Abstract
The natural history and epidemiology of Pseudomonas aeruginosa infections in non-cystic fibrosis (non-CF) bronchiectasis is not well understood.
As such it was our intention to determine the evolution of airway infection and the transmission potential of P. aeruginosa in patients with non-CF bronchiectasis.
A longitudinal cohort study was conducted from 1986–2011 using a biobank of prospectively collected isolates from patients with non-CF bronchiectasis. Patients included were ≥18 years old and had ≥2 positive P. aeruginosa cultures over a minimum 6-month period. All isolates obtained at first and most recent clinical encounters, as well as during exacerbations, that were morphologically distinct on MacConkey agar were genotyped by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). A total of 203 isolates from 39 patients were analysed. These were compared to a large collection of globally epidemic and local CF strains, as well as non-CF isolates.
We identified four patterns of infection in non-CF bronchiectasis including: 1) persistence of a single strain (n=26; 67%); 2) strain displacement (n=8; 20%); 3) temporary disruption (n=3; 8%); and 4) chaotic airway infection (n=2; 5%). Patterns of infection were not significant predictors of rates of lung function decline or progression to end-stage disease and acquisition of new strains did not associate with the occurrence of exacerbations. Rarely, non-CF bronchiectasis strains with similar pulsotypes were observed in CF and non-CF controls, but no CF epidemic strains were observed. While rare shared strains were observed in non-CF bronchiectasis, whole-genome sequencing refuted patient–patient transmission.
We observed a higher incidence of strain-displacement in our patient cohort compared to those observed in CF studies, although this did not impact on outcomes.
Short abstract
Pseudomonas aeruginosa demonstrates distinct infection patterns in non-cystic fibrosis bronchiectasis http://ow.ly/PnvA30jvZDi
Introduction
Chronic airway infections are a hallmark of suppurative lung diseases, including non-CF bronchiectasis. Bronchiectasis is characterised by the abnormal dilation and thickening of the bronchial walls, enabling chronic airway infections, and manifests clinically with a persistent productive cough [1]. Affected individuals may additionally experience acute pulmonary exacerbations (PEx), which present with increased respiratory symptomology, worsened lung function and systemic symptoms [2].
The presence of Pseudomonas aeruginosa in the airways has been associated with accelerated decline in lung function, worsened quality of life and increased morbidity and mortality in non-CF bronchiectasis [3, 4]. Whilst both the evolution and patho-adaptation of P. aeruginosa infections have been extensively studied in cystic fibrosis (CF), our understanding of these processes lags in non-CF bronchiectasis [5, 6]. Despite notable advancements in non-CF bronchiectasis research, much of our current understanding of airway infections in non-CF bronchiectasis is extrapolated from CF. Specifically, the following observations have been made regarding P. aeruginosa infections in CF: 1) changes in clinical status such as PEx are not solely due to the acquisition of new infections but arise from perturbations affecting the original colonising strains [7]; 2) adult P. aeruginosa airway infections are generally stable with the exception of rare events of acquisition and eventual replacement or co-infection of epidemic (ePA) strains [7–10]; 3) transmission of P. aeruginosa infections is possible and may be associated with adverse clinical consequences [11–13].
Whilst recent studies [14, 15] have started to investigate these trends, continued research will contribute to a better understanding of non-CF bronchiectasis. Therefore, we set out to characterise the epidemiology, transmission characteristics and clinical outcomes of P. aeruginosa infection in non-CF bronchiectasis using a longitudinal collection of biobanked isolates. Given the similarities between CF and non-CF bronchiectasis lung disease, we hypothesised that similar trends in the epidemiology and outcomes of P. aeruginosa infections would be observed between both diseases.
