Significance
We designed a synthetic heterotrimeric triple helix by jointly considering stability of a target abc association of three unique chains and the energy gap between the target and 26 competing states. The critical balance of electrostatic and hydrogen-bonding interactions is dramatically revealed in an atomic-resolution structure of the design. Mutations in multibody electrostatic interactions uncover cooperative networks of salt bridges. This work advances our understanding of the role of surface electrostatics and hydrogen bonding in protein stability and fold specificity and provides computational tools for modeling collagen.
Keywords: protein design, self-assembly, triple helix, cooperativity, molecular dynamics
Abstract
One-quarter of the 28 types of natural collagen exist as heterotrimers. The oligomerization state of collagen affects the structure and mechanics of the extracellular matrix, providing essential cues to modulate biological and pathological processes. A lack of high-resolution structural information limits our mechanistic understanding of collagen heterospecific self-assembly. Here, the 1.77-Å resolution structure of a synthetic heterotrimer demonstrates the balance of intermolecular electrostatics and hydrogen bonding that affects collagen stability and heterospecificity of assembly. Atomistic simulations and mutagenesis based on the solved structure are used to explore the contributions of specific interactions to energetics. A predictive model of collagen stability and specificity is developed for engineering novel collagen structures.
The three chains comprising the collagen triple helix primarily associate as homotrimers, although one-quarter—7 of 28—exist biologically as heterotrimers (1, 2). Fibril-forming type I collagen found in bone and skin is formed by the association of two α1 and one α2 chains. Likewise, the nonfibrillar type IV collagen is composed of three different chains (3, 4). The oligomerization state of collagen in the extracellular matrix can provide essential cues to modulate cell behavior, direct tissue morphogenesis, and, in some cases, promote fibrosis pathologies (5, 6). Although noncollagenous prodomains primarily dictate specificity of collagen assembly (7–9), it is intriguing to explore how the triple-helical domains may also modulate collagen self-assembly.
Understanding the intermolecular forces underlying heterospecific assembly is also important for molecular-scale engineering. Compositional control of multicomponent assemblies can be used to modulate material properties for a diverse array of molecular systems, from metal-organic ligand complexes (10, 11) to DNA origami (12, 13) and protein nanocages (14, 15). A major challenge in such systems is the exponential increase in complexity as more components are incorporated. In the case of collagen, three unique components can nominally form 33 = 27 trimeric assemblies. Computational approaches have proved to be powerful for exploring this large combinatorial space on oligomers of various secondary structure types (16–18).
The triple-helix structure imposes constraints on the sequence space available for synthetic collagen design. Collagen sequences are composed of tandem arrays of Gly-X-Y triplets, where frequently X = Pro and Y = (4R)-hydroxyproline (Hyp or O). Instead of a hydrophobic core, the collagen triple helix is a zipper of backbone hydrogen bonds, which are critical for stability and folding, but do not control heterospecificity (19). Instead, specificity may be engineered by introducing networks of side-chain interactions along the protein surface (20–22). A strategy of engineering complementary electrostatic interactions has proved successful in directing the formation of synthetic heterotrimers (23–27). However, the extent to which the complexity of self-assembly may be controlled is limited by our understanding of the energetics of polar interactions on the highly solvent-exposed surface of collagen.
Stabilizing effects of salt bridges have been studied by rationally substituting X and Y positions with charged residues in homotrimeric (POG)n or (PPG)n host sequences (28–32). There are currently only a few structures of collagen-like proteins in the Protein Data Bank (PDB) (33) and even fewer examples of designed structures held together by electrostatic interactions (34). Discrete sequence-based electrostatic scoring functions have been used to predict collagen stability from sequences (35, 36). Multiobjective optimization algorithms have been applied to simultaneously promote the formation of the specific target association and disfavor the competing states, leading to the design of an abc heterotrimer capable of specific assembly (37) and an obligate abc heterotrimer where folding requires all three chains (18). However, the lack of high-resolution structural information critically limits our ability to control stability and specificity, particularly under conditions of increasing the complexity (38–40).
To address this structural knowledge gap, we pursued the atomic resolution crystal structure of a registry-specific collagen heterotrimer, designed by optimizing salt-bridge networks on the surface of a target triple helix. The structure was used to select specific residues for mutagenesis and molecular modeling to better understand detailed energetic contributions of electrostatic interactions. This work advances our understanding of the role of surface electrostatics and hydrogen bonding in protein stability and fold specificity and provides computational tools for modeling collagen.
Results and Discussion
Backbone Topology.
The pitch of the triple helix has an established sequence dependence, with imino-acid-rich sequence forming a tighter 7 residues per 2 turns (7/2) helical repeat and imino-acid poor sequences forming a looser 10 residues per 3 turns (10/3) repeat (41, 42). Geometric analysis of the abc structure using a six-step parameter model adapted from DNA (42) showed that the heterotrimer formed a 10/3 helix where the twist parameter was −107.6°, close to the ideal 10/3 helix value of 108° (SI Appendix, Table S2). Consistent with previous high-resolution structures of collagen peptides (34, 43, 44), a repeated water-backbone side-chain network was observed along the face of the triple helix (SI Appendix, Fig. S1).
Specific Registry.
