Significance
Gram-positive sortase enzymes represent two broad functional categories—those that cross-link proteins to the cell wall and those that can catalyze this reaction and polymerize proteins to build adhesive pilus fibers. Here we report an in vitro reproduction of a robust pilus polymerization reaction using a variant of a corynebacterial pilus-specific sortase in which the catalytic center is unmasked. By molecular modeling, we uncovered a conserved structural element of pilus-specific sortases critical for protein ligation in vitro and further demonstrated that the activated sortase ligates the isolated domains of the pilin harboring the donor and acceptor motifs for ligation. Besides enabling future molecular studies and antibiotic development, our system provides a powerful platform for bioconjugation and protein engineering.
Keywords: Corynebacterium diphtheriae, sortase, pilus polymerization, protein ligation, transpeptidation
Abstract
Covalently cross-linked pilus polymers displayed on the cell surface of Gram-positive bacteria are assembled by class C sortase enzymes. These pilus-specific transpeptidases located on the bacterial membrane catalyze a two-step protein ligation reaction, first cleaving the LPXTG motif of one pilin protomer to form an acyl-enzyme intermediate and then joining the terminal Thr to the nucleophilic Lys residue residing within the pilin motif of another pilin protomer. To date, the determinants of class C enzymes that uniquely enable them to construct pili remain unknown. Here, informed by high-resolution crystal structures of corynebacterial pilus-specific sortase (SrtA) and utilizing a structural variant of the enzyme (SrtA2M), whose catalytic pocket has been unmasked by activating mutations, we successfully reconstituted in vitro polymerization of the cognate major pilin (SpaA). Mass spectrometry, electron microscopy, and biochemical experiments authenticated that SrtA2M synthesizes pilus fibers with correct Lys–Thr isopeptide bonds linking individual pilins via a thioacyl intermediate. Structural modeling of the SpaA–SrtA–SpaA polymerization intermediate depicts SrtA2M sandwiched between the N- and C-terminal domains of SpaA harboring the reactive pilin and LPXTG motifs, respectively. Remarkably, the model uncovered a conserved TP(Y/L)XIN(S/T)H signature sequence following the catalytic Cys, in which the alanine substitutions abrogated cross-linking activity but not cleavage of LPXTG. These insights and our evidence that SrtA2M can terminate pilus polymerization by joining the terminal pilin SpaB to SpaA and catalyze ligation of isolated SpaA domains in vitro provide a facile and versatile platform for protein engineering and bio-conjugation that has major implications for biotechnology.
Adhesive protein polymers, called “pili” or “fimbriae,” are expressed on the cell envelope by many Gram-negative and Gram-positive bacteria, and they are critical for bacterial virulence (1). Many types of Gram-negative pili have been reported, including the well-studied retractable type IV, conjugative, and chaperone-assisted pili (2). These pili are formed by distinct pathways (2, 3); however, none of these pili are covalently linked polymers, unlike the sortase-catalyzed pili found in many Gram-positive bacteria, including Actinomyces oris, Enterococcus faecalis, Bacillus cereus, and numerous species of streptococci and lactobacilli (4–6).
One of the well-studied sortase-mediated pilus assembly systems involves Corynebacterium diphtheriae (7), the causative agent of pharyngeal diphtheria (8). C. diphtheriae produces three distinct pilus types (7, 9, 10), each comprised of a pilus tip adhesin, a pilus shaft made of the major pilin, and a base pilin that is covalently anchored to the cell wall (11). The archetypal SpaA-type pilus, which mediates adherence to the pharyngeal epithelium (12), consists of the tip pilin SpaC, shaft pilin SpaA, and pilus base SpaB (13). A pilus-specific sortase named “SrtA” is required for pilus polymerization (13), performing a repetitive, irreversible transpeptidation reaction that covalently links the pilin subunits via an isopeptide bond (14). Although each Spa pilin harbors a cell wall sorting signal (CWSS), which starts with a conserved LPXTG motif, followed by a stretch of hydrophobic amino acids and a positively charged tail (15), SpaA contains a pilin motif with the Lys residue K190 acting as a nucleophile for the aforementioned transpeptidation reaction (13). According to the current model (16), SrtA cleaves the LPXTG motif of Spa pilins between Thr and Gly, forming acyl-enzyme intermediates between the Thr residue and the SrtA catalytic Cys residue. This intermediate is then nucleophilically attacked by the reactive K190 of an incoming SpaA subunit. In pilus biogenesis, the SpaC–SrtA acyl-enzyme intermediate forms first, resulting in the joining of the ε-amine group of K190 to the Thr carbonyl carbon atom in the LPXT of SpaC. Pilus polymerization ensues when additional SpaA protomers are joined progressively to the pilus base by the SrtA enzyme via the same Lys-mediated transpeptidation reaction. Polymerization is terminated with the entry of SpaB into the pilus base (11), which is then anchored to the cell wall by the housekeeping sortase SrtF (17). This cell wall anchoring of pilus polymers is likely similar to that of surface proteins catalyzed by the prototype SrtA enzyme from Staphylococcus aureus (18, 19). While most of this biphasic model of pilus assembly in Gram-positive bacteria (6), i.e., pilus polymerization followed by cell wall anchoring, has been validated experimentally, the molecular determinants that make up a pilus-specific sortase and enable the enzyme to join proteins together remain unknown.
The SrtA enzyme is classified as a member of the class C sortase subgroup within the sortase superfamily that has the unique ability to cross-link proteins via Lys–Thr isopeptide bonds (20, 21). Although all sortases share a canonical β-barrel sortase superfamily fold (22, 23), class C enzymes are distinguished by the presence of a conserved N-terminal region that forms a “lid” that covers the active site structurally and functionally (24–26). In Streptococcus pneumoniae, X-ray crystallographic evidence originally suggested that the lid region was flexible, possibly modulating substrate binding; however, subsequent studies in solution utilizing NMR showed this region to be relatively rigid in the SrtC1 enzyme (24, 27–29). Mutations of the lid region in A. oris SrtC2 or Streptococcus agalactiae SrtC1 did not alter the pilus polymerizing activities in vivo (30, 31); nonetheless, the mutations caused enzyme instability and increased hydrolytic activity in S. agalactiae SrtC1 (30), supporting a regulatory role for the N-terminal lid. However, the unique structural properties that enable class C sortase enzymes to cross-link proteins have not been identified.
We report here the crystal structures of the C. diphtheriae class C sortase SrtA lacking the signal peptide and transmembrane domain (referred to as “SrtAWT”) and a mutant of this protein that has substitutions in the lid interface which normally masks the catalytic pocket (SrtA2M). Using these recombinant enzymes and an SpaA substrate that lacks the signal peptide and transmembrane domain, we succeeded in reconstituting the SpaA pilus shaft polymerization reaction in vitro, demonstrating that the removal of SrtA’s lid not only unmasks the catalytic center structurally but also enables the polymerizing activity in vitro. Subsequently, by structural modeling and phylogenic and mutational analyses, we identified two structural elements that enable SrtA to cross-link proteins. Importantly, we showed that the activated sortase can ligate the isolated pilin domains, thus defining the donor and acceptor motifs for the ligation reaction. The system we report provides a platform for in vitro mechanistic investigations of Gram-positive pilus assembly, antibiotic development, and biotechnological applications of protein modification and conjugation via a unique transpeptidation reaction.
Results and Discussions
Structure of the C. diphtheriae Pilus-Specific Sortase.
