Significance
Aberrant or unwanted transcripts can be degraded by the RNA exosome with the help of the nuclear exosome-targeting (NEXT) complex. NEXT, composed of RNA-binding protein RBM7, scaffold ZCCHC8, and helicase MTR4, is implicated in stress response, neurodegeneration, and viral ribogenesis. Here, we characterize the activities of NEXT that support its role in exosome-mediated decay. NEXT catalyzes 3′→5′ helicase activity and disrupts RNA:RNA and DNA:RNA duplexes more efficiently than MTR4. Optimal activity is observed when substrates include a uridine-rich motif, for interactions with RBM7, and a 3′ poly(A) tail. The ZCCHC8 C-terminal domain binds the helicase core and can stimulate MTR4 helicase/ATPase activities. Our results highlight the interplay among NEXT subunits to ensure effective targeting of substrates.
Keywords: RNA, RBM7, NEXT complex, R-LOOP, RNA decay
Abstract
The nuclear exosome-targeting (NEXT) complex functions as an RNA exosome cofactor and is involved in surveillance and turnover of aberrant transcripts and noncoding RNAs. NEXT is a ternary complex composed of the RNA-binding protein RBM7, the scaffold zinc-knuckle protein ZCCHC8, and the helicase MTR4. While RNA interactions with RBM7 are known, it remains unclear how NEXT subunits collaborate to recognize and prepare substrates for degradation. Here, we show that MTR4 helicase activity is enhanced when associated with RBM7 and ZCCHC8. While uridine-rich substrates interact with RBM7 and are preferred, optimal activity is observed when substrates include a polyadenylated 3′ end. We identify a bipartite interaction of ZCCHC8 with MTR4 and uncover a role for the conserved C-terminal domain of ZCCHC8 in stimulating MTR4 helicase and ATPase activities. A crystal structure reveals that the ZCCHC8 C-terminal domain binds the helicase core in a manner that is distinct from that observed for Saccharomyces cerevisiae exosome cofactors Trf4p and Air2p. Our results are consistent with a model whereby effective targeting of substrates by NEXT entails recognition of elements within the substrate and activation of MTR4 helicase activity.
A central player in the processing, quality control, and turnover of nearly every class of RNA produced in eukaryotic nuclei is the RNA exosome, a conserved multisubunit ribonuclease complex (1–3). The RNA exosome interacts with a variety of cofactors and protein complexes. Early work in Saccharomyces cerevisiae identified cellular compartment-specific RNA helicases and associated cofactors that engage RNA substrates to promote efficient threading into the catalytic sites of the RNA exosome through posttranscriptional modifications and/or disruption of structured RNA. In the cytoplasm, the exosome interacts with the cofactor Ski7p, which, in turn, interacts with the Ski complex that consists of the tetratricopeptide repeat-containing protein Ski3p, the β-propeller protein Ski8p, and the Ski2p helicase (4). The Ski complex and Ski7p recruit the exosome to a stalled 80S ribosome undergoing nonstop decay (5). In the nucleus, the Mtr4p helicase associates with the S. cerevisiae RNA exosome via interactions with Mpp6p (6, 7) and Rrp47p (8). Mtr4p is involved in the maturation of the large subunit 5.8S ribosomal RNA (9). Mtr4p is also part of the Trf4p/Trf5p-Air1p/Air2p-Mtr4p polyadenylation (TRAMP) complex in which the Trf4p/Trf5p subunit catalyzes nontemplated 3′ oligoadenylation to prepare RNA substrates for degradation (10). TRAMP participates in the turnover of RNA substrates that include hypomodified tRNAs and cryptic unstable transcripts (CUTs) in the yeast nuclei (11, 12).
Related exosome-associated RNA helicases have been identified in human: namely, SKIV2L (13) and MTR4 (also known as SKIV2L2) (14). MTR4 is present in the nucleus and is a central subunit of several protein complexes, including the nuclear exosome-targeting (NEXT) complex (15), the poly(A) exosome-targeting (PAXT) connection (16), and the polysome protector complex (PPC) (17). NEXT is a nucleoplasmic ternary complex consisting of RBM7, ZCCHC8, and MTR4 (15). RBM7 is a 31-kDa protein with a polyuridine-specific RNA recognition motif (RRM) (18), while the zinc-knuckle protein ZCCHC8 acts as a scaffold, mediating the interaction between RBM7 and MTR4 (16). NEXT is involved in exosome-mediated surveillance and decay of noncoding RNAs, such as enhancer RNAs (eRNAs) and aberrant 3′-extended transcripts from small nuclear RNA (snRNA), telomerase RNA, and replication-dependent histone genes (15, 19, 20). It ensures a unidirectional output from pervasive transcription of bidirectional promoters by targeting promoter upstream transcripts (PROMPTs) (21) or upstream antisense RNAs (uaRNAs) (22) for decay. NEXT interacts with various transcript-associated complexes: namely, the nuclear cap-binding complex (23), the spliceosome (24), and the pre-mRNA 3′ processing complex (25). Given its wide range of substrates and interactions, NEXT is an important node in regulating nuclear exosome activities and takes part in several biological processes, including DNA damage response (26), stress response (27), and viral ribogenesis (28). Consequently, its functional inactivation can lead to altered RNA metabolism and is implicated in the development of spinal muscular atrophy-like disorder (29).
The biochemical activities of NEXT have not been characterized to date. It is assumed to play a role in disrupting RNA structures and ultimately delivering RNA substrates to the exosome. How each NEXT subunit contributes to this function remains elusive. In this study, we reconstitute the NEXT core complex and demonstrate its ability to catalyze 3′-to-5′ RNA unwinding. We show that the helicase activity of NEXT is enhanced relative to MTR4 alone. NEXT exhibits sequence preferences for binding and/or unwinding RNA substrates. By mapping an RNA path in NEXT, we determine that optimal substrates include a 3′ poly(A) tail and a polyuridine motif upstream of the 3′ end for interaction with RBM7. We also characterize interactions of ZCCHC8 with MTR4 and identify two MTR4-binding regions within ZCCHC8. One of these regions includes the highly conserved C-terminal domain of ZCCHC8 that binds the helicase core of MTR4 and is required for enhancing MTR4 helicase and ATPase activities. Together, this work provides a functional dissection of interactions and interplay between NEXT subunits and RNA that are consistent with specific substrate targeting to the nuclear RNA exosome.
