcript isoforms with truncated leader sequences from their precursor mRNAs (10, 14). In addition, endonucleolytic cleavages within multigene operons can lead to the differential stability of the upstream and downstream products, allowing cells to tune the stoichiometries of mRNAs and their protein products among cotranscribed genes. An example of a multigene transcript that undergoes endoribonuclease-dependent maturation is the glycolysis operon cggR-gapA in B. subtilis (9, 16, 17). The first gene, cggR, encodes a transcriptional repressor of itself and the entire operon. CggR is needed by the cell at a lower level than the product of the second gene, the glycolytic enzyme GAPDH (GapA) (18). Cleavage by an endoribonuclease, RNase Y in this case, near the end of cggR destabilizes the cggR portion, while leaving the gapA portion stable, resulting in higher levels of the gapA mRNA (17, 19, 20). In contrast to canonical mRNA turnover, operon mRNA processing by RNA endonucleases provides a posttranscriptional mechanism for differentiating expression among cotranscribed genes. However, beyond a few documented examples, there have been few systematic surveys of endonuclease-mediated mRNA isoform maturation across a transcriptome. It is also unknown whether other factors participate and potentially regulate these mRNA processing events.
In this work, we focus on RNase Y and its potential regulators. RNase Y is the major RNA endonuclease responsible for mRNA decay in B. subtilis. RNase Y cleavages of several mRNAs have been shown to lead to rapid degradation, such as for sinR (21, 22), rpsO (23), and those containing S-box riboswitches (8). Depletion of RNase Y affects the abundance of ≈1,000 mRNAs and increases the bulk mRNA half-life by more than twofold (16, 24, 25). It has been suggested that RNase Y interacts with other proteins important for mRNA turnover, such as the 3′-to-5′ exoribonuclease polynucleotide phosphorylase (PNPase), and the DEAD box RNA helicase CshA, forming a complex called the degradosome (9, 16, 26, 27). RNase Y localizes to the cell periphery via a single-pass transmembrane domain (28–30). Whether RNase Y’s activity is regulated is unknown.
It was recently reported that the complex of YlbF, YmcA, and YaaT (the Y-complex) physically interacts with RNase Y and is required for processing of the cggR-gapA transcript in B. subtilis (22). Previously investigated as a regulator of biofilm formation (31, 32), the Y-complex was found to destabilize the transcript encoding the negative regulator of biofilm formation, SinR. Modestly elevated sinR mRNA levels in mutants of the Y-complex lead to a block in biofilm formation due to its hypersensitivity to levels of the transcriptional repressor (22, 33, 34). The Y-complex is reported to be required for other types of developmental processes in B. subtilis (35), such as genetic competence (36), and is coconserved with RNase Y in other gram-positive bacteria, raising the possibility of a broad functional association. Whether the Y-complex is required for RNase Y activity outside of cggR-gapA and sinR was previously unknown.
To examine the global role of the Y-complex in mRNA processing, we turned to a technique developed by one of our laboratories for RNA end-enrichment sequencing (Rend-seq) (37). This approach not only allows us to identify the 5′ and 3′ ends of overlapping RNA isoforms with nucleotide resolution but also enables quantification of transcript abundance. Comparing the transcript isoforms present in strains lacking each component of the Y-complex with wild-type cells revealed the loss of shorter isoforms that have truncated regions of either the 5′ UTR or even entire cistrons. We found that these shorter mRNA isoforms are processed from longer isoforms via RNase Y cleavage, which leads to unstable upstream fragments and stable downstream fragments, analogous to the cggR-gapA operon. Furthermore, analyses of both novel and previously known RNase Y targets of noncoding RNAs show that the Y-complex has limited contribution to their maturation. In addition, we show that the function of the Y-complex toward mRNA processing is conserved in Staphylococcus aureus, with the ortholog of YlbF being required for the processing of a conserved operon. This work suggests that the Y-complex plays a broad role in mRNA processing by acting as an accessory factor for RNase Y, and that RNase Y directs two classes of processing events: those that are Y-complex–dependent and those that are independent.
Results
Deletion of the Y-Complex Leads to Altered Operon mRNA Isoforms.
To globally assess the role of the Y-complex in polycistronic mRNA processing, we sought to determine the differences in transcript isoforms between wild-type B. subtilis and mutants lacking each component of the Y-complex. We carried out Rend-seq experiments to both identify and quantify operon mRNA isoforms. Rend-seq achieves end-enrichment by sparse fragmentation of cellular RNAs, which generates fragments containing the original RNA ends and, to a much lesser extent, fragments that start at internal positions (37). The fragments are then converted into a cDNA library for short-read, high-throughput sequencing and mapped to the B. subtilis genome. For each position on the genome, the fragment counts for the 5′-mapped reads and 3′-mapped reads are plotted separately. With a greater than 40-fold end-enrichment, the positions of original 5′ and 3′ ends of transcripts show single-nucleotide peaks in the 5′-mapped reads and 3′-mapped reads, respectively. Furthermore, each peak in 5′-mapped (or 3′-mapped) reads is accompanied by a proportional increase (or decrease) in density downstream. As a result, overlapping mRNA isoforms can be readily resolved and quantified. We have previously demonstrated that the RNA ends identified by Rend-seq are consistent with results from hundreds of independent 5′ and 3′ mapping experiments (37). We have also shown that Rend-seq reproduces the mRNA isoform architecture reported for the hbs locus in B. subtilis, which consists of two transcription start sites (TSSs) and a processed 5′ end due to RNase Y (14) (see Fig. 3H).
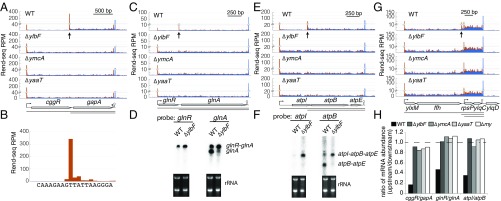
By applying Rend-seq to RNA from wild-type cells and strains lacking components of the Y-complex, we confirmed that the Y-complex is required for RNase Y processing of the cggR-gapA operon. Rend-seq from wild-type cells (B. subtilis strain 3610 with an epsH knockout to avoid biofilm formation; see SI Appendix, Extended Materials and Methods) confirmed two major transcript isoforms that differ in the position of their 5′ ends, as indicated by the two peaks in 5′-mapped reads (orange) with corresponding stepwise increases of read density (Fig. 1 A and H). The positions of the TSS and processed 5′ end agree with previous evidence based on primer extension experiments (38) (Fig. 1B). Consistent with the Y-complex being required for the generation of the shorter isoform, Rend-seq for mutants lacking the Y-complex showed only the full-length transcript, as evidenced by the disappearance of both the peak that corresponds to the 5′ end of the shorter isoform and the associated stepwise change in read density (Fig. 1A). Furthermore, the unprocessed full-length transcript becomes more abundant, as evidenced by the increases in both the TSS peak height and the read density for cggR. As a result, the absence of RNase Y processing in Y-complex mutants leads to nearly equal expression of cggR and gapA transcripts, increasing the relative abundance from 0.15:1 in wild-type cells to 0.93:1 in mutants of the Y-complex (Fig. 1H).
Fig. 1.