Materials and methods
Patients and strains
Patients were identified from a retrospective review of the microbiologic records of the Calgary Bronchiectasis Clinic (CBC) Biobank (1980–2015). This biobank contains prospectively collected and inventoried bacterial pathogens recovered in real time from the sputum of non-CF bronchiectasis patients upon submission of a quantitative sputum culture as part of their routine care. From each sputum sample, plated on MacConkey agar, every individual morphotype of suspected P. aeruginosa was confirmed as P. aeruginosa using standard methodologies in real time. Morphotypes were defined according to a priori definitions (see supplemental table S1). Individual isolates were then subsequently transferred into skim milk stock and frozen at −80 °C and entered into the CBC Biobank. For inclusion in to our study, patients had a diagnosis of bronchiectasis with the exclusion of CF and two or more sputum samples within the biobank spanning ≥6 months where P. aeruginosa was isolated. Bronchiectasis was confirmed radiographically [16]. From each patient, every individual P. aeruginosa morphotype from their first encounter (FE; the first isolate within the biobank), their most recent encounter (RE; the most recent isolate for active and inactive patients) and, where available, serial alternate year isolates and exacerbation encounters (EE; isolates identified at the time of PEx) were assessed.
The prevalence of shared strains of P. aeruginosa and the potential for cross-infection amongst non-CF bronchiectasis patients was assessed using a control cohort consisting of 812 isolates obtained from CF patients over the last 30 years, 22 isolates obtained from local environmental sources (both natural environments and hospital facilities) and 35 strains obtained from community-acquired blood stream infections (CA-BSI). Of the 812 CF-derived isolates, 65 representatives of globally distributed ePA strains known to commonly exist within patients with suppurative lung infections were used [9, 17].
Molecular typing
The pattern of P. aeruginosa infections, including whether patients were chronically infected with the same strain longitudinally or whether serial infections with different isolates occurred, was primarily assessed using a pulsed-field gel electrophoresis (PFGE) protocol described by Parkins et al. [9]. Strains conforming to the criteria of Tenover et al. [18] were considered related. PFGE profiles were compared using BioNumerics Version 7.0 (Applied Maths, Austin, TX, USA). The resulting dendrograms were generated at 2.0% position tolerance and 1.5% optimisation using the unweighted pair-group method with arithmetic mean (UPGMA) and the Sørensen–Dice similarity coefficient. Clusters of P. aeruginosa strains derived from three or more unrelated patients were a priori considered clonal [9].
Following PFGE, multilocus sequence typing (MLST) was used to confirm the relatedness of the FE and the RE isolates for each patient [19]. The resulting MLST profiles were then referenced to an online P. aeruginosa MLST database (http://pubmlst.org/paeruginosa/) in order to identify allele and sequence types. Isolates were considered putatively clonally related if the MLST profiles had less than two loci altered but were assigned a novel sequence type if strains were one or more loci different than identified in the online database [9].
Isolates with similar MLST profiles from unrelated patients were sequenced using whole-genome sequencing (WGS) to investigate the potential for patient-to-patient transmission as described previously [20]. Contigs (sets of overlapping DNA segments that combined represent a consensus region of DNA) were assembled using the A5-MiSeq pipeline [21], annotated with PROKKA [22] and subjected to genome comparison using Roary [23].
Clinical information and definitions
In order to determine whether the different patterns of infection were associated with particular patient characteristics, acute PEx events, or the rate of lung function decline, a detailed chart review was performed [24]. Chronic P. aeruginosa colonisation at baseline was defined as per the Leeds criteria adapted from CF, whereby patients were defined as chronically infected if more than 50% of cultures obtained over the preceding year were positive for P. aeruginosa [25]. The aetiology of bronchiectasis was classified as: post-infectious, immunodeficiency, idiopathic and other. PEx events were clinician-defined based on documentation of increasing respiratory symptoms or radiographic changes resulting in new antimicrobial therapy. The pattern of infection was characterised using molecular typing and defined based on similarity of isolates. If all isolates were found to be clonally related then the patient was defined as a “stable” infection. If isolates collected from a patient were dissimilar, they were classified within the strain displacement category. Patients attending the CBC are seen on a consultation basis but continue to receive the bulk of their care from primary care physicians and local respirologists. With respect to infection prevention standards, we enforce strict cough and hand hygiene and actively discourage patient contact. Patients do not share clinic rooms, waiting rooms or inpatient rooms. However, we do not utilise universal contact precautions nor ask patients to wear surgical masks. The study was approved by the local ethics board (REB16-0035).