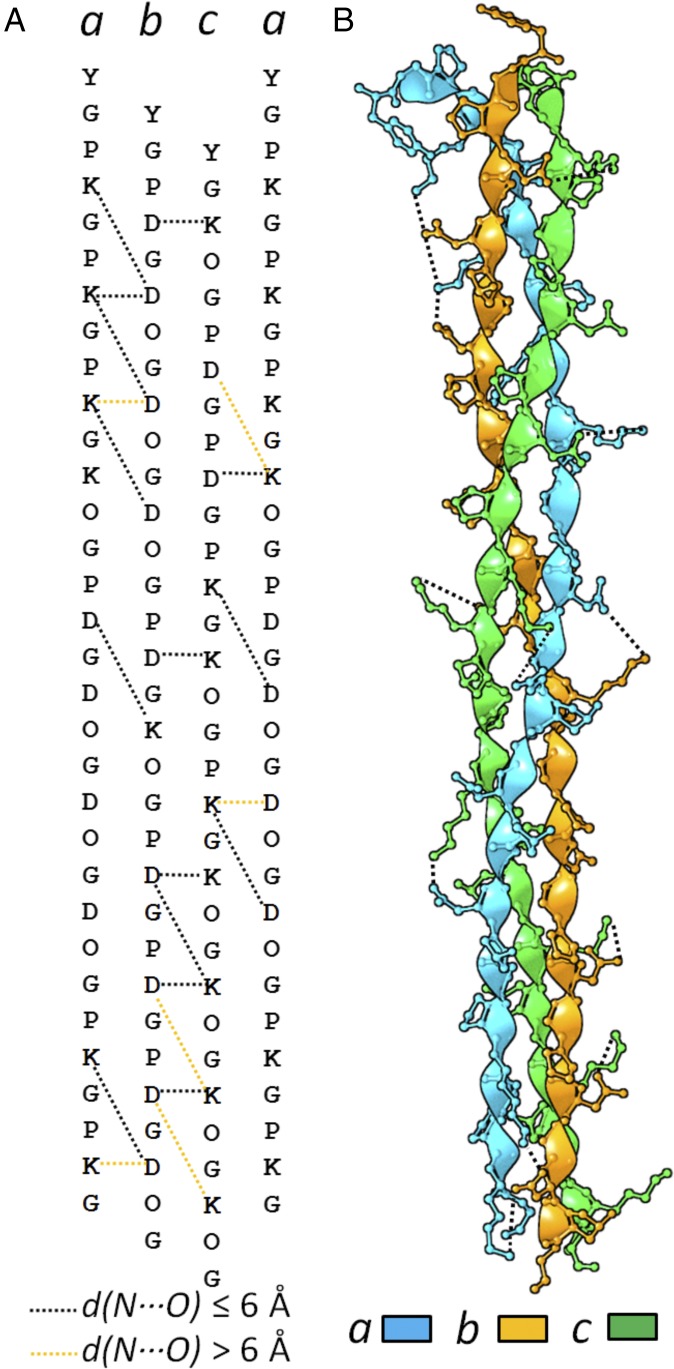
In prior studies, we knew only that abc assembled with the correct stoichiometry, but could not discriminate between the six registries: abc, acb, bac, bca, cab, and cba (18). The discrete sequence-based model predicted that, of the six registries, abc would contain the most favorable charge pairs and fewest charge repulsions (SI Appendix, Table S3). After multiple unsuccessful attempts to determine a structure, a tyrosine–glycine was added at the N terminus of each chain to facilitate peptide concentration measurements. This modification did not affect the stoichiometry of assembly and increased the melting temperature (Tm) of the 1:1:1 mixture from the previously observed 29 °C to 34 °C. The improved stability may have facilitated crystals of sufficient quality for structure determination. abc clearly adopted the intended registry in both triple helices of the asymmetric unit and formed the majority of predicted salt-bridge interactions (Fig. 1).
Fig. 1.
Sequence-based design (18) (A) and atomic-resolution structure (B) of abc (PDB ID code 5YAN). Salt bridges with distance d between Asp side-chain carboxylate oxygen and Lys side-chain amine nitrogen d <6 Å are denoted by black dashed lines. Salt bridges that are possible from a discrete sequence model, but are found to be weak (d > 6 Å) in the structure are denoted with yellow dashed lines. See SI Appendix for the crystallography statistics and structure details.
Side-Chain Interactions.
The triple-helix fold constrained interactions between neighboring chains to two geometric classes: axial (Y-X′), where the Y position on one chain sat above the X′ position of the neighboring chain in a manner coincident with the helical axis; and lateral (Y-X), where the two neighboring chains interacted in a direction perpendicular to the helical axis (Fig. 2). The notation for interacting pairs throughout will be the Y position residue followed by the X or X′ position residue. For Y-X and Y-X′ arrangements, there are four combinations of Lys and Asp, resulting in a total of eight types of charge–pair interaction. In the structure, there were 21 interchain salt bridges between Asp and Lys (Fig. 1), including both axial and lateral interactions. Based on the separation between interresidue charged groups, two of the four opposite charge-pair types, axial DK (7.05 ± 1.43 Å) and lateral KD (6.3 ± 0.62 Å), were consistently quite weak in the structure. The other favorable charge pairs, axial KD (2.9 ± 0.19 Å) and lateral DK (4.6 ± 0.76 Å), were consistently stronger. Reported homotrimer structures (30) contained primarily axial KD pairs with geometries very similar to those in abc.
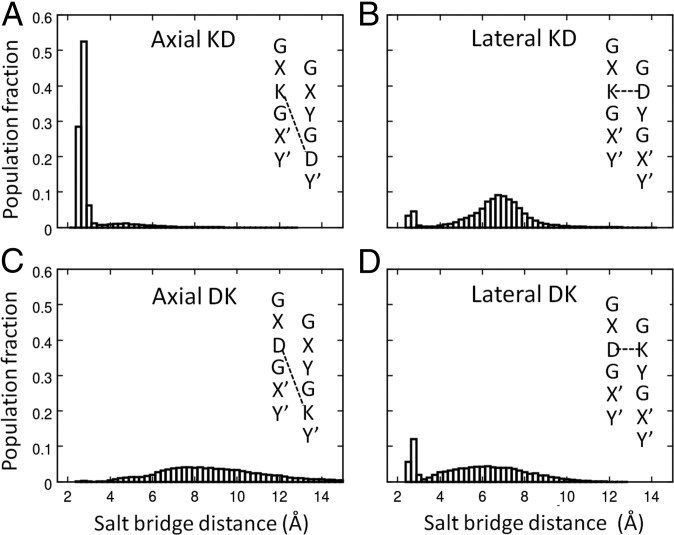
Fig. 2.