Absence of short polycistronic mRNA isoforms in B. subtilis lacking components of the Y-complex. Rend-seq data show 5′-mapped (orange) and 3′-mapped (blue) read counts plotted for the cggR-gapA (A), glnR-glnA (C), atpI-atpB-atpE (E), and ylxM-ffh-rpsP-ylqD (G) regions of the B. subtilis genome from wild-type (WT) cells (strain 3610, ∆epsH background) and mutants lacking ylbF, ymcA, and yaaT, respectively. Horizontal lines below the gene annotations indicate potential isoforms predicted by Rend-seq for WT cells. Longer isoforms due to partial read-through of transcription terminators after gapA, atpE, and ylqD are not depicted. Vertical arrows point to positions of the 5′ ends of isoforms that disappear in mutants lacking components of the Y-complex. Peak shadows are removed from Rend-seq data (SI Appendix, Extended Materials and Methods) (37). For each position, the number of reads mapped is normalized by the number of million reads for that sequencing sample that align to the B. subtilis genome outside of rRNA and tRNAs. (B) Rend-seq 5′-mapped reads surrounding the 5′ end of the gapA-only isoform (±10 nt) in WT cells, with the base corresponding to each position below. Northern blot analysis shows the identity of operonic mRNA isoforms in WT cells and a mutant lacking YlbF for the glnR-glnA (D) and atpI-atpB (F) operons, with rRNA used as a loading control. The 5′ ends predicted for the ylxM-ffh-rpsP-ylqD operon were previously verified by 5′ RACE (37). (H) Ratio of mRNA levels between neighboring genes at either side of Y-complex–dependent 5′ ends. The mRNA levels are quantified using Rend-seq. RPM, reads per million.
More broadly, we identified several other operons that showed a similar absence of shorter mRNA isoforms in mutants lacking components of the Y-complex. For example, the two-gene operon of the transcriptional repressor glnR and the glutamine synthetase glnA (Fig. 1C) was present as both a full-length glnR-glnA transcript and a glnA-only transcript in wild type. The latter was absent in Y-complex mutants, causing the mRNA levels of glnA and glnR to be equal (Fig. 1H). Northern blot analysis confirmed the loss of the shorter isoform and the increased level of the longer isoform in cells lacking YlbF (Fig. 1D). Similarly, in the ATP synthase operon, Rend-seq data showed that a shorter isoform starting downstream from the first gene, atpI, is absent in Y-complex mutants (Fig. 1E), leading to similar mRNA levels between atpI and atpB (Fig. 1H). Again, Northern blot analysis confirmed the presence of the shorter isoform in the wild-type cells (Fig. 1F). The positions of Y-complex–dependent 5′ ends for these and 10 other examples [e.g., the ffh-rpsP operon (Fig. 1G) and the tgt-yrbF operon (SI Appendix, Fig. S1A)] are summarized in Dataset S1. The differential expression of operon genes is either reduced or completely lost when the Y-complex is disabled. The Y-complex is therefore required to achieve, and potentially regulate, the stoichiometry of cotranscribed proteins.
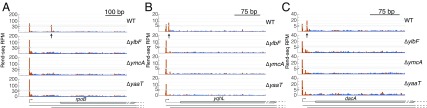
In addition to differentiating expression of multicistronic mRNAs, the Y-complex has a dramatic effect on alternative 5′ UTRs for the first gene of operons. Several operons that have alternative leader sequences in wild type showed a loss of the isoforms with shorter leaders in mutants. These included the 5′ UTRs for operons encoding the β-subunit of RNA polymerase, RpoB (Fig. 2A); a protein with unknown function, YqhL (Fig. 2B); the penicillin-binding protein DacA (Fig. 2C); and five other examples (SI Appendix, Fig. S1A and Dataset S1). The identity of these altered 5′ ends was shown with nucleotide resolution (SI Appendix, Fig. S1B) and confirmed by 5′ RACE and Sanger sequencing (SI Appendix, Fig. S1C) for rpoB. Taken together, deleting any components of the Y-complex alters mRNA isoforms for at least 21 operons that include 73 genes across the B. subtilis transcriptome, expanding its known effects beyond the cggR-gapA operon.
Fig. 2.
Absence of short 5′ UTRs in B. subtilis lacking components of the Y-complex. Rend-seq 5′-mapped (orange) and 3′-mapped (blue) read counts are plotted for the rpoB (A), yqhL (B), and dacA (C) regions of the B. subtilis genome from wild-type (WT) cells (strain 3610, ∆epsH background) and mutants lacking ylbF, ymcA, and yaaT. Horizontal lines below gene annotations indicate potential isoforms predicted by Rend-seq for WT cells. For rpoB and dacA, transcription continues past the regions shown into downstream genes. Vertical arrows point to positions of the 5′ ends of isoforms that disappear in mutants lacking components of the Y-complex. Peak shadows are removed from Rend-seq data (SI Appendix, Extended Materials and Methods) (37). RPM, reads per million.
Cleavage by RNase Y Is Required for the Y-Complex–Dependent Maturation of Operon mRNAs.
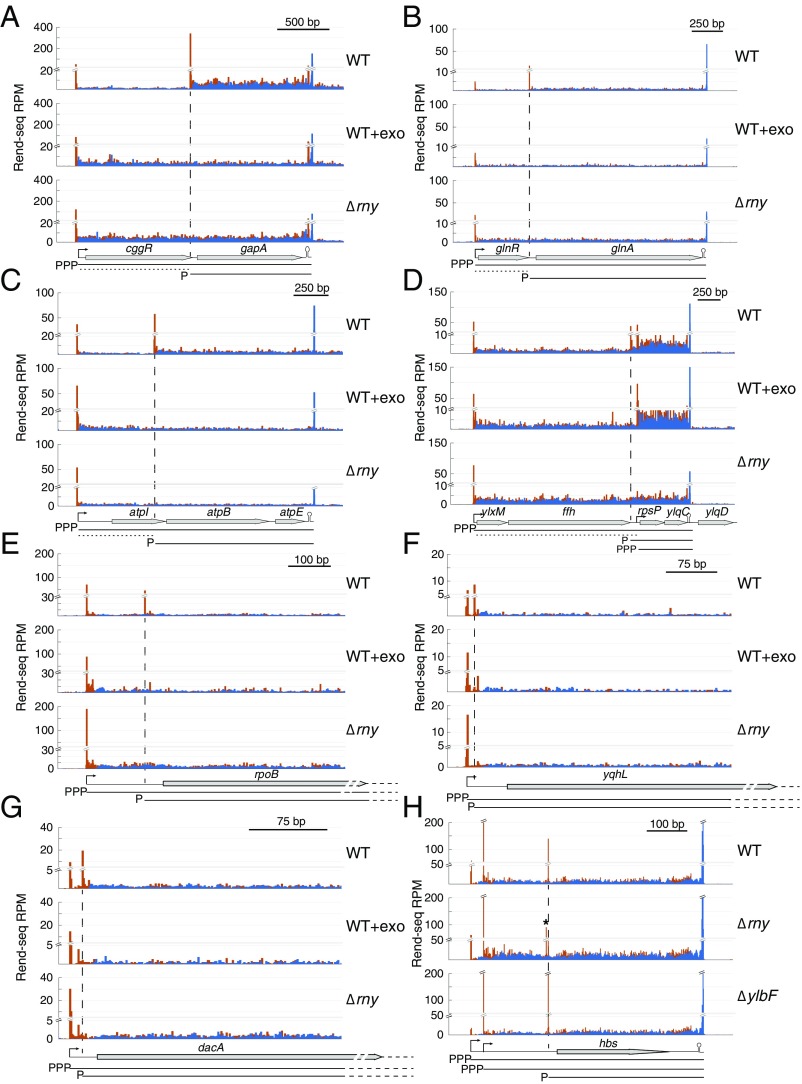
Since the Y-complex directly interacts with RNase Y and affects the maturation of the cggR-gapA operon mRNA (22), we sought to determine if the short mRNA isoforms that are absent in Y-complex mutants are derived from RNase Y-dependent processing events. Rend-seq data from cells lacking RNase Y show that the shortened transcript isoforms for cggR-gapA (Fig. 3A), glnR-glnA (Fig. 3B), atpI-atpB (Fig. 3C), and ffh-rpsP (Fig. 3D) were indeed absent: the peaks corresponding to their 5′ ends disappear. Similarly, the short alternative 5′ UTRs were absent in mutants lacking RNase Y for rpoB (Fig. 3E), yqhL (Fig. 3F), and dacA (Fig. 3G) (the complete list of operons whose shortened isoforms require both RNase Y and the Y-complex is provided in Dataset S1). Accompanying the disappearance of the shorter isoforms, the longer isoforms showed increased abundance in the RNase Y mutant in a manner similar to mutants of the Y-complex (Fig. 1H). These results are consistent with a model in which the Y-complex–dependent shorter isoforms are the products of RNase Y cleavage. However, it is also possible that the shorter isoforms represent internal TSSs whose synthesis relative to the upstream TSSs is indirectly affected by RNase Y.