Results
MTR4 Helicase Activity Is Activated When Incorporated into NEXT.
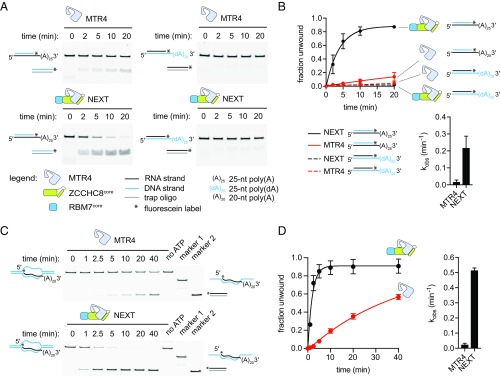
Reconstitution of NEXT using full-length proteins proved to be challenging. RBM7 and ZCCHC8 were prone to aggregation and degradation during purification, as observed by others (30), resulting in low yield. After probing various truncations, we identified a soluble RBM7 construct (residues 6 to 94; RBM7core) containing the RNA Recognition Motif (RRM) and a minimal ZCCHC8 construct (ZCCHC8core) with deletion of a low complexity region (residues 416 to 507) that reduced degradation and improved yield. Full-length MTR4 and these variants of RBM7 and ZCCHC8 were used to reconstitute a soluble NEXT core complex (Fig. 1A) (denoted as “NEXT” hereafter for simplicity). The helicase activities of MTR4 and NEXT were examined using a strand displacement gel-shift assay in which the release of a fluorescently labeled RNA strand from a double-stranded substrate was monitored upon duplex unwinding. The reaction was supplemented with a trap oligo to capture and prevent reannealing of the displaced strand. Each substrate consists of a 16-bp duplex region with or without a poly(A) overhang on either the 5′ or 3′ end. MTR4 and NEXT displayed unwinding activities for the 3′ tailed RNA duplex but not for substrates with blunt ends or a 5′ overhang, consistent with a 3′-to-5′ helicase polarity (Fig. 1B). Neither AMPPNP nor ADP was able to substitute for ATP in unwinding reactions (Fig. 1C). Furthermore, a mutation (E253Q) in the DExH motif involved in ATP hydrolysis resulted in undetectable MTR4 and NEXT helicase activities, indicating a requirement for ATP hydrolysis in the unwinding process (Fig. 1D). Interestingly, MTR4 helicase activity appears stimulated in NEXT. At identical enzyme concentration, NEXT was more efficient than MTR4 in unwinding the 3′ tailed substrate (Fig. 1 E and F; also, see SI Appendix, Table S1). Despite the low yield, we compared the activities of NEXT reconstituted using the full-length proteins to our optimized NEXT minimal core complex and observed comparable activities (SI Appendix, Fig. S1). Hence, we proceeded to use the NEXT minimal core complex for subsequent experiments. The strand displacement rate extrapolated to enzyme saturation (Vmax) for NEXT was >30-fold higher than that of MTR4 alone (Fig. 1G). These data suggest that MTR4 helicase activity is stimulated within the NEXT complex.
Fig. 1.
RNA helicase activities of MTR4 and NEXT. (A) Coomassie-stained SDS/PAGE gel showing recombinant MTR4 and the reconstituted NEXT core complex. The legend applies to all panels in the figure. (B) End point helicase assay of MTR4 and NEXT on RNA duplex substrates with 3′ A25 overhang, 5′ A25 overhang, or blunt ends. Unwinding reactions were quenched after 40 min. No ATP lanes represent unwinding reactions with 2 µM MTR4 or 0.5 µM NEXT in the absence of ATP. Marker lanes indicate the expected band for the unwound fluorescein-labeled (*) RNA strand captured by the trap oligo (gray line). (C) Dependence of MTR4 and NEXT helicase activity on ATP hydrolysis. Assays were carried out using a 3′ A25 tailed RNA substrate with 2 µM MTR4 or 0.5 µM NEXT and stopped after 40 min. (D) Inactivation of MTR4 and NEXT helicase activity due to a mutation (E253Q; denoted by EQ superscript) in the DExH-box motif. The end point helicase assays were performed as in C. (E) Representative native PAGE gel showing the time courses of MTR4 (800 nM) and NEXT (800 nM) unwinding reactions on 3′ A25 tailed RNA duplex. (F) Graph showing the time courses of MTR4 and NEXT helicase activity on 3′ A25 tailed RNA duplex. Data points show mean ± SD (n = 9). Rate constants (mean ± SD) shown in the bar graph were determined by fitting the data to the integrated first-order rate law. (G) Graph showing the dependence of strand displacement rate on protein concentration for MTR4 and NEXT using the 3′ A25 tailed RNA substrate. Data points show mean ± SD (n = 3). Curves represent best fit to the equations v0 = Vmax [E]/K1/2 for MTR4 and v0 = Vmax*[E]/(K1/2 + [E]*/(1 + [E]/K′1/2)) for NEXT. The bar graph shows a comparison of maximal strand displacement rate Vmax (mean ± SD) for MTR4 and NEXT.
NEXT Resolves DNA:RNA Hybrids with an RNA-Tracking Strand.
The RNA exosome and MTR4 participate in the resolution of DNA:RNA hybrids produced during asymmetric DNA strand mutagenesis in antibody diversification in B cells. Genetic ablation of exosome subunits and cellular depletion of MTR4 resulted in accumulation of R-loops and reduced Ig class switch recombination efficiency (14, 31). Interestingly, MTR4 was isolated with NEXT subunits from B cells (14). Thus, we sought to determine if MTR4 could resolve DNA:RNA hybrids in vitro in the context of NEXT. Unwinding of a DNA:RNA heteroduplex with a 3′ poly(A) RNA tracking strand was observed for both MTR4 and NEXT, with the latter exhibiting 13-fold higher activity (Fig. 2 A and B; see also SI Appendix, Table S1). In contrast, a DNA:RNA hybrid with a DNA tracking strand was a poor substrate (Fig. 2 A and B). NEXT and MTR4, albeit to a weaker extent, were also capable of releasing RNA from a three-stranded substrate designed to mimic R-loops (32) (Fig. 2 C and D). These results suggest that MTR4, given its weaker helicase activity in isolation, may be incorporated into NEXT for effective resolution of DNA:RNA hybrids.
