Significance
Membrane fusion is a fundamental process of eukaryotic cells required for subcellular organization and cell–cell communication, involving SNARE proteins and regulatory Sec1/Munc18 (SM) proteins. In plant cytokinesis, membrane vesicles delivered to the cell-division plane fuse with one another to form the partitioning membrane, which requires cytokinesis-specific Qa-SNARE KNOLLE and interacting SM protein KEULE in Arabidopsis. KEULE has a paralog named SEC1B, which originated through gene duplication and subsequent functional diversification during angiosperm evolution. Biochemical interaction studies and analysis of relevant double mutants reveal that a predominant interaction of SEC1B with Qa-SNARE SYP132 entailed its preferential role in secretion, whereas KEULE acquired a unique role in cytokinesis through its interaction with cytokinesis-specific Qa-SNARE KNOLLE.
Keywords: membrane traffic, cell-plate formation, secretion, Qa-SNAREs, SEC1/Munc18
Abstract
Sec1/Munc18 (SM) proteins contribute to membrane fusion by interacting with Qa-SNAREs or nascent trans-SNARE complexes. Gymnosperms and the basal angiosperm Amborella have only a single SEC1 gene related to the KEULE gene in Arabidopsis. However, the genomes of most angiosperms including Arabidopsis encode three SEC1-related SM proteins of which only KEULE has been functionally characterized as interacting with the cytokinesis-specific Qa-SNARE KNOLLE during cell-plate formation. Here we analyze the closest paralog of KEULE named SEC1B. In contrast to the cytokinesis defects of keule mutants, sec1b mutants are homozygous viable. However, the keule sec1b double mutant was nearly gametophytically lethal, displaying collapsed pollen grains, which suggests substantial overlap between SEC1B and KEULE functions in secretion-dependent growth. SEC1B had a strong preference for interaction with the evolutionarily ancient Qa-SNARE SYP132 involved in secretion and cytokinesis, whereas KEULE interacted with both KNOLLE and SYP132. This differential interaction with Qa-SNAREs is likely conferred by domains 1 and 2a of the two SM proteins. Comparative analysis of all four possible combinations of the relevant SEC1 Qa-SNARE double mutants revealed that in cytokinesis, the interaction of SEC1B with KNOLLE plays no role, whereas the interaction of KEULE with KNOLLE is prevalent and functionally as important as the interactions of both SEC1B and KEU with SYP132 together. Our results suggest that functional diversification of the two SEC1-related SM proteins during angiosperm evolution resulted in enhanced interaction of SEC1B with Qa-SNARE SYP132, and thus a predominant role of SEC1B in secretion.
Membrane fusion in eukaryotes is mediated by the formation of trans-complexes between SNARE proteins C-terminally anchored in adjacent membranes. Sec1/Munc18 (SM) proteins assist in SNARE complex formation (1). The family of SM proteins comprises four members that act at different subcellular locations [SEC1, plasma membrane; SLY1, endoplasmic reticulum (ER), and Golgi; VPS45, trans-Golgi network (TGN)-early endosome; VPS33, late endosome-lysosome/vacuole] and are evolutionarily conserved across the eukaryotes (2). VPS45 and VPS33 have also been studied in the flowering plant Arabidopsis (3–6). The plasma membrane-localized Sec1p of yeast has counterparts in multicellular eukaryotes, with the latter often occurring in several isoforms. For example, Munc18 isoforms in mammals are specialized toward specific tasks or are expressed in tissue-specific ways (7). The Arabidopsis genome codes for three variants of Sec1p-related SM proteins, named KEULE (KEU), SEC1A, and SEC1B (8). KEULE has been studied in some detail, whereas SEC1A and SEC1B have been barely touched. KEULE is essential for vesicle fusion by which the partitioning membrane named “cell plate” is formed in cytokinesis (8–12). KEULE has recently been proposed also to coordinate that membrane fusion with the dynamics of phragmoplast microtubules supporting cell plate formation (13). KEULE (named SEC11) has also been implicated in trafficking to the plasma membrane in interphase by interacting with PEN1 (also known as SYR1 or SYP121), a Qa-SNARE protein involved in ABA response, pathogen attack, and programmed stomatal closure (14, 15). It is not known whether the two other SEC1-related proteins play different roles than KEULE or whether there is substantial functional overlap between the three SEC1 isoforms.
Here we analyze the Arabidopsis SEC1 isoform SEC1B in regard to functional requirement, subcellular localization, and Qa-SNARE interaction, in comparison with the cytokinesis-essential isoform KEULE. SEC1B overlapped functionally with KEULE in general secretion and, to a lesser extent, in cytokinesis. However, unlike KEULE, SEC1B almost failed to interact with Qa-SNARE KNOLLE (KN) in cytokinesis but interacted strongly with the ancient Qa-SNARE SYP132, indicating a prominent role for SEC1B in secretory traffic to the plasma membrane.
Results
SM Protein SEC1B Is Nonessential Because of Functional Overlap with Its Paralog KEULE.