Favorable charge–pair distance distributions determined by MD simulations of abc for (A) axial KD interactions, (B) lateral KD interactions, (C) axial DK interactions, and (D) lateral DK interactions. Distributions are averaged over all occurrences of a particular class of interactions. Distances are between the closest side-chain carboxylate oxygen on Asp and the terminal side-chain amine on Lys. Residue-pair-specific distributions are presented in SI Appendix, Fig. S2.
Geometries of the various salt-bridge types were consistent across abc and between the two triple-helices asymmetric unit. To assess whether charge–pair interactions were biased by lattice contacts or other potential crystallization artifacts, we performed molecular dynamics (MD) simulations on individual triple helices from the abc asymmetric unit. Analyses of the resulting MD ensemble supported that the electrostatic interactions were dominating side-chain conformations (Fig. 2). The axial KD interaction was constrained between a narrow range of 2–3 Å, while axial DK and lateral KD showed little evidence of short salt-bridge formation. Lateral DK can exist in both states in simulation (SI Appendix, Fig. S3) and adopted intermediate distance in the experimental structure.
Deconstructing Ion Pair Energetics.
Unlike substitution studies on homotrimers, where mutations affect energetics across all three chains, abc offers a platform to pinpoint specific ion pairs and characterize their contribution to stability. Two types of mutations were introduced into abc: charged residue to alanine to disrupt the salt bridge and charge reversal substitutions (i.e., K → D or D → K) to assess destabilizing effects of the repulsive interactions. It should be noted that the primary method of characterizing stability is thermal unfolding, monitored by CD at the characteristic positive ellipticity band at 225 nm. We did not perform refolding experiments and thus have not demonstrated true equilibrium melting temperatures. Instead, we followed a gradual temperature schedule as developed (45). Consequently, the apparent melting temperatures do not represent direct measurements of stability and must be interpreted as apparent stabilities.
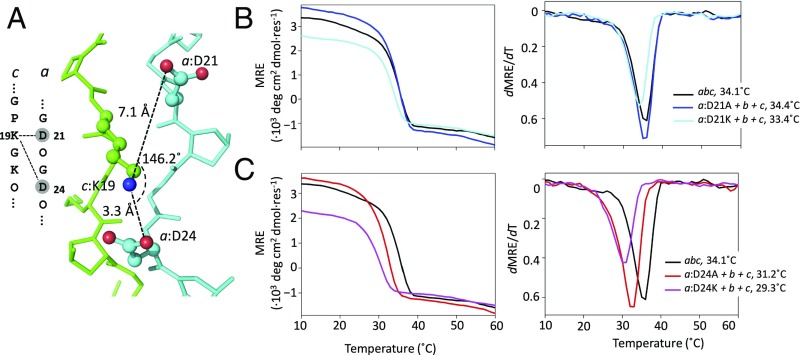
Based on simulations and geometric analysis of the abc structure, a KD interaction was predicted to be more favorable in the axial than the lateral arrangement. For example, c:K19 simultaneously participated in a tight axial salt bridge with a:D24 and a weak lateral interaction with a:D21 (Fig. 3A). Mutating a:D24A disrupted the strong axial KD salt bridge, reducing the Tm by 2.9 °C (Fig. 3C). In contrast, a:D21A had little effect on stability (Fig. 3B). Charge reversal substitutions at these positions followed a similar pattern. The a:D24K mutation reduced Tm by 1.9 °C relative to a:D24A, whereas a smaller destabilization of 0.9 °C was observed for a:D21K relative to a:D21A.
Fig. 3.
Mutagenesis of the three-residue network c:K19-a:D21/c:K19-a:D24 between chains c and a, comprising both lateral and axial KD salt bridges. (A) Three-residue network depicted in both sequence and structure. Thermal denaturation observed by CD spectroscopy at 225 nm for abc, a:D21 (B) or a:D24 (C) substituted with Ala or Lys, and combined with peptides b and c.
An isolated axial KD pair, where neither side chain was in proximity to interact with additional charged groups, appeared to be stronger than one in the complex salt-bridge network. Disrupting the axial KD interaction between c:K13 and a:D18 led to a much larger destabilization (5.5 °C) (Table 1 and SI Appendix, Fig. S4). Comparison of the axial KD interactions in the two contexts suggested anticooperativity in complex salt bridges for this type of interaction.
Table 1.
Effects of Ala substitutions on stability
| Interaction | Location | Tm, °C | ΔTm, °C* |
| Lateral DK | b:D16 c:K15A | 30.8 | 3.3 |
| c:D10A a:K12 | 31.3 | 2.8 | |
| Lateral KD | c:K19 a:D21A | 34.4 | −0.3 |
| Axial DK | a:D16 b:K18A | 33.5 | 0.6 |
| c:D7A a:K12 | 35.0 | −0.9 | |
| Axial KD | c:K19 a:D24A | 31.2 | 2.9 |
| c:K13 a:D18A | 28.6 | 5.5 |
Isolated charge-pair networks are highlighted in bold.