Fig. 2.
MTR4 and NEXT unwind DNA:RNA hybrids with an RNA tracking strand. (A) Time courses of unwinding reactions of MTR4 (800 nM) and NEXT (800 nM) on DNA:RNA hybrids (10 nM). DNA and RNA strands are depicted as blue and black lines, respectively. Legend applies to all panels in the figure. (B) Quantitation of unwinding experiment in A with average values (n = 5 for substrate with RNA tracking strand and n = 3 for substrate with DNA tracking strand) plotted and fitted to a pseudo first-order rate law. Error bars represent ±SD. The bar graph shown compares the observed rate constants of MTR4 and NEXT on a 3′ A25 tailed DNA:RNA heteroduplex substrate with an RNA tracking strand. (C) MTR4 (800 nM) and NEXT (800 nM) facilitate displacement of fluorescein-labeled (*) RNA with a 3′ A20 extension from a three-stranded substrate (10 nM) that mimics R-loops. No ATP lanes represent unwinding reactions in the absence of ATP quenched after 40 min. Markers 1 and 2 indicate the expected bands for the fluorescein-labeled (*) RNA annealed to either a DNA strand (blue line) of the R-loop mimic or the trap oligo (gray line), respectively. Legends for substrate schematics as in A. (D) Quantitation of MTR4 and NEXT unwinding of the R-loop mimic. Fractions unwound (mean ± SD) are plotted against time. Best fit curve to pseudo first-order rate law is derived from data (n = 3) collected from three separate reactions. The bar graph shown compares the observed rate constants (mean ± SD) of MTR4 and NEXT for unwinding the R-loop mimic.
NEXT Exhibits Preference for Binding and Unwinding a 3′ Poly(U) Tailed RNA Substrate.
RBM7 binds RNA promiscuously but exhibits some preference for polyuridine sequences (18, 19). Indeed, NEXT bound a 3′ poly(U) tailed duplex with 18-fold lower dissociation constant (Kd) compared with a 3′ poly(A) tailed duplex (Fig. 3A). No binding preferences were observed for MTR4 alone or a binary complex of ZCCHC8 and MTR4, underscoring a role of RBM7 in providing poly(U) specificity. Comparable MTR4 unwinding kinetics was observed for a 3′ poly(U) tailed substrate relative to 3′ poly(A) tailed substrates (SI Appendix, Fig. S2). We determined whether the observed preference for polyuridine sequences influence unwinding by NEXT. Using a molecular beacon helicase assay (33), we compared the initial velocity of strand displacement at various concentrations of NEXT, using substrates with either poly(A) or poly(U) 3′ overhangs (Fig. 3B). While strand displacement rates extrapolated to enzyme saturation (Vmax) were comparable between 3′ poly(U) tailed and 3′ poly(A) tailed substrates (Fig. 3C; see also SI Appendix, Table S1), the half-maximal rate concentration (K1/2) for the 3′ poly(U) tailed substrate was ∼30-fold lower than that for the 3′ poly(A) tailed substrate (Fig. 3C and SI Appendix, Table S1). The rate-limiting step for unwinding has not been determined, but differences in K1/2 might reflect the contribution of RBM7 to binding to the 3′ poly(U) tailed substrate within NEXT. Together, these results corroborate the reported preferential targeting of NEXT to U-rich elements within NEXT targets in vivo (18, 19).
Fig. 3.
Sequence specificity of NEXT. (A) Bar graph showing the equilibrium dissociation constant (Kd) of MTR4, ZCCHC8core–MTR4, and NEXT on RNA duplexes with either 3′ A20 overhang or 3′ U20 overhang. Dissociation constants were determined using EMSA. Error bars show ±SD (n = 3). (B) Plot of initial velocity (v0) of strand displacement versus NEXT concentration. Curves represent best fit to the equations v0 = Vmax [E]/K1/2 for 3′ A20 tailed RNA substrate and v0 = Vmax*[E]/(K1/2 + [E]*/(1 + [E]/K′1/2)) for 3′ U20 RNA substrate. Data (n = 3) were collected from three separate reactions; error bars represent ±SD. (C) Bar graph comparing the maximal strand displacement rate (Vmax) and half-maximal rate concentration (K1/2) of NEXT on 3′ tailed A20 and 3′ tailed U20 substrates.
Mapping NEXT Subunit Contacts to RNA.
We sought to map the interactions between NEXT and RNA using a site-directed 4-thiouridine cross-linking assay. NEXT was incubated with a nonhydrolyzable ATP analog AMPPNP and fluorescently labeled 20-nt RNA that contained a single internal 4-thiouridine incorporated at several positions. RNA-protein cross-linking was induced by exposure to long-wavelength UV. The 3′ end of the RNA substrate forms cross-links most efficiently with MTR4, followed by cross-links to ZCCHC8 when 4-thiouridine was positioned −8 and −12 (Fig. 4A). Cross-links to RBM7 occurred within a broader region of RNA centered at position −16 (Fig. 4A). While it was previously known that RBM7 and MTR4 bound RNA, the RNA cross-linking pattern (depicted in Fig. 4B) supports an RNA path that involves contacts to ZCCHC8 in a manner that is consistent with NEXT subunit organization reported previously (16).
Fig. 4.
Interaction of NEXT with RNA. (A) Site-directed 4-thiouridine cross-linking assay of NEXT with an A20 substrate. (B) Diagram showing the proposed RNA binding path in NEXT. (C) Molecular beacon RNA duplex substrates I to III used to test the effect of the position of uridine sequences on RNA affinity and helicase activity of NEXT. (D) Bar graph showing the equilibrium dissociation constant (Kd) of NEXT on substrates I to III. Dissociation constants were measured using EMSA. Error bars represent ±SD from three experiments (n = 3). (E) Molecular beacon helicase assay of NEXT. Reactions were carried out at 30 °C using 50 nM NEXT and 10 nM RNA. Data (n = 3) were fitted to the integrated pseudo first-order rate law. Error bars show ±SD. (F) Bar graph showing the observed rate constants and reaction amplitudes obtained for substrates I to III, 3′ A20 tailed duplex, and 3′ U20 tailed duplex. Error bars represent ±SD.