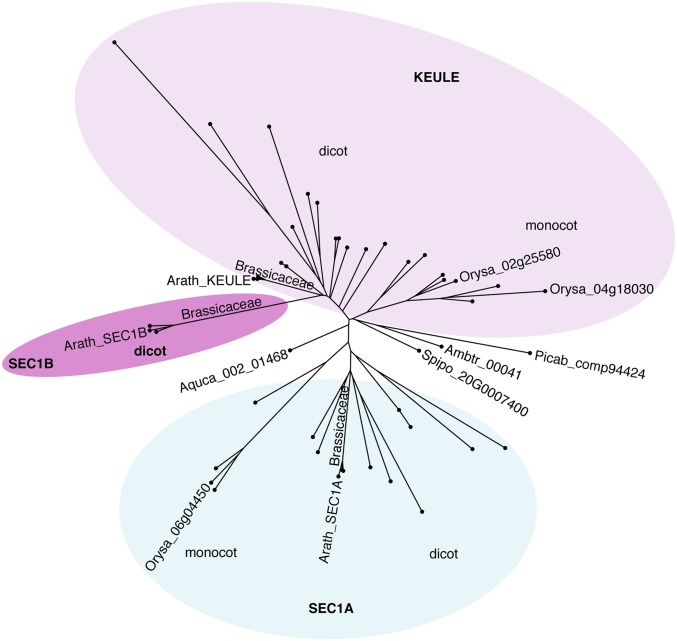
From an evolutionary perspective, gymnosperms like Norway spruce (Picea abies), the basal angiosperm Amborella, the basal dicot Aquilegia, and the secondarily simplified monocot Spirodela all encode only a single SEC1 isoform (Fig. 1 and SI Appendix, Fig. S1) (16–23). Most dicot and monocot angiosperms have three SEC1 isoforms. Two copies of KEULE/SEC1B appear to have arisen by gene duplication independently in the two angiosperm lineages, whereas the gene duplication giving rise to SEC1A and KEULE/SEC1B may have occurred before the monocot–dicot split some 140 million y ago (24). The two copies of the KEULE/SEC1B gene are most strongly diverged by sequence in the Brassicaceae and allied species.
Fig. 1.
Simplified phylogenetic tree of plant SEC1 proteins. The phylogenetic tree was generated using the neighbor-joining method in the CLC workbench program (abridged). Note that KEULE and SEC1B evolved differentially from SEC1A and that SEC1B is only present in dicot plants. Ambtr, Amborella trichopoda (basal dicot); Aquca, Aquilegia caerulea (basal angiosperm); Arath, Arabidopsis thaliana (Brassicaceae); Orysa, Oryza sativa (Monocot); Picab, Picea abies (gynmosperm); Spipo, Spirodela polyrhiza (basal monocot). See SI Appendix, Fig. S1 for a detailed phylogenetic tree.
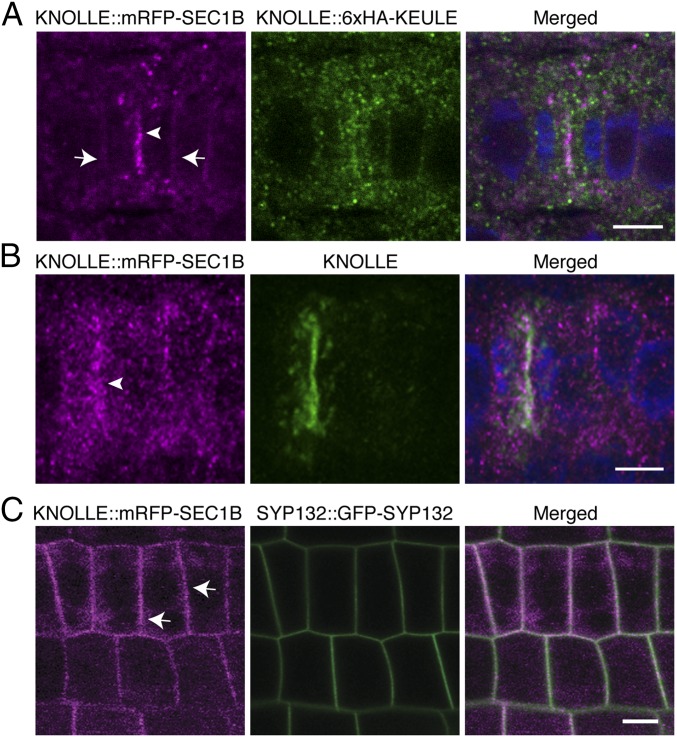
In Arabidopsis, the two SM proteins, KEULE and SEC1B, are closely related by sequence (69% identity, 82% similarity) (SI Appendix, Fig. S2) and might thus perform similar functions. One criterion is the subcellular localization of the protein. HA-tagged KEULE accumulates at the forming cell plate of dividing seedling root cells, in addition to cytosolic staining in both dividing and nondividing root cells (Fig. 2A) (25). mRFP-tagged SEC1B was detected in the cytosol, at the plasma membrane, and the forming cell plate in seedling root cells (Fig. 2 and SI Appendix, Fig. S3A). SEC1B colocalized with cytokinesis-specific Qa-SNARE KNOLLE at the forming cell plate (Fig. 2B). At the plasma membrane, SEC1B also colocalized with the Qa-SNARE SYP132 (Fig. 2C). SEC1B localization was insensitive to brefeldin A (BFA), a fungal toxin interfering with exchange factors for ARF small GTPases (ARF-GEFs) (SI Appendix, Fig. S3B) (26). This observation suggests that SEC1B, like KEULE, localizes to the cell plate and the plasma membrane independently of membrane traffic from the Golgi/TGN (25). In contrast, the more distantly related SEC1A was only detected in the cytosol of seedling root cells (SI Appendix, Fig. S3C).