Fig. 3.
RNase Y cleavage generates the truncated mRNA isoforms. (A–G) Rend-seq 5′-mapped (orange) and 3′-mapped (blue) read counts plotted for the cggR-gapA, glnR-glnA, atpI-atpB-atpE, ylxM-ffh-rpsP-ylqD, rpoB, yqhL, and dacA regions of the B. subtilis genome. For each region, Rend-seq data are shown for RNA extracted from wild-type (WT) cells (strain 3610, ∆epsH background), WT cells treated with a 5′-exo (WT+exo) (strain 168 background), and a mutant lacking RNase Y (Δrny). Horizontal lines below the gene annotations indicate potential isoforms predicted by Rend-seq for WT cells, with the 5′ moiety marked. Horizontal dashed lines indicate potential cleavage products that are rapidly degraded. Longer isoforms due to partial read-through of transcription terminators after gapA, atpE, and ylqD are not depicted. For rpoB and dacA, transcription continues past the regions shown into downstream genes. Vertical dashed lines indicate positions of the 5′ ends of the truncated isoforms that are absent in the mutant of RNase Y and are sensitive to 5′-exo treatment in vitro. Peak shadows are removed from Rend-seq data (SI Appendix, Extended Materials and Methods). (H) Rend-seq data plotted for the hbs region of the B. subtilis genome for RNA extracted from WT cells and mutants lacking RNase Y (Δrny) or a component of the Y-complex (ΔylbF). The asterisk indicates an alternative 5′ end that appears in the mutant of RNase Y, which is consistent with the isoform labeled as “M + 4” in the study by Braun et al. (14). P, monophosphate; PPP, triphosphate; RPM, reads per million.
To test whether the shorter isoforms are the product of RNA cleavage events and not TSSs, we first examined whether their 5′ nucleotides carry 5′-monophosphates, as expected from RNase Y cleavage and/or potential subsequent exonuclease activities, instead of the 5′-triphosphates that come from transcription initiation (39). Total RNA from wild-type cells was treated with a 5′ monophosphate-specific exonuclease (5′-exo) to degrade RNAs whose 5′ ends are the product of RNases. The RNA products of this reaction were then assessed by Rend-seq. The Y-complex–dependent shortened transcript isoforms were all absent in the sample treated with the 5′-exo, suggesting that they are not the product of internal TSSs (Fig. 3 A–G). We next examined whether the regions upstream of the shorter isoforms were more susceptible to 3′-to-5′ exonucleolytic decay, which would be a consequence of RNase Y cleavage. Consistently, we found that in a mutant lacking PNPase, the major 3′-to-5′ exonuclease, the upstream regions showed increased read density even though the shorter isoforms were still present (SI Appendix, Fig. S2). Together, these lines of evidence suggest that the shortened isoforms arise from RNase Y cleavage, whereas the upstream cleavage products are rapidly degraded by PNPase. The sequential action by RNase Y and PNPase results in differential stability for different regions of the cotranscribed mRNA, creating alternative isoforms as previously seen for the cggR-gapA operon.
We found that all but one instance of RNase Y-dependent mRNA maturation required all three components of the Y-complex. Using Rend-seq data for the wild type, the rny mutant, and wild type treated with 5′-exo, we identified 22 high-confidence operons that have truncated mRNA isoforms due to RNase Y cleavage (SI Appendix, Extended Materials and Methods). Among these operons, the only maturation event that does not require the Y-complex is hbs, which was previously reported to depend on RNase Y and also confirmed by our Rend-seq data (Fig. 3H) (14).
The Y-Complex Is Required for Some, but Not All, RNase Y Processing Sites in Noncoding RNAs.
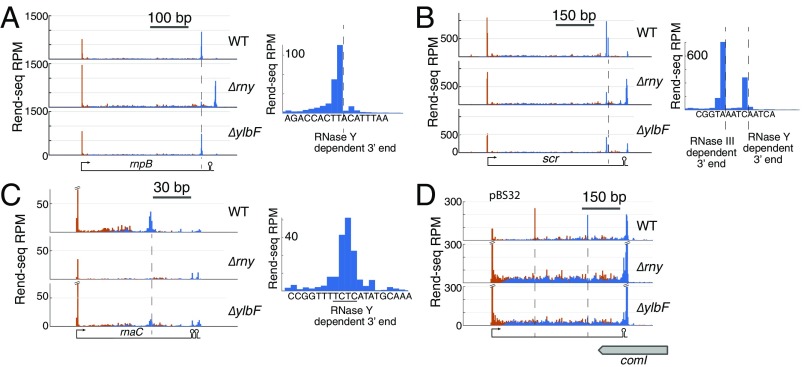
Given that the Y-complex is broadly required for mRNA maturation by RNase Y, we next sought to determine if the Y-complex also participates in RNase Y-dependent 3′ maturation of noncoding RNAs, such as the RNA components of RNase P (rnpB) and the signal recognition particle (scr) (7). Consistent with published Northern blot analyses, Rend-seq data showed that both rnpB and scr are predominantly present in their mature, cleaved forms in wild-type cells and present mainly in premature forms in cells lacking RNase Y (Fig. 4A). In contrast to the RNase Y mutant in which cleavages were absent or reduced, both rnpB and scr were still processed in mutants of the Y-complex, as evidenced by the peaks in 3′-mapped reads that correspond to RNase Y cleavage sites. In the case of scr, the fraction of processed RNA was reduced but still present in mutants of the Y-complex (Fig. 4B and SI Appendix, Fig. S3). These results indicate that, unlike most mRNA maturation events, these noncoding RNA processing sites do not strictly require the accessory Y-complex.
Fig. 4.
Noncoding RNA maturation by RNase Y and its dependency on the Y-complex. Rend-seq 5′-mapped (orange) and 3′-mapped (blue) read counts are plotted for the rnpB (A), scr (B), and rnaC (C) regions of the B. subtilis genome from wild-type (WT) cells (∆epsH background) and mutants lacking rny and ylbF. Mutants lacking YmcA or YaaT behave similarly (SI Appendix, Fig. S3). Dotted lines indicate the positions of mature 3′ ends. Zoomed-in views of 3′-mapped reads at the positions surrounding the mature 3′ ends are shown. (D) Rend-seq data from WT cells and mutants lacking rny and ylbF mapped to the B. subtilis 3610 plasmid pBS32 for the region antisense to comI. Dotted lines indicate the position of RNA ends that depend on the presence of RNase Y. RPM, reads per million.
Rend-seq data also revealed previously uncharacterized instances of noncoding RNA maturation that depended on RNase Y but with limited contribution of the Y-complex. One example is the maturation of the small regulatory RNA rnaC. Comparison between wild type and cells lacking RNase Y showed that rnaC was transcribed past the mature 3′ end and into a sequence that resembles factor-independent transcription terminators. As indicated by the absence of mature product in cells lacking RNase Y, RNase Y was required for the shortening of rnaC by ∼39 nucleotides from the 3′ end (Fig. 4C). Interestingly, in mutants of the Y-complex, both mature and pre-RNAs were present, suggesting that the cleavage is only partially dependent on the Y-complex. Other examples include two unannotated RNAs encoded on the plasmid, pBS32, in B. subtilis strain 3610 that are present in altered forms in mutants lacking the Y-complex or RNase Y. One is an antisense RNA to the competence inhibitor comI (Fig. 4D), and the other is an antisense RNA to the extracytoplasmic function (ECF)-like sigma factor zpdN (SI Appendix, Fig. S3). That these are noncoding RNAs is supported by the lack of ribosome density from published ribosome profiling data on these RNAs (34). Taken together, these results show that RNase Y processes several noncoding RNAs with various dependencies on the Y-complex.