The archetypal SpaA pilus polymer produced by corynebacteria is built by the dedicated pilus-specific sortase SrtA (7, 13). To gain insight into the mechanism of pilus polymerization, we determined the structure of SrtA by X-ray crystallography. We performed crystallization screens using a soluble fragment encompassing the catalytic domain of SrtA (residues 37–257, SrtAWT), which was cloned, expressed, and purified from Escherichia coli. SrtAWT crystallized as a homodimer in the P61 2 2 space group. Diffraction data were collected to 2.1-Å resolution and were phased by molecular replacement (Tables 1 and 2). The electron density for residues 37–248 was well defined, enabling their structure to be modeled, while density for the remaining C-terminal residues is missing, presumably due to a disordered state.
Table 1.
Crystal data collection statistics
| Statistics | SrtAWT | SrtA2M |
| X-ray wavelength, Å | 0.9792 | 0.9792 |
| Space group | P 6122 | P 21 |
| Unit cell dimensions | a = b = 77.7 Å, c = 202.3 Å, α = β = 90°, γ = 120° | a = 65.2 Å, b = 45.5 Å, c = 74.9 Å, α = γ = 90°, β = 96.4 |
| Resolution, Å | 38.9–2.1 (2.14–2.1) | 38.9–1.85 (1.88–1.85) |
| No. of unique reflections | 21,872 (1,050) | 36,705 (1,431) |
| Completeness, % | 99.7 (100) | 97.7 (77.4) |
| R-merge | 0.142 (0.916) | 0.087 (0.410) |
| CC1/2, Å2 | −(0.939) | −(0.741) |
| I/σ | 30.7 (6.4) | 16.5 (1.95) |
| Redundancy | 15.4 (15.2) | 3.5 (2.2) |
| Molecules per asymmetric unit | 1 | 2 |
| No. of protein residues | 215 | 430 |
Numbers in parenthesis are shown for the highest-resolution shell.
Table 2.
Structure refinement statistics
| Statistics | SrtAWT | SrtA2M |
| Resolution range, Å | 38.9–2.1 (2.157–2.1) | 38.9–1.85 (1.898–1.85) |
| Reflections | 21,846 (1,533) | 36,692 (2,195) |
| σ cutoff | None | None |
| R-value, all, % | 16.22 | 17.26 |
| R-value (R-work), % | 16.05 (17.4) | 17.12 (23.7) |
| Free R-value, % | 19.56 (24.0) | 20.08 (23.7) |
| Rms deviations from ideal geometry | ||
| Bond length, Å | 0.017 | 0.012 |
| Angle, ° | 1.76 | 1.60 |
| Chiral, Å | 0.101 | 0.088 |
| No. of atoms | ||
| Protein | 1,737 | 2,974 |
| Sulfate | 5 | — |
| Water | 167 | 258 |
| Mean B-factor, Å2 | ||
| All atoms | 32.0 | 30.7 |
| Protein atoms | 31.2 | 30.3 |
| Protein main chain | 28.6 | 29.0 |
| Protein side chain | 33.9 | 31.6 |
| Sulfate | 66.0 | — |
| Water | 38.5 | 35.9 |
| MolProbity summary | ||
| Ramachandran outliers, % | 0.0 | 0.0 |
| Ramachandran favored, % | 97.18 | 98.89 |
| Rotamer outliers, % | 2.02 | 0.89 |
| C-beta deviations | 0.0 | 0.0 |
| Clash score | 1.73 | 1.34 |
| MolProbity score | 1.31 | 0.86 |
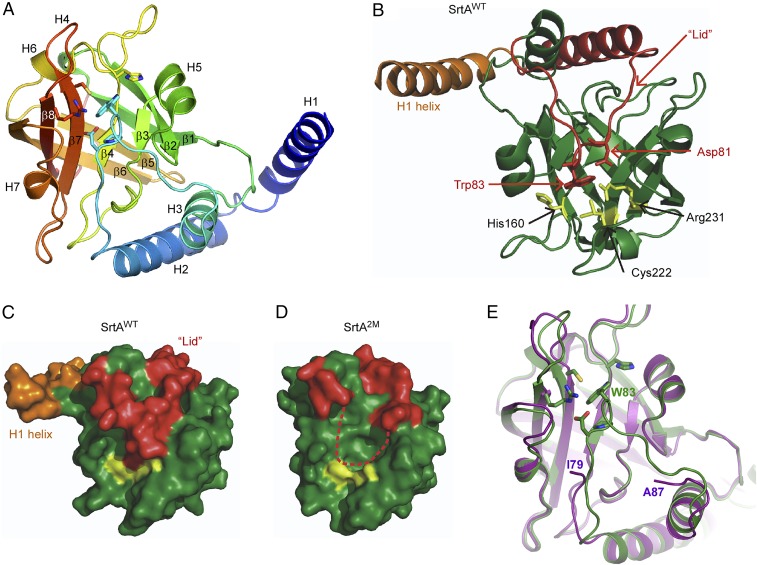
The overall structure of SrtAWT conforms to the typical sortase fold described previously (22), containing an eight-stranded β-barrel core flanked by several 310 and α-helices (Fig. 1 A and B). Three additional α-helices are located at the N terminus of SrtAWT (Fig. 1 A and B) and contain the distinguishing lid structure that occludes the enzyme’s active site in a class C sortase (Fig. 1B). Interestingly, the H1 helix mediates homodimerization in the crystal structure and is generally removed from the body of the enzyme (Fig. 1 A and B), while helices H2 and H3 are positioned immediately adjacent to the active site and are connected by a loop that contains the highly conserved DPW lid motif that interacts with the active site (Fig. 1 A and B). W83 in the lid participates in aromatic stacking interactions with the active site C222 and nearby H160 residues. In addition, D81 within the motif interacts with the active site R231 residue, suggesting its regulatory role in lid positioning and pilin polymerization. Importantly, residues within the catalytic triad His–Cys–Arg are well resolved, and C222 can be modeled in two distinct positions with 50% occupancy, pointing both toward and away from the active site (Fig. 1 A and B).
Fig. 1.
Structural analysis of the C. diphtheriae pilus-specific sortase SrtA. (A) The crystal structure of SrtA was determined to 2.1-Å resolution, with the overall protein fold presented as rainbow coloring from blue to red corresponding to the N- to C-terminal positions. The helices are marked as H1–H7, and the β-strands are marked as β1–β8. (B) The lid region is marked in red with conserved lid residues D81 and W83 and catalytic residues C222, H160, and R231 in yellow. The C222 residue is shown only in the main conformation. (C and D) Hydrophobic surface renderings of WT SrtA (SrtAWT) (C) and the lid mutant (SrtA2M) structures (D) with the H1 helix and lid loop structures in red. The H1 helix of SrtA2M is absent, and its lid structure is not visible, as indicated by a red dashed line. (E) Superposition of the SrtAWT (green) and the lid mutant SrtA2M (pink) structures was generated by PyMOL.
To investigate the functional importance of the lid in polymerization, we next generated a recombinant SrtA mutant protein in which the DPW lid motif (residues 81–83) was mutated to GPG, hereafter referred to as “SrtA2M.” We succeeded in determining the crystal structure of SrtA2M at 1.85-Å resolution using crystallization conditions that differed from those used for the WT protein (Materials and Methods). In the electron density map for SrtA2M, residues 80–86 that represent the lid were invisible. Presumably, the lid residue substitutions prevented contacts with the active site, causing the mutant lid to adopt a range of conformations. Remarkably, a second major difference between the two structures is the absence of interpretable electron density for the H1 helix in the SrtA2M lid mutant, which might be caused by flexibility around the hinge between helices H1 and H2 and by the absence of stabilizing interactions with neighboring molecules in the crystals of SrtA2M.