Fig. 2.
Subcellular localization of KNOLLE::mRFP-SEC1B. Immunofluorescence (A and B) and live-imaging (C) of KNOLLE::mRFP-SEC1B in seedling roots: (A) KNOLLE::mRFP-SEC1B (magenta) and KNOLLE::6xHA-KEULE (green) labeled with anti-HA antibody in fixed seedling root. (B) KNOLLE::mRFP-SEC1B (magenta) and KNOLLE (green) labeled with anti-KNOLLE antiserum in fixed seedling root. (C) Live imaging of KNOLLE::mRFP-SEC1B (magenta) and SYP132::GFP-SYP132 (green) in seedling roots. Note that mRFP-SEC1B locates at the cell division plane (arrowheads in A and B) and plasma membrane (arrows in A and C) (see also SI Appendix, Fig. S3A). Note also that the punctate signal of mRFP-SEC1B becomes more prominent after fixation (compare also Right and Left panels in SI Appendix, Fig. S3A). DAPI was used for staining nuclei (blue). (Scale bars: 10 µm.)
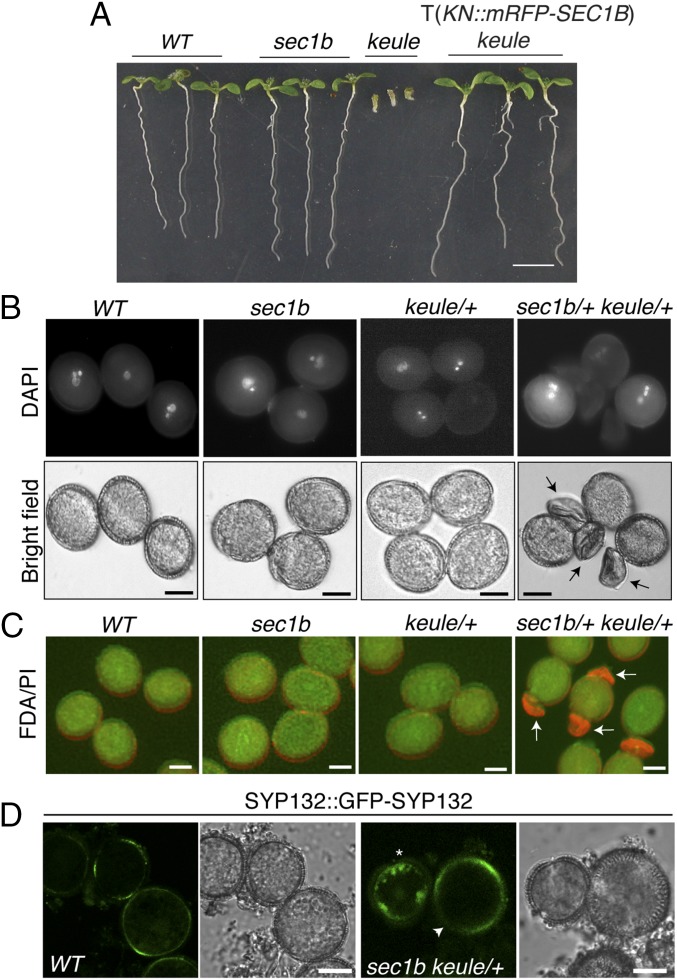
The functional relatedness of SEC1B to KEULE was also revealed by the ability of SEC1B when strongly expressed from the KNOLLE cassette (KNOLLE::mRFP-SEC1B) to rescue the KEULE deletion mutant keuleMM125 (Fig. 3A and SI Appendix, Fig. S3 D and F and Table S1) (8). The same rescuing effect had been demonstrated for KNOLLE::6xHA-KEULE (25). In contrast, KNOLLE::mRFP-SEC1A failed to rescue keuleMM125 (SI Appendix, Fig. S3 E and G and Table S1).
Fig. 3.
Analysis of sec1b single and sec1b keule double mutant. (A) Mutant phenotype. Note that the sec1b single mutant is indistinguishable from wild-type (WT), unlike the keule single mutant, which is fully rescued with a KNOLLE(KN)::mRFP-SEC1B transgene (T) (see SI Appendix, Table S1 for genetic analysis). (B and C) Mature pollen of sec1b keule double mutant. Pollen were stained with DAPI (B) or with fluorescein diacetate (FDA) (green) and propidium iodide (PI) (red) (C). Note that sec1b keule double-mutant pollen are collapsed and not labeled with DAPI or FDA (arrows in B and C; see SI Appendix, Fig. S5B for images in separate channels; SI Appendix, Table S2C for quantification). (D) SYP132::GFP-SYP132 expression in developing pollen of sec1b keule/+ plants. Note the aggregates of GFP-SYP132 in the sec1b keule double mutant (asterisk, Right) compared with the accumulation of GFP-SYP132 in a sec1b single mutant at the plasma membrane (arrowhead, Right) as in wild type (Left). See also SI Appendix, Fig. S7 for overview and line intensity profiles and SI Appendix, Table S3 for quantitative measurement. (Scale bars: 2 mm in A and 10 µm in B–D.)
