Significance
Obesity has become a major epidemic around the globe, with fatal comorbidities such as type 2 diabetes, heart disease, and cancer that have led to alarming health concerns in modern medicine. Although excess adipose tissue is associated with negative metabolic outcomes, the physiological functions of epigenetic regulators in adipose tissue and metabolism are unclear. Here we show that KDM4B in adipose tissues epigenetically controls energy expenditure, oxidation, lipolysis, and thermogenesis. Loss of Kdm4b impairs energy expenditure, adaptive thermogenesis, and adipose tissue lipolysis, resulting in obesity and associated metabolic dysfunction. Our results provide insights into control of obesity and suggest that modulation of KDM4B levels or activity may be a potential therapeutic target for human obesity.
Keywords: obesity, epigenetics, KDM4B, metabolism
Abstract
Although significant progress has been made in understanding epigenetic regulation of in vitro adipogenesis, the physiological functions of epigenetic regulators in metabolism and their roles in obesity remain largely elusive. Here, we report that KDM4B (lysine demethylase 4B) in adipose tissues plays a critical role in energy balance, oxidation, lipolysis, and thermogenesis. Loss of KDM4B in mice resulted in obesity associated with reduced energy expenditure and impaired adaptive thermogenesis. Obesity in KDM4B-deficient mice was accompanied by hyperlipidemia, insulin resistance, and pathological changes in the liver and pancreas. Adipocyte-specific deletion of Kdm4b revealed that the adipose tissues were the main sites for KDM4B antiobesity effects. KDM4B directly controlled the expression of multiple metabolic genes, including Ppargc1a and Ppara. Collectively, our studies identify KDM4B as an essential epigenetic factor for the regulation of metabolic health and maintaining normal body weight in mice. KDM4B may provide a therapeutic target for treatment of obesity.
Obesity has become a major epidemic around the globe, with fatal comorbidities such as type 2 diabetes, heart disease, and cancer that have led to alarming health concerns in modern medicine (1). The condition develops when the balance between energy intake and energy expenditure is chronically disrupted (2). To address obesity beyond simply reducing intake, vigorous research is aimed at augmenting energy output in metabolic tissues, including adipose tissue. Although excess adipose tissue is associated with negative metabolic outcomes, adipocytes are well known to play critical roles in health and disease as an endocrine organ and a master regulator of energy homeostasis and metabolic health (3, 4). Brown adipose tissue (BAT) is notable for its regulatory function in metabolism and adaptive thermogenesis (4). The traditional role of white adipose tissue (WAT), which is stored in both visceral and s.c. depots, is to serve as a primary energy reservoir. Alterations in fatty acid oxidation in WAT also contribute to adiposity (2, 4). At this time, endeavors to better understand adipose tissue biology in an effort to alleviate obesity are extensive and rapidly evolving.
Modification of chromatin architecture and compaction by histone methylation manipulates the accessibility of regulatory transcriptional factors and thereby controls the activation and repression of gene expression. This process allows for dynamic coordination of gene activation, as the methylating action of histone methyl transferases can be rapidly reversed by histone demethylation. Connections between histone methylation and metabolic disease have recently been revealed. For example, Nicotinamide N-methyltransferase (NNMT) knockdown in white fat (WAT) and liver protects against diet-induced obesity by augmenting cellular energy expenditure. NNMT1 deficiency modifies methylated H3K4 levels at gene promoters for ornithine decarboxylase and spermidine-spermine N1-acetyltransferase (encoded by Sat1), leading to altered S-adenosyl methionine and NAD+ levels (5). Adipose tissue-specific deletion of euchromatic histone-lysine N-methyltransferase (EHMT1) leads to a marked reduction of BAT-mediated adaptive thermogenesis, obesity, and systemic insulin resistance. Inhibition of EHMT1 in BAT results in severe loss of BAT characteristics and induces muscle differentiation in vivo through demethylation of H3K9me2/me3 at muscle-selective gene promoters (6).
KDM4B , also known as JMJD2B, is a histone demethylase that is involved in the demethylation of repressive H3K9 trimethylation marks to promote gene activation (7, 8). Recently, our group found that KDM4B epigenetically regulates osteogenic differentiation of human mesenchymal stem/stromal cells from bone marrow in vitro (9, 10). To elucidate the functional role of KDM4B in vivo, we generated Kdm4b knockout (KO) mice. Unexpectedly, we found that KDM4B is an epigenetic modulator of energy homeostasis and metabolic health in mice. Loss of Kdm4b impairs energy expenditure, adaptive thermogenesis, and adipose tissue lipolysis, resulting in obesity and associated metabolic dysfunction. Mechanistically, we found that KDM4B directly controls the transcriptional activation of multiple metabolic genes by erasing H3K9me3 marks.
Results
Kdm4b-Deficient Mice Exhibit Age- and Diet-Induced Obesity.
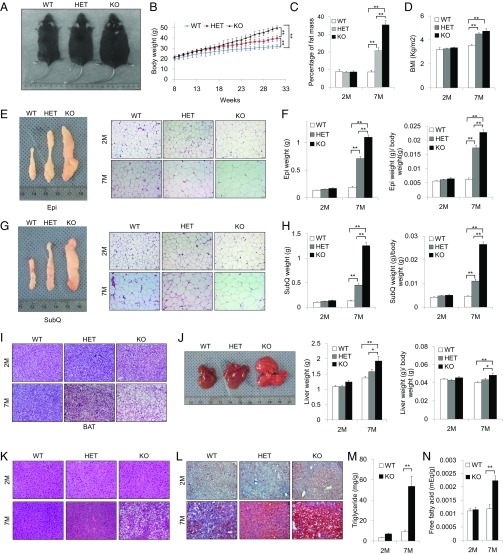
To explore the biological functions of KDM4B in vivo, we generated global Kdm4b KO mice (KO or Kdm4b−/−). This was accomplished using a Kdm4b allele with loxP sites flanking exon 5 encoding the JmjC domain, which is essential for iron chelating function and demethylase activity. A cytomegalovirus-Cre (CMV-Cre) recombinase transgene was used to remove exon 5, resulting in a frame-shift and premature translation termination of Kdm4b. The deletion of Kdm4b was confirmed by Western blotting (SI Appendix, Fig. S1A). KO mice were viable at birth, but with a birth rate of about 16% (78/495), which was lower than the Mendelian ratio (25%), indicating possible embryonic lethality in a portion of Kdm4b–/– embryos. In addition to the lower birth rate, all KO mice died 1–2 d after weaning them 21 d after birth. However, the postponement of weaning to 28 d improved the survival rate of these animals. Unexpectedly, KO mice on a normal chow diet became noticeably obese with age compared with their WT littermates. Interestingly, heterozygous (HET) mice had body weights intermediate between WT and KO mice, suggesting that the effect of KDM4B on obesity is potentially dose dependent (Fig. 1A). To assess the kinetics of weight gain in these animals, we measured body weight every week starting at 2 mo of age. Body weights of the three genotypes were similar at 2 mo, but with increasing age, mice having reduced Kdm4b gene dosage gained weight more rapidly (Fig. 1B). By 7 mo of age, body weight and body mass index (BMI) were significantly higher in KO mice, with HET mice having an intermediate phenotype. The differences in body weight were, at least in part, a result of greater fat mass, as determined by NMR (Fig. 1 C and D).