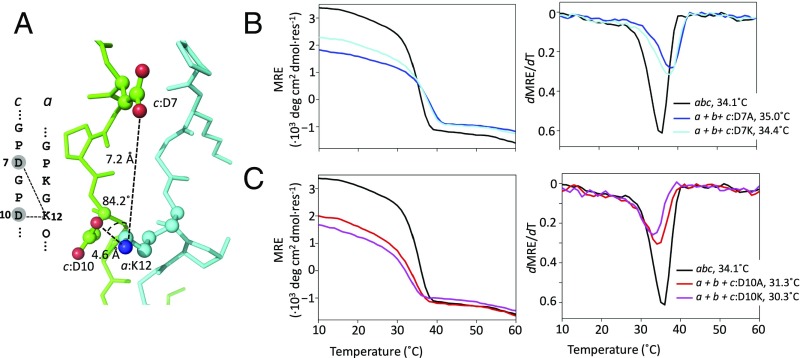
Similarly, the DK pairs interacted more tightly in lateral over axial arrangements, as seen in structure and simulation. a:K12 at an X position formed a tight lateral salt bridge with c:D7 and a weak axial one with c:D10 in the abc structure (Fig. 4A). Disrupting this interaction with a c:D10A mutation decreased Tm by 2.8 °C (Fig. 4C). In contrast, when the axial interaction was disrupted with the c:D7A mutation, Tm increased slightly (Fig. 4B). The charge-reversal mutations followed a similar pattern, with c:D10K having a greater impact on stability than c:D7K (Table 2). Disrupting the isolated interaction between b:D16 and c:K15 was more destabilizing than in the complex salt-bridge network (SI Appendix, Fig. S5), indicating weak, if any, anticooperativity.
Fig. 4.
Three-residue c:D7-a:K12/c:D10-a:K12 network containing axial and lateral DK salt bridges. (A) Salt bridges are depicted in both sequence and structure. (B and C) c:D7 (B) and c:D10 (C) were mutated to Ala and Lys, mixed with peptides a and b, and characterized by thermal denaturation, monitored at 225 nm by CD spectroscopy.
Table 2.
Effects of charge reversals on stability
| Interaction | Location | Tm, °C | ΔTm, °C* |
| Lateral DD | b:D16 c:K15D | 28.2 | −2.6 |
| Lateral KK | c:D10K a:K12 | 30.0 | −1.3 |
| c:K19 a:D21K | 33.4 | −1.0 | |
| Axial DD | a:D16 b:K18D | 29.0 | −4.5 |
| Axial KK | c:D7K a:K12 | 34.4 | −0.6 |
| c:K19 a:D24K | 29.3 | −1.9 |
Isolated charge-pair networks are highlighted in bold.
Corresponding values are in Table 1.
Based on the interactions probed in abc, complex salt-bridge networks involving three charged groups showed anticooperativity, with the strongest effect on residues participating in an axial KD interaction. For proteins in general, it has been proposed that the extent and direction of cooperativity in multibody salt-bridge networks is dependent on the angle, θ, between the central charged group and two interacting partners (46) (Figs. 3A and 4A). When θ < 90°, the central charged residue can interact tightly with both partners, resulting in positive cooperativity. When θ > 90°, the central charge must adopt different conformations to optimally interact with one of the other partners, resulting in anticooperativity (46). For abc, the average θ for lateral DK interaction was ∼85.4 ± 13.5°, on the cusp of anticooperativity. The axial KD θ = 132.2 ± 11.9°. In either case, salt-bridge networks appeared to be anticooperative, and isolated charge pairs contributed greater stability.
Lys–Lys repulsions had less impact on stability than Asp–Asp repulsions (Table 2), consistent with previous observations (47). This supports the assumptions of previously used discrete sequence-based scoring functions (18, 35, 48), where repulsions between acidic residues were weighted more strongly, presumably because the shorter side chains and decreased conformational freedom of Asp or Glu relative to Lys or Arg increased the effective strength of such repulsions.
Prediction of Collagen Self-Assembly.
It is challenging to model electrostatic interactions on protein surfaces. Given that all nonglycine positions in collagen are on the surface, such calculations are highly dependent on how well electrostatics, polar interactions, and solvation are modeled. Leveraging structural information from abc and accompanying atomistic simulations, we extended previous discrete sequence-based scoring functions by adding terms for isolated charge pairs and those involved in multibody networks. The contributions of side-chain flexibility were estimated from MD simulation and used to weight contributions of interactions to thermal stability:
| [1] |
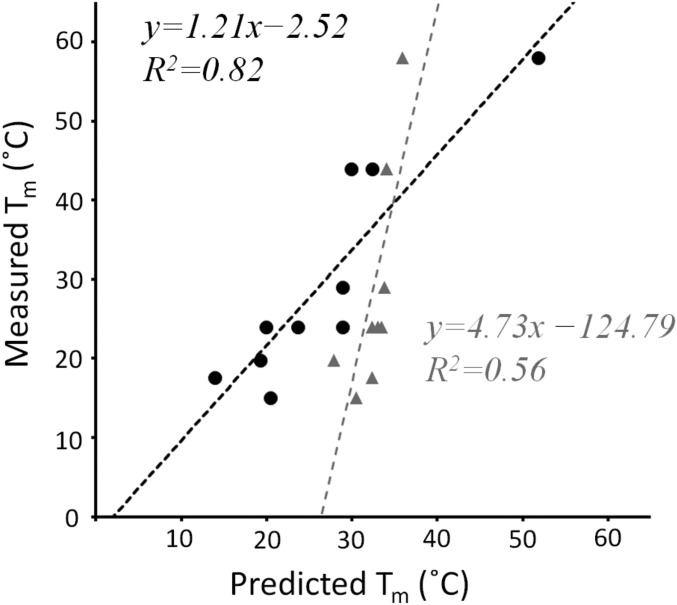
For a salt-bridge type i, ni is the number of observations, ΔTm,i is the contribution to stability based on mutagenesis, and pi is a weight obtained from molecular simulations (SI Appendix, Table S4). This scoring function was evaluated on a set of heterotrimers of known or modeled structures that exclusively utilized Lys–Asp interactions to promote stability and specificity. The dataset included circular permutations of peptides a, b, and c (39) and a collagen mimetic peptides designed by Fallas and Hartgerink (37). The updated scoring function showed improved performance relative to that used for the original design of abc (Fig. 5).