Statistical analysis
Non-normally distributed data were represented using median values with interquartile range (IQR). Categorical variables were analysed using the two-tailed Fisher's exact test. Continuous data were analysed using the two-tailed Mann–Whitney U-test. For our primary clinical outcome analysis, yearly lung function decline (% predicted forced expiratory volume in 1 s (FEV1)) between chronic and strain-displacement groups was modelled through the use of generalised estimating equations (GEEs) with exchangeable correlation structures and robust standard errors. Analyses were conducted with STATA Version 13.1 (StataCorp, College Station, TX, USA) and Prism 5.0 (Graphpad Software, La Jolla, CA, USA).
Results
Patient demographics
The inclusion criteria were met by 39 patients with non-CF bronchiectasis (table 1). The aetiology of non-CF bronchiectasis was primarily post-infective (51%) and idiopathic (44%), although there were single cases of immunodeficiency (common variable immunodeficiency) and toxic inhalation. Median age at enrolment was 58 (IQR 23–81). Baseline % predicted FEV1 at cohort inclusion was 53% (IQR 39.8–73%) and % predicted forced vital capacity (FVC) was 67% (IQR 51–83%). Eleven patients (28%) had a history of lung resection prior to enrolment, with one patient (2.6%) receiving a lung transplant during his time at the non-CF bronchiectasis clinic. One patient died during the study and 43% of patients eventually required long-term oxygen supplementation. Thirty-one patients (79%) had chronic infection at the FE [25]. Notably, chronic infection at baseline did not differ between the strain-displacement and stable groups (p=1.00), nor did length of microbiological follow-up.
TABLE 1.
Baseline patient demographics
| Selected patient demographics | Stable (n=26) | Strain displacement (n=13) |
| Age at enrolment years | 66.7 (47.9, 72.7) | 61.46 (41, 71) |
| Duration of follow-up years | 2.8 (1, 7) | 4.8 (1, 10.4) |
| FEV1 % predicted | 51 (39, 65) | 58 (40.5, 78.5) |
| Respiratory comorbidities | ||
| Sinusitis | 11 (42) | 6 (46) |
| Reactive airway disease | 7 (27) | 3 (23) |
| COPD | 8 (31) | 1 (8) |
| Aetiology | ||
| Idiopathic | 13 (50) | 4 (31) |
| Post-infective | 13 (50) | 7 (54) |
| Immunodeficiency | 1 (8) | |
| Other | 1 (8) | |
| Recorded antibiotics used | ||
| Inhaled tobramycin | 6 (23) | 3 (23) |
| Azithromycin | 6 (23) | 1 (8) |
| Ciprofloxacin | 3 (12) | 3 (23) |
| Additional therapies | ||
| Inhaled β2-agonist | 23 (88) | 10 (77) |
| Inhaled CS | 16 (62) | 5 (38) |
| Ipratropium bromide | 2 (8) | 6 (46) |
| Systemic CS | 5 (19) | 2 (15) |
| Spiriva | 6 (23) | 2 (15) |
| Long-term oxygen therapy | 12 (46) | 5 (38) |
Data are presented as n (%) or median (interquartile range). FEV1: forced expiratory volume in 1 s; COPD: chronic obstructive pulmonary disease; CS: corticosteroid.