Mutants Lacking the Y-Complex and RNase Y Have Elevated Levels of Riboswitches.
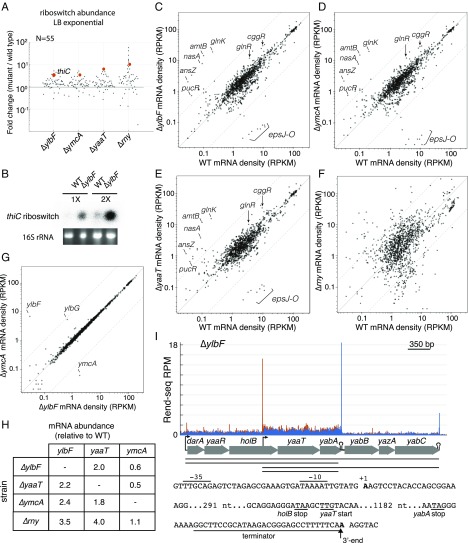
In addition to its roles in the maturation of operon mRNAs and noncoding RNAs, RNase Y is known to initiate the turnover of several S-box and T-box riboswitches in B. subtilis (8, 24). To determine if the Y-complex has a potential role in riboswitch turnover, we globally assessed the abundance of annotated riboswitches and RNA leader sequences in wild-type cells and cells lacking the Y-complex using the Rend-seq data. To increase the number of riboswitches whose abundance is high enough to be quantified, we carried out Rend-seq from cells grown in three conditions: exponential growth in a rich medium [Luria–Bertani (LB) medium] and exponential growth and stationary phase growth in the minimal salts glutamate glycerol medium (MSgg) (Fig. 5A, SI Appendix, Fig. S4 A and B, and Dataset S2). Of the 61 annotated riboswitches and RNA leader sequences that were produced under at least one condition, 36 showed a greater than twofold increase in abundance in Y-complex mutants in at least one condition (41 in the RNase Y mutant). For each condition, the riboswitches and RNA leader sequences that showed the highest increase in abundance in the RNase Y mutant were also more abundant in all of the Y-complex mutants. The riboswitches that are affected include previously reported ones, such as S-box and T-box riboswitches (8, 24), as well as others, such as Thi-box riboswitches (e.g., tenA, thiC), gcvT, pbuG, pbuE, lysC, and ribD, as well as the protein-dependent RNA leader sequences for infC and trpE. The increase in thiC riboswitch abundance was independently confirmed by Northern blotting (Fig. 5B). These results suggest that the Y-complex also has an effect on the stability of most riboswitches and RNA leader sequences.
Fig. 5.
Changes in RNA abundance in mutants lacking the Y-complex. (A) Fold-change in riboswitch and RNA leader abundances in mutants lacking ylbF, ymcA, yaaT, and rny compared with wild type. The number of riboswitches and RNA leaders whose expression levels are quantified in LB medium is indicated. Red dots indicate the thiC riboswitch. Only a few leaders showed greater than twofold repression, which are all regulated by CcpA (blgF, glpD, glpF, and glpT) and likely a consequence of transcriptional regulation. (B) Northern blot analysis showing the abundance of the thiC riboswitch in wild-type (WT) cells and cells lacking YlbF. Two dilutions of total RNA (1× and 2×) are shown with the 16S rRNA used as a loading control. The mRNA abundances for all annotated and expressed genes (>100 reads) in B. subtilis are plotted for their levels in the WT versus strains lacking ylbF (C), ymcA (D), yaaT (E), and rny (F). RNA was collected from cells in exponential phase in LB medium. Dashed lines indicate a 10-fold difference from the diagonal. RPKM, reads per 1,000 bases of gene per million bases of total reads that map to the B. subtilis genome, excluding rRNAs and tRNAs. (G) mRNA abundances for all annotated and expressed (read count >100) genes in B. subtilis are plotted for their levels in the ∆ylbF versus ∆ymcA. All other combinations are shown in SI Appendix, Fig. S5. (H) mRNA levels of ylbF, ymcA, and yaaT in WT and strains lacking components of the Y-complex. (I) Rend-seq 5′-mapped (orange) and 3′-mapped (blue) read counts plotted for the yaaT region of the genome from cells lacking ylbF. In addition to the known TSS upstream of darA and the TSS in holB reported here, small additional peaks in 5′ reads of unknown origin are present. The sequence of the 5′ and 3′ UTRs of yaaT and yabA are shown. Putative −10 and −35 elements of the σA consensus sequences upstream of the peak in 5′-mapped reads internal to holB and a putative terminator sequence upstream of the peak in 3′-mapped reads downstream of yabA are indicated. RPM, reads per million.
RNase Y Has Broad Effects on the Transcriptome and Phenotypes.
The only previously reported role of the Y-complex in RNA turnover is for the mRNA for sinR, the master regulator of biofilm formation, which is a known target of RNase Y (21, 22). Based on sinR message abundance and its promoter activities in Y-complex mutants, it was shown that the mRNA is stabilized in the absence of YlbF or YmcA under biofilm-inducing conditions, resulting in a small increase in sinR mRNA and protein levels (∼2.8-fold and ∼1.8-fold, respectively). The RNA-sequencing (RNA-seq) results here confirmed that there is no measurable change in sinI mRNA abundance but that there is an increase in sinR message abundance in mutants of the Y-complex and RNase Y in the stationary phase in MSgg medium (1.6-fold) (SI Appendix, Fig. S4C and Dataset S3). Dubnau et al. (40) recently reported qRT-PCR results that mutants of the Y-complex have reduced sinI and sinR mRNA abundance in comparison to a reference transcript, fusA. We note that, in our transcriptome data, the level of fusA is also increased relative to most mRNAs (Dataset S3), which explains the apparent inconsistency between qRT-PCR results and those from Northern blots and RNA-seq. Nevertheless, for both sinR mRNA and riboswitches, the effects of deleting the genes for the Y-complex were smaller than the all-or-none effects seen for mRNA maturation.
Given that the Y-complex mainly affects operon mRNA maturation and not noncoding RNAs, we propose that the Y-complex is a specificity factor for RNase Y activity. Consistent with this model, mRNA abundances in mutants lacking the Y-complex differ from the wild type only for a specific set of genes, whereas the strain lacking RNase Y showed global remodeling of the transcriptome (Fig. 5 C–F and Dataset S3). For strains lacking components of the Y-complex, the genes with elevated transcript levels mainly consisted of those whose polycistronic mRNAs are matured by RNase Y or those involved in nitrogen utilization, presumably due to misregulation of the glnR-glnA operon. [The increased abundance for the eps genes in wild-type cells was a consequence of a polar effect from the use of a deletion of the epsH gene to prevent biofilm formation, and not due to the absence of the Y-complex (see SI Appendix, Extended Materials and Methods).] Interestingly, among the three components of the Y-complex, deleting ylbF and ymcA yielded strikingly similar effects on the transcriptome (Fig. 5G), whereas cells lacking YaaT exhibited an additional induction of prophage genes (Dataset S3). In contrast to the Y-complex, many more genes had altered mRNA levels in the RNase Y mutants (R = 0.72 compared with wild type vs. R = 0.94 between ∆ylbF and wild type with log-transformed data). The large shifts in the transcriptome are likely due both to direct and indirect effects of RNase Y-dependent RNA turnover, which does not require the Y-complex. Together with the observation that mutants lacking RNase Y showed more severe phenotypes than mutants lacking the Y-complex (41), these data suggest that there are at least two branches of RNase Y activities: one that requires the Y-complex and one that does not.
Negative Feedback Regulation of the Y-Complex via RNase Y.