To reveal functional requirements of the SEC1B gene, we analyzed a knockout allele caused by T-DNA insertion (GABI-KAT_601G09) (SI Appendix, Fig. S4 A and B). The level of mRNA accumulation for both SEC1A and KEULE appeared not to be altered in the sec1b knockout mutant (SI Appendix, Fig. S4B). Thus, there was no compensatory up-regulation of KEULE expression in response to the loss of SEC1B protein. The sec1b mutant plants were homozygous viable, fertile, and phenotypically normal and thus indistinguishable from wild-type plants (Fig. 3A). Moreover, sec1b mutant seedlings displayed normal root hairs (SI Appendix, Fig. S4C), in contrast to keule mutant seedlings (8). Thus, SEC1B appeared not to play an essential role in Arabidopsis development. It should be noted, however, that in all tissues and developmental stages analyzed, KEULE was expressed about 10-fold more strongly than SEC1B (SI Appendix, Fig. S4D) (27).
Unlike the sec1b knockout mutant, elimination of KEULE gene function is zygotically lethal, resulting in morphologically abnormal seedlings (Fig. 3A) (8). The keule mutant embryos display characteristic cytokinesis defects, including cell-wall stubs and unfused vesicles in the plane of cell division (10). Interestingly, no sec1b keule double homozygotes were detected among the seedling progeny of the selfed double heterozygotes. Instead, many unfertilized ovules were observed in the siliques of selfed plants (SI Appendix, Fig. S5A and Table S2A), suggesting that reproduction was compromised. Reciprocal crosses with wild type revealed that both embryo sac and pollen were affected. Strongly reduced transmission through pollen (2%) and the embryo sac (7%) indicated that the sec1b keule double mutant was nearly gametophytically lethal (SI Appendix, Table S2B). Moreover, ∼25% of the pollen produced by doubly heterozygous plants were collapsed (Fig. 3 B and C and SI Appendix, Fig. S5B and Table S2C). Because the asymmetric division of the microspore was not obviously impaired (SI Appendix, Fig. S6 K and L), the pollen phenotype might rather reflect defects during the growth phase of the pollen. To analyze this further, we examined the subcellular localization of Qa-SNARE SYP132 fused to GFP. While GFP-SYP132 was detected at the plasma membrane in pollen of wild type as well as sec1b mutant or keule-heterozygous plants, aggregates were detected in the cytoplasm in ∼47% of the microspores produced by sec1b keule/+ plants (Fig. 3D; see SI Appendix, Fig. S7 for line intensity profiles, SI Appendix, Table S3 for quantitative measurement, and Movie S1). In conclusion, KEULE and SEC1B were functionally sufficiently similar to one another to largely compensate for each other’s absence in the single mutants, except for the main role of KEULE in cytokinesis.
Physical Interactions of SEC1-Like SM Proteins with SYP1 Qa-SNAREs.
SM proteins exert their effects by interacting with Qa-SNAREs or incipient trans-SNARE complexes (1). KEULE has been shown to interact with the open form of monomeric KNOLLE, which requires the sequence-specific linker separating the N-terminal helices from the SNARE domain (25). We performed several assays to determine whether and to what extent KEULE and SEC1B can interact with the two SYP1 Qa-SNAREs involved in membrane fusion during cytokinesis: KNOLLE and SYP132 (25, 28, 29).
In semiquantitative yeast two-hybrid assays, KEULE interacted with both KNOLLE and SYP132 in their constitutively open forms to similar extents (SI Appendix, Fig. S9 A, E, and F). In contrast, SEC1B interacted with SYP132 very strongly but only 10-fold less with KNOLLE (SI Appendix, Fig. S9 B, E, and F). SEC1B also tended to interact with the constitutively open form of SYP132 more strongly than with the normal form. Thus, unlike KEULE, SEC1B had a clear preference to interact with Qa-SNARE SYP132 involved in both cytokinesis and secretion (29).