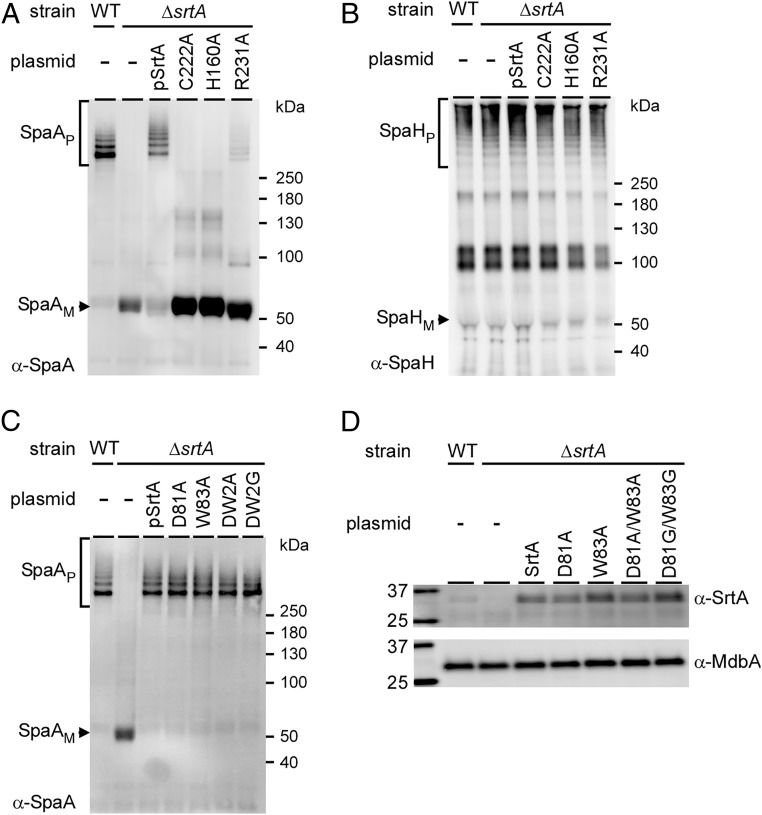
To evaluate the involvement of the predicted catalytic residues and the lid in pilus assembly, corynebacterial cells harboring WT and its isogenic mutants were subjected to cell fractionation, and protein samples were immunoblotted with specific antibodies against SpaA (α-SpaA), the cognate substrate of SrtA that forms the pilus shaft (7, 13). As shown in Fig. 2A, SpaA polymers (P) were observed in cell wall fractions of the WT strain, but they were absent in the srtA deletion mutant, as previously reported (11). Ectopic expression of SrtA rescued the pilus assembly defect of the ∆srtA mutant (Fig. 2A, third lane), and Ala substitution of the catalytic residues C222, H160, and R231 abrogated pilus assembly (Fig. 2A, last three lanes). In control experiments, we demonstrated that none of these mutations affected the assembly of the SpaH-type pili, as expected (Fig. 2B) (9). Strikingly, the lid mutants are catalytically active in pilus polymerization. Like the WT and the complementing strains, strains expressing mutations in the DPW motif still produced pilus polymers (Fig. 2C), and immunoblotting analysis of the membrane fractions revealed no changes in the SrtA protein level when the lid was mutated (Fig. 2D).
Fig. 2.
Catalytic residues are required for pilus assembly in vivo. Cells of C. diphtheriae strains in equivalent numbers were subjected to cell fractionation. Protein samples from cell wall fractions (A–C) and protoplasts (D) were analyzed by immunoblotting with specific antibodies against SpaA (α-SpaA) (A and C), SpaH (α-SpaH) (B), and SrtA (α-SrtA) (D), with α-MdbA as a membrane-loading control. Pilus monomers (subscript M), polymers (subscript P), and molecular mass markers are indicated.
To visualize these SpaA polymers, corynebacterial cells were immobilized on carbon-coated nickel grids, washed with water, and stained with 0.75% uranyl formate before viewing with an electron microscope. Because the parental strain NCTC 13129 produced short pili that were hardly detected (SI Appendix, Fig. S1A, WT), we constructed a multicopy vector expressing both SpaA and SrtA. Using this vector as a template, SrtA2M was generated by site-directed mutagenesis (Materials and Methods). The generated vectors were introduced into a corynebacterial double mutant lacking both spaA and srtA (ΔspaA/ΔsrtA). Compared with the WT strain, overexpression of SpaA and SrtA resulted in increased production of long pili, as expected (SI Appendix, Fig. S1A, ΔspaA/ΔsrtA/pSpaA-SrtA). Consistent with the above results, mutations in the DPW motif did not affect pilus assembly (SI Appendix, Fig. S1A, ΔspaA/ΔsrtA/pSpaA-SrtA2M). To confirm that these long pilus fibers are SpaA pili, the same set of strains was subjected to immunoelectron microscopy (IEM) (32), whereby immobilized cells were stained with α-SpaA, followed by staining with gold particles conjugated with IgG, before washing and staining with uranyl acetate. As shown in SI Appendix, Fig. S1B, SpaA-stained pili were detected in both strains producing WT SrtA and SrtA2M, whereas short pili were observed in the WT strain, and no pili were observed in the ΔspaA/ΔsrtA and ΔspaA/ΔsrtA/pSpaAK190A-SrtA2M strains.
Thus, the overall structure of the C. diphtheriae SrtAWT enzyme resembles class C sortases, or pilus-specific sortases, which possess a distinguishing feature of this class of enzymes, the lid region (16, 21). In agreement with previous studies (30, 31), the elimination of the lid’s interaction at the catalytic pocket does not dramatically affect pilus assembly in vivo.
In Vitro Reconstitution of Archetypal C. diphtheriae SpaA Pilus Polymerization.
Previous structural and biochemical studies of pilus-specific sortase enzymes in several streptococcal species indicate that the lid may modulate substrate entry into the active site (24, 25, 28, 30). We envisioned that a loss of lid closure might increase the accessibility of the active site. To test this hypothesis, we used the thiol-reactive reagent 4,4′-dithiodipyridine (DTDP) (33, 34) to probe the solvent accessibility of the catalytic Cys residue (C222). Disulfide exchange between thiol side chains of Cys residues and DTDP gives rise to 4-thiopyridone, which shows strong absorption at 324 nm (34). The recombinant proteins SrtAWT or SrtA2M (0.6 mg/mL) were rapidly mixed with 0.32 mM DTDP, and the rate of reaction between DTDP and C222 was monitored as an increase in absorbance at 324 nm. Time-dependent changes in absorbance were fit to single or double exponential equations to derive rates, as described in Materials and Methods. As shown in Fig. 3A, data for the SrtAWT enzyme best fit an equation with a single exponential rate of 2.17 ± 0.02/min. In contrast, data for the lid anchor mutant best fit an equation with two much faster exponential rates, 228 ± 7/min and 16 ± 3/min (Fig. 3A, Inset), which indicates that the catalytic C222 was readily accessible in this mutant. The two different rates may be due to slow exchange between two conformations in the mutant protein. If so, the conformation with the faster rate is the dominant form, since it represents 80% of the total change in absorbance.
Fig. 3.