Fig. 5.
Stability measurements for 10 experimentally characterized synthetic collagen peptide heterotrimer systems were taken from various studies (18, 37, 39). Corresponding predicted stabilities were calculated by using Eq. 1 and the original scoring function used to design abc (18, 35). Where registry of a heterotrimer was not determined, the most stable computed association state was assumed. A detailed breakdown of favorable and unfavorable electrostatic interactions is presented in SI Appendix, Table S5.
A 10 °C computed stability gap exists between abc and the next most stable species, cab and bca (SI Appendix, Table S3), indicating that the specific assembly was likely due to disparities in charged residue networks between the target and competing states. Similarly, for the alanine and charge-reversal substitutions, the abc registry had the highest computed stability (SI Appendix, Figs. S7 and S8 and Table S6), and the next most stable states were bca and cab, indicating that single substitutions did not significantly perturb the association state energy landscape.
For type I collagen (COL1), the registry of the α1:α1:α2 heterotrimer has not been unequivocally determined. Previous computational, model peptide, and structural studies have variously placed the single α2 chain in the leading, middle, or trailing position (36, 49–53). Although collagen assembly is largely directed by globular prodomains (54), the fibrillar domain also showed preferences for specific association states. By using Eq. 1 on the ∼1,000-residue-long triple-helical regions of rat COL1A1 and COL1A2—UniProtKB IDs P02454 and P02466 (55) —the α2α1α1 association state with α2 in the leading position had the most favorable score (SI Appendix, Fig. S9). The next most stable state was an α1 homotrimer which has been observed to form, albeit with poor efficiency (56, 57). The α2 homotrimer had a poor assembly score, and its formation was not observed (58). The high computed stability of α2α1α1 conflicts with a structural analysis of COL1 complexed with von Willebrand factor, which requires a α1α1α2 association state (53). It is proposed that collagen stability is marginal at physiological temperatures to facilitate matrix remodeling in vivo (59).
In many cases within the COL1 sequence, regions showed preference for different association states and/or stoichiometries (SI Appendix, Fig. S10). The regions that most clearly discriminated the α2α1α1 stoichiometry occurred toward the center of the sequence, rather than near the N or C termini, suggesting that prodomains may determine specificity of assembly at the ends that is then facilitated through processive folding of the center by heterospecific salt-bridge networks. This analysis was parameterized on peptides and only considered K/D interactions leading to unrealistic melting temperatures for longer sequences. The landscape may change when considering the energetic features of other types of residue interactions. abc provides a useful platform for exploring position- and residue-specific pairwise interactions to address this question in a more complete manner.
Higher-Order Assembly.
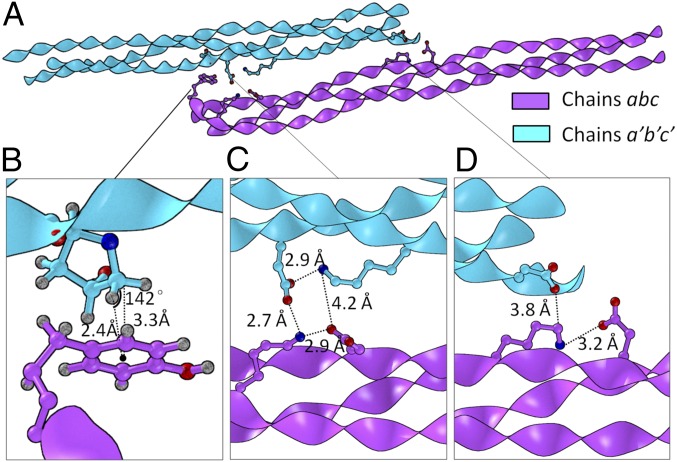
Two molecules of abc associated in an antiparallel configuration in the asymmetric unit (Fig. 6A). Although abc exists in solution as a single triple helix (39), interhelical and lattice contacts may provide insight into how supramolecular assembly is controlled. The helix–helix interaction was primarily mediated by solvent, but several direct interhelical interactions were observed. There was a CH-π contact interaction between the Cδ hydrogen of a′:Hyp19 and a:Tyr1. The geometry of this contact, donor-to-ring-center distance, and angle were well within expected cutoffs for a CH-π contact (Fig. 6B) (60). Notably, peptides without the tyrosine were difficult to crystalize and did not diffract to sufficient resolution for structure determination. CH-π contacts have been observed between Pro and Phe in the α2β1 integrin structure (61) and have been implicated in telopeptide-mediated collagen self-assembly (62).
Fig. 6.
(A) Relative orientation of two triple helices in an asymmetric unit. (B) CH-π interaction between a’:Hyp19 and a:Tyr1. The geometric center of the aromatic ring is represented with a dot. (C and D) Two complex salt-bridge networks. To show atomic-level details of the CH-π interaction, the stick representations in B are further enlarged compared with those in C and D.
Multiple examples were found in abc of Lys simultaneously forming intrahelical and interhelical salt bridges with Asp (Fig. 6 C and D). The participation of Lys in multiple simultaneous interactions on the triple helix is rare (63) and appears to be limited to structures with significant Lys and Asp content (30). Weak lattice contacts between asymmetric units were also observed (SI Appendix, Fig. S11). Notably, these contacts did not significantly bias charge pair geometries relative to distributions observed in molecular simulations, indicating that interactions that effectively mediate triple-helix assembly do not come at the cost of intrahelical stability (63).