Having defined an RNA binding path, we designed additional substrates (I to III) containing a pentauridine sequence at defined positions within the 3′ overhang (Fig. 4C) and determined the ability of NEXT to bind and unwind these substrates. Based on cross-linking (Fig. 4B), substrates I to III include pentauridine sequences at positions expected to bind RBM7 and/or ZCCHC8. These substrates displayed equilibrium binding and observed rate constants comparable with a 3′ U20 tailed substrate (Fig. 4 D–F). However, the reaction amplitudes for substrates I to III indicated that unwinding proceeded to near completion (Fig. 4 E and F). These results suggest that the sequence of the RNA substrate can influence the recruitment and activities of NEXT and that an optimal substrate includes a polyuridine motif upstream of a 3′ adenylate tail.
ZCCHC8 C-Terminal Domain Interacts with MTR4 and Is Necessary for Activating MTR4 Helicase Activity.
Acting as an adaptor, ZCCHC8 is proposed to bridge interactions between RBM7 and MTR4 (16, 30). While the proline-rich domain in spliceosome-associated protein (PSP) (residues 283 to 327) of ZCCHC8 recruits the RRM module of RBM7 (30), determinants underlying interactions of ZCCHC8 with MTR4 remain unclear. To identify regions in ZCCHC8 important for binding MTR4, we performed pull-down assays using copurified complexes between Strep-tagged RBM7core and various constructs of ZCCHC8 (Z1 to Z4) as bait (Fig. 5A). Consistent with previous reports, RBM7 was not sufficient for direct interaction with MTR4 (Fig. 5B, lane 11). In contrast, MTR4 precipitates in the presence of full-length ZCCHC8 (Z1) (Fig. 5B, lane 7), as well as the ZCCHC8 fragment ZCCHC81–327 (Z2) (Fig. 5B, lane 8). While ZCCHC8283–658 (Z3) failed to interact with MTR4 (Fig. 5B, lane 9), inclusion of the C-terminal domain (CTD) in the Z4 construct (ZCCHC8283–415 + 508–707) precipitates MTR4 (Fig. 5B, lane 10). Additionally, Strep-tagged ZCCHC8CTD alone (Z5; ZCCHC8659–707) precipitates MTR4 (Fig. 5B, lane 12). From these results, we infer a bipartite interaction of ZCCHC8 with MTR4. The first appears to involve the N-terminal region (residues 1 to 282) that contains a predicted coiled-coil region and a zinc-knuckle motif, and the second involves the C-terminal domain ZCCHC8CTD.
Fig. 5.
Mapping of ZCCHC8 interaction with MTR4 uncovers a role for the ZCCHC8 C-terminal domain in stimulating MTR4 helicase activity. (A) ZCCHC8 constructs (Z1 to Z5) were used for the pull-down assays. (B) Coomassie-stained SDS/PAGE gel showing the input and eluate of the pull-down assay. Various ZCCHC8 constructs were copurified with Strep-RBM7core and used as bait, except for lanes 6 and 12 wherein Strep-Z5 was used as bait. (C) Plot of strand displacement rate (mean ± SD, n = 3) at varying protein concentrations for a NEXT variant with deletion of CTD in ZCCHC8 (NEXTΔCTD), and binary complexes of ZCCHC8CTD–MTR4 and ZCCHC8core–MTR4. Assays were performed using 3′ A25 tailed RNA substrate. Maximal rates (mean ± SD) shown in the bar graph were obtained by fitting the data to the equation: v0 = Vmax [E]/K1/2.
The sequence of ZCCHC8CTD is conserved across homologs of ZCCHC8 in vertebrates (SI Appendix, Fig. S3), and deletion of the ZCCHC8CTD led to a substantial loss of unwinding activity (Fig. 5C). While ZCCHC8CTD appears to be required for optimal activity, it is not sufficient as neither ZCCHC8CTD–MTR4 nor ZCCHC8core–MTR4 binary complexes displayed activities comparable with NEXT (Figs. 1G and 5C and SI Appendix, Table S1). Furthermore, maximal strand displacement rates for ZCCHC8CTD–MTR4 and ZCCHC8core–MTR4 were ∼sevenfold lower compared with NEXT (Figs. 1G and 5C; also see SI Appendix, Table S1). These results underscore requirements for both RBM7 and ZCCHC8 to achieve maximal activity.
Crystal Structure of ZCCHC8CTD Bound to MTR4.
To determine the structural basis for MTR4 interactions with ZCCHC8CTD, we isolated a complex between ZCCHC8CTD and MTR4∆N∆Arch that lacks the N-terminal residues 1 to 70 and the arch domain. The complex crystallized in space group P61 with a single complex in the asymmetric unit. The structure was solved by molecular replacement using S. cerevisiae Mtr4p (PDB ID code 2XGJ) (34) as search model. Electron density for the ZCCHC8CTD peptide was evident. The structure was rebuilt based on the human sequence and refined at 2.2 Å resolution with Rwork/Rfree of 0.19/0.22 (see SI Appendix, Table S2 for data collection and refinement statistics).
The helicase core of MTR4 features a globular fold consisting of a pair of RecA-like domains (RecA1 and RecA2), a winged helix (WH) domain, and a helical bundle (HB) domain (Fig. 6A). Although most regions are highly similar to the structure of yeast Mtr4p, the N-terminal β-hairpin is shorter and does not latch onto RecA2 (Fig. 6A and SI Appendix, Fig. S4A). RecA1 and RecA2 are juxtaposed and form a solvent-exposed cleft where motifs responsible for binding and hydrolyzing ATP are located. While at low occupancy (refined to 0.70), an ADP molecule resides in this cleft with the adenine nucleobase recognized by the Q-motif residue Gln144 and stacked between the side chains of Phe138 and Arg527 (Fig. 6A). The WH domain packs against RecA1 and the C-terminal HB domain that encloses the RNA binding cavity formed by the RecA1–RecA2 interface.
Fig. 6.