Involvement of the lid in pilus polymerization in vitro. (A) Accessibility of the thiol group of the active site C222 residue in SrtAWT (main panel) or SrtA2M (Inset) enzymes was determined by stopped-flow experiments, whereby the reaction between the thiol-reactive reagent DTDP and C222 was monitored by absorbance at 324 nm. The experiments were performed in triplicate. (B) In vitro reconstitution of SpaA pilus polymerization was carried out at room temperature using various forms of recombinant SrtA and SpaA proteins at the molar ratio of 1:3. The reaction samples were analyzed by SDS/PAGE and Coomassie staining after 0, 24, and 48 h of incubation. Additional samples after 72 h of incubation were taken for the SrtA2M reactions (black triangles). SpaA monomers (M), polymers (P), and molecular mass markers are indicated. An SrtA–SpaA intermediate is marked with asterisks. (C) Protein samples from pilus polymerization reactions were analyzed by electron microscopy with negative staining using 0.75% uranyl formate. For comparison, recombinant SrtA2M and SpaA K190A proteins were included. (D) The isopeptide bond between residue T494 of the LPLTG motif and Lys residue K190 of the pilin motif in SpaA polymers in B was examined by MS/MS. Shown is the m/z tandem mass spectrum of the linked peptide (sequence shown in the Inset). (E) Fusion proteins between SUMO and the N-terminal SpaA domain (NSpaA; residues 30–194) and between maltose-binding protein (MBP) and the C-terminal SpaA domain (CSpaA; residues 350–500) were used with the SrtA2M enzyme in the in vitro pilus polymerization assay as described in B. The reaction samples were analyzed by SDS/PAGE and Coomassie staining after 24 h. The reactive Lys residue K190 and the LPXTG motif in the two domains are indicated.
The increased DTDP reactivity of the active site Cys residue in SrtA2M described above raises the possibility that the mutant enzyme may be able to assemble pili in vitro, which has been difficult to reconstitute so far for pilus-assembling sortases. We therefore sought to reconstitute pilus polymerization in vitro using various recombinant sortase enzymes and a soluble form of SpaA (residues 30–500), which is devoid of the N-terminal signal peptide and C-terminal membrane anchor domain (see diagram in Fig. 3E). Sortases were mixed with SpaA at a 1:3 molar ratio, and aliquots were removed for SDS/PAGE analysis and Coomassie staining at 0, 24, and 48 h. In the SrtAWT samples, a few new high molecular mass (HMM) bands were weakly observed after 24 and 48 h of incubation, one migrating between the 50 and 100 kDa markers and the others around 100 kDa (Fig. 3B, lanes SrtAWT). Remarkably, with the SrtA2M enzyme, HMM SpaA polymers (SpaAP) were abundantly formed within 24 h and increased further after 48 and 72 h (Fig. 3B, lanes SrtA2M). Consistent with the results in Fig. 2A, the catalytically inactive enzyme in which C222 was replaced by Ala, SrtAC222A, failed to produce any SpaA polymers (Fig. 3B, lanes SrtAC222A). Intriguingly, removal of the H1 helix in the SrtA2M enzyme also abrogated pilus polymerization (Fig. 3B, lanes ΔSrtA2M). The significance of this helix in the transpeptidation activity of sortase is discussed below.
To visualize HMM SpaA polymers formed by SrtA2M, the reaction mixtures after 72 h of incubation were subjected to electron microscopy, whereby aliquots were applied to nickel grids; bound proteins were washed and stained with 0.75% uranyl formate before viewing with an electron microscope. As shown in Fig. 3C, strands of SpaA polymers were observed in the reaction with the SrtA2M enzyme but not with an SpaAK190A mutant substrate defective in the nucleophilic attack, thus authenticating the visualization of Gram-positive pilus polymers synthesized in vitro. The synthesized pilus polymers had a width of ∼10 nm and a length ranging from ∼200–500 nm, equivalent to 25–62 subunits, with each protomer measuring about 8 nm (35). For comparison, pili produced by SrtA2M in vivo had widths ranging from 7.6–9.3 nm and lengths up to 2 μm (SI Appendix, Fig. S1A).
To determine if the recombinant SrtA2M enzyme faithfully catalyzes the pilus transpeptidation reaction, we determined whether the SpaA subunits in the HMM SpaA polymers were linked together via covalent Lys isopeptide bonds in which the Thr residue of the LPLT sorting signal was joined to the Lys residue within the pilin motif (13). Indeed, MS analysis of excised HMM SpaA polymer SDS/PAGE bands revealed the presence of an isopeptide bond between the carbonyl carbon of T494 and the sidechain amine of K190 (Fig. 3D and Table 3), as was observed in the native SpaA pili assembled in vivo (36). In line with the role of Lys190 in pilus polymerization, the SpaA mutant substrate in which K190 was replaced by Ala was unable to form polymers with the active SrtA2M enzyme (Fig. 3B, lanes SpaAK190A). Remarkably, the MS data revealed the presence of SrtA2M and SpaA in the marked band migrating between the 50 and 100 kDa markers in both SrtAWT and SrtA2M samples (Fig. 3B, asterisks), suggesting that the enzyme is joined to the SpaA substrate via a labile thioacyl bond forming an acyl-enzyme heterodimer intermediate. To our astonishment, the MS analysis also revealed the presence of SrtA2M in the HMM SpaA polymer bands migrating at and above the 100-kDa marker (Fig. 3B, lanes SrtA2M, bracket). The results are in agreement with our previous identification of the native acyl-enzyme intermediates formed between SrtA and SpaA polymers in vivo in C. diphtheriae as demonstrated by immunoblotting (37).
Table 3.
MS analysis of synthetic SpaA pilus polymers
| HPLC retention time, min | Calculated MH+ | Predicted peptides | Mass accuracy, ppm | Pilus assembly reaction |
| 24.4 | 1947.0334 | DVHVYPKHQALS :: ELPLT | 4.0 | Reaction with SpaA30–500 |
| 26.5 | 1410.7627 | DVHVYPK* :: ELPLT | 2.8 | Reaction with SpaA30–500 |
| 22.9 | 1675.8802 | DVHVYPKHQ* :: ELPLT | 3.6 | Reaction with SpaA30–500 |
| 29.0 | 2336.2034 | DVHVYPKHQALS :: NAGFELPLT | 4.7 | Reaction with SpaA30–500 |
| 23.3 | 2400.2922 | DVHVYPKHQALSEPVK :: ELPLT | 2.9 | Reaction with SpaA30–500 |
| ND | 2485.2756 | DQITLITCTPYAVNSHR :: ELPLT | ND | Reaction with SpaA30–500 |
| ND | 2874.4455 | DQITLITCTPYAVNSHR :: NAGFELPLT | ND | Reaction with SpaA30–500 |
| ND | 1657.8717 | DQITLITCTP* :: ELPLT | ND | Reaction with SpaA30–500 |
| ND | 2047.0416 | DQITLITCTP* :: NAGFELPLT | ND | Reaction with SpaA30–500 |
| 24.5 | 1947.0334 | DVHVYPKHQALS :: ELPLT | 4.5 | Reaction with NSpaA and CSpaA |
| 26.5 | 1410.7627 | DVHVYPK :: ELPLT | 3.3 | Reaction with NSpaA and CSpaA |
| 22.9 | 1675.8802 | DVHVYPKHQ :: ELPLT | 3.5 | Reaction with NSpaA and CSpaA |
| ND | 2336.2034 | DVHVYPKHQALS :: NAGFELPLT | ND | Reaction with NSpaA and CSpaA |
| ND | 2400.2922 | DVHVYPKHQALSEPVK :: ELPLT | ND | Reaction with NSpaA and CSpaA |
| ND | 2485.2756 | DQITLITCTPYAVNSHR :: ELPLT | ND | Reaction with NSpaA and CSpaA |
| ND | 2874.4455 | DQITLITCTPYAVNSHR :: NAGFELPLT | ND | Reaction with NSpaA and CSpaA |
| ND | 1657.8717 | DQITLITCTP* :: ELPLT | ND | Reaction with NSpaA and CSpaA |
| ND | 2047.0416 | DQITLITCTP* :: NAGFELPLT | ND | Reaction with NSpaA and CSpaA |
| 36.5 | 1023.6448 | PKLI :: ELPLT | 11.9 | Reaction with CSpaA and SpaB |
MH+, the mass of the singly protonated species; ND, not determined.
Not expected cleavage sites.