Conclusions
The crystal structure of abc confirms a single registry of an obligate heterotrimer mediated by a complementary network of surface salt bridges. Their energetic contributions to stability can be ranked as axial KD > lateral DK > axial DK ∼ lateral KD. Complex salt bridges involving multiple charged residues can exhibit anticooperativity due to geometric constraints imposed by the triple helix. With structure-based constraints on computational modeling, collagen-folding stability and chain registry can be modeled with improved accuracy. These scoring functions will be used to enhance stability and specificity of collagen assembly, targeting conditions of increasing the system complexity (39, 64). The heterotrimer also provides a powerful platform to study collagen function and pathological mutations with structural precision.
Materials and Methods
Crystallization.
Peptides a, b, and c were dissolved in a 20 mM Tris⋅HCl buffer at pH 7.5 with 100 mM NaCl making the solutions of 5 mM concentration. a, b and c peptide solutions were mixed at a 1:1:1 ratio, incubated at 4 °C overnight, and then set for crystallization by using the hanging-drop method at 4 °C. After ∼2 mo, the best crystals were obtained under 60% (vol/vol) (+/−)-2-methyl-2,4-pentanediol, 40 mM sodium cacodylate trihydrate at pH 7.0, 80 mM potassium chloride, and 12 mM spermine tetrahydrochloride. The diffraction data were collected at 100 K, and the best one was diffracted to 1.77 Å. The space group was P212121, indexed by the hkl2000 software (HKL Research).
Structure Determination and Refinement.
The structures were solved by molecular replacement with the Phenix software suite (65) by using a fragment (residues 6–21) of human type III collagen (PDB ID code 3DMW) (31). This structure was chosen because of its low imino-acid containing sequence and 10/3 helical conformation (SI Appendix, Table S2), as would also be expected for abc. Initial phases were improved by rigid body refinement, followed by rounds of simulated annealing and anisotropic B-factor refinement using the Phenix suite. Model rebuilding was done in COOT (66). The refinement was performed by autoBUSTER (67). Water picking was started at 1.77 Å, at which point simulated annealing was replaced by atomic position refinement. The crystal structure has been deposited in the PDB (PDB ID code 5YAN). Refer to SI Appendix, Table S1 for crystallography statistics.
MD.
The coordinates from the abc structure were used as the initial structure for MD simulation. The structure was placed in a truncated dodecahedron periodic box of explicit TIP3P water (68) with 39,291 water molecules. The distance from the surface of the box to the closest atom of the solute was set to 10 Å. The simulation was carried out in the Amber99sb*-ILDN (69) force field with GROMACS (70). The lengths of bonds involving hydrogens were constrained, allowing for a 2-fs time step. Long-range electrostatic interactions were evaluated in reciprocal space by using the particle-Mesh Ewald method (71) with a maximum spacing for the fast Fourier transform grid of 1.2 Å and an interpolation via a sixth-order polynomial. The minimal cutoff distance for electrostatic and van der Waals interactions was set to 12 Å. The system was relaxed to a local energy minimum by using the steepest descent method (72). Subsequently, a 10-ns NPT and a 100-ns NVT simulation were conducted. A temperature of 297 K was maintained via the velocity rescaling algorithm (0.1 ps relaxation time), and the pressure P = 1 bar was controlled by using the weak coupling method of Berendsen et al. (73).
Peptide Synthesis.
The peptides were synthesized by using solid-phase Fmoc chemistry, purified to 95% purity by reverse-phase HPLC with mass spectrometry at GL Biochem Ltd. N and C termini were uncapped. See SI Appendix for peptide sequences, mass spectrometric analyses, and HPLC chromatograms for all of the peptides (SI Appendix, Fig. S12).
Supplementary Material
Acknowledgments
We thank Jiawei Wang at Tsinghua University and Helen Berman at Rutgers University for useful discussions. MD simulations were performed at the National Supercomputing Center in Wuxi, China. This work was supported by 1000 Plan of China Grant K2069999 (to F.X.), National Natural Science Foundation of China (NSFC) Grants 51603089 (to F.X.) and 21603088 (to H.Z.), and Natural Science Foundation of Jiangsu Province, China Grants BK20151126 (to F.X.) and BK20161066 (to H.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5YAN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802171115/-/DCSupplemental.