Fig. 1.
Kdm4b KO mice exhibit obesity phenotypes on normal chow diet. (A) Representative images of male 7-mo-old WT, HET, and KO mice. (B) Growth curve of WT, HET, and KO littermates (n = 8). (C) Determination of percentage of fat mass of WT, HET, and KO mice using NMR imaging at 2 mo and 7 mo (n = 8). (D) Determination of BMI of WT, HET and KO mice starting at 28 d for 2-mo-old and 7-mo-old mice (n = 8). (E) Representative images of Epi WAT from WT, HET, and KO mice at 7 mo and H&E staining of Epi from WT, HET, and KO mice. (Scale bars, 50 µm.) (F) Determination of Epi weights from WT, HET, and KO mice at 2 and 7 mo (n = 8). (G) Representative images of SubQ WAT from WT, HET, and KO mice at 7 mo and H&E staining of SubQ from WT, HET, and KO mice at 2 and 7 mo. (Scale bars, 50 µm.) (H) Determination of SubQ from WT, HET, and KO mice at 2 and 7 mo (n = 8). (I) H&E staining of BAT for WT, HET, and KO mice at 2 and 7 mo. (Scale bars, 50 µm.) (J) Representative images of livers of WT, HET, and KO mice at 7 mo, and determination of liver weights (n = 8). (K) H&E staining of livers for WT, HET, and KO mice at 2 and 7 mo. (Scale bars, 50 µm.) (L) Oil Red O staining of livers for WT, HET, and KO mice at 2 and 7 mo. (Scale bars, 50 µm.) (M and N) Determination of percentage triglyceride (M) and free fatty acids (N) in liver tissues of WT and KO mice (n = 6). *P < 0.05; **P < 0.01. All data are presented as the mean ± SEM.
At 7 mo of age, both Epi WAT and SubQ WAT fat pads were larger in KO and HET compared with WT mice (Fig. 1 E–H). In both WAT depots, tissue weight was increased in KO compared with WT (∼sixfold) and HET mice (three- to fourfold; Fig. 1 F and H). Furthermore, H&E staining of Epi and SubQ adipose tissues revealed significantly increased adipocyte diameter in KO and HET mice compared with WT animals (Fig. 1 E and G). This increase in adipocyte size was also observed in BAT sections of KO and HET mice and increased lipid storage in BAT was evident in KO mice (Fig. 1I). In addition, KO animals experienced significant liver enlargement and were more prone to hepatic steatosis, as demonstrated by H&E staining and Oil Red O staining (Fig. 1 J–L). Intrahepatic triglyceride and free fatty acid levels were significantly increased in the livers of 7-mo-old KO animals compared with their WT littermates (Fig. 1 M and N). Plasma total triglyceride and cholesterol, as well as LDL and very LDL cholesterol levels, were higher in 7-mo-old KO animals compared with their WT littermates (Table 1). As no differences in these lipids were seen in 2-mo-old animals, this hyperlipidemia is likely caused by obesity after the loss of KDM4B function.
Table 1.
Lipid panel for WT and KO mice at 2 and 7 mo
| 2 mo | 7 mo | |||||
| Metabolic parameters | WT | KO | P value | WT | KO | P value |
| Triglyceride, mg/dL | 57.9 ± 6.7 | 62.3 ± 11.1 | 0.36 | 53.6 ± 14.3 | 69.7 ± 12.5 | 0.011 |
| Total cholesterol, mg/dL | 79.9 ± 9.2 | 77.8 ± 11.9 | 0.70 | 52.4 ± 22.6 | 143 ± 17.0 | 0.00038 |
| LDL cholesterol, mg/dL | 16.7 ± 3.4 | 16.2 ± 4.75 | 0.81 | 15.2 ± 14.1 | 55.2 ± 8.1 | 0.0043 |
| VLDL | 11.6 ± 1.27 | 12.5 ± 2.32 | 0.35 | 10.8 ± 3.1 | 14 ± 2.4 | 0.015 |
The differences between WT and KO animals became more dramatic when animals were subjected to a high-fat diet (HFD) beginning at 2 mo of age. After 8 wk of the HFD, KO mice gained 15 g more weight than WT animals (SI Appendix, Fig. S1 B–E). In addition, the 4-mo-old KO mice on the HFD had larger epi and subQ fat pads and contained larger adipocytes (SI Appendix, Fig. S1 F–I). Intrascapular BAT from KO mice was characterized by larger adipocytes with increased lipid content compared with WT mice (SI Appendix, Fig. S1J). Greater accumulation of lipids and development of hepatic steatosis occurred in livers from KO mice on HFD compared with WT mice, as demonstrated by Oil Red O staining and H&E (SI Appendix, Fig. S1 K and L).
Kdm4b Deficiency Induces Insulin Resistance.
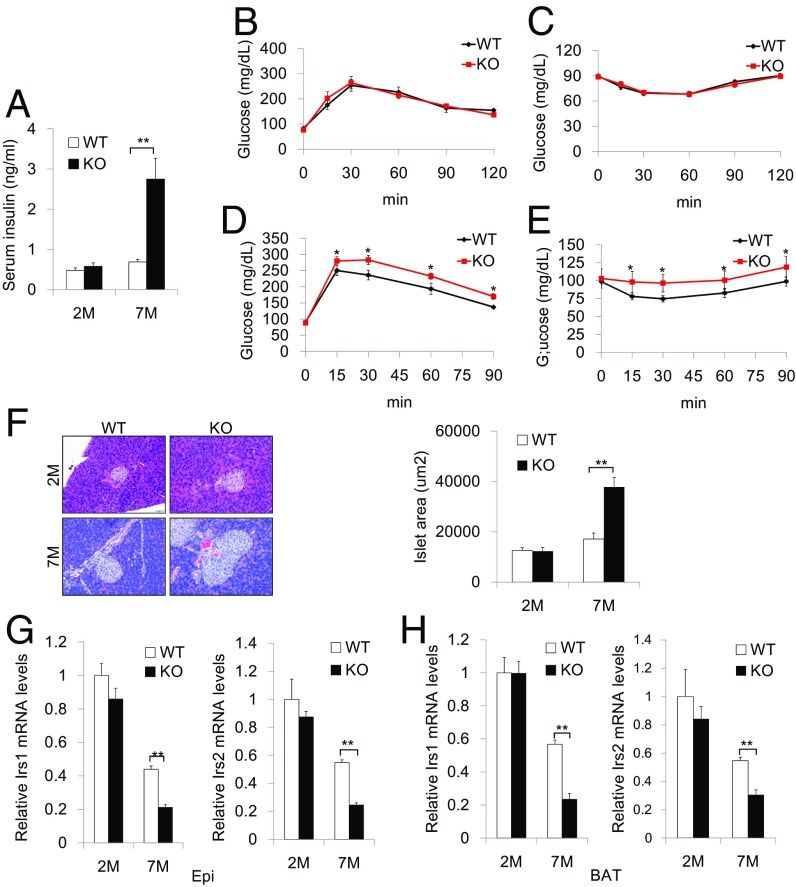
To assess the metabolic effects of Kdm4b deficiency-induced obesity, we assessed glucose homeostasis. When fed a chow diet, there was no difference in fasting plasma insulin concentration and blood glucose levels between 2-mo-old KO and WT littermates. However, at 7 mo of age, chow-fed KO mice exhibited a fivefold increase in basal serum insulin concentration (Fig. 2A). Glucose tolerance and insulin tolerance tests at 2 mo of age showed no impairment in KO mice (Fig. 2 B and C). However, at 7 mo of age, after KO mice had accumulated substantially more adipose tissue, Kdm4b ablation led to impaired glucose tolerance and insulin sensitivity (Fig. 2 D and E). In addition, serum triglyceride and cholesterol concentrations were increased in 7-mo-old KO mice, but not in 2-mo-old animals (Table 1).