Proteins involved in RNA degradation are sometimes negatively autoregulated through the stability of their own mRNAs (42–45). In cells lacking RNase Y or components of the Y-complex, the mRNA for two of its components, YlbF and YaaT, was increased to levels ranging from approximately fourfold in cells lacking RNase Y to approximately twofold in mutants lacking other components of the Y-complex (Fig. 5H and SI Appendix, Fig. S6A). Differing from ylbF and yaaT, the abundance of ymcA was similar between wild type and mutants lacking RNase Y, YlbF, or YaaT. Therefore, the synthesis of at least two of the three members of this complex appears to be regulated by their own activity, although the mode of regulation remains to be determined.
In addition to yaaT’s negative feedback regulation, Rend-seq revealed a previously unrecognized yaaT transcript isoform. The yaaT gene has been thought to be transcribed only as part of the eight-gene operon of darA-yaaR-holB-yaaT-yabA-yabB-yazA-yabC (46). Rend-seq results confirmed the presence of this eight-gene polycistronic mRNA and also revealed the presence of two additional transcript isoforms: a two-gene transcript of only yaaT and yabA as well as a minor five-gene transcript of yaaT-yabA-yazA-yazC-yabC (Fig. 5I). The presence of these two additional isoforms, as well as the increase in yaaT transcript abundance in cells lacking YlbF, was confirmed by Northern blot analysis (SI Appendix, Fig. S6B). The 5′ end of these transcripts, upstream of yaaT, was not sensitive to 5′-exo treatment and also conformed to the canonical −10 and −35 promoter motifs, indicating that it likely corresponds to a TSS that is internal to the upstream holB ORF (Fig. 5I). The 3′ end of the yaaT-yabA transcript was preceded by a transcription termination sequence just after the yabA ORF (Fig. 5I). Together, these findings suggest that ylbF and yaaT transcripts are particularly sensitive to Y-complex–dependent degradation through RNase Y, providing a mechanism for maintaining the activity of the Y-complex.
YaaT-GFP Localizes to the Cell Periphery in a Manner Dependent on RNase Y.
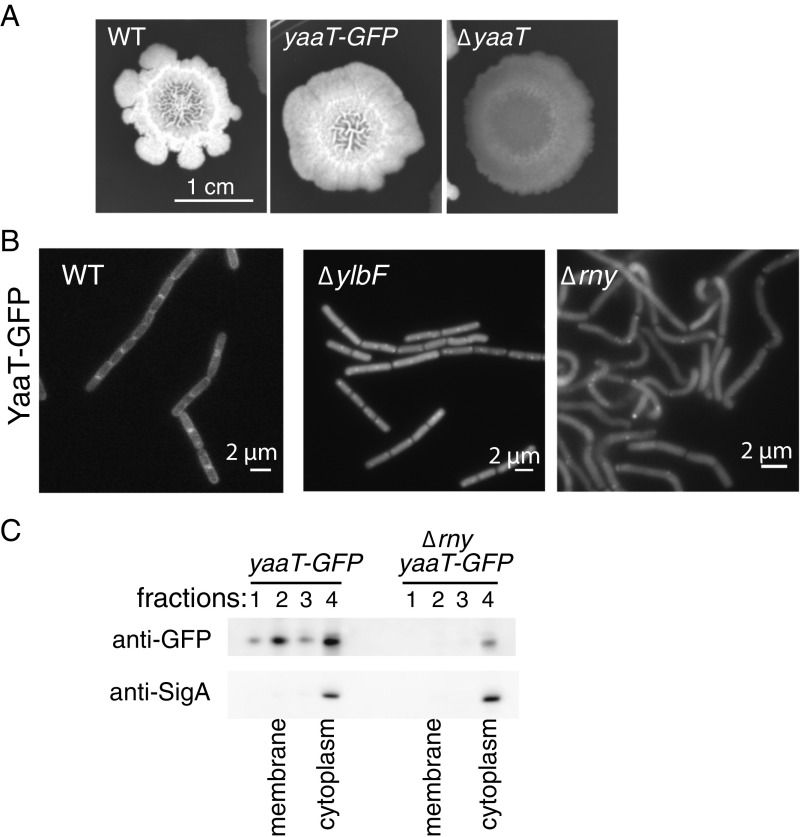
Although RNase Y is known to localize to the cell membrane with an enrichment at the division septum (29, 30), whether or not the Y-complex also localizes to the cell periphery is debated. In contrast to RNase Y, the components of the Y-complex do not contain predicted transmembrane domains. In 2002, Hosoya et al. (35) reported that a C-terminal GFP fusion of YaaT localizes to the cell periphery with enrichment at the site of septation. In contrast, Dubnau et al. (40) showed that an N-terminal YFP-YaaT fusion localizes to the cytoplasm. We revisited the question of YaaT localization using the endogenous promoter to drive the synthesis of a YaaT-GFP C-terminal fusion at its native locus in a markerless manner. The strain harboring YaaT-GFP was able to produce wild-type biofilms, indicating that the YaaT-GFP fusion was functional (Fig. 6A). Fluorescence microscopy showed that YaaT-GFP was indeed enriched at the cell periphery and sites of septation (Fig. 6B). Interestingly, the localization of YaaT-GFP to the cell periphery was dependent on the presence of YlbF (Fig. 6B): In cells lacking YlbF, YaaT-GFP localized to the cytoplasm with little signal at the division septum. In mutants lacking RNase Y, YaaT-GFP localization also appeared to be cytoplasmic, although the cell morphology was substantially altered for this slow-growing strain (Fig. 6B).
Fig. 6.
Subcellular localization of YaaT-GFP. (A) Colony biofilms produced by wild-type (WT; strain 3610 with intact eps operon), yaaT-GFP, and ∆yaaT strains grown on LB glycerol manganese, with the structured wrinkles reflecting matrix production. (B) Micrographs showing the localization of YaaT-GFP in WT (strain 3610 with intact eps operon), ∆ylbF, and ∆rny cells. (C) Western blot against YaaT-GFP (anti-GFP antibody) and SigA for fractions from a membrane flotation experiment. Cleared lysates were centrifuged in an OptiPrep medium gradient, and fractions were collected from the top. Cytoplasmic proteins remain in fraction 4, while membrane-associated proteins migrate to fraction 2. The band shown is of the approximate size of YaaT-GFP and is absent in a WT whole-cell lysate lacking GFP. The full Western blots are shown in SI Appendix, Fig. S6C.
Independently, we tested the localization of YaaT-GFP to the cell membrane using a membrane flotation assay that separates the membrane from cytoplasm (47) (Fig. 6C). An antibody against GFP showed a clear enrichment of YaaT-GFP in the membrane fraction, whereas a cytoplasmic protein (SigA), as a control, was absent. This localization to the membrane fraction was completely dependent on the presence of RNase Y (Fig. 6C). This experiment provides independent evidence supporting the results of fluorescence microscopy that one component of the Y-complex localizes to the cell periphery in a manner that depends on RNase Y.
Conservation of Y-Complex Function.