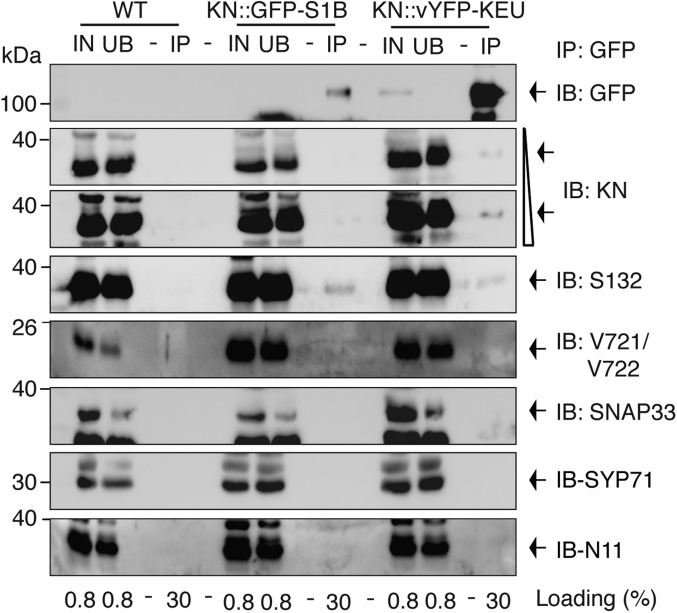
To assess the interaction between SYP1 Qa-SNAREs and SEC1-related SM proteins in planta, we performed coimmunoprecipitation assays with extracts from seedlings expressing KNOLLE::vYFP-KEULE or KNOLLE::GFP-SEC1B (Fig. 4) (25, 30). The anti-GFP immunoprecipitates were examined for the presence of the two Qa-SNAREs, KNOLLE and SYP132. In addition, the presence of their SNARE partners VAMP721/722, SNAP33, SYP71, and NPSN11 was also analyzed to clarify whether SEC1B interacted with the monomeric Qa-SNARE, as shown for the KEULE–KNOLLE interaction, or with the assembled SNARE complex, as shown for the interaction of Munc18-1 with the mammalian neuronal SNARE complex (25, 31). Endogenous SYP132 was coimmunoprecipitated with SEC1B. In contrast, virtually no KNOLLE, nor any of their SNARE partners, were detected in the immunoprecipitate (Fig. 4). KEULE interacted with both Qa-SNAREs but not with any of their SNARE partners VAMP721/722, SNAP33, SYP71, and NPSN11 in seedlings (Fig. 4) (25, 32). The interaction of KEULE with SYP132 was comparatively weak, as evidenced by the fact that in another coimmunoprecipitation assay with KNOLLE::6xHA-KEULE, only Myc-tagged SYP132, but not endogenous SYP132, was detected in the precipitate (SI Appendix, Fig. S8). In conclusion, both KEULE and SEC1B interact with monomeric Qa-SNAREs but apparently not with assembled SNARE complexes. Furthermore, only KEULE but not SEC1B showed detectable interaction with KNOLLE. Conversely, SEC1B interacted more strongly than KEULE with endogenous SYP132, taking their relative protein levels into account. These results suggest that the yeast two-hybrid interaction assays reflect the in situ conditions. Thus, SEC1B might only make a small contribution to cytokinesis compared with KEULE.
Fig. 4.
Coimmunoprecipitation analysis of SEC1-like SM proteins and SYP1 Qa-SNAREs. Protein extracts from KNOLLE::GFP-SEC1B (KN::GFP-S1B) and KNOLLE::vYFP-KEULE (KN::vYFP-KEU) seedlings were subjected to immunoprecipitation with anti-GFP beads. Wild-type (WT, Col) seedlings were used as control. Immunoprecipitates (IP) were immunoblotted (IB) with the antisera indicated: GFP, anti-GFP; KN, anti-KNOLLE; S132, anti-SYP132; V721/V722, anti-VAMP721/V722; S33, anti-SNAP33; S71, anti-SYP71; N11, anti-NPSN11. Note that KNOLLE is barely detected in GFP-SEC1B precipitates in longer exposure, but similar to the background level of control. IN, input; M, molecular markers; UB, unbound (size in kilodaltons on the Left). Loading (%), relative loading volume to total volume.
Domains of SEC1-Related SM Proteins Conferring Differential Interaction with Qa-SNAREs.
SEC1 proteins comprise five domains—1, 2a, 3a, 3b, and 2b—as revealed by crystal structure analysis (33). In neuronal SEC1 (nSEC1), largely conserved amino acid residues in domains 1 and 3a make contact with specific amino acid residues of the Qa-SNARE syntaxin 1a (33). To examine whether the differential interaction of the two closely related SM proteins with the Qa-SNAREs KNOLLE and SYP132 is mediated by specific domains, we generated chimeric proteins by swapping domains 1 and 2a between KEULE and SEC1B: one chimera comprised domains 1 and 2a of KEULE and the other domains of SEC1B (KEULE1–2aSEC1B3a–2b, KS), whereas the complementary chimera comprised domains 1 and 2a of SEC1B and the other domains of KEULE (SEC1B1–2aKELE3a–2b, SK) (SI Appendix, Fig. S9C; see SI Appendix, Fig. S2 for the sequences). Semiquantitative yeast two-hybrid interaction assay revealed that the SK chimera interacted strongly with SYP132 but not with KNOLLE, whereas the complementary KS chimera weakly interacted with both SYP132 and KNOLLE as did KEULE (SI Appendix, Fig. S9 D and E). These results suggest that domains 1 and 2a of the closely related SM proteins KEULE and SEC1B might contain features conferring differential interaction with Qa-SNAREs.
Genetic Interactions of SYP1 Qa-SNAREs and SEC1-Like SM Proteins.