Fig. 2.
Whole-body Kdm4b KO mice develop impaired glucose and insulin tolerance. (A) Measurement of serum insulin level for WT and KO mice at 2 and 7 mo (n = 8). (B and C) Measurement of plasma glucose during GTT (B) and ITT (C) for 2-mo-old WT and KO mice (n = 5). (D and E) Measurement of plasma glucose during GTT (D) and ITT (E) for 7-mo-old WT and KO mice (n = 5). (F) Representative H&E staining images of pancreases and determination of islet size for WT and KO mice at 2 and 7 mo (n = 5). (Scale bar, 50 µm.) (G and H) Expression levels of markers Irs1 and Irs2 in WAT (G) and BAT (H) for WT and KO mice at 2 and 7 mo (n = 6). All data are presented as the mean ± SEM. *P < 0.05; **P < 0.01.
Histological examination of the pancreas revealed a substantial increase in the size of the pancreatic islets of 7-mo-old global KO mice fed a chow diet (Fig. 2F), and a more exaggerated threefold increase in 4-mo-old KO mice after HFD feeding (SI Appendix, Fig. S1M). These changes in pancreatic morphology are likely indicative of compensatory enlargement of β-cell mass and hypertrophy of islets in response to insulin resistance (11). qRT-PCR revealed that insulin receptor substrate 1 (Irs1) and insulin receptor substrate 2 (Irs2) were significantly down-regulated in both WAT and BAT in 7-mo-old KO animals (Fig. 2 G and H). Together, these results demonstrate that deletion of Kdm4b leads to obesity, which results in insulin resistance, aberrant pancreatic β cell mass, and insulin signaling gene expression.
Kdm4b Deficiency Leads to Impairments in Energy Expenditure.
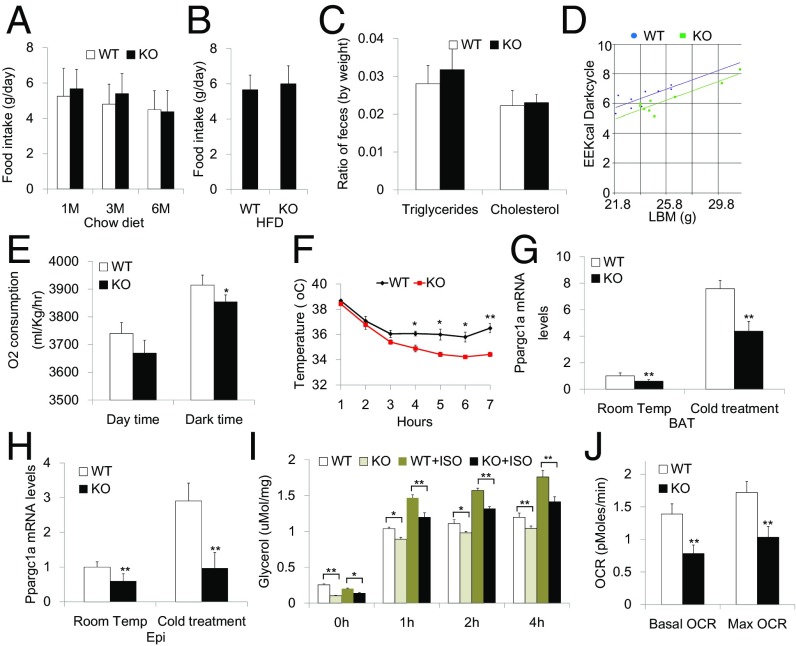
Obesity occurs when the delicate balance between energy intake and expenditure is violated. To evaluate the etiology of obesity in Kdm4b-deficient mice, we first examined caloric intake. There were no differences between WT and KO mice in their daily food intake of chow diet and HFD and the nutrition absorption from food, as demonstrated by similar fecal lipid content of triglyceride and cholesterol (Fig. 3 A–C). In contrast, Kdm4b deficiency led to significant decreases in energy expenditure and reduced oxygen consumption during the dark phase of the circadian cycle (Fig. 3 D and E). Kdm4b-deficient mice also exhibited impaired adaptive thermogenesis when placed at 4 °C (Fig. 3F). This was associated with reduced Ppargc1a gene expression in mice maintained at room temperature, and blunted Ppargc1a induction on cold exposure in both BAT and Epi WAT (Fig. 3 G and H). Additional aberrations in WAT metabolism were also observed, with reduced rates of lipolysis of Epi WAT isolated from Kdm4b-deficient mice under basal conditions and after β-adrenergic stimulation using isoproterenol (Fig. 3I). Defective lipolysis is consistent with the increased adipocyte diameter observed in adipose tissue of Kdm4b-deficient mice, and could contribute to impaired thermogenesis through reduction in fatty acid supply to BAT. The oxygen consumption rate of Epi WAT measured ex vivo showed reductions in KO fat tissue under basal conditions and after trifluorocarbonylcyanide phenylhydrazone (FCCP) stimulation, suggesting reduced oxidative metabolism in WAT after Kdm4b deletion (Fig. 3J).
Fig. 3.
Whole-body Kdm4b KO mice exhibit impairments in energy expenditure levels, adaptive thermogenesis, lipolysis, and oxidation. (A) Determination of food intake for WT and KO mice at 1, 3, and 6 mo (n = 6). (B) Determination of high-fat food intake for WT and KO mice at 3 mo (n = 6). (C) Determination of fecal content of triglycerides and cholesterol for WT and KO mice at 2 mo (n = 8). (D) Determination of energy expenditure for WT and KO mice at 2 mo (n = 9). (E) Determination of oxygen consumption for WT and KO mice at 2 mo (n = 9). (F) Measurement of body temperature for WT and KO mice under cold treatment over the course of 6 h (n = 4). (G and H) Ppargc1a gene expression in BAT (G) and Epi (H) after cold treatment. (I) Determination of lipolysis ratio in Epi fat of WT and KO mice (n = 5). (J) Determination of oxygen consumption rate (OCR) in Epi fat of WT and KO mice measured in Seahorse Extracellular Flux Analyzer (n = 3). *P < 0.05; **P < 0.01. All data are presented as the mean ± SEM.
Adipose-Specific Deletion of Kdm4b Results in Obesity with Metabolic Impairment.