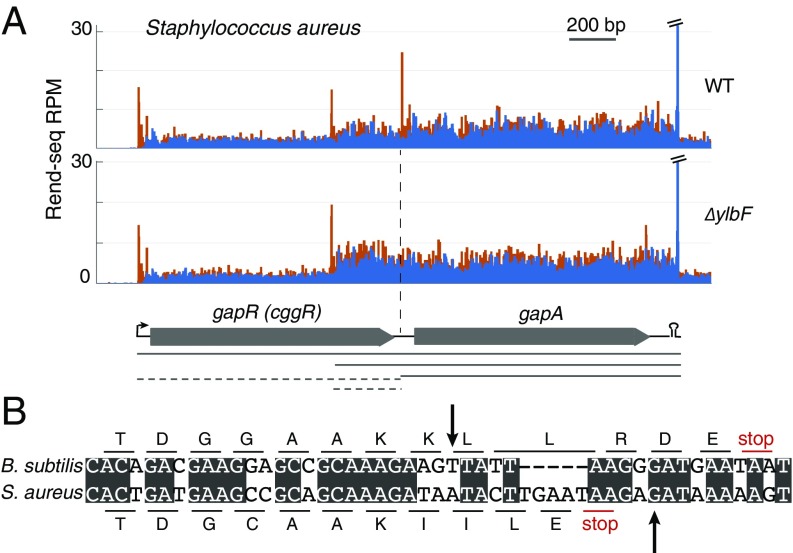
RNase Y, YlbF, YmcA, and YaaT are all highly conserved among Firmicutes. As we have previously noted, rny and yaaT show an additional pattern of cooccurrence extending to other gram-positive bacteria (22). To determine if the role of the Y-complex in RNA processing is conserved, we carried out a Rend-seq experiment in S. aureus to compare wild-type cells [HG003, a virulent derivative of multilocus sequence type ST8 strain NCTC8325 repaired for two regulatory genes (48)] with a mutant lacking the ortholog of ylbF (SAOUHSC_01070). These results showed that generation of one of the three mRNA isoforms of the cggR(gapR)-gapA operon also requires the YlbF ortholog in S. aureus (Fig. 7A). Despite the similarity in the genes included on the processed isoform to that of B. subtilis, the 5′ nucleotide sequence of the mature downstream transcript is not conserved (Fig. 7B). This 5′ end was previously shown to be dependent on RNase Y (49). The exact position of the mature 5′ end might be either the direct RNase Y cleavage site or the product of further 5′-to-3′ exonuclease activity after RNase Y cleavage upstream. Therefore, it remains a possibility that the endonucleolytic cleavage site is conserved between B. subtilis and S. aureus. In addition to the cggR-gapA operon, we observed that the known cleavages of two riboswitches (serS and valS) by RNase Y in S. aureus (49) also require the ortholog YlbF (SI Appendix, Fig. S6 D and E). The requirement of YlbF for the processing of at least three mRNAs in S. aureus, together with the evolutionary cooccurrence of the entire Y-complex with RNase Y, suggests that the function of these proteins in mRNA processing is conserved among Firmicutes.
Fig. 7.
Ortholog of YlbF in S. aureus is required for cleavage of cggR-gapA. (A) Rend-seq 5′-mapped (orange) and 3′-mapped (blue) read counts plotted for the cggR(gapR)-gapA region of the S. aureus HG003 genome in wild-type (WT) cells and mutants lacking the ortholog of YlbF. RPM, reads per million. (B) Nucleotide sequence alignment of the regions surrounding the 5′ end of truncated gapA transcripts in S. aureus and B. subtilis.
Discussion
In this work, we determined the global role of the Y-complex in RNase Y activities for the maturation of mRNAs and noncoding RNAs throughout the B. subtilis transcriptome. Using Rend-seq to identify and quantify RNA isoforms, we defined the set of mRNAs and noncoding RNAs matured by RNase Y. Interestingly, although most mRNA maturation events also require the presence of the Y-complex, a subset of RNAs can be matured without the Y-complex. Together with the drastic phenotypic differences between mutants lacking RNase Y and lacking the Y-complex, these data suggest that the Y-complex acts as a specificity factor for the endonuclease.
How might the Y-complex influence RNase Y specificity? Although our study provides a list of Y-complex–dependent and –independent target sequences near the 5′ ends of the processed isoforms, we have not identified consensus primary or secondary structural motifs that might specify the Y-complex activity. Biochemical studies using purified RNase Y lacking the N-terminal transmembrane domain have shown that the cleavage at the yitJ riboswitch, without the Y-complex, is sensitive to the phosphorylation state of the 5′ end and the secondary structural context of the cleavage site (8). It is possible that the Y-complex allows RNase Y to bypass some of these requirements in vivo, making it possible to access additional sites. For instance, cleavages internal to polycistronic mRNAs occur far away from the TSS, and the 5′ monophosphate requirement may need to be alleviated. By comparison, RNase E, the functional analog of RNase Y in E. coli, can cleave both by binding to monophosphorylated 5′ ends and via “direct entry” (50, 51). Interestingly, the yitJ riboswitch is among the 36 riboswitches whose abundance increases in the absence of the Y-complex in vivo, suggesting that the Y-complex may also contribute to its degradation. Biochemical reconstitution of the Y-complex may help elucidate its substrate specificity.
Consistent with previous evidence that the Y-complex affects RNase Y activity via a physical association (22), we confirmed that YaaT-GFP, produced from its endogenous locus, is localized to the cell periphery both by fluorescence microscopy and by a membrane-floatation experiment. We also showed that this localization depends on both YlbF and RNase Y. These results differ from those of Dubnau et al. (40), but they are in agreement with the previous report of Hosoya et al. (35). RNase Y-dependent localization to the cell membrane is fully consistent with previous two-hybrid evidence that YlbF and YmcA interact with RNase Y (22) and with our current evidence that the function of RNase Y is strongly dependent on the Y proteins.
Our transcriptome data also support a previous view that not all components of the Y-complex are functionally equivalent. Although the defects in RNA maturation are consistent across all three single deletions of the complex, mRNA abundances are markedly different in ∆yaaT compared with ∆ylbF and ∆ymcA (SI Appendix, Fig. S5). This observation is consistent with the fact that a strain lacking YaaT has a different biofilm phenotype than mutants of YlbF and YmcA (22). We cannot distinguish whether the difference in transcriptome is due to YaaT’s potentially unique effects on RNA decay, which are not directly reported in our data, or due to other functions of YaaT outside the Y-complex. Ribosome profiling data for cells under steady-state growth suggest that the rates of protein synthesis for these three components are proportional to the reported stoichiometry of the complex (37, 52). Therefore, the majority of YaaT likely resides in the complex (52). However, it is feasible that there exists a small fraction of YaaT outside the complex that could potentially influence biofilm formation.
Are there biological consequences to the perturbations in multigene mRNA processing events in mutants lacking RNase Y or the Y-complex? It is common for bacteria to transcribe multiple contiguous genes as part of the same mRNA message, even when the protein products are needed in different quantities. We found several incidences in which the correct stoichiometry among these transcripts is set through RNase Y cleavage and the subsequent destabilization of different portions of the transcript. In addition to intergenic cleavages, mRNA processing in the 5′ UTR may affect translation of the first gene, and not others, in polycistronic mRNA, hence also leading to stoichiometric imbalance between cotranscribed genes. One of the most noticeable differences in Y-complex mutants is that such polycistronic mRNA maturation is abolished. Operons like cggR-gapA and glnR-glnA share a common feature in which the transcriptional repressor of the operon (CggR and GlnR) is kept at a lower level than the metabolic enzyme (GapA and GlnA) due to RNase Y and the Y-complex. Absence of the Y-complex therefore leads to overproduction of the repressors, and potentially aberrant regulation. For cggR-gapA, the aberrant regulation might explain the defect in growth transition that could arise from carbon limitation (22). For glnR-glnA, we see extensive changes in the abundance of nitrogen metabolism genes in mutants lacking the Y-complex, with many of the most increased mRNAs falling into this category. It would be interesting to explore the possibility that the Y-complex is a regulator for carbon and nitrogen metabolism through its interaction with RNase Y, especially in light of recent evidence that the Y-complex carries sulfur-iron clusters (53).
Dubnau and coworkers (52) have reported that the Y-complex is a regulator of the phosphorelay that is responsible for activation of the master regulator Spo0A that governs entry into sporulation. Could the Y proteins function both as part of a membrane-associated ribonuclease complex, as we have demonstrated here, and as activators of relay proteins? We note that the Y proteins are widely conserved among Firmicutes, including many species that have RNase Y but lack a phosphorelay, such as S. aureus, for which we have shown that the ortholog of YlbF is required for proper processing of mRNAs. We also note that none of the three Y protein genes surfaced in classic screens for mutants defective in sporulation and that our past efforts to detect a defect in Spo0A activation in Y protein mutants were unsuccessful. Nonetheless, it is possible that such mutations do impair Spo0A activation under certain specific conditions. However, a straightforward explanation for such effects, if they exist, is that they are an indirect consequence of the global effects of Y gene mutations on metabolism rather than a direct effect on the Spo0F and Spo0B relay proteins as claimed by Dubnau and coworkers (52). Thus, it seems unlikely that the Y-complex is both serving as a specificity factor for RNase Y and also moonlighting as a regulator for the sporulation phosphorelay. In toto, these results reinforce the view that the Y proteins are in a functional complex with RNase Y, and they are in keeping with our previous suggestion that YlbF, YmcA, and YaaT be named RcsA, RcsB, and RcsC, respectively, for RNase Y-containing complex subunits A, B, and C (22).