Structure of the ZCCHC8CTD–MTR4∆N∆Arch complex. (A) Overall structure of ZCCHC8CTD (lemon green) bound to the helicase core of MTR4. NT and CT denote the N and C termini of ZCCHC8CTD. (Inset) A snapshot of a simulated annealing omit map (gray mesh) from the modeled ADP, contoured at 0.9 σ. (B) Superposition of ZCCHC8CTD–MTR4∆N∆Arch model (color scheme as in A) with the structure of S. cerevisiae Mtr4p (light gray) in complex with Trf4p (magenta) and Air2p (teal) peptides (PDB ID code 4U4C). (C–F) Magnified views of the interaction of the ZCCHC8CTD peptide with MTR4. (G) Unwinding assays of NEXT reconstituted with the indicated ZCCHC8 variant. Reactions were performed using a 3′ A25 tailed duplex substrate and 500 nM protein. Assays were performed in triplicate with a representative gel for each sample shown.
ZCCHC8CTD adopts an extended conformation via interactions with every domain of the helicase core (Fig. 6A and SI Appendix, Fig. S4B). It is situated at the base of the helicase core, opposite to the presumed location of the arch domain, a position that is distinct from the binding sites of TRAMP subunits Trf4p and Air2p (Fig. 6B) (35). The interface between ZCCHC8CTD and MTR4 buries >2,000 Å2 of total solvent-accessible surface area. Amino acid side chains within the interface are highly conserved in both MTR4 and ZCCHC8 (SI Appendix, Figs. S3 and S4C). ZCCHC8CTD contains a short N-terminal helix αA (residues 663 to 667) that is positioned in a crevice formed by the RecA1 and WH domains (Fig. 6C). This helix is preceded by a salt bridge between ZCCHC8CTD Asp662 and RecA1 Arg244 (Fig. 6C). On the nonpolar surface of αA, Phe666 is within van der Waals distance of RecA1 residues Tyr237 and Leu272 and WH residues Leu573, Val576, and Ile579. Arg575 and Arg875 within the WH domain are within hydrogen bonding distance to the main chain carbonyl oxygen atoms of Phe666 and Thr668, and Gly669, respectively (Fig. 6C). Superposition of the ZCCHC8CTD–MTR4∆N∆Arch structure with a complex of Hel308 with partially unwound DNA (SI Appendix, Fig. S4D) (36) suggests that ZCCHC8CTD is adjacent to the putative RNA exit channel of MTR4 with ZCCHC8CTD residues Phe673, Glu674, Phe675, and Glu676 located near the rim of the channel (Fig. 6D). Main chain amides within ZCCHC8CTD are within hydrogen bonding distance to the C-terminal carboxylate of MTR4 (Fig. 6E). ZCCHC8CTD forms another helix (αB, residues 684 to 691) that contacts RecA1 through interactions that include a parallel cation-π interaction between RecA1 Arg200 and ZCCHC8CTD Tyr685 (Fig. 6E) while its aliphatic surface contacts RecA2 (Fig. 6F).
To determine if any of the aforementioned contacts between ZCCHC8CTD and MTR4 were important for NEXT activity, we performed site-directed mutagenesis in three regions of the ZCCHC8CTD–MTR4 interface and reconstituted NEXT with ZCCHC8core mutants. A pair of mutations (D662A/F666K) designed to perturb the interaction of ZCCHC8CTD helix αA with RecA1 and WH did not alter helicase activity of NEXT (Fig. 6G). We then tested if residues adjacent to the putative RNA exit channel impacted unwinding. Neither alanine substitutions of Phe673 and Phe675 nor Glu674 and Glu676 diminished helicase activity of NEXT (Fig. 6G). Finally, mutations (I688E/L692E) designed to disrupt the hydrophobic interface between ZCCHC8CTD helix αB and RecA2 resulted in loss of unwinding. Furthermore, deletion of residues 681 to 707 also compromised duplex unwinding, demonstrating the contribution of this region to NEXT helicase activity (Fig. 6G).
ZCCHC8CTD Stimulates the RNA-Dependent ATPase Activity of MTR4.
ZCCHC8CTD makes extensive contacts with the MTR4 ATPase cassettes (RecA1 and RecA2), and mutations that disrupt ZCCHC8CTD interaction with RecA2 reduced NEXT helicase activity, so we asked if ZCCHC8CTD contributes to stimulation of MTR4 helicase activity by activating its ATPase activity. In the absence of RNA, both MTR4 and NEXT exhibited low intrinsic ATPase activity (Fig. 7 and SI Appendix, Fig. S5). Addition of RNA stimulated ATP hydrolysis by MTR4 and, to a greater extent, NEXT. NEXT containing a ZCCHC8 variant with truncation of the CTD displayed reduced RNA-induced ATPase activity comparable with MTR4 alone. Likewise, NEXT with ZCCHC8 mutations (I688E/L692E) designed to impair CTD interactions with RecA2 yielded a weaker RNA-dependent ATPase activity in comparison with WT NEXT. These data suggest a role for ZCCHC8CTD in promoting ATPase activity. Interestingly, an MTR4 complex with ZCCHC8CTD recapitulated the same kinetics of ATPase activity as NEXT in the presence of RNA. These results suggest that ZCCHC8CTD contributes to MTR4 helicase activity, in part, by enhancing its RNA-dependent ATPase activity.
Fig. 7.
ZCCHC8CTD stimulates RNA-dependent ATPase activity of MTR4. Time courses of ATP hydrolysis in the presence or absence of the 3′ A25 tailed RNA duplex. An expanded view of ATPase activities in the absence of RNA can be found in SI Appendix, Fig. S5. Data show mean ± SD (n = 6 for reactions with RNA, n = 3 for reactions without RNA).