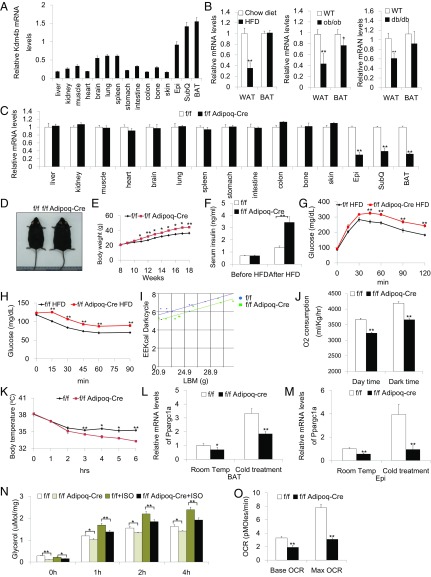
To identify potential sites of KDM4B action, we profiled Kdm4b expression in various murine tissues. Interestingly, Kdm4b expression was most highly enriched in major fat depots, including SubQ and Epi WAT, as well as BAT (Fig. 4A), suggesting that KDM4B might have an important physiological role in adipose tissues. Furthermore, a survey of Kdm4b expression in mice with HFD-induced obesity as well as genetic obesity caused by deficiency of leptin (ob/ob) or leptin receptor (db/db) revealed that Kdm4b mRNA abundance was attenuated in obesity. This includes suppression of Kdm4b expression in WAT across all obesity mouse models, as well as in BAT of ob/ob mice (Fig. 4B).
Fig. 4.
Kdm4b adipose-specific KO mice have a similar phenotype with whole-body Kdm4b KO mice. (A) Determination of Kdm4b expression in different tissues using qRT-PCR. Hprt was used as an internal control. (B) Determination of Kdm4b mRNA levels in diet-induced obesity model, ob/ob and db/db transgenic mice (n = 6). (C) Assessment of adipose-specific deletion of Kdm4b by qRT-PCR. f/f, Kdm4bf/f; f/f Adipoq-Cre, Kdm4bf/fAdipoq-Cre (n = 5). (D) Representative images of Kdm4bf/f and Kdm4bf/fAdipoq-Cre littermates after HFD feeding. (E) Growth curve of Kdm4bf/f and Kdm4bf/fAdipoq-Cre littermate on a HFD (n = 8). (F) Serum insulin in Kdm4bf/f and Kdm4bf/fAdipoq-Cre mice before and after HFD feeding (n = 8). (G and H) Plasma blood glucose during GTT and ITT in HFD-fed Kdm4bf/f and Kdm4bf/fAdipoq-Cre mice (n = 5). (I) Determination of energy expenditure for Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice, using metabolic cages at 2 mo (n = 9). (J) Determination of oxygen consumption in day and dark time for Kdm4bf/f and Kdm4bf/f Adipoq-Cre 2-mo mice (n = 9). (K) Measurements of body temperature of Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice under cold treatment over the course of 6 h (n = 4). (L and M) Ppargc1a gene expression in BAT (L) and Epi (M) after cold treatment of control Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice (n = 4). (N) Determination of lipolysis ratio in Epi of Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice (n = 5). (O) Measurements of oxidation ratio of Epi WAT in Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice analyzed by Seahorse (n = 3). *P < 0.05; **P < 0.01. All data are presented as the mean ± SEM. OCR, oxygen consumption rate.
To investigate the role of KDM4B in adipose tissues in vivo, Kdm4bf/f mice were crossed with transgenic mice expressing Cre recombinase under the control of the adipocyte-specific adiponectin (Adipoq) gene promoter, allowing generation of fat-specific Kdm4b KO mice (12). Kdm4b expression levels were evaluated in various tissues, and the results confirmed the deletion of Kdm4b specifically in WAT and BAT in Kdm4bf/fAdipoq-Cre mice (Fig. 4C). The Kdm4bf/fAdipoq-Cre mice were born at the expected Mendelian ratio and had normal morphology at birth. When these animals were exposed to HFD, progressively and significantly greater body weight gain in Kdm4bf/fAdipoq-Cre mice was observed (Fig. 4 D and E). After 10 wk of HFD, NMR-analyzed fat mass composition and BMI were significantly elevated in Kdm4bf/fAdipoq-Cre mice (SI Appendix, Fig. S2 A and B). In addition, Kdm4bf/fAdipoq-Cre mice displayed increased adiposity with larger and heavier Epi and SubQ fat, as well as greater adipocyte diameter in both Epi and SubQ fat after HFD feeding (SI Appendix, Fig. S2 C–F). The H&E staining of BAT revealed a marked increase in adipocyte size and lipid storage of brown adipocytes in Kdm4bf/fAdipoq-Cre mice compared with Kdm4bf/f mice (SI Appendix, Fig. S2G). These obesity phenotypes of adipose-specific Kdm4b KO mice closely mirrored the findings with whole-body Kdm4b KO mice, suggesting that the disruption of KDM4B function in adipose tissue is essential for Kdm4b deficiency-induced obesity. The obese phenotype with its associated metabolic dysfunction was also observed in an independent conditional strain generated using aP2-Cre (Kdm4bf/faP2-Cre mice), which expresses Cre in adipocytes as well as in additional cell types such as macrophages (SI Appendix, Fig. S3). In contrast, deletion of Kdm4b in liver driven by albumin-Cre produced mice of normal size and body composition on both chow and HFD, revealing that hepatic KDM4B does not play a role in the obesity phenotype (SI Appendix, Fig. S4).
Adipose tissue-specific Kdm4b-deficient mice exhibited impaired glucose homeostasis. After HFD feeding, fasting insulin levels were greatly elevated in Kdm4bf/fAdipoq-Cre mice, and glucose and insulin tolerance tests revealed that the specific deletion of Kdm4b in adipose tissue leads to glucose intolerance and reduced insulin sensitivity (Fig. 4 F–H). Such metabolic deficiencies were not present before diet-induced obesity (SI Appendix, Fig. S2 H and I). Plasma total triglyceride and cholesterol, as well as LDL and very LDL cholesterol levels, were higher in HFD-feeding Kdm4bf/fAdipoq-Cre mice compared with their WT littermates (Table 2). No differences in these lipids were seen before HFD feeding (Table 2). Enlarged pancreatic islets were also observed in adipose-specific Kdm4b-deficient mice fed a HFD (SI Appendix, Fig. S2 J and K). The HFD adipose-specific KO mice also had significantly reduced expression of Irs1 and Irs2 in both Epi WAT and BAT (SI Appendix, Fig. S2 L and M).
Table 2.
Lipid panel for Kdm4bf/f (f/f) and Kdm4bf/fAdipoq-Cre (f/f Adipoq-Cre) mice before and after HFD feeding
| Before HFD | After HFD | |||||
| Metabolic parameters | f/f | f/f Adipoq-Cre | P value | f/f | f/f Adipoq-Cre | P value |
| Triglyceride, mg/dL | 56.4 ± 9.3 | 65.0 ± 9.9 | 0.07 | 55.2 ± 10.0 | 66.7 ± 15.8 | 0.034 |
| Total cholesterol, mg/dL | 81.8 ± 10.8 | 83.7 ± 10.6 | 1.00 | 141.9 ± 34.6 | 173.9 ± 30.0 | 0.017 |
| LDL cholesterol, mg/dL | 18.7 ± 3.8 | 17.6 ± 6.8 | 0.29 | 68.5 ± 25.0 | 90.8 ± 24.1 | 0.025 |
| VLDL | 11.2 ± 2.0 | 12.8 ± 2.1 | 0.09 | 11.1 ± 2.0 | 13.7 ± 3.1 | 0.019 |
We assessed whether the specific deletion of Kdm4b in adipose tissues impairs energy expenditure, as observed in the global Kdm4b-deficient mice. Similar to the whole-body KO model, Kdm4b deficiency, specifically in the adipose tissues, was sufficient to cause impaired energy expenditure and oxygen consumption (Fig. 4 I and J). The adipose tissue-specific Kdm4b-deficient mice also exhibited impaired adaptive thermogenesis when exposed to 4 °C (Fig. 4K). Blunted Ppargc1a induction on cold exposure in both BAT and Epi WAT was also exhibited in adipose tissue-specific Kdm4b-deficient mice (Fig. 4 L and M). Isolated Kdm4b-deficient Epi WAT exhibited impaired basal and isoproterenol-stimulated lipolysis and reduced oxygen consumption compared with that from control Kdm4bf/f mice (Fig. 4 N and O). SubQ fat is the major site of cold-induced adipose tissue browning (13). We also tested whether KDM4B was required for cold-induced browning in SubQ fat. Histological examination showed that there was no difference in SubQ fat browning between WT and KO mice or Kdm4bf/f and Kdm4bf/fAdipoq-Cre mice on cold stimulation (SI Appendix, Fig. S5 A and B).