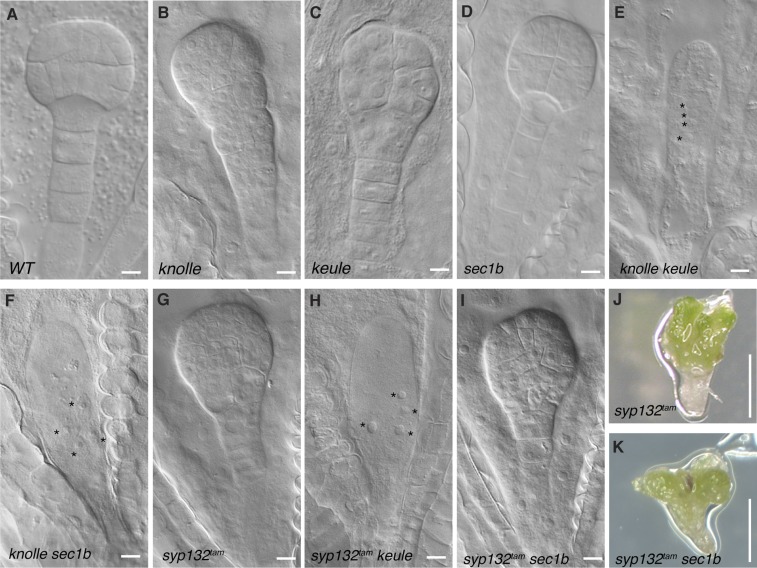
In addition to the physical interaction assays, all possible double mutants of SYP1 Qa-SNAREs and SEC1-like SM proteins were generated and analyzed phenotypically (Fig. 5). The knolle keule double mutant clearly showed an embryo-lethal phenotype in which cytokinesis was completely blocked from the zygote stage of embryogenesis, as reported previously (10). Consequently, the mutant embryo was a single growing cell in which the number of nuclei increased over time (Fig. 5E). This result suggested that the single mutants, knolle and keule, were not embryo-lethal but displayed abnormal seedling phenotypes because functionally overlapping related genes enabled completion of embryogenesis. The easiest explanation would be that functionally overlapping SYP1 Qa-SNARE and SEC1-like SM proteins might interact with KEULE and KNOLLE, respectively, such that in the knolle keule double mutant, keule knockout would render a KNOLLE-redundant SYP1 Qa-SNARE inactive and knolle knockout would render a KEULE-redundant SEC1-like SM protein inactive. Consistent with this idea, our recent study has revealed that SYP132 is the relevant redundant SYP1 Qa-SNARE and that syp132tam mutant, a combined mutant of two alleles of syp132T-DNA and RPS5A>>amiR(SYP132), in a knolle mutant background results in lethal embryos nearly resembling the knolle keule double mutant (29). Furthermore, a syp132tam keule double mutant also showed a strong embryo-lethal phenotype, approaching the knolle keule phenotype (Fig. 5H; compare with the syp132tam mutant alone in Fig. 5G and SI Appendix, Table S4A). Thus, KEULE interacts genetically with both KNOLLE and SYP132 in cytokinesis. Regarding the genetic interactions of SEC1B, a knolle sec1b double mutant also gave a very similar embryo-lethal phenotype to knolle keule (Fig. 5F; compare with Fig. 5E and SI Appendix, Table S4B). These data suggested that both KEULE and SEC1B interact with SYP132 in cytokinesis. In contrast to syp132tam keule, however, a syp132tam sec1b double mutant did not die as an embryo that completely failed to undergo cytokinesis. Instead, the syp132tam sec1b double mutant completed embryogenesis, displaying a seedling-lethal phenotype that closely resembled the syp132tam single-mutant phenotype (Fig. 5 I and K; compare with Fig. 5 G and J and SI Appendix, Table S4C). Additionally, reciprocal crosses of syp132T-DNA/SYP132 s1b/SEC1B doubly heterozygous plants with wild-type plants revealed that the transmission frequency of the syp132T-DNA sec1b double mutant was reduced via the pollen (SI Appendix, Table S5B). This result supported the conclusion that the interaction of SEC1B with SYP132 is involved in a general secretory pathway. Thus, unlike KEULE, SEC1B did not play an essential role in KNOLLE activation during embryogenesis, which suggests functional diversification among the SEC1-like SM proteins.
Fig. 5.
Analysis of double mutants revealing interaction between SEC1-like SM proteins and SYP1 Qa-SNARE proteins. (A–I) Embryo images of wild type (WT; A), knolle (B), keule (C), sec1b (D), knolle keule (E), knolle sec1b (F), syp132tam (G), syp132tam keule (H), and syp132tam sec1b (I). Note that knolle keule, knolle sec1b and syp132tam keule double mutants form a single-celled embryo with multiple nuclei, due to almost completely blocking cytokinesis (asterisks in E, F, and H). In contrast, the syp132tam sec1b double mutant is phenotypically indistinguishable from the syp132tam single mutant at both embryo (I, compare with G) and seedling (K, compare with J) stages. See SI Appendix, Table S4 for genetic analysis. (Scale bars: 10 µm in A–I; 5 mm in J and K.)
Discussion
The two paralogs of Arabidopsis SEC1 protein, KEULE and SEC1B, appear to be closely related to each other, displaying substantial functional overlap. This is clearly demonstrated by the keule sec1b double mutant being nearly gametophytically lethal, whereas sec1b mutant plants are homozygous viable and keule knockout mutants are only seedling-lethal. Our data did not give any evidence for a primary defect in pollen cytokinesis, whereas the growth defect and the eventual collapse of the developing pollen grain suggested that secretory trafficking might be seriously impaired in the keule sec1b double mutant, which is consistent with the compromised delivery of GFP-SYP132 to the plasma membrane. The simplest interpretation of the data would be that KEULE and SEC1B are stable proteins such that carryover from the meiocyte would suffice for the asymmetric division of the microspore (and possibly for the division of the generative cell as well) but not for the subsequent substantial growth of the tricellular pollen before maturation.