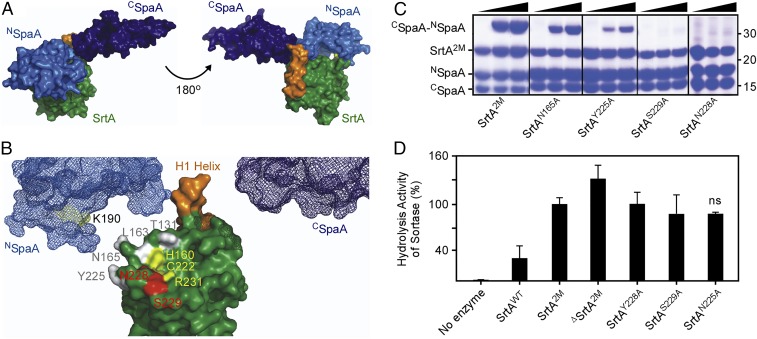
To further probe the mechanism of SpaA pilus assembly, we dissected the SpaA molecule into two components: the N-terminal domain (NSpaA, residues 30–194, encompassing the pilin motif with the K190 nucleophile) and the C-terminal domain (CSpaA, residues 350–500, containing the CWSS with the LPLTG motif) (Fig. 3E). The recombinant proteins were expressed in E. coli and purified (SI Appendix). When the two isolated domains were mixed at equal concentrations (300 µM) in a reaction with the lid-substituted SrtA2M enzyme (100 µM), a di-polypeptide conjugate was readily formed (Fig. 3E). Significantly, the presence of the expected Thr-Lys isopeptide in this conjugate was confirmed by MS (Table 3). Furthermore, control reactions demonstrated that SrtA containing the WT lid is markedly inactive in catalyzing cross-linking of the isolated domains (SI Appendix, Fig. S2). Together, our results support the concept that the lid in class C sortase functions in the molecular gating of substrate entry to the enzyme active site. Thus, we have demonstrated pilus polymerization in Gram-positive Actinobacteria in an in vitro reaction.
Structural Elements in a Sortase Required for Protein Polymerization.
To gain insight into how SrtA joins the SpaA proteins together during polymerization, we performed molecular modeling of the NSpaA–SrtA–CSpaA ternary complex in which the isopeptide bond is modeled using our previously determined crystal structures of SpaA [Protein Data Bank (PDB) ID code 3HR6] (36) and the isolated SrtA (PDB ID code 5K9A) proteins. We first generated a model of the SrtA–SpaA acyl-intermediate, juxtaposing the C terminus of the C-terminal SpaA domain with the active site C222 residue in SrtA. Because the crystal structure of SpaA lacks the CWSS that forms the acyl-intermediate with SrtA, we modeled the acyl-intermediate by placing the C-terminal domain of SpaA ∼25 Å away from the active site Cys to accommodate the nine missing C-terminal residues that contain the CWSS. To construct the ternary complex, we then positioned the coordinates of the SpaA N-terminal domain near the acyl-intermediate to juxtapose the reactive Lys K190 of the pilin motif with the active site C222 residue (Fig. 4A). The resulting model of the ternary complex makes it readily obvious that the β7/β8 loop near the active site Cys residue and the N-terminal H1 helix in SrtA are in contact with the SpaA N-terminal domain, raising the possibility that these elements might play a role in recognizing the region of SpaA that houses the reactive Lys nucleophile. Strikingly, a primary sequence alignment of SrtA and other class C sortases indicates that they all contain a conserved TP(Y/L)XIN(S/T)H motif within the β7/β8 loop (SI Appendix, Fig. S3). This motif is clearly absent in other types of sortases that are known to attach proteins to the cell wall (class A, B, D, and E enzymes) but are unable to polymerize proteins. We thus postulated that the β7/β8 loop may play a role in conferring the polymerization activity in the class C enzymes.
Fig. 4.
Structural modeling reveals SrtA residues critical for transpeptidation activities. (A) An SrtA–SpaA pilin polymerase attack complex was visualized and assembled using PyMOL. Shown is an SpaA molecule splitting into its two domains, NSpaA (light blue) and CSpaA (dark blue). The SrtA enzyme is shown in green, and the H1 helix potentially bridging interactions between the two SpaA domains is seen in orange. (B) The detailed locations of the SrtA catalytic triad (marked in yellow) and the surrounding residues at the active site of the pilin polymerase attack complex are shown. The N-terminal H1 helix bridges the two reactive domains of SpaA and potentially facilitates interactions for the formation of the SrtA–SpaA polymerization attack complex. (C) Transpeptidation activity of SrtA2M and its variants (N165A, Y225A, N228A, and S229A) was determined in the pilus polymerization assay described in Fig. 3E, using domain substrates NSpaA and CSpaA. Protein samples were analyzed by SDS/PAGE and Coomassie staining after 24 h. The ligated product CSpaA–NSpaA, sortase enzymes, substrates, and molecular markers are indicated. (D) Hydrolysis activity of SrtA enzymes was determined by an HPLC-based assay. WT or mutant SrtA (50 μM) was incubated with 500 µM KNAGFELPLTGGSGRI (SpaApep) in a 100-μL assay at 37 °C for 48 h. Reaction products were monitored and separated using HPLC at an absorbance of 215 nm. The peak fractions were collected and identified by MALDI-TOF-MS. The hydrolysis activity by SrtA2M is set as 100%. The results are presented as the average of three independent experiments; error bars indicate SDs; ns, not significant.
In our model of the ternary reaction intermediate, the side chains of Y225, N228, and S229 within the TP(Y/L)XIN(S/T)H motif extend from the enzyme’s surface in a position to contact NSpaA. To explore their possible roles in catalysis, we constructed a series of mutants of the lid-opened SrtA2M mutant enzyme in which each of these residues was individually replaced by Ala. The purified S229A and N228A mutant SrtA2M proteins were each defective in transpeptidation in vitro, as no isopeptide-linked SpaA–SpaA product was produced even after 48 h; the Y225A mutant protein had impaired transpeptidation activity as well, but to a lesser extent than the S229A and N228A mutants (Fig. 4C). Recall that the removal of the H1 helix in the SrtA2M enzyme also abrogates pilus polymerization (Fig. 3B, lanes ΔSrtA2M). We have determined that the absence of the H1 helix does not cause the protein to unfold, since the 1H-15N heteronuclear single-quantum correlation (HSQC) spectra of SrtA2M and ΔSrtA2M are generally similar (SI Appendix, Fig. S4). We conclude that specific residues within the β7/β8 loop and the presence of the H1 helix form a functionally important contact surface with NSpaA. This is supported by experiments with an SrtA2M mutant harboring the N165A substitution in the proximal β4/β5 loop, which showed that this mutant retained nearly WT activity (Fig. 4C; lanes SrtAN165A).
The model of the ternary complex raises the possibility that critical residues in the β7/β8 loop and the H1 helix may be required only for nucleophile recognition during the transpeptidation reaction but not for the other step of catalysis in which the LPXTG sorting motif is cleaved to form the thioacyl enzyme–substrate intermediate (4, 38). To test this hypothesis, we determined the importance of these structural elements in thioacyl-intermediate formation, using an established HPLC-based assay (39, 40) and an SpaA-derived peptide KNAGFELPLTGGSGRI (SpaApep) as the substrate. The enzymes and the SpaApep substrate were mixed at a 1:10 molar ratio, and the loss of the intact peptide was monitored by HPLC with the hydrolysis activity of SrtA2M set as 100%. Consistent with a selective role in nucleophile recognition, none of the mutants exhibited any significant defect in cleaving the LPXTG motif (Fig. 4D). Importantly, the ΔSrtA2M enzyme, which was inactive in the pilus polymerization assay (Fig. 3B), cleaved the SpaApep substrate with an efficiency comparable to that of the activated SrtA2M mutant enzyme (Fig. 4D). Under these conditions, the hydrolysis kinetics of the SrtA2M and ΔSrtA2M enzymes displayed a comparable Vmax of 2.3 ± 0.2 and 3.3 ± 0.8 µM/h, respectively, unlike that of SrtAWT, which was significantly reduced (0.6 ± 0.1 µM/h). These results prompt us to propose that the conserved TP(Y/L)XIN(S/T)H motif within the β7/β8 loop is a hallmark feature of the class C sortases that enables molecular recognition of the pilin motif Lys nucleophile in their cognate substrates. The H1 helix appears to play a similar role; however, it is not well conserved in class C sortases.