KDM4B Controls Metabolic Gene Expression.
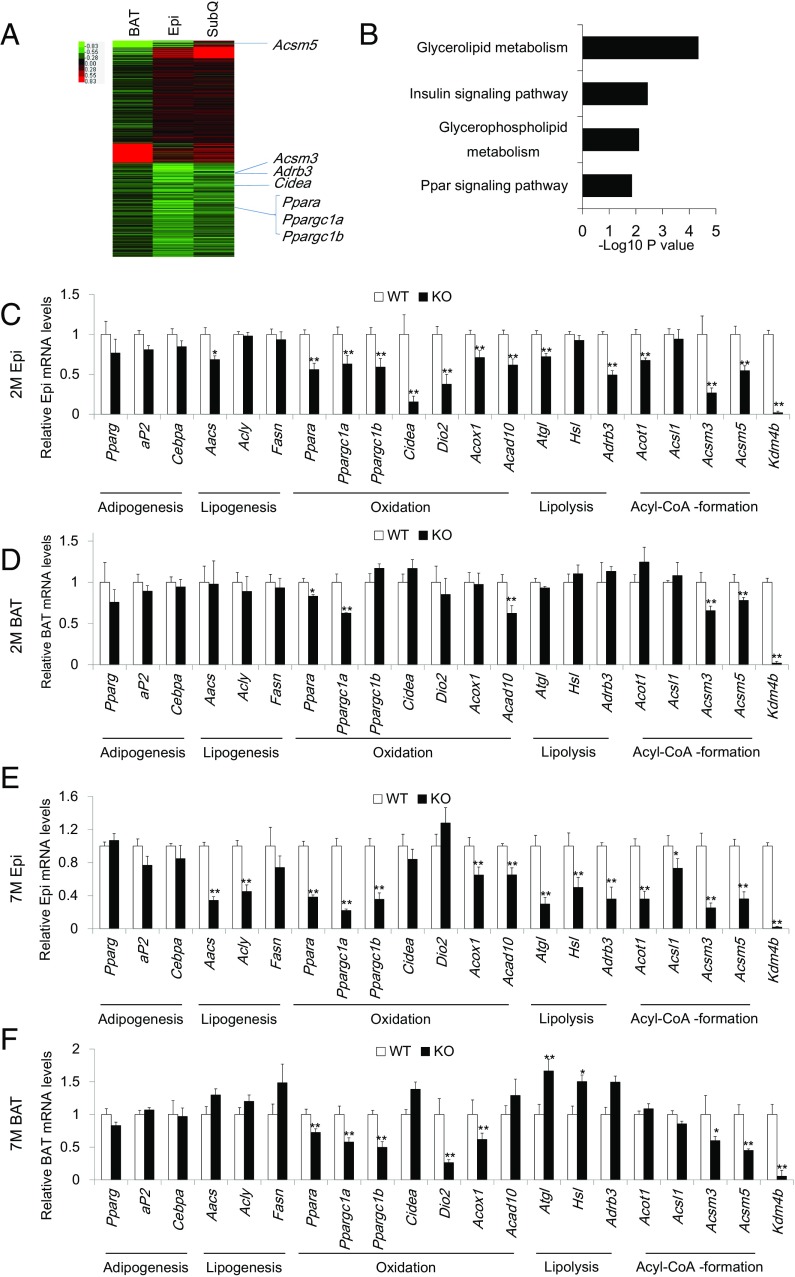
To explore the molecular mechanisms of Kdm4b deficiency-induced obesity, we performed a microarray analysis on the Epi, SubQ, and BAT of 2-mo-old WT and whole body Kdm4b KO mice. Gene expression profiles identified 1,508 genes that were down-regulated and 547 genes that were up-regulated in Epi fat of KO mice compared with WT mice (Fig. 5A). In SubQ fat, 1,276 genes were down-regulated and 885 genes were up-regulated. In BAT, 1,396 genes were down-regulated and 580 genes were up-regulated with Kdm4b deficiency. We found that many metabolic-related genes were reduced in Epi, SubQ, and BAT depots, with Epi fat exhibiting the most dramatic down-regulation in gene expression in KO mice compared with WT mice (Fig. 5A). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of Epi WAT indicated that down-regulated genes with Kdm4b deficiency were enriched in pathways associated with PPAR and insulin signaling, as well as glycerolipid and glycerophospholipid metabolism (Fig. 5B).
Fig. 5.
KDM4B regulates metabolic genes involved in oxidation, lipolysis, and acyl-CoA formation. (A) Heat map showing the expression levels of genes in BAT, Epi WAT, SubQ WAT in KO mice compared with WT mice. (B) Gene ontology enrichment analysis for down-regulated genes in Epi WAT. Bars represent −log10 of P values. (C and D) Real-time RT-PCR analysis of metabolic marker genes in Epi (C) and BAT (D) for WT and KO mice at 2 mo (n = 8). (E and F) Real-time RT-PCR analysis of metabolic marker genes in Epi (E) and BAT (F) for WT and KO mice at 7 mo (n = 8). *P < 0.05; **P < 0.01. All data are presented as the mean ± SEM.
The genes influenced by Kdm4b levels included genes encoding two transcription factors that are known to be critical for energy expenditure and oxidation in vivo: PPARα and PGC-1α. qRT-PCR confirmed that loss of KDM4B reduced the expression of Ppara and Ppargc1a in Epi WAT of KO mice (Fig. 5C). In addition, qRT-PCR confirmed that Ppargc1b and genes involved in lipolysis (Atgl and Adrb3), acyl-CoA formation (Acot1, Acsm3, and Acsm5), and oxidation/thermogenesis (Cidea, Dio2, Acox1, and Acad10) are down-regulated in WAT from KO animals compared with their WT littermates (Fig. 5C). A subset of these genes also exhibited reduced expression in BAT of KO mice (Fig. 5D). In contrast, no significant changes in marker genes for adipogenesis (Pparg, Fabp4, and Cebpa) and lipogenesis (Aacs, Acly, and Fasn) were detected in either WAT or BAT (Fig. 5 C and D). Also, there was no significant difference in Ucp1 mRNA expression from Epi, SubQ, and BAT of WT and KO mice (SI Appendix, Fig. S6A). We further tested the expression of UCP1 under cold stimulation and found that there was no difference in UCP1 protein levels between WT and KO mice (SI Appendix, Fig. S6B).