Natural history of P. aeruginosa in non-CF bronchiectasis and clinical outcomes
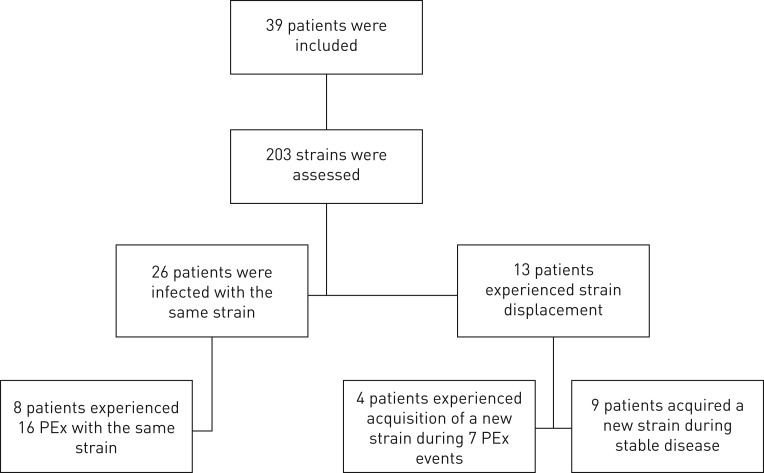
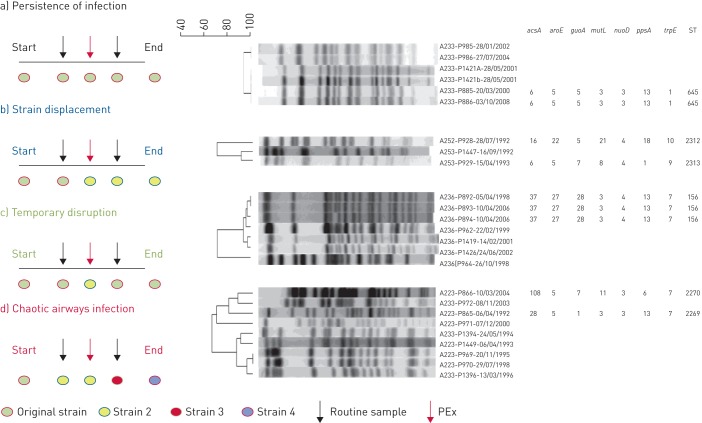
A total of 203 isolates (range: two to eight per patient), representing eight distinct morphotypes out of a possible 34 which were defined a priori (see supplementary table S1 and figure S1), were genotyped and patterns of colonisation were distinguished based on the PFGE profiles for each patient. In only one instance were multiple morphotypes apparent at a single time point. The median duration between the earliest and most recent isolates was 3.2 years (IQR 1.1–8.3) and ranged from 0.5–21 years. If all isolates characterised for each patient over the study period were clonally related, the patient was defined to have a “stable” infection. Conversely, if the initial isolate was no longer observed but a new isolate was, the intermediate isolates for each patient were used to determine the overall history of infection. Using this classification, we observed four patterns of P. aeruginosa colonisation: 1) absolute persistence of the original infecting strain without invasion by an extraneous isolate (n=26; 67%); 2) strain displacement where the original strain was no longer observed (n=8; 20%); 3) temporary acquisition of a new strain type eventually reverting back to the original strain (n=3; 8%); and 4) chaotic airways colonisation where new strains were continually identified (n=2; 5%) (figure 1). To assess if unstable airways infections associated with a different aetiology and subsequent clinical course, those with stable infections were compared to those where more than one strain type was identified (combining the three other groups) (figure 2).
FIGURE 1.
Flow chart of study design including the total number of non-cystic fibrosis bronchiectasis patients enrolled. PEx: pulmonary exacerbation.
FIGURE 2.
Displaying the diversity in the natural history of infections of Pseudomonas aeruginosa in patients with non-cystic fibrosis bronchiectasis including: a) patients with stable infection by a single strain of P. aeruginosa (persistence of infection; n=26); b) strain displacement (n=8); c) temporary disruption (n=3); d) chaotic airways infection (n=2). The dendrograms were generated at 2.0% position tolerance and 1.5% optimisation using the unweighted pair-group method with arithmetic mean (UPGMA) and the Sørensen–Dice similarity coefficient. Isolates were named following the format “A(patient)-P(isolate)-(date isolated)”. ST: sequence typing; PEx: pulmonary exacerbation.
Patterns of infection analysis revealed no association with patient demographics, including aetiology of non-CF bronchiectasis, comorbidities, sex, age and duration of culture follow-up. Antibiotic therapies received during the study, including inhaled tobramycin and azithromycin, had no association with stable or unstable infections. Similarly, the rate of FEV1 decline between patients with stable strains −0.7% per year (95% CI −1 to −0.5%) and those with strain displacement +0.2% per year (95% CI −0.2 to –0.6%) was not found to be different (p=0.37). Additionally, 12 patients experienced 23 PEx events during clinic visits, of which 16 events (70%) were associated with the same chronically colonising strain being isolated (p=0.01) (figure 2). Four of the 12 patients experienced seven PEx events (30%) which were associated with a new strain, different from the chronically infecting isolate. Of the seven PEx events associated with different strains, four were transient infections, one persisted after strain-displacement and two were identified in individuals with chaotic airways infection where they were subsequently displaced by a further new strain.
Prevalence of clonal isolates and patient-to-patient transmission in non-CF bronchiectasis