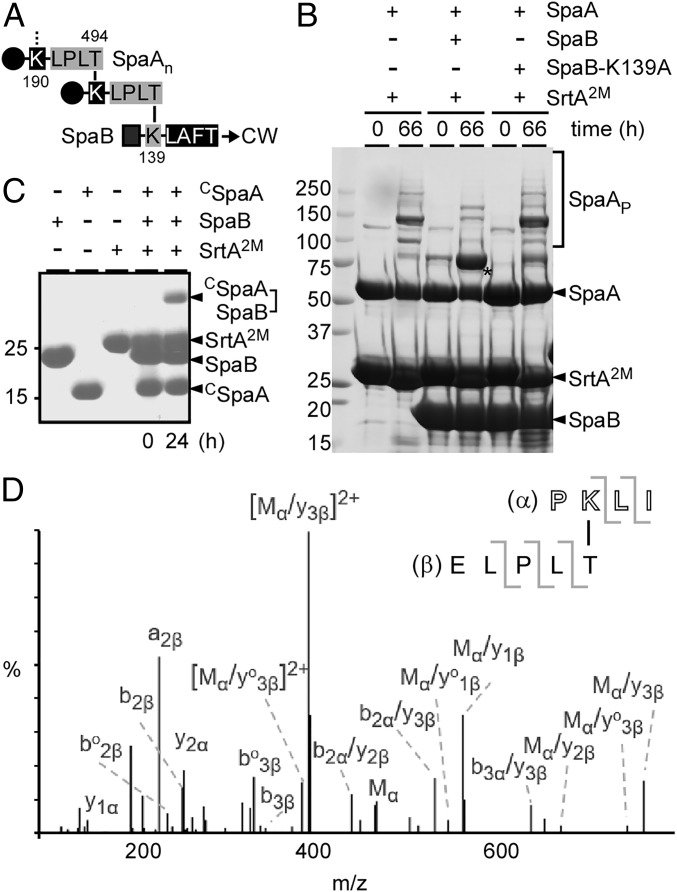
SrtA-Catalyzed Pilus Polymerization Is Terminated by SpaB.
Our previous studies suggest that SpaB acts as a molecular switch that terminates pilus polymerization by incorporating into the pilus polymer as the terminal subunit, and this reaction requires the Lys residue K139 present on SpaB (11), which is then anchored to the cell wall by the housekeeping sortase SrtF (17). The SpaA polymer is presumed to be linked to the terminal SpaB via an isopeptide bond formed between the Thr residue of the SpaA LPXTG motif and K139 (Fig. 5A). To examine if this is the case, we produced a recombinant SpaB protein (residues 25–180), which lacked the N-terminal signal peptide and the hydrophobic domain and the C-terminal charged tail of the CWSS but contained the LAFTG motif. As a control, an SpaB mutant protein with the K139A substitution mutation was also generated. These recombinant proteins, expressed in and purified from E. coli, were then used in the pilus polymerization assay and analyzed by SDS/PAGE and Coomassie staining as described in Fig. 3B. In the presence of the SrtA2M enzyme, recombinant SpaA protein was polymerized into HMM species as expected (Fig. 5B, first two lanes). Remarkably, when SpaB was added into this reaction, the formation of SpaA polymers was significantly reduced, and the SpaA–SpaB dimer accumulated (Fig. 5B, third and fourth lanes, asterisk); it is important to note that while some trimeric forms of SpaA were observed, SrtA2M was unable to further polymerize SpaA pilins in the presence of WT SpaB, although SpaA substrates were abundantly available (Fig. 5B, lane 4). This suggests that the SpaB K139 may be more nucleophilic than the SpaA K190 or that SpaB K139 may have a higher affinity and the ability to outcompete SpaA for pilin cross-linking reactions. The fact that SpaA pilus polymerization catalyzed by SrtA2M was not affected in the presence of the SpaB K139A mutant protein is consistent with K139 as the nucleophile in the cross-linking reaction (Fig. 5B, last two lanes).
Fig. 5.
SrtA-catalyzed pilus polymerization is terminated by the pilus base SpaB pilin. (A) Depicted is an SpaA polymer (SpaAn) with individual subunits cross-linked by an isopeptide bond between K190 and T494. The SpaA polymer is linked to the terminal SpaB via an isopeptide bond between the T494 residue of SpaA and the K139 residue of SpaB, which in turn is covalently attached to the bacterial peptidoglycan (CW). (B) A pilus polymerization termination assay was performed with SrtA2M enzyme and SpaA substrate (2:1 molar ratio) in the presence (same concentration as SpaA) or absence of SpaB or SpaB with the K139A mutation. The reactions were stopped after 66 h by the addition of SDS-containing sample buffer, and protein samples were analyzed by SDS/PAGE and Coomassie staining. The asterisk indicates an SpaA–SpaB dimer. Molecular mass markers (kDa) are shown. (C) CSpaA (residues 350–500), SpaB, and SrtA2M were used in the pilus polymerization termination assay, with substrates and enzyme at a 3:1 molar ratio. Protein samples were analyzed by SDS/PAGE and Coomassie staining after 24-h incubation. (D) A gel band corresponding to a CSpaA–SpaC dimer was excised for tryptic digestion and MS/MS. Shown is the MS/MS spectrum, which revealed the presence of the isopeptide bond formed between T494 and K139.
The observation that the SpaA–SpaB dimer is the predominant form of pilin conjugates produced by SrtA2M in the presence of SpaB (Fig. 5B, lane 4) prompted us to test whether the pilin motif is dispensable for SrtA-catalyzed SpaA–SpaB conjugation. To examine if this is the case, the recombinant protein CSpaA (see Fig. 4C), lacking the pilin motif, was used in place of recombinant SpaA. The reaction was performed as described in Fig. 4C. Indeed, after 24 h a band corresponding to the size of a truncated SpaA–SpaB conjugate was observed (Fig. 5C). To corroborate this and analyze the linkage between SpaA and SpaB, this band was excised for MS analysis (see Fig. 3D for methods). MS/MS data confirmed the isopeptide bond between the SpaA T494 residue and the SpaB K139 residue (Fig. 5D and Table 3). Clearly, our results demonstrate that SpaB is a termination factor for pilus polymerization.
In conclusion, we report here the high-resolution crystal structures of the C. diphtheriae pilus-specific sortase SrtA enzyme (SrtAWT) and a mutant form of the enzyme with mutations in the lid region (SrtA2M) and through these illuminate some of the basic features of the sortase that functions to polymerize pilus proteins in Gram-positive bacteria. The structure of the WT enzyme displayed a characteristic “closed” configuration of a class C sortase with its catalytic site occluded by a molecular lid (Fig. 1). By introducing specific amino acid substitutions within the lid, we were able to generate an enzyme whose catalytic pocket displayed an open conformation with no other major perturbations detected in the atomic structure. The functional importance of these two states of the enzyme was demonstrated by our ability to reconstitute a robust pilus polymerization reaction in vitro using the cognate shaft pilin. While the WT form could not polymerize the shaft pilin in a reaction, the opened-lid version of the enzyme is highly active. We showed that the activated enzyme was able to recognize the sorting signal, form the relevant acyl-enzyme intermediate, recognize the pilin motif Lys residue, and catalyze isopeptide bond formation conjugating Lys of one pilin protomer with Thr of another protomer (Fig. 3). We then utilized structural modeling to identify specific structural elements conserved in a pilus-specific sortase which are important for catalyzing the transpeptidation reaction in vitro (Fig. 4). We also provide additional in vitro evidence in support of SpaB being a terminator of pilus polymerization (Fig. 5).