Discussion
The RNA exosome and its cofactors work together to surveil and ultimately degrade a large repertoire of eukaryotic transcripts to maintain a healthy and functional transcriptome. In the nucleus, the MTR4 helicase is part of several protein complexes that deliver and prime substrates for processing or degradation by the exosome. S. cerevisiae Mtr4p exhibits 3′-to-5′ helicase activity (12, 37, 38). We have shown that the human homolog MTR4 also catalyzes unwinding of RNA duplexes with a single-stranded 3′ RNA extension. NEXT displays enhanced helicase activity compared with MTR4, demonstrating a functional advantage in forming a complex with RBM7 and ZCCHC8. Interestingly, isolated yeast Mtr4p displays weaker helicase activity compared with its activities when associated with the TRAMP subunits Trf4p and Air2p (39). It remains unclear if this is part of a regulatory mechanism to tune helicase activities for particular functions. We also provide in vitro evidence that supports a role for NEXT in resolving DNA:RNA hybrids. DNA:RNA heteroduplexes in the form of R-loops are involved in various cellular processes, including gene regulation and Ig diversification (14, 31, 40). Paradoxically, R-loops are also potential sources of genomic instability (40). The RNA exosome and MTR4 appear to play important roles to keep these antagonistic functions for R-loops in check (14, 31). By recruiting and activating MTR4, NEXT appears poised to mediate turnover of these structures independently or in conjugation with the exosome.
The RRM module of RBM7 is reported to display a binding specificity for uridine-rich sequences (18). Consistent with this model, NEXT exhibits an RBM7-dependent preference for substrates with polyuridine sequences. The position of a polyuridine tract within the single-stranded 3′ extension of the duplex also contributes to effective binding and unwinding by NEXT. Optimal activity can be achieved when a polyuridine stretch is placed upstream of the 3′ end at positions predicted to interact with RBM7. RNA-seq and iCLIP analyses have revealed a preference for polypyrimidine tracks in NEXT substrates that include PROMPTs (19, 41), snRNAs (19), and introns of pre-mRNA (19). These U-rich elements are likely recognized by RBM7 to recruit NEXT and, in turn, the exosome. In vivo substrates of NEXT are typically polyadenylated (21), but it does not appear to be a strict requirement for degradation (41). However, this latter report (41) measured transcript levels under steady-state conditions. It is plausible that polyadenylation could increase the rate of decay or that the presence of a polyuridine motif takes precedence over having poly(A) tails. Polyadenylation may otherwise be required for substrates that lack polyuridine elements near the 3′ end. Nevertheless, we posit that efficient exosome targeting by NEXT is governed by multiple features of the substrate that are recognized by distinct elements from each NEXT subunit. The temporal interaction of NEXT with other protein complexes that decorate the nascent transcript may also influence the fate of the RNA (19). Together, these unique features of NEXT and substrates provide some insights to the ability of NEXT to discriminate between RNA substrates that are destined for degradation by the exosome.
The role of ZCCHC8 in NEXT appears not to be limited to its scaffolding function as its C-terminal domain promotes RNA-dependent ATP hydrolysis and is necessary for enhancing the helicase activity of MTR4 within NEXT. Binding of either ZCCHC8CTD or ZCCHC8core to MTR4 is not sufficient to attain the same level of unwinding activity as NEXT, underscoring the role of RBM7 for productive unwinding. RBM7 potentially assists in substrate binding during the course of unwinding or, similar to TRAMP, increases the rate of unwinding initiation (42).
A common theme has emerged regarding subunit organization within MTR4-containing complexes whereby RNA-binding adaptors [RBM7 in NEXT, PABPN1 in Poly(A) exosome targeting (PAXT) connection] are linked to MTR4 via a zinc knuckle-containing protein (ZCCHC8 in NEXT or ZFC3H1 in PAXT) (16). We confirm this binding mode and resolve an RNA binding path from site-directed 4-thiouridine cross-linking patterns that involves all three subunits of NEXT. In addition, we identify two regions of ZCCHC8 that bind MTR4. One is located in the N-terminal segment of ZCCHC8 that includes a predicted coiled-coil region and zinc-knuckle motif, proximal to the PSP domain that recruits the RBM7 RRM. MTR4 also binds the C-terminal domain of ZCCHC8. Specifically, ZCCHC8CTD interacts with a highly conserved surface at the base of the helicase core modules, unique from the binding sites reported for TRAMP subunits Trf4p and Air2p. While NEXT and TRAMP subunits contact MTR4 through the helicase core, several exosome partners are recruited to MTR4 via its arch domain (43). Hence, in addition to being an RNA helicase, MTR4 serves as an interaction hub, providing a platform for multiple exosome accessory factors to access the exosome.
Materials and Methods
Cloning, Protein Expression, and Purification.
RBM7 and MTR4 coding sequences were cloned into pET-28a with an N-terminal decahistidine Smt3 (His10-Smt3) tag. ZCCHC8 constructs were prepared as N-terminal Smt3 fusions in pRSF-Duet1. All truncations and mutations were cloned using USER cloning (44) or a QuikChange site-directed mutagenesis kit following the manufacturer’s protocol. Recombinant proteins were produced in Escherichia coli BL21-CodonPlus(DE3) RIL (Novagen) grown in superbroth. Protein expression was induced by addition of 0.25 mM isopropyl-β-d-thiogalactoside and further incubation with shaking at 18 °C for 16 to 20 h. Cells were resuspended in lysis buffer [20 mM Tris⋅HCl, pH 8.0, 350 mM NaCl, 0.1 mM Tris(2-carboxyethyl)phosphine (TCEP)], lysed by sonication, and clarified by centrifugation. The soluble protein extracts were applied onto an Ni2+-NTA column (Qiagen). After several washes, the bound recombinant proteins were eluted with a buffer containing 20 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, and 250 mM imidazole, and purified further using a HiTrap Heparin HP column (GE Healthcare). The Smt3 tags were cleaved by overnight incubation with the Ulp protease (45). Finally, proteins were subjected to size exclusion chromatography in a buffer containing 20 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, and 0.1 mM TCEP, concentrated, and stored at −80 °C until further use. Reconstitution of the NEXT complex was achieved by incubating a copurified complex of RBM7–ZCCHC8 with molar excess of MTR4, followed by size exclusion chromatography.
Preparation of RNA Substrates.
RNA oligonucleotides used in the helicase assays were synthesized and purified by Integrated DNA Technologies. 4-Thiouridine-labeled RNAs were obtained from Dharmacon Inc. The sequences of all RNA used are listed in SI Appendix, Table S3. Lyophilized RNA were resuspended in annealing buffer containing 20 mM Tris, pH 7.0, and 100 mM potassium acetate. RNA duplexes were prepared by heating oligonucleotide mixtures to 95 °C for 5 min, followed by incubation at 16 °C for at least an hour. Oligonucleotide-based R-loop mimic was prepared as described previously (32).