We also examined the gene expression profiles of 7-mo-old WT and KO mice. Although the trend was similar to the gene profiles for 2-mo-old mice, more accentuated down-regulation of genes for lipolysis and acyl-coA formation was observed in Epi-WAT, and additional marker genes for lipolysis (Hsl) and acyl-coA formation (Acsl1) were significantly down-regulated (Fig. 5E). Interestingly, lipogenesis marker genes, including Aacs and Acly, were also significantly suppressed in Epi WAT of KO mice, which could represent a secondary effect resulting from age-induced obesity at 7 mo (Fig. 5E). We also confirmed the gene expression in BAT from 7-mo-old Kdm4b KO mice (Fig. 5F). Ppara and Ppargc1a are both down-regulated in BAT.
To confirm that KDM4B does not play a role in adipogenesis, preadipocytes of WT and KO mice were induced to differentiate into adipocytes and their adipogenic potential was compared. Oil Red O staining and expression levels of adipogenic marker genes revealed no differences between the two genotypes (SI Appendix, Fig. S5 C and D).
To determine whether gene expression changes in adipose tissues result from local effects of reduced KDM4B activity within these tissues, we examined metabolic gene expression levels in Epi WAT and BAT of 2-mo-old Kdm4bf/f Adipoq-Cre mice. Consistent with the expression profile of 2-mo-old whole-body Kdm4b KO mice, Ppara and Ppargc1a were significantly down-regulated in Epi WAT of Kdm4bf/f Adipoq-Cre mice compared with Kdm4bf/f mice. A set of genes involved in lipolysis (Atgl and Adrb3), acyl-CoA formation (Acsm3 and Acsm5), and oxidation/thermogenesis (Ppargc1b, Cidea, and Acad10) were also found to be down-regulated in Epi-WAT of Kdm4bf/f Adipoq-Cre mice (SI Appendix, Fig. S7A). In BAT of Kdm4bf/f Adipoq-Cre mice, Ppargc1a, Dio2, Acox1, and Ascm3 were significantly decreased (SI Appendix, Fig. S7B). In mice fed a HFD, down-regulation of genes associated with lipolysis (Atgl, Hsl, and Adrb3) and acyl-CoA formation (Acsl1, Ascm3, and Ascm5) was even more pronounced than in chow-fed mice (SI Appendix, Fig. S7 C and D). In addition, adipose tissue-specific Kdm4b deficiency led to decreased BAT expression of Ppargc1a and Ppara and reduced Epi WAT expression of lipogenesis genes, including Aacs and Fasn (SI Appendix, Fig. S7 C and D). We also test those genes in fully differentiated Kdm4b WT and KO adipocytes. Consistent with data in adipose tissues, those metabolic genes were also significantly reduced in Kdm4b KO adipocytes, further indicating that the down-regulation of those genes is adipocyte-intrinsic in Kdm4b KO mice (Fig. 6A).
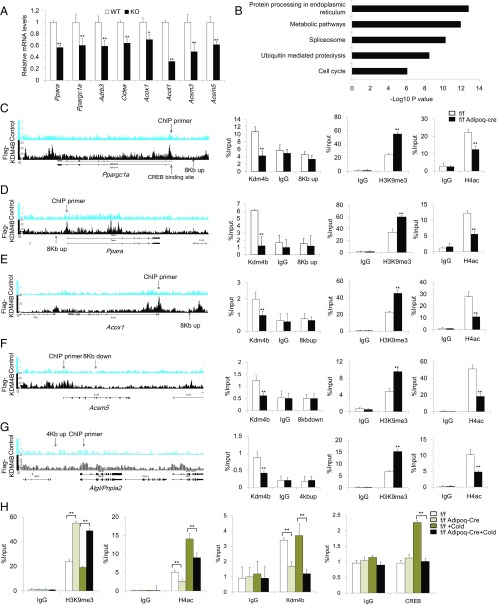
Fig. 6.
Loss of KDM4B leads to increased H3K9me3 and decreased H4ac to the metabolic genes promoter. (A) Relative mRNA levels of Ppara, Ppargc1a, Adrb3, Cidea, Acox1, Acot1, Acsm3, and Acsm5 in differentiated Kdm4b WT and KO adipocytes. Data are presented as the mean ± SEM. (B) KEGG pathway analysis of Kdm4b direct targets identified by ChIP-Seq. (C–G) The ChIP-Seq result, location of ChIP primers, determination of Kdm4b occupancy, the level of H3K9me3, and the levels of H4ac on the promoters of Ppargc1a, Ppara, Acox1, Acsm5, and Atgl in Epi WAT of 2-mo-old Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice measured by ChIP-qPCR. Error bars represent mean ± SD for triplicate experiments. (H) The level of H3K9me3, H4ac, KDM4B occupancy, and CREB binding on the promoter of Ppargc1a before and after cold treatment in 2-mo-old Kdm4bf/f and Kdm4bf/f Adipoq-Cre mice. *P < 0.05; **P < 0.01. Error bars represent mean ± SD for triplicate experiments.
KDM4B Directly and Epigenetically Controls Metabolic Genes in Adipose Tissues.
KDM4B epigenetically promotes gene transcription by erasing the repressive H3K9me3 marks. To elucidate whether KDM4B in adipose tissues directly regulated metabolic regulatory genes identified from microarray analyses, we performed chromatin immunoprecipitation (ChIP)-sequencing assays (ChIP-Seq). Unfortunately, although several commercial anti-mouse KDM4B antibodies were able for detecting locus specific enrichment using ChIP-qPCR, they were not suitable for ChIP-Seq studies because of substantial background noise. To overcome this problem, we isolated preadipocytes from Kdm4b KO mice and transduced them with the retroviruses expressing Flag-KDM4B or the empty vector. Subsequently, Kdm4b−/− preadipocytes expressing Flag-KDM4B or the empty vector were induced to differentiate, and differentiated adipocytes were used for ChIP-Seq with Flag antibodies.