Although the keule single mutant displays seemingly specific cytokinesis defects, the root hairs are stunted or absent in keule seedlings, in contrast to knolle and sec1b seedlings, suggesting an additional, noncytokinetic function for KEULE (8). Interestingly, estradiol-inducible amiRNA(SYP132) expression in root hair cells caused reduced root hair growth (34), which might be KEULE-dependent. Nonetheless, KEULE appears to be primarily involved in cytokinesis, whereas SEC1B makes its major contribution to secretory traffic. Thus, like their interacting SYP1 Qa-SNAREs, KNOLLE, and SYP132, these SM proteins appear to have specialized to some extent. However, the single mutants of keule and sec1b suggested a nonreciprocal relationship between the two proteins: KEULE could replace SEC1B completely, whereas SEC1B could not replace KEULE in cytokinesis even though strongly overexpressed SEC1B from the KNOLLE promoter can fulfill the function of KEULE in the keule knockout mutant by interacting with KNOLLE, which in turn mediates membrane fusion in cytokinesis. This was clearly demonstrated by the analysis of Qa-SNARE SM-protein double mutants. In combination with either knolle or syp132, the keule mutant blocked cytokinesis completely, revealing the ability of KEULE to interact with both Qa-SNAREs. In contrast, sec1b inhibited cytokinesis completely only in the knolle mutant background, whereas the sec1b syp132 double mutant essentially resembled the syp132 single mutant seedling, suggesting preferential interaction of SEC1B with the ancient Qa-SNARE SYP132 in both secretion and cytokinesis. This conclusion is also supported by the physical interaction analyses involving yeast two-hybrid and coimmunoprecipitation assays.
It is important to note that strong overexpression of SEC1B from the KNOLLE promoter in cytokinesis can rescue keule mutant seedlings, suggesting that the two proteins, KEULE and SEC1B, are functionally sufficiently similar to replace each other. The yeast two-hybrid data suggest that both KEULE and SEC1B can interact with KNOLLE, although SEC1B interacts much more strongly with SYP132 compared with KNOLLE and the interaction of KEULE with SYP132. This difference, together with the 10-fold lower level of SEC1B expression compared with KEULE, might explain why the lack of SEC1B is not deleterious, whereas absence of KEULE impairs cytokinesis profoundly. In a highly simplified view, KNOLLE–KEULE interaction might account for most of membrane-fusion activity in cytokinesis, with SYP132–KEULE, SYP132–SEC1B and, possibly, KNOLLE–SEC1B interactions each making minor contributions (SI Appendix, Fig. S10). In contrast, fusion of secretory vesicles with the plasma membrane involving SYP132 appears to be equally well supported by both KEULE and SEC1B.
The yeast two-hybrid assay revealed that domains 1 and 2a of the two SM proteins KEULE and SEC1B influence the way the SM proteins interact with the Qa-SNAREs KNOLLE and SYP132. Our results are consistent with the structural analysis of nSec1–syntaxin 1a interaction, which identified several conserved amino acid residues in the domain 1 of nSec1 that play a prominent role in the interaction with the N-terminal helices (Habc domain) or the SNARE domain (H3 domain) of syntaxin 1a (33).
Our results suggest that functional diversification of KEULE and SEC1B only started in early angiosperm evolution. Considering that SEC1B predominantly interacted with the ancient Qa-SNARE SYP132, KEULE (or its precursor, the not yet duplicated SEC1-related protein) might have acquired an additional function after KNOLLE had arisen in the early angiosperms. Nonetheless, the retention of a KNOLLE gene in the secondarily simplified duckweed Spirodela polyrhiza suggests that the single remaining SEC1-related protein is not only related to KEULE by sequence but also able to interact with KNOLLE like KEULE in Arabidopsis. This raises the possibility that membrane fusion in angiosperm cytokinesis was made more efficient by the coevolution of a cytokinesis-specific Qa-SNARE KNOLLE, as opposed to its nonspecialized precursor SYP132, and an interacting SM protein KEULE, as opposed to the mainly secretory SM protein SEC1B.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana plants were grown either on soil or on agar plates with MS medium (2.15 g/L Murashige and Skoog, 0.5 g/L MES, 1% sucrose, pH 5.6) at 23 °C in continuous light. Transgenic plants were generated with the floral-dip method of Agrobacterium tumefaciens-mediated transformation (35). T1 plants were selected by spraying with 1:1,000 diluted BASTA (183 g/L glufosinate; AgrEvo) or hygromycin (20 µg/mL; Duchefa). T-DNA insertional sec1b (GABI-KAT_601G09, sulfonamide-resistant) mutant seed was purchased from the European Arabidopsis Stock Centre (NASC).
The following transgenic lines were used: KNOLLE::6xHA-KEULE (25), KNOLLE::Myc-SYP132 (28), SYP132::GFP-SYP132 (29, 36), KNOLLE::Myc-KNOLLE (25), and VHA-a1-GFP (37).
In Silico Analysis.
SEC1-related sequences were obtained from genome database sources, such as Phytozome v12 (https://phytozome.jgi.doe.gov/pz/portal.html) and others (https://www.cacaogenomedb.org; spirodelagenome.org/jgi_csp; congenie.org; banana-genome-hub.southgreen.fr/organism/Musa/acuminata). Peptide sequences were aligned in the CLC workbench (v7.8.1) program. The unrooted phylogenetic tree was generated using the neighbor-joining method together with the bootstrap test (1,000 replicates) in the CLC workbench (v7.8.1) program.