Gel-Shift Helicase Assays.
Unwinding reactions were carried out at 30 °C in 20 mM 3-(N-morpholino)propanesulfonic acid (Mops)-NaOH, pH 6.5, 50 mM NaCl, 0.5 mM MgCl2, 5% (vol/vol) glycerol, 5 mM β-mercaptoethanol (BME), and 0.4 U/µL murine RNase inhibitor (New England Biolabs). Proteins were incubated with 10 nM RNA substrate for 5 min, and the reactions were initiated by simultaneous addition of 2 mM ATP (pH 7.0), 2 mM MgCl2, and 400 nM DNA capture strand. Reactions were quenched using 0.5% (wt/vol) SDS, 5 mM EDTA, and 10% (vol/vol) glycerol. Samples were electrophoresed in Novex 20% Tris base, boric acid, EDTA (TBE) gel (Thermo Fisher Scientific). Fluorescent bands were imaged using a Typhoon FLA 9500 laser scanner and were quantified using ImageJ (46). The amount of unwound substrate was estimated by taking the fraction of the band intensity of the unwound strand over the sum of the intensities of the unreacted and unwound substrate. Data were analyzed using Graphpad Prism version 7.
Molecular Beacon Helicase Assay.
A real-time molecular beacon helicase assay was performed based on the methods reported previously (33). In this assay, the helicase-dependent displacement of the RNA strand labeled with 5′ Iowa Black RQ and 3′ Cy5 leads to stem-loop formation that puts the fluorophore in close proximity to the quencher, resulting in a decrease in fluorescence signal. Reactions were conducted in 20 mM Tris⋅HCl, pH 7.0, 50 mM NaCl, 0.5 mM MgCl2, 5% (vol/vol) glycerol, 5 mM BME, and 0.4 U/µL murine RNase inhibitor (New England Biolabs). Proteins were preincubated with 10-nM substrates for 10 min, and the reactions were started by addition of 2 mM ATP and 2 mM MgCl2. Fluorescence was recorded at 30 °C every 30 s using a SpectraMax M5 microplate reader (Molecular Devices) with 643/667 nm as excitation/emission wavelengths. The fraction of unwound substrate was calculated using data normalized to the fluorescence at the start of the reaction and the quenched molecular beacon single-stranded RNA. Data were obtained from three replicates and were analyzed using Graphpad Prism version 7.
Electrophoretic Mobility-Shift Assay.
Various concentrations of protein were mixed with 20 nM fluorescein-labeled RNA duplex in 20 mM Tris, pH 7.0, 50 mM NaCl, 2 mM adenosine 5′-(β,γ-imido)triphosphate (AMPPNP), 2.5 mM MgCl2, 5% (vol/vol) glycerol, 5 mM BME, and 0.4 U/µL murine RNase inhibitor (New England Biolabs). The binding mixture was incubated at 22 °C for an hour. Samples were applied to a Novex 4 to 20% TBE gel (Thermo Fisher Scientific) for native polyacrylamide gel electrophoresis. Gel bands were visualized using a Typhoon FLA 9500 laser scanner and were quantified using ImageJ (46). Data from three replicates were analyzed using Graphpad Prism version 7. Dissociation constants (Kds) were obtained by fitting the data in the equation: fraction bound = (Bmax[P]h)/(Kdh + [P]h) where [P] is the protein concentration, h is the Hill’s coefficient, and Bmax is the maximal fraction bound.
Site-Directed 4-Thiouridine Cross-Linking Assay.
Proteins (200 nM) were incubated with 100 nM RNA (A20) labeled with internal 4-thiouridine (4SU) in the indicated position in a buffer containing 20 mM Tris, pH 7.0, 50 mM NaCl, 5 mM BME, 1 U/µL RNase inhibitor, human placenta (New England Biolabs), 1.5 mM MgCl2, and 1 mM AMPPNP (pH 7.0) at 22 °C for 30 min. Cross-linking was induced by exposure of the protein–RNA mixture to 365 nm UV light using a 4-W handheld UV lamp (UVP) for 20 min at 4 °C. Samples were quenched with NuPAGE lithium dodecyl sulfate sample buffer (Thermo Fisher Scientific) and subjected to SDS/PAGE analysis using NuPAGE 4 to 12% Bis-Tris protein gels (Thermo Fisher Scientific). Gels were scanned using a Typhoon FLA 9500 laser scanner. The 5′ fluorescein-labeled oligonucleotides used included the following 5′-to-3′ sequences: AAA AAA AAA AAA AAA AAA (4SU)A, AAA AAA AAA AAA AAA A(4SU)A AA, AAA AAA AAA AAA (4SU)AA AAA AA, AAA AAA AA(4SU) AAA AAA AAA AA, AAA A(4SU)A AAA AAA AAA AAA AA, and (4SU)AA AAA AAA AAA AAA AAA AA.
Pull-Down Assays.
The Strep-tagged RBM7 and ZCCHC8CTD constructs were prepared as N-terminal His10-Smt3-Strep fusions in pET-28a. Soluble protein extracts of His10-Smt3-Strep-RBM7 and Smt3-ZCCHC8 variants were mixed and bound to Streptactin beads (IBA Lifesciences) in the presence of the Ulp protease. After several wash steps, the beads were incubated with MTR4 in binding buffer [20 mM Tris, pH 8.0, 175 mM NaCl, 0.01% (vol/vol) octylphenoxy poly(ethyleneoxy)ethanol, branched, and 0.1 mM TCEP] and washed, and the bound proteins were eluted with the binding buffer supplemented with 5 mM desthiobiotin. Protein samples were applied onto NuPAGE 4 to 12% Bis-Tris protein gels (Thermo Fisher Scientific) and visualized by Coomassie staining.
ATPase Assay.
ATPase assays were performed using an ADP-Glo kit (Promega) using the manufacturer’s protocol. Proteins (1 μM) were preincubated with 200 nM RNA in a solution containing 20 mM Mops, pH 6.5, 50 mM NaCl, 2.5 mM MgCl2, 5% (vol/vol) glycerol, 5 mM BME, and 0.4 U/µL murine RNase inhibitor (New England Biolabs). ATP was added to a final concentration of 1 mM, and the reaction mixture was incubated at 30 °C for various time points. Luminescence was measured using the SpectraMax M5 microplate reader with 250-ms integration time.