We identified 11,867 significantly enriched binding peaks. These peaks distributed to the intron (35%), distal intergenic (23.8%), promoter (26.6%), coding exon (3.1%), downstream of gene (5,000 bp; 4.3%), 5′UTR (6.1%), and 3′UTR (1.1%). The cis-regulatory element annotation system analysis revealed that KDM4B showed very high relative enrichment in the promoter regions (26.6% compared with 1% of the genome background). Consistently, the average bindings of KDM4B for all refseq gene promoters, spanning ±5 kb of the closest transcription starting sites, showed that KDM4B is significantly enriched within 1 kb of the transcription starting site. There was no binding in Kdm4b−/− cells expressing the empty vector, which confirmed that Flag-KDM4B binding is specific (SI Appendix, Fig. S8A). KEGG pathway analysis indicated that KDM4B targets are highly associated with metabolic pathway, protein processing in endoplasmic reticulum, splicesome, ubiquitin-mediated proteolysis, and cell cycle (Fig. 6B). We compared and overlapped KDM4B-regulated genes in Epi fat with the genes containing KDM4B-binding peaks, and found that 697 KDM4B-regulated genes, including Ppara, Ppargc1a, Acox1, Acsm5, and Atgl, have the KDM4B-binding peaks (SI Appendix, Fig. S8B). To further confirm our results, we performed individual ChIP-qPCR, using Epi WAT of 2-mo-old Kdm4bf/f and Kdm4bf/fAdipoq-Cre mice. ChIP-qPCR validated that KDM4B was present in the promoters of Ppara, Ppargc1a, Acox1, Acsm5, and Atgl in WT adipose tissues, but this occupancy was absent in Kdm4b-deficient adipose tissues (Fig. 6 C–G). Moreover, ChIP-qPCR showed that H3K9me3 levels were significantly increased on the promoters of Ppara, Ppargc1a, Acox1, Acsm5, and Atgl in Kdm4b-deficient adipose tissues compared with WT adipose tissues (Fig. 6 C–G). H3K9me3 levels did not change on the promoters of genes (aP2 and Acly) in Kdm4b-deficient mice (SI Appendix, Fig. S8 C and D). Histone H4 acetylation (H4ac) is a general active mark of gene transcription. Therefore, we performed ChIP-qPCR to examine the levels of H4ac on the promotors of Ppargc1a, Ppara, Acox1, Acsm5, and Atgl. ChIP-qPCR revealed that the levels of H4ac were significantly reduced on these gene promoters in Kdm4b-deficient adipose tissues compared with WT adipose tissues (Fig. 6 C–G).
We further explored how KDM4B epigenetically regulated Ppargc1a induction. ChIP assays showed that KDM4B was constitutively present on the Ppargc1a promoter regardless of cold treatment in control Epi WAT tissues. KDM4B could not be detected on the Ppargc1a promoter in Kdm4b-deficient Epi WAT tissues (Fig. 6H). Consistently, the levels of H3K9me3 marks on the Ppargc1a promoter were significantly lower in control Epi WAT tissues than in Kdm4b-deficient Epi WAT tissues (Fig. 6H), suggesting that easing H3K9me3 might play a priming role in Ppargc1a transcription. The active H4ac marks on the Ppargc1a promoter were higher in WT Epi WAT tissues than in Kdm4b-deficient Epi WAT tissues (Fig. 6H). Because cold stimulation induces the cAMP-responsive element-binding protein (CREB) to the Ppargc1a promoter to activate its transcription (14, 15), we further examined whether loss of KDM4B affected the recruitment of CREB to the Ppargc1a promoter. ChIP assays revealed that CREB was induced to bind to the Ppargc1a promoter in WT Epi WAT tissues on cold stimulation. On the contrary, the occupancy of CREB on the Ppargc1a promoter was completely absent after cold treatment in Kdm4b-deficient Epi WAT tissues (Fig. 6H).
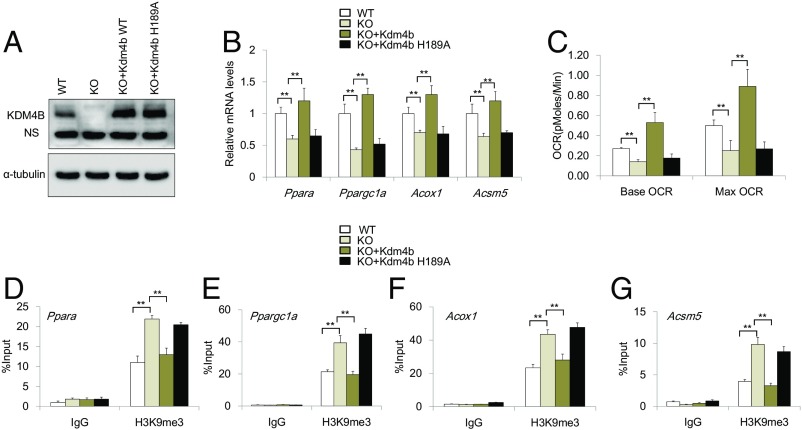
To test whether the enzymatic activity of KDM4B was required for KDM4B-mediated energy homeostasis, we generated a catalytically inactive mutant of Flag-KDM4B-H189A (16). KO preadipocytes were transduced with retroviruses expressing Flag-KDM4B or Flag-KDM4B-H189A and subsequently differentiated into adipocytes (Fig. 7A). RT-qPCR showed that the restoration of KDM4B, but not KDM4B-H189A, was able to rescue the expression of Ppara, Ppargc1a, Acox1, and Acsm5 in KO adipocytes (Fig. 7B). Oxygen consumption rate assays found that the restoration of KDM4B, but not KDM4B-H189A, was able to restore the oxygen consumption of KO adipocytes under basal conditions and after FCCP stimulation (Fig. 7C). ChIP-qPCR revealed that the restoration of KDM4B, but not KDM4B-H189A, significantly reduced H3K9me3 levels on the promoters of Ppargc1a, Ppara, Acox1, and Acsm5 in KO adipocytes, respectively (Fig. 7 D–G).
Fig. 7.
The enzymatic activity of KDM4B is required for KDM4B-mediated gene expression and energy homeostasis. (A) Western blotting confirmed that Kdm4b and Kdm4b-H189A were restored in KO adipocytes. (B) The expression of Ppara, Ppargc1a, Acox1, and Acsm5 was examined by RT-qPCR. (C) The oxygen consumption of KO adipocytes expressing KDM4B or KDM4B-H189A under basal conditions and after isoproterenol stimulation. OCR, oxygen consumption rate. (D–G) The levels of H3K9me3 on the promoters of Ppargc1a, Ppara, Acox1, and Acsm5 by ChIP-qPCR, respectively. WT, WT adipocytes; KO, KO adipocytes; KO+Kdm4b, KO adipocytes expressing Flag-KDM4B; KO+Kdm4b-H189A, KO adipocytes expressing Flag-KDM4B-H189A.
Discussion
As the incidence of obesity continues to rise at an alarming rate, finding a solution to address this global public health problem is imperative. As part of this effort, adipocytes have been the target of intense investigation for their known role in obesity and metabolic diseases. Here, we identified an epigenetic modulator, KDM4B, that directly regulates essential metabolic genes in adipose tissues to maintain energy homeostasis. Both whole-body and adipose-specific Kdm4b deletion resulted in increased susceptibility to obesity. The onset of obesity was not caused by excessive caloric intake but, rather, was the result of impairments in metabolic processes leading to a reduction in energy expenditure and impaired adaptive thermogenesis. Kdm4b-deficient mice also developed insulin resistance, hyperlipidemia, and morphologic alterations in the liver and pancreas. Further studies will be required to determine whether all these effects are secondary to obesity or, rather, reflect specific roles for KDM4B in these tissues.
Our findings using conditional KO animals for liver and adipose tissue revealed that liver-specific Kdm4b deletion was not associated with obesity, whereas adipose-specific deletion produced striking phenotypes mirroring findings from whole-body KO animals. Moreover, the phenotypes in Kdm4bf/f Alb-Cre mice indicated that the liver defects seen in Kdm4b-deficient animals were not caused by alterations in hepatic biology but, rather, were induced by obesity, and specifically by the accumulation of Epi fat, which could indirectly increase hepatic lipid. Conversely, Kdm4bf/f Adipoq-Cre mice developed obesity and exhibited impaired glucose metabolism and insulin resistance as a result of decreased insulin signaling in adipose tissue. Tissue-specific deletion allowed us to identify BAT and WAT adipose tissues as the main sites of KDM4B action and highlighted the specific role of adipose KDM4B in regulating whole-body energy homeostasis. Both whole-body and tissue-specific conditional KOs revealed a causal relationship between Kdm4b deficiency and metabolic malfunction. Furthermore, Kdm4b expression levels were reduced in diet-induced and genetically obese mouse models, suggesting that Kdm4b dysregulation may occur in obesity resulting from multiple causes. The link between Kdm4b levels and obesity highlights KDM4B as a potential therapeutic target for obesity.