References
- 1.Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heino J. The collagen family members as cell adhesion proteins. Bioessays. 2007;29:1001–1010. doi: 10.1002/bies.20636. [DOI] [PubMed] [Google Scholar]
- 3.Tolstoshev P, Haber R, Crystal RG. Procollagen alpha2 mRNA is significantly different from procollagen alpha1(I) mRNA in size or secondary structure. Biochem Biophys Res Commun. 1979;87:818–826. doi: 10.1016/0006-291x(79)92031-x. [DOI] [PubMed] [Google Scholar]
- 4.Johansson C, Butkowski R, Wieslander J. The structural organization of type IV collagen. Identification of three NC1 populations in the glomerular basement membrane. J Biol Chem. 1992;267:24533–24537. [PubMed] [Google Scholar]
- 5.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hynes RO, Naba A. Overview of the matrisome—an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol. 2012;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudko SP, Bächinger HP. Structural insight for chain selection and stagger control in collagen. Sci Rep. 2016;6:37831. doi: 10.1038/srep37831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudko SP, Engel J, Bächinger HP. The crucial role of trimerization domains in collagen folding. Int J Biochem Cell Biol. 2012;44:21–32. doi: 10.1016/j.biocel.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Bourhis JM, et al. Structural basis of fibrillar collagen trimerization and related genetic disorders. Nat Struct Mol Biol. 2012;19:1031–1036. doi: 10.1038/nsmb.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sepehrpour H, Saha ML, Stang PJ. Fe-Pt twisted heterometallic bicyclic supramolecules via multicomponent self-assembly. J Am Chem Soc. 2017;139:2553–2556. doi: 10.1021/jacs.6b11860. [DOI] [PubMed] [Google Scholar]
- 11.Fujita D, et al. Self-assembly of tetravalent Goldberg polyhedra from 144 small components. Nature. 2016;540:563–566. doi: 10.1038/nature20771. [DOI] [PubMed] [Google Scholar]
- 12.Han D, et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 13.Mao C, LaBean TH, Relf JH, Seeman NC. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature. 2000;407:493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 14.Padilla JE, Colovos C, Yeates TO. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci USA. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher JM, et al. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson AR, et al. Computational design of water-soluble α-helical barrels. Science. 2014;346:485–488. doi: 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- 17.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu F, Zahid S, Silva T, Nanda V. Computational design of a collagen A:B:C-type heterotrimer. J Am Chem Soc. 2011;133:15260–15263. doi: 10.1021/ja205597g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salem G, Traub W. Conformational implications of amino acid sequence regularities in collagen. FEBS Lett. 1975;51:94–99. doi: 10.1016/0014-5793(75)80861-1. [DOI] [PubMed] [Google Scholar]
- 21.Traub W, Fietzek PP. Contribution of the α2 chain to the molecular stability of collagen. FEBS Lett. 1976;68:245–249. doi: 10.1016/0014-5793(76)80446-2. [DOI] [PubMed] [Google Scholar]
- 22.Hulmes DJS, Miller A, Parry DAD, Piez KA, Woodhead-Galloway J. Analysis of the primary structure of collagen for the origins of molecular packing. J Mol Biol. 1973;79:137–148. doi: 10.1016/0022-2836(73)90275-1. [DOI] [PubMed] [Google Scholar]
- 23.Fallas JA, Lee MA, Jalan AA, Hartgerink JD. Rational design of single-composition ABC collagen heterotrimers. J Am Chem Soc. 2012;134:1430–1433. doi: 10.1021/ja209669u. [DOI] [PubMed] [Google Scholar]
- 24.Gauba V, Hartgerink JD. Surprisingly high stability of collagen ABC heterotrimer: Evaluation of side chain charge pairs. J Am Chem Soc. 2007;129:15034–15041. doi: 10.1021/ja075854z. [DOI] [PubMed] [Google Scholar]
- 25.Gauba V, Hartgerink JD. Self-assembled heterotrimeric collagen triple helices directed through electrostatic interactions. J Am Chem Soc. 2007;129:2683–2690. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- 26.Jalan AA, Demeler B, Hartgerink JD. Hydroxyproline-free single composition ABC collagen heterotrimer. J Am Chem Soc. 2013;135:6014–6017. doi: 10.1021/ja402187t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Leary LER, Fallas JA, Hartgerink JD. Positive and negative design leads to compositional control in AAB collagen heterotrimers. J Am Chem Soc. 2011;133:5432–5443. doi: 10.1021/ja111239r. [DOI] [PubMed] [Google Scholar]
- 28.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry. 2005;44:1414–1422. doi: 10.1021/bi048216r. [DOI] [PubMed] [Google Scholar]
- 29.Venugopal MG, Ramshaw JA, Braswell E, Zhu D, Brodsky B. Electrostatic interactions in collagen-like triple-helical peptides. Biochemistry. 1994;33:7948–7956. doi: 10.1021/bi00191a023. [DOI] [PubMed] [Google Scholar]
- 30.Fallas JA, Dong J, Tao YJ, Hartgerink JD. Structural insights into charge pair interactions in triple helical collagen-like proteins. J Biol Chem. 2012;287:8039–8047. doi: 10.1074/jbc.M111.296574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boudko SP, et al. Crystal structure of human type III collagen Gly991-Gly1032 cystine knot-containing peptide shows both 7/2 and 10/3 triple helical symmetries. J Biol Chem. 2008;283:32580–32589. doi: 10.1074/jbc.M805394200. [DOI] [PubMed] [Google Scholar]
- 32.Kramer RZ, et al. Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair. J Mol Biol. 2000;301:1191–1205. doi: 10.1006/jmbi.2000.4017. [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer Green R, Berman HM. An overview of structural studies of the collagen triple helix. In: Bansal M, Srinivasan N, editors. Biomolecular Forms and Functions: A Celebration of 50 Years of the Ramachandran Map. World Scientific; Singapore: 2013. [Google Scholar]
- 35.Xu F, Zhang L, Koder RL, Nanda V. De novo self-assembling collagen heterotrimers using explicit positive and negative design. Biochemistry. 2010;49:2307–2316. doi: 10.1021/bi902077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanda V, Zahid S, Xu F, Levine D. Computational design of intermolecular stability and specificity in protein self-assembly. Methods Enzymol. 2011;487:575–593. doi: 10.1016/B978-0-12-381270-4.00020-2. [DOI] [PubMed] [Google Scholar]
- 37.Fallas JA, Hartgerink JD. Computational design of self-assembling register-specific collagen heterotrimers. Nat Commun. 2012;3:1087. doi: 10.1038/ncomms2084. [DOI] [PubMed] [Google Scholar]