Crystallization, Structure Determination, and Analysis.
Crystals of ZCCHC8CTD in complex with MTR4 with truncations of the N terminus (residues 1 to 70) and the arch domain (residues 601 to 842 replaced with a single serine linker) were obtained by the hanging-drop vapor diffusion technique at 20 °C. The complex (15 mg/mL) was preincubated with 2 mM ADP and 2 mM MgCl2 and mixed with equal volume of the reservoir solution containing 1.6 M sodium/potassium (L)-tartrate and 0.1 M Bis-Tris, pH 6.5. Crystals were cryoprotected by serial transfer to 50% (vol/vol) sodium malonate with 0.1 M Hepes, pH 7.5. Diffraction data were collected at the Advanced Photon Source 24-ID-C beamlines [Northeastern Collaborative Access Team (NE-CAT)] and processed using HKL2000 (47). The structure was solved using Phaser (48) in the Phenix suite (49) with the structure of the S. cerevisiae Mtr4p (PDB ID code 2XGJ) used as search model. The model was built using Coot (50) and refined using Phenix (49). Molprobity (51) was used to assess model geometry. A composite omit map was generated using Phenix (49). Surface conservation was analyzed using Consurf (52). Structures were rendered using PyMol (Schrodinger, LLC).
Supplementary Material
Acknowledgments
We thank the members of the C.D.L. laboratory for helpful discussions. We also thank Devanshi Jain, Kurt Januszyk, Eva-Maria Weick, and Dimitrios Zattas for their critical review of the manuscript. Work was based in part upon research conducted at NE-CAT beamlines [P41 GM103403, NIH National Institute of General Medical Sciences (NIGMS), S10 RR029205, NIH-Office of Research Infrastructure Programs (ORIP) High-End Instrumentation (HEI) grant]. Beamline research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. This research was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Awards R01GM079196 (to C.D.L.), R35GM118080 (to C.D.L.), and P30 CA008748 [NIH National Cancer Institute (NCI)-Cancer Center Support Grant]. C.D.L. is an investigator of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6C90).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803530115/-/DCSupplemental.
References
- 1.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 2.Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17:227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- 3.Zinder JC, Lima CD. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017;31:88–100. doi: 10.1101/gad.294769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: Crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 6.Wasmuth EV, Zinder JC, Zattas D, Das M, Lima CD. Structure and reconstitution of yeast Mpp6-nuclear exosome complexes reveals that Mpp6 stimulates RNA decay and recruits the Mtr4 helicase. eLife. 2017;6:e29062. doi: 10.7554/eLife.29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk S, Bonneau F, Ebert J, Kögel A, Conti E. Mpp6 incorporation in the nuclear exosome contributes to RNA channeling through the Mtr4 helicase. Cell Rep. 2017;20:2279–2286. doi: 10.1016/j.celrep.2017.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuch B, et al. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 2014;33:2829–2846. doi: 10.15252/embj.201488757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckard SC, et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol. 2014;15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim J, et al. Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell. 2017;169:523–537.e15. doi: 10.1016/j.cell.2017.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lubas M, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Meola N, et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell. 2016;64:520–533. doi: 10.1016/j.molcel.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Ogami K, et al. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017;31:1257–1271. doi: 10.1101/gad.302604.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrossova D, et al. RBM7 subunit of the NEXT complex binds U-rich sequences and targets 3′-end extended forms of snRNAs. Nucleic Acids Res. 2015;43:4236–4248. doi: 10.1093/nar/gkv240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubas M, et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 2015;10:178–192. doi: 10.1016/j.celrep.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 20.Tseng C-K, et al. Human telomerase RNA processing and quality control. Cell Rep. 2015;13:2232–2243. doi: 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 21.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 22.Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci USA. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen PR, et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarov EM, Owen N, Bottrill A, Makarova OV. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res. 2012;40:2639–2652. doi: 10.1093/nar/gkr1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blasius M, Wagner SA, Choudhary C, Bartek J, Jackson SP. A quantitative 14-3-3 interaction screen connects the nuclear exosome targeting complex to the DNA damage response. Genes Dev. 2014;28:1977–1982. doi: 10.1101/gad.246272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiedje C, et al. p38MAPK/MK2-mediated phosphorylation of RBM7 regulates the human nuclear exosome targeting complex. RNA. 2015;21:262–278. doi: 10.1261/rna.048090.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rialdi A, et al. The RNA exosome syncs IAV-RNAPII transcription to promote viral ribogenesis and infectivity. Cell. 2017;169:679–692.e14. doi: 10.1016/j.cell.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giunta M, et al. Altered RNA metabolism due to a homozygous RBM7 mutation in a patient with spinal motor neuropathy. Hum Mol Genet. 2016;25:2985–2996. doi: 10.1093/hmg/ddw149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falk S, et al. Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors. Nat Commun. 2016;7:13573. doi: 10.1038/ncomms13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pefanis E, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Belon CA, Frick DN. Monitoring helicase activity with molecular beacons. Biotechniques. 2008;45:433–442. doi: 10.2144/000112834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falk S, et al. The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol Cell. 2014;55:856–867. doi: 10.1016/j.molcel.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Büttner K, Nehring S, Hopfner K-P. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein J, Patterson DN, Wilson GM, Toth EA. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3′->5′ helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–4942. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- 38.Jackson RN, et al. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia H, Wang X, Anderson JT, Jankowsky E. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc Natl Acad Sci USA. 2012;109:7292–7297. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preker P, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patrick EM, Srinivasan S, Jankowsky E, Comstock MJ. The RNA helicase Mtr4p is a duplex-sensing translocase. Nat Chem Biol. 2017;13:99–104. doi: 10.1038/nchembio.2234. [DOI] [PubMed] [Google Scholar]
- 43.Losh JS, van Hoof A. Gateway arch to the RNA exosome. Cell. 2015;162:940–941. doi: 10.1016/j.cell.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Bitinaite J, et al. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K, Cr IU. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.