Statistical Analysis.
The dataset was analyzed using R software (https://www.r-project.org/) and performing ANOVA (single-factor for SI Appendix, Fig. S9 A and B; two-way for SI Appendix, Fig. S9D) and a posteriori Tukey test. F value = variance of the group means/mean of the within group variances. P = the significance probability associated with the F value.
See SI Appendix, Materials and Methods, for details on molecular cloning, genetic and transcript analysis, chemical treatment, pollen staining, coimmunoprecipitation and immunoblot analyses, immunofluorescence analysis, and yeast analysis.
Supplementary Material
Acknowledgments
We thank Anton A. Sanderfoot, Masa H. Sato, and Paul Schulze-Lefert for sharing published materials; Sonja Touihri for technical support; and Christopher Grefen, Sandra Richter, and Farid El-Kasmi for discussion and critical reading of the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft through Grant Ju179/19-1 (to G.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.C.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722611115/-/DCSupplemental.
References
- 1.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell. 2000;11:2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zouhar J, Rojo E, Bassham DC. AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 2009;149:1668–1678. doi: 10.1104/pp.108.134361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka H, et al. Cell polarity and patterning by PIN trafficking through early endosomal compartments in Arabidopsis thaliana. PLoS Genet. 2013;9:e1003540. doi: 10.1371/journal.pgen.1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rojo E, Zouhar J, Kovaleva V, Hong S, Raikhel NV. The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol Biol Cell. 2003;14:361–369. doi: 10.1091/mbc.E02-08-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, et al. Comparative studies of Munc18c and Munc18-1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2013;110:E3271–E3280. doi: 10.1073/pnas.1311232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assaad FF, Huet Y, Mayer U, Jürgens G. The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet. 1996;253:267–277. doi: 10.1007/pl00008594. [DOI] [PubMed] [Google Scholar]
- 10.Waizenegger I, et al. The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr Biol. 2000;10:1371–1374. doi: 10.1016/s0960-9822(00)00775-2. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, et al. Regulation of cytokinesis by exocyst subunit SEC6 and KEULE in Arabidopsis thaliana. Mol Plant. 2013;6:1863–1876. doi: 10.1093/mp/sst082. [DOI] [PubMed] [Google Scholar]
- 12.Fendrych M, et al. The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell. 2010;22:3053–3065. doi: 10.1105/tpc.110.074351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steiner A, et al. The membrane-associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis. Mol Plant. 2016;9:528–540. doi: 10.1016/j.molp.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Karnik R, et al. Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell. 2015;27:675–694. doi: 10.1105/tpc.114.134429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnik R, et al. Arabidopsis Sec1/Munc18 protein SEC11 is a competitive and dynamic modulator of SNARE binding and SYP121-dependent vesicle traffic. Plant Cell. 2013;25:1368–1382. doi: 10.1105/tpc.112.108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birol I, et al. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics. 2013;29:1492–1497. doi: 10.1093/bioinformatics/btt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nystedt B, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- 18.Project AG. Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat Commun. 2014;5:3311. doi: 10.1038/ncomms4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegrzyn JL, et al. Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics. 2014;196:891–909. doi: 10.1534/genetics.113.159996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamala S, et al. Assembly and validation of the genome of the nonmodel basal angiosperm Amborella. Science. 2013;342:1516–1517. doi: 10.1126/science.1241130. [DOI] [PubMed] [Google Scholar]
- 22.Neale DB, et al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 2014;15:R59. doi: 10.1186/gb-2014-15-3-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimin A, et al. Sequencing and assembly of the 22-gb loblolly pine genome. Genetics. 2014;196:875–890. doi: 10.1534/genetics.113.159715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 25.Park M, Touihri S, Müller I, Mayer U, Jürgens G. Sec1/Munc18 protein stabilizes fusion-competent syntaxin for membrane fusion in Arabidopsis cytokinesis. Dev Cell. 2012;22:989–1000. doi: 10.1016/j.devcel.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 27.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 28.Reichardt I, et al. Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis. Traffic. 2011;12:1269–1280. doi: 10.1111/j.1600-0854.2011.01222.x. [DOI] [PubMed] [Google Scholar]
- 29.Park M, et al. Concerted action of evolutionarily ancient and novel SNARE complexes in flowering-plant cytokinesis. Dev Cell. 2018;44:500–511.e4. doi: 10.1016/j.devcel.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 30.Müller I, et al. Syntaxin specificity of cytokinesis in Arabidopsis. Nat Cell Biol. 2003;5:531–534. doi: 10.1038/ncb991. [DOI] [PubMed] [Google Scholar]
- 31.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Kasmi F, et al. SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol Biol Cell. 2013;24:1593–1601. doi: 10.1091/mbc.E13-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 34.Ichikawa M, et al. Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:790–800. doi: 10.1093/pcp/pcu048. [DOI] [PubMed] [Google Scholar]
- 35.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 36.Enami K, et al. Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:280–289. doi: 10.1093/pcp/pcn197. [DOI] [PubMed] [Google Scholar]
- 37.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.