- 38.Nanda V, Belure SV, Shir OM. Searching for the Pareto frontier in multi-objective protein design. Biophys Rev. 2017;9:339–344. doi: 10.1007/s12551-017-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu F, Silva T, Joshi M, Zahid S, Nanda V. Circular permutation directs orthogonal assembly in complex collagen peptide mixtures. J Biol Chem. 2013;288:31616–31623. doi: 10.1074/jbc.M113.501056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belure SV, Shir OM, Nanda V. The Genetic and Evolutionary Computation Conference. Association for Computing Machinery; New York: 2017. Protein design by multiobjective optimization: Evolutionary and non-evolutionary approaches; pp. 1081–1088. [Google Scholar]
- 41.Bella J. A new method for describing the helical conformation of collagen: Dependence of the triple helical twist on amino acid sequence. J Struct Biol. 2010;170:377–391. doi: 10.1016/j.jsb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Xu F, et al. Parallels between DNA and collagen - comparing elastic models of the double and triple helix. Sci Rep. 2017;7:12802. doi: 10.1038/s41598-017-12878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer RZ, Bella J, Mayville P, Brodsky B, Berman HM. Sequence dependent conformational variations of collagen triple-helical structure. Nat Struct Biol. 1999;6:454–457. doi: 10.1038/8259. [DOI] [PubMed] [Google Scholar]
- 44.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 45.Persikov AV, Xu Y, Brodsky B. Equilibrium thermal transitions of collagen model peptides. Protein Sci. 2004;13:893–902. doi: 10.1110/ps.03501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acevedo-Jake AM, Clements KA, Hartgerink JD. Synthetic, register-specific, AAB heterotrimers to investigate single point glycine mutations in Osteogenesis imperfecta. Biomacromolecules. 2016;17:914–921. doi: 10.1021/acs.biomac.5b01562. [DOI] [PubMed] [Google Scholar]
- 47.Parmar AS, Joshi M, Nosker PL, Hasan NF, Nanda V. Control of collagen stability and heterotrimer specificity through repulsive electrostatic interactions. Biomolecules. 2013;3:986–996. doi: 10.3390/biom3040986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Summa CM, Rosenblatt MM, Hong J-K, Lear JD, DeGrado WF. Computational de novo design, and characterization of an A(2)B(2) diiron protein. J Mol Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 49.Piez KA, Trus BL. Sequence regularities and packing of collagen molecules. J Mol Biol. 1978;122:419–432. doi: 10.1016/0022-2836(78)90419-9. [DOI] [PubMed] [Google Scholar]
- 50.Bender E, Silver FH, Hayashi K, Trelstad RL. Type I collagen segment long spacing banding patterns. Evidence that the alpha 2 chain is in the reference or A position. J Biol Chem. 1982;257:9653–9657. [PubMed] [Google Scholar]
- 51.Ottl J, Musiol HJ, Moroder L. Heterotrimeric collagen peptides containing functional epitopes. Synthesis of single-stranded collagen type I peptides related to the collagenase cleavage site. J Pept Sci. 1999;5:103–110. doi: 10.1002/(SICI)1099-1387(199902)5:2<103::AID-PSC188>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 52.Perumal S, Antipova O, Orgel JP. Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis. Proc Natl Acad Sci USA. 2008;105:2824–2829. doi: 10.1073/pnas.0710588105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brondijk TH, Bihan D, Farndale RW, Huizinga EG. Implications for collagen I chain registry from the structure of the collagen von Willebrand factor A3 domain complex. Proc Natl Acad Sci USA. 2012;109:5253–5258. doi: 10.1073/pnas.1112388109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma U, et al. Structural basis of homo- and heterotrimerization of collagen I. Nat Commun. 2017;8:14671. doi: 10.1038/ncomms14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.UniProt Consortium T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moro L, Smith BD. Identification of collagen alpha1(I) trimer and normal type I collagen in a polyoma virus-induced mouse tumor. Arch Biochem Biophys. 1977;182:33–41. doi: 10.1016/0003-9861(77)90280-6. [DOI] [PubMed] [Google Scholar]
- 57.Jimenez SA, Bashey RI, Benditt M, Yankowski R. Identification of collagen alpha1(I) trimer in embryonic chick tendons and calvaria. Biochem Biophys Res Commun. 1977;78:1354–1361. doi: 10.1016/0006-291x(77)91441-3. [DOI] [PubMed] [Google Scholar]
- 58.Lees JF, Tasab M, Bulleid NJ. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J. 1997;16:908–916. doi: 10.1093/emboj/16.5.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brandl M, Weiss MS, Jabs A, Sühnel J, Hilgenfeld R. C-H...π-interactions in proteins. J Mol Biol. 2001;307:357–377. doi: 10.1006/jmbi.2000.4473. [DOI] [PubMed] [Google Scholar]
- 61.Emsley J, Knight CG, Farndale RW, Barnes MJ. Structure of the integrin alpha2beta1-binding collagen peptide. J Mol Biol. 2004;335:1019–1028. doi: 10.1016/j.jmb.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 62.Kar K, et al. Aromatic interactions promote self-association of collagen triple-helical peptides to higher-order structures. Biochemistry. 2009;48:7959–7968. doi: 10.1021/bi900496m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parmar AS, James JK, Grisham DR, Pike DH, Nanda V. Dissecting electrostatic contributions to folding and self-assembly using designed multicomponent peptide systems. J Am Chem Soc. 2016;138:4362–4367. doi: 10.1021/jacs.5b10304. [DOI] [PubMed] [Google Scholar]
- 64.Xu F, et al. Compositional control of higher order assembly using synthetic collagen peptides. J Am Chem Soc. 2012;134:47–50. doi: 10.1021/ja2077894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams PD, Mustyakimov M, Afonine PV, Langan P. Generalized X-ray and neutron crystallographic analysis: More accurate and complete structures for biological macromolecules. Acta Crystallogr D Biol Crystallogr. 2009;65:567–573. doi: 10.1107/S0907444909011548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bricogne G. Direct phase determination by entropy maximization and likelihood ranking: Status report and perspectives. Acta Crystallogr D Biol Crystallogr. 1993;49:37–60. doi: 10.1107/S0907444992010400. [DOI] [PubMed] [Google Scholar]
- 68.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 69.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 72.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 73.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.