Kdm4b deficiency did not affect adipocyte differentiation, but altered expression of many metabolic genes in adipose tissues. Notable among these was the gene encoding PGC-1α, a key player in oxidative metabolism and mitochondrial biogenesis that promotes energy expenditure mainly through thermogenesis (17–20). Robust activation of PGC-1α occurs in adipose tissue in response to a variety of environmental cues (19). In our study, marked induction of Ppargc1a expression in response to cold temperature in Kdm4bf/f mice vanished with the loss of Kdm4b in adipose tissues. Given the critical role of PGC-1α in metabolism, the expression of Ppargc1a is tightly regulated. Our studies identified a role for KDM4B in the epigenetic regulation of Ppargc1aby erasing H3K9me3 marks. It is somewhat surprising that fat-specific Kdm4b KO mice exhibited decreased energy expenditure without UCP1 inhibition. Recent studies have demonstrated that beige fat engages several UCP1-independent mechanisms to generate heat, including creatine-driven substrate cycle and sarcoplasmic/endoplasmic reticulum calcium ATPases 2b-mediated calcium cycling (21, 22). It is therefore possible that Kdm4b deficiency may lead to the inhibition of these non-UCP1 thermogenic pathways to attenuate energy expenditure. In summary, our findings indicate that KDM4B has an important role in metabolic homeostasis by regulation of gene expression in BAT and WAT, with subsequent effects on energy metabolism. The observation that heterozygous Kdm4b mice have reduced adiposity suggests that modulation of KDM4B levels or activity may be a potential therapeutic target for human obesity.
Materials and Methods
Mice.
The Kdm4bflox/flox mice were purchased from Sanger (UK) with a C57BL6 genetic background. The flox site flanked Kdm4b exon 5, which encodes the jumonji domain. The deletion of exon 5 resulted in a frame shift and early translation termination. First, the neo-LacZ cassette was deleted by Flp-FRT recombination (Jackson Laboratory, catalog no. 009086) and Flippase was crossed out by mating with C57BL6 WT mice. Then Kdm4bflox/+ mice were crossed with CMV-Cre mice (Jackson Laboratory, catalog no. 006054) to obtain Kdm4b heterozygotes. Heterozygotes were intercrossed to obtain WT, heterozygous, and homozygous genotypes. Tissue-specific Kdm4b-deficient mice were generated by crossing Kdm4bf/f mice with mice expressing Alb-Cre (Jackson Laboratory, catalog no. 003574) or Adipoq-Cre (Jackson Laboratory, catalog no. 010803). All mice were housed in pathogen-free facilities under 12-h light/12-h dark cycle. All protocols were approved by the Division of Laboratory Animal Medicine of the University of California, Los Angeles, and were in accordance with National Institutes of Health guidelines.
Body Weight, Food Intake, and Body Composition.
Body weight was measured weekly on a normal chow diet, where the mice were fed for a period of 7 mo. Body weights were measured weekly until the end of the experimental protocol. For the HFD feeding model, 8-wk-old mice were fed a HFD (60% fat, 20% protein, and 20% carbohydrate by kcal; D12492, Research diets) for ∼10–12 wk. Then WAT and BAT were harvested for RNA extraction. Ob/ob and db/db mice models were described before (23). All experiments were performed in male mice.
For food intake, mice were single housed for 1 wk before measurement. The food intake data were collected for a 2-wk period. Body composition was analyzed by a rodent NMR scanner (Bruker Biospin), as described previously (24). The mice placed in the scanner for measurement of body composition were analyzed for fat mass (g), muscle mass (g), and free fluid (g). The percentage of fat mass was normalized by total body weight. The fat mass ratio was calculated by fat mass divided by total body weight. BMI was calculated by BMI formula by dividing body weight (g) by body length (mm) squared (BMI = body weight/body length2).
Statistical Analysis.
Statistical analysis between two groups was carried out with Student’s t test. For comparison between groups involving two genotypes of mice along with procedures or times, the differences were assessed using two-way ANOVA. ANCOVA was used for energy expenditure experiment. P ≤ 0.05 was considered a statistically significant difference.
Study Approval.
All mouse studies were performed under the approved University of California, Los Angeles, Chancellor’s Animal Research Committee (ARC) ARC#2013–002-11B, and ARC#2007–058-08A and were in accordance with National Institutes of Health guidelines.
Extended method and information about histology, lipids and insulin measurement, glucose tolerance test (GTT) and insulin tolerance test (ITT), indirect calorimetry and oxygen consumption rate, lipolysis, cold treatment, RNA extraction, qRT-PCR and gene expression study, and ChIP-qPCR and ChIP-Seq assays are described in SI Appendix, Materials and Methods. The microarray and ChIP-Seq datasets have been deposited to the NCBI Gene Expression Omnibus (GEO) database under accession numbers GSE102289 and GSE102148, respectively.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01AR063089 and R01DE16513; a Core Voucher Award from the University of California, Los Angeles, Clinical and Translational Science Institute; the Shapiro Family Charitable Foundation; and NIH Grants U24DK092993 and DK076169 of Mouse Metabolic Phenotyping Center program in the University of California, Davis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The ChIP-Seq datasets have been submitted to the NCBI Gene Expression Omnibus (GEO) database under the accession no. GSE102148.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721814115/-/DCSupplemental.
References
- 1.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao RR, et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell. 2014;157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y. Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 8.Deng P, Chen QM, Hong C, Wang CY. Histone methyltransferases and demethylases: Regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int J Oral Sci. 2015;7:197–204. doi: 10.1038/ijos.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye L, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HL, Yu B, Deng P, Wang CY, Hong C. Transforming growth factor-β-induced KDM4B promotes chondrogenic differentiation of human mesenchymal stem cells. Stem Cells. 2016;34:711–719. doi: 10.1002/stem.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimura A, et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature. 2012;483:350–354. doi: 10.1038/nature10798. [DOI] [PubMed] [Google Scholar]
- 12.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen P, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Spiegelman BM, Puigserver P, Wu Z. Regulation of adipogenesis and energy balance by PPARgamma and PGC-1. Int J Obes Relat Metab Disord. 2000;24(Suppl 4):S8–S10. doi: 10.1038/sj.ijo.0801492. [DOI] [PubMed] [Google Scholar]
- 21.Kazak L, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. 2015;163:643–655. doi: 10.1016/j.cell.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda K, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017;23:1454–1465. doi: 10.1038/nm.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GX, et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med. 2014;20:1436–1443. doi: 10.1038/nm.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhasin KK, et al. Maternal low-protein diet or hypercholesterolemia reduces circulating essential amino acids and leads to intrauterine growth restriction. Diabetes. 2009;58:559–566. doi: 10.2337/db07-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.