Given that the SrtA2M enzyme is able to catalyze pilus polymerization and SpaB incorporation in vitro, although pilus assembly in vivo is not apparently altered when the mutant enzyme is present, we surmise that the SrtA lid might play some form of a modulatory role in pilus polymerization and termination in vivo. For instance, as SpaB is a preferred substrate of the housekeeping sortase SrtF (17), charging SrtF with SpaB triggers an SrtF–SrtA interaction. This interaction could potentially alter the configuration of the SrtA lid, allowing SrtA-mediated entry of SpaB to the pilus base and subsequently transferring the pilus polymer to SrtF to complete the cell wall-anchoring step. Future experiments may be designed to examine the lid dynamics in the presence or absence of SpaB or SrtF.
Importantly, we have shown here that the separated domains of the pilin, one containing the pilin motif and the other containing the sorting motif, could be ligated efficiently to produce a di-polypeptide conjugate containing the Lys–Thr isopeptide bond. This provides a powerful protein ligation platform for engineering designer proteins that is mechanistically different from the “sortagging” technology developed with the archetypal S. aureus sortase which normally functions to cross-link surface proteins to the bacterial cell wall but does not polymerize proteins (41, 42). We envision that the surface display of protein polymers, protein labeling of living cells, and protein ligation are a few examples of many potential biotechnological and biological applications of this enzyme.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
Bacterial strains and plasmids used in this study are listed in SI Appendix, Table S1. SI Appendix, SI Materials and Methods contains information regarding recombinant plasmids, protein purification, protein crystallization, and structure determination. C. diphtheriae strains were grown in Heart Infusion (HI) broth (Becton Dickinson) or on HI agar plates at 37 °C. When needed, kanamycin was added at a concentration of 25 μg/mL. E. coli DH5α and BL21 (DE3), used for cloning and protein expression and purification, respectively, were grown in either Luria–Bertani or 2× YT broth (Sigma-Aldrich) at 37 °C in the presence of ampicillin at 100 μg/mL.
In Vitro Reconstitution of Pilus Polymerization.
In vitro reactions were carried out at room temperature, and proteins were dissolved in assay buffer [50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 1 mM DTT]. All reactions used a fixed 100-µM concertation of SrtA enzyme and 300-µM SpaA substrate (either full length or each individual domain). Reactions were stirred gently by continuous rotation. Aliquots were taken at 0 h, 24 h, 48 h, and 72 h, and reactions were quenched by the addition of two volumes of SDS loading dye. For a pilus termination assay, SpaB was used at the same concentration as SpaA, whereas the molar concentrations of SrtA2M enzyme and SpaA substrate followed 3:1 or 2:1 ratios. The reactions were performed in 24 or 66 h, respectively.
Probing Accessibility of the SrtA Active Site.
Reaction rates of DTDP and SrtA proteins via the Cys C222 residue were determined by stopped-flow experiments, which were performed at 23 °C using an Applied Photophysics Ltd. Model SX.18 MV sequential stopped-flow spectrofluorimeter with a 150 W Xe/Hg lamp and a dead time of 1.7 ms. All triplicate reactions were carried out in reaction buffer [50 mM 3-(N-morpholino)propanesulfonic acid (Mops), 200 mM KCl, 1 mM EDTA, pH 7.5]. Absorbance was monitored at 324 nm after solutions were rapidly mixed in syringe A, containing 0.6 mg/mL protein, and syringe B, containing 0.32 mM DTDP. Reaction rates (k) were derived by fitting data to the following equations with one (Eq. 1) or two (Eq. 2) rates:
| [1] |
or
| [2] |
where A is absorbance at 324 nm at time t, and Amax is the maximum absorbance.
Cell Fractionation and Western Blotting.
Cell fractionation and Western blotting were performed according to published procedures with some modifications (43, 44). Briefly, midlog-phase cultures of C. diphtheriae strains grown at 37 °C were normalized to an OD600 of 1.0 and were subjected to cell wall protein extraction using mutanolysin (300 U/mL). Protein samples obtained from culture medium (S) and cell wall (W) were trichloroacetic acid precipitated and acetone washed. The protoplasts after the cell wall extraction were used for analysis of cell membrane-bound proteins. Protein samples were resuspended in SDS sample buffer containing 3% urea and were heated at 100 °C for 10 min before SDS/PAGE analysis using 3–12% or 3–20% Tris-Gly gradient gels. Detection of proteins was performed by immunoblotting with specific antibodies (1:20,000 for α-SpaA; 1:4,000 for α-SpaH; 1:5,000 for α-MdbA; and 1:4,000 for α-SrtA).
MS of Pilus Polymers.
Protein digestion and isopeptide bond identification were performed according to previous protocols (36, 45). Specifically, proteins entrapped in gel bands were reduced with 10 mM DTT (Sigma) at 60 °C for 1 h and then were alkylated with 50 mM iodoacetamide (Sigma) at 45 °C for a few minutes in the dark. These reduction and alkylation steps were skipped for the acyl-intermediate samples. Samples were digested with 200 ng trypsin (Thermo Scientific) at 37 °C overnight. At the end of trypsin digestion, 200 ng of Asp-N endoproteinase (Thermo Scientific) was added for another overnight incubation. Digested peptides were extracted from the gel bands in 50% acetonitrile/49.9% water/0.1% TFA and were cleaned with C18 StageTip (46) before MS analysis.
Digested peptides were separated on an EASY-Spray column (25 cm × 75 μm i.d., PepMap RSLC C18, 2 μm; Thermo Scientific) connected to an EASY-nLC 1000 nUPLC (Thermo Scientific) using a gradient of 5–35% acetonitrile in 0.1% formic acid and a flow rate of 300 nL/min for 30 min. Tandem mass spectra were acquired in a data-dependent manner with an Orbitrap Q Exactive mass spectrometer (Thermo Fisher Scientific) interfaced to a nanoelectrospray ionization source.
The raw MS/MS data were converted into MGF format by Thermo Proteome Discoverer 1.4 (Thermo Scientific). In-house programs to search for the isopeptides were used for two different approaches. The first approach was performed as previously described (36) and was used to calculate the masses of predicted peptides containing the isopeptide linkage to guide the search. The second approach was based on published (36) and our own observations of the presence of ions specific for the fragments of ELPLT (m/z 215.138, 225.122, 243.132, 294.180, 312.190, 322.174, and 340.186). The in-house programs sifted through tens of thousands of mass spectra searching specifically for this information and extracted MS/MS spectra for further analyses.
Determination of SrtA Hydrolysis by an HPLC-Based Assay.
In vitro hydrolysis reactions were performed based on the method developed by Kruger et al. (47). WT or mutant SrtA (50 µM) was incubated with 500 µM KNAGFELPLTGGSGRI (SpaApep) in 100-µL reactions at 37 °C for 24 h. The reactions were quenched by adding 50 µL of 1 M HCl and were injected onto a Waters XBridge Peptide BEH C18 reversed-phase HPLC column. Peptides were eluted by applying a gradient from 5–51% acetonitrile (in 0.1% TFA) over 25 min at a flow rate of 1 mL/min. Elution of the peptides was monitored by absorbance at 215 nm. Peak fractions were collected, and their identities were confirmed by MALDI-TOF-MS.