More broadly, differential mRNA stability among operon-encoded genes presents a general strategy for defining and regulating the stoichiometry of functionally related proteins. A critical element of this strategy is the ability to specify the sites for initiating RNA processing. Analogous to the previously characterized examples of sRNA guiding RNase E in trans and stem-loop guiding RNase III in cis (12, 13, 45), we found here that the Y-complex is specifically required for RNase Y in processing events that lead to differential isoform abundance. Being a potential specificity factor similar to small RNAs (sRNAs) and structured RNAs, the physiological roles of the Y-complex may evolve rapidly. By precisely resolving and quantifying complex mRNA isoforms, Rend-seq will provide an opportunity for elucidating the posttranscriptional control of gene expression in diverse organisms.
Materials and Methods
Extended materials and methods are provided in SI Appendix.
Bacterial Strains and Media.
All Rend-seq experiments except for one were carried out on B. subtilis strain 3610 and its derivatives. For the Rend-seq sample treated with 5′-exo, B. subtilis 168 was used. B. subtilis PY79 was used during strain construction. B. subtilis was grown in LB medium or MSgg medium. S. aureus was grown in tryptic soy broth (TSB). Biofilms were grown on LB glycerol manganese 1.5% agar (LB, 1% glycerol, 10 mM MnCl2) (54) at room temperature for 5 d.
Growth Conditions for RNA-Seq and RNA Extraction.
Overnight cultures were back-diluted to an OD600 of 0.001 in LB or MSgg medium. Cells from exponential cultures were collected at an OD600 of 0.25–0.3. Stationary phase cells from MSgg medium were collected at an OD600 of ≈2.5. S. aureus was grown in TSB, and cells were collected at an OD600 of 0.5. RNA was extracted using TRIzol (ThermoFisher) and cleaned up and treated with DNase on Rneasy columns (Qiagen).
Rend-Seq and Library Preparation.
RNA was collected from B. subtilis strain 3610 and its mutants except for the exonuclease-treated sample (B. subtilis 168). Total RNA (after DNase treatment) was depleted of rRNA using MICROBExpress (ThermoFisher). The RNA was fragmented by adding RNA fragmentation buffer (1×; ThermoFisher) for 30 s at 95 °C and quenched by addition of RNA fragmentation stop buffer (ThermoFisher). Fragmented RNAs between 20 and 40 bp were isolated by PAGE and converted to a cDNA library by 3′ ligation of a DNA linker, RT, circularization, and amplification by PCR. For the exonuclease-treated samples, total RNA from B. subtilis strain 168 exponential growth in LB medium was treated with 5′-exo (Illumina) before fragmentation.
Sequencing and Data Analysis.
Sequencing was performed on an Illumina HiSeq 2000 instrument at the BioMicroCenter (Massachusetts Institute of Technology). Reads stripped of the linker sequence were aligned using Bowtie v. 1.2.1.1 (options -v 1 -k 1) to the reference genomes NC 000964.3 (B. subtilis chromosome), KF365913.1 (B. subtilis plasmid pBS32), and NC 007795.1 (S. aureus). The 5′ and 3′ ends of mapped reads were counted separately at genomic positions to produce WIG files, which were normalized per million non-rRNA and non-tRNA reads for each sample. Shadows were removed from WIG files first by identifying the position of peaks and then by reducing the other end of the aligned reads by the peak’s enrichment factor to produce the final normalized and shadow-removed WIG files. Read counts were plotted with MATLAB (MathWorks) for the gene positions listed in Dataset S4. For each position, the number of reads is normalized to the total number of reads for that sample that align to the B. subtilis genome outside of rRNAs and tRNAs.
Datasets.
A list of datasets with information is available in Dataset S4. Raw and processed data are available at the Gene Expression Omnibus (accession no. GSE108295). The data are available for reviewers at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108295.
Microscopy.
Micrographs of YaaT-GFP in the wild-type and ∆ylbF backgrounds were acquired on an Olympus BX61 upright fluorescence microscope and in the ∆rny background on a Nikon Ti-E inverted microscope. Cells grown in LB medium to exponential phase were collected and immobilized on 2.5% agarose pads.
Flotation and Western Blot.
For YaaT-GFP strains in a wild-type background and lacking RNase Y, cells were collected at an OD600 of ≈0.5. Cleared lysates were applied to an OptiPrep density gradient medium (Sigma). Samples were spun at 213,600 × g in Optima Max-XP Ultracentrifuge (Beckman) for 3 h. For Western blots, rabbit anti-SigA polyclonal antibodies were used at a concentration of 1:40,000 and rabbit anti-GFP polyclonal antibodies were used at a concentration of 1:3,000 (ThermoFisher). They were visualized with goat anti-rabbit antibodies conjugated to horseradish peroxidase at a concentration of 1:10,000 (ThermoFisher).
Supplementary Material
Acknowledgments
We thank L. Foulston and A. DeFrancesco for creating the ylbF deletion in S. aureus and collecting cells for RNA-seq; N. Bradshaw for advice on the flotation experiment; and M. Anderson, L. Schons-Fonesca, and A. Grossman for access to and help with their microscope. We thank Byoung-Mo Koo for the rny and pnpA B. subtilis strains. We thank C. Condon and colleagues and B.-M. Koo for valuable feedback and comments. We thank Y. Chai and members of the Losick, Grossman, and G.-W.L. laboratories for discussions and the MIT BioMicro Center for sequencing. This work was supported by NIH Grant R00GM105913, NIH Grant R35GM124732, the Pew Biomedical Scholars Program, the Searle Scholars Program, the Sloan Research Fellowship, Smith Family Awards (to G.-W.L.), a Natural Sciences and Engineering Research Council Fellowship (to J.-B.L.), a Howard Hughes Medical Institute International Student Research Fellowship (to J.-B.L.), and NIH Grant GM18568 (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108295 (accession no. GSE108295).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803283115/-/DCSupplemental.