Electron Microscopy.
For visualization of in vitro pilus polymers, pilus polymerization reactions were diluted in half with water and 7-μL aliquots were applied onto carbon-coated nickel grids, washed five times with distilled water, and stained with 0.75% uranyl formate for 2 min before viewing by a JEOL JEM-1400 electron microscope.
For visualization of pili produced by corynebacterial cells, electron microscopy and IEM were performed according to a published protocol (48). Briefly, corynebacterial cells grown on HI agar plates were washed and suspended in PBS. Seven-microliter aliquots of the cell suspension were applied onto nickel grids, washed, and stained with 0.75% uranyl formate before viewing by an electron microscope. For IEM, cells were stained with α-SpaA (1:100 dilution), followed by staining with 12-nm gold particles conjugated to IgG, before staining with 1% uranyl acetate.
To estimate the dimension of pili, ImageJ (https://imagej.nih.gov/ij/) was employed. Twenty-five measurements were performed at different locations of pili for each strain. Statistical analysis was performed by GraphPad Prism.
Supplementary Material
Acknowledgments
We thank members of the R.T.C. and H.T.-T. laboratories for critical review and discussion of the manuscript and members of the Structural Biology Center at Argonne National Laboratory for help in conducting X-ray diffraction data collection. This work was supported by Department of Health and Human Services, NIH National Institute of Allergy and Infectious Diseases Award AI52217 (to R.T.C.) and in part by Center of Structural Genomics of Infectious Diseases Contracts HHSN272201200026C and HHSN272201700060C (to A.J.), National Institute of General Medical Sciences Grant GM103479 (to J.A.L.), and National Institute of Dental and Craniofacial Research Awards DE017382 and DE025015 (to H.T.-T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5K9A and 6BWE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800954115/-/DCSupplemental.
References
- 1.Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: Assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol. 2017;15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 3.Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: Diversity in structure and function. FEMS Microbiol Rev. 2012;36:1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 6.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: Assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 8.Rogers EA, Das A, Ton-That H. Adhesion by pathogenic corynebacteria. Adv Exp Med Biol. 2011;715:91–103. doi: 10.1007/978-94-007-0940-9_6. [DOI] [PubMed] [Google Scholar]
- 9.Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc Natl Acad Sci USA. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 14.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 15.Navarre WW, Schneewind O. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel SD, Liu J, Ton-That H. Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol. 2016;34:31–37. doi: 10.1016/j.mib.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaminathan A, et al. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 19.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: A nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobitz AW, Kattke MD, Wereszczynski J, Clubb RT. Sortase transpeptidases: Structural biology and catalytic mechanism. Adv Protein Chem Struct Biol. 2017;109:223–264. doi: 10.1016/bs.apcsb.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khare B, V L Narayana S. Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly. Protein Sci. 2017;26:1458–1473. doi: 10.1002/pro.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manzano C, Izoré T, Job V, Di Guilmi AM, Dessen A. Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of Gram-positive pathogens. Biochemistry. 2009;48:10549–10557. doi: 10.1021/bi901261y. [DOI] [PubMed] [Google Scholar]
- 25.Khare B, Fu ZQ, Huang IH, Ton-That H, Narayana SV. The crystal structure analysis of group B Streptococcus sortase C1: A model for the “lid” movement upon substrate binding. J Mol Biol. 2011;414:563–577. doi: 10.1016/j.jmb.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persson K. Structure of the sortase AcSrtC-1 from Actinomyces oris. Acta Crystallogr D Biol Crystallogr. 2011;67:212–217. doi: 10.1107/S0907444911004215. [DOI] [PubMed] [Google Scholar]
- 27.Neiers F, et al. Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity. J Mol Biol. 2009;393:704–716. doi: 10.1016/j.jmb.2009.08.058. [DOI] [PubMed] [Google Scholar]
- 28.Jacobitz AW, et al. The “lid” in the Streptococcus pneumoniae SrtC1 sortase adopts a rigid structure that regulates substrate access to the active site. J Phys Chem B. 2016;120:8302–8312. doi: 10.1021/acs.jpcb.6b01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manzano C, et al. Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae. Structure. 2008;16:1838–1848. doi: 10.1016/j.str.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Cozzi R, et al. Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. FASEB J. 2011;25:1874–1886. doi: 10.1096/fj.10-174797. [DOI] [PubMed] [Google Scholar]
- 31.Wu C, et al. Structural determinants of Actinomyces sortase SrtC2 required for membrane localization and assembly of type 2 fimbriae for interbacterial coaggregation and oral biofilm formation. J Bacteriol. 2012;194:2531–2539. doi: 10.1128/JB.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez BC, Chang C, Wu C, Tran B, Ton-That H. Electron transport chain is biochemically linked to pilus assembly required for polymicrobial interactions and biofilm formation in the Gram-positive actinobacterium Actinomyces oris. MBio. 2017;8:e00399-17. doi: 10.1128/mBio.00399-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putkey JA, et al. Fluorescent probes attached to Cys 35 or Cys 84 in cardiac troponin C are differentially sensitive to Ca(2+)-dependent events in vitro and in situ. Biochemistry. 1997;36:970–978. doi: 10.1021/bi9617466. [DOI] [PubMed] [Google Scholar]
- 34.Epps DE, Vosters AF. The essential role of a free sulfhydryl group in blocking the cholesteryl site of cholesteryl ester transfer protein (CETP) Chem Phys Lipids. 2002;114:113–122. doi: 10.1016/s0009-3084(01)00187-6. [DOI] [PubMed] [Google Scholar]
- 35.Echelman DJ, et al. CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks. Proc Natl Acad Sci USA. 2016;113:2490–2495. doi: 10.1073/pnas.1522946113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttilla IK, et al. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in Gram-positive bacteria. J Bacteriol. 2009;191:5603–5612. doi: 10.1128/JB.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Aulabaugh A, et al. Development of an HPLC assay for Staphylococcus aureus sortase: Evidence for the formation of the kinetically competent acyl enzyme intermediate. Anal Biochem. 2007;360:14–22. doi: 10.1016/j.ab.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Ton-That H, Mazmanian SK, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 41.Antos JM, et al. Site-specific protein labeling via sortase-mediated transpeptidation. Curr Protoc Protein Sci. 2017;89:15.13.11–15.13.19. doi: 10.1002/cpps.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proft T. Sortase-mediated protein ligation: An emerging biotechnology tool for protein modification and immobilisation. Biotechnol Lett. 2010;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- 43.Chang C, Mandlik A, Das A, Ton-That H. Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol Microbiol. 2011;79:1236–1247. doi: 10.1111/j.1365-2958.2010.07515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reardon-Robinson ME, et al. A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol. 2015;98:1037–1050. doi: 10.1111/mmi.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thevis M, Ogorzalek Loo RR, Loo JA. In-gel derivatization of proteins for cysteine-specific cleavages and their analysis by mass spectrometry. J Proteome Res. 2003;2:163–172. doi: 10.1021/pr025568g. [DOI] [PubMed] [Google Scholar]
- 46.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 47.Kruger RG, Dostal P, McCafferty DG. Development of a high-performance liquid chromatography assay and revision of kinetic parameters for the Staphylococcus aureus sortase transpeptidase SrtA. Anal Biochem. 2004;326:42–48. doi: 10.1016/j.ab.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Chang C, Huang IH, Hendrickx AP, Ton-That H. Visualization of Gram-positive bacterial pili. Methods Mol Biol. 2013;966:77–95. doi: 10.1007/978-1-62703-245-2_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.