References
- 1.Hui MP, Foley PL, Belasco JG. Messenger RNA degradation in bacterial cells. Annu Rev Genet. 2014;48:537–559. doi: 10.1146/annurev-genet-120213-092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohanty BK, Kushner SR. Regulation of mRNA decay in bacteria. Annu Rev Microbiol. 2016;70:25–44. doi: 10.1146/annurev-micro-091014-104515. [DOI] [PubMed] [Google Scholar]
- 3.Cohen SN, McDowall KJ. RNase E: Still a wonderfully mysterious enzyme. Mol Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- 4.Bandyra KJ, Luisi BF. Licensing and due process in the turnover of bacterial RNA. RNA Biol. 2013;10:627–635. doi: 10.4161/rna.24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao Y, et al. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell. 2017;65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aït-Bara S, Carpousis AJ. RNA degradosomes in bacteria and chloroplasts: Classification, distribution and evolution of RNase E homologs. Mol Microbiol. 2015;97:1021–1135. doi: 10.1111/mmi.13095. [DOI] [PubMed] [Google Scholar]
- 7.Gilet L, DiChiara JM, Figaro S, Bechhofer DH, Condon C. Small stable RNA maturation and turnover in Bacillus subtilis. Mol Microbiol. 2015;95:270–282. doi: 10.1111/mmi.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Commichau FM, et al. Novel activities of glycolytic enzymes in Bacillus subtilis: Interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruscella P, Shahbabian K, Laalami S, Putzer H. RNase Y is responsible for uncoupling the expression of translation factor IF3 from that of the ribosomal proteins L35 and L20 in Bacillus subtilis. Mol Microbiol. 2011;81:1526–1541. doi: 10.1111/j.1365-2958.2011.07793.x. [DOI] [PubMed] [Google Scholar]
- 11.Burton ZF, Gross CA, Watanabe KK, Burgess RR. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983;32:335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- 12.Régnier P, Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 13.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun F, Durand S, Condon C. Initiating ribosomes and a 5′/3′-UTR interaction control ribonuclease action to tightly couple B. subtilis hbs mRNA stability with translation. Nucleic Acids Res. 2017;45:11386–11400. doi: 10.1093/nar/gkx793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochat T, Bouloc P, Repoila F. Gene expression control by selective RNA processing and stabilization in bacteria. FEMS Microbiol Lett. 2013;344:104–113. doi: 10.1111/1574-6968.12162. [DOI] [PubMed] [Google Scholar]
- 16.Lehnik-Habrink M, et al. The RNA degradosome in Bacillus subtilis: Identification of CshA as the major RNA helicase in the multiprotein complex. Mol Microbiol. 2010;77:958–971. doi: 10.1111/j.1365-2958.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu B, et al. Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains. Mol Microbiol. 2014;94:41–55. doi: 10.1111/mmi.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meinken C, Blencke HM, Ludwig H, Stülke J. Expression of the glycolytic gapA operon in Bacillus subtilis: Differential syntheses of proteins encoded by the operon. Microbiology. 2003;149:751–761. doi: 10.1099/mic.0.26078-0. [DOI] [PubMed] [Google Scholar]
- 19.Gerwig J, Stülke J. Caught in the act: RNA-seq provides novel insights into mRNA degradation. Mol Microbiol. 2014;94:5–8. doi: 10.1111/mmi.12769. [DOI] [PubMed] [Google Scholar]
- 20.Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehnik-Habrink M, et al. RNA processing in Bacillus subtilis: Identification of targets of the essential RNase Y. Mol Microbiol. 2011;81:1459–1473. doi: 10.1111/j.1365-2958.2011.07777.x. [DOI] [PubMed] [Google Scholar]
- 22.DeLoughery A, Dengler V, Chai Y, Losick R. Biofilm formation by Bacillus subtilis requires an endoribonuclease-containing multisubunit complex that controls mRNA levels for the matrix gene repressor SinR. Mol Microbiol. 2016;99:425–437. doi: 10.1111/mmi.13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao S, Bechhofer DH. Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J Bacteriol. 2010;192:3279–3286. doi: 10.1128/JB.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laalami S, et al. Bacillus subtilis RNase Y activity in vivo analysed by tiling microarrays. PLoS One. 2013;8:e54062. doi: 10.1371/journal.pone.0054062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durand S, Gilet L, Bessières P, Nicolas P, Condon C. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8:e1002520. doi: 10.1371/journal.pgen.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux CM, DeMuth JP, Dunman PM. Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J Bacteriol. 2011;193:5520–5526. doi: 10.1128/JB.05485-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvo E, Alabi S, Liu B, Schlessinger A, Bechhofer DH. Interaction of Bacillus subtilis polynucleotide phosphorylase and RNase Y: Structural mapping and effect on mRNA turnover. J Biol Chem. 2016;291:6655–6663. doi: 10.1074/jbc.M115.711044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehnik-Habrink M, et al. RNase Y in Bacillus subtilis: A natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol. 2011;193:5431–5441. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cascante-Estepa N, Gunka K, Stülke J. Localization of components of the RNA-degrading machine in Bacillus subtilis. Front Microbiol. 2016;7:1492. doi: 10.3389/fmicb.2016.01492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bürmann F, Sawant P, Bramkamp M. Identification of interaction partners of the dynamin-like protein DynA from Bacillus subtilis. Commun Integr Biol. 2012;5:362–369. doi: 10.4161/cib.20215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Branda SS, et al. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 33.Norman TM, Lord ND, Paulsson J, Losick R. Memory and modularity in cell-fate decision making. Nature. 2013;503:481–486. doi: 10.1038/nature12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramaniam AR, et al. A serine sensor for multicellularity in a bacterium. eLife. 2013;2:e01501. doi: 10.7554/eLife.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosoya S, Asai K, Ogasawara N, Takeuchi M, Sato T. Mutation in yaaT leads to significant inhibition of phosphorelay during sporulation in Bacillus subtilis. J Bacteriol. 2002;184:5545–5553. doi: 10.1128/JB.184.20.5545-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tortosa P, Albano M, Dubnau D. Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol Microbiol. 2000;35:1110–1119. doi: 10.1046/j.1365-2958.2000.01779.x. [DOI] [PubMed] [Google Scholar]
- 37.Lalanne JB, et al. Evolutionary convergence of pathway specific enzyme expression stoichiometry. Cell. 2018;173:749–761.e38. doi: 10.1016/j.cell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig H, et al. Transcription of glycolytic genes and operons in Bacillus subtilis: Evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol. 2001;41:409–422. doi: 10.1046/j.1365-2958.2001.02523.x. [DOI] [PubMed] [Google Scholar]
- 39.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 40.Dubnau EJ, et al. A protein complex supports the production of Spo0A-P and plays additional roles for biofilms and the K-state in Bacillus subtilis. Mol Microbiol. 2016;101:606–624. doi: 10.1111/mmi.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figaro S, et al. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol. 2013;195:2340–2348. doi: 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain C, Belasco JG. Autoregulation of RNase E synthesis in Escherichia coli. Nucleic Acids Symp Ser. 1995;114:85–88. [PubMed] [Google Scholar]
- 43.Robert-Le Meur M, Portier C. Polynucleotide phosphorylase of Escherichia coli induces the degradation of its RNase III processed messenger by preventing its translation. Nucleic Acids Res. 1994;22:397–403. doi: 10.1093/nar/22.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bardwell JC, et al. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989;8:3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiChiara JM, Liu B, Figaro S, Condon C, Bechhofer DH. Mapping of internal monophosphate 5′ ends of Bacillus subtilis messenger RNAs and ribosomal RNAs in wild-type and ribonuclease-mutant strains. Nucleic Acids Res. 2016;44:3373–3389. doi: 10.1093/nar/gkw073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolas P, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 47.Bradshaw N, Losick R. Asymmetric division triggers cell-specific gene expression through coupled capture and stabilization of a phosphatase. eLife. 2015;4:e08145. doi: 10.7554/eLife.08145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herbert S, et al. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khemici V, Prados J, Linder P, Redder P. Decay-initiating endoribonucleolytic cleavage by RNase Y is kept under tight control via sequence preference and sub-cellular localisation. PLoS Genet. 2015;11:e1005577. doi: 10.1371/journal.pgen.1005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clarke JE, Kime L, Romero A D, McDowall KJ. Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 2014;42:11733–11751. doi: 10.1093/nar/gku808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrey SM, Mackie GA. Roles of the 5′-phosphate sensor domain in RNase E. Mol Microbiol. 2011;80:1613–1624. doi: 10.1111/j.1365-2958.2011.07670.x. [DOI] [PubMed] [Google Scholar]
- 52.Carabetta VJ, et al. A complex of YlbF, YmcA and YaaT regulates sporulation, competence and biofilm formation by accelerating the phosphorylation of Spo0A. Mol Microbiol. 2013;88:283–300. doi: 10.1111/mmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanner AW, et al. The RicAFT (YmcA-YlbF-YaaT) complex carries two [4Fe-4S]2+ clusters and may respond to redox changes. Mol Microbiol. 2017;104:837–850. doi: 10.1111/mmi.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shemesh M, Chai Y. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol. 2013;195:2747–2754. doi: 10.1128/JB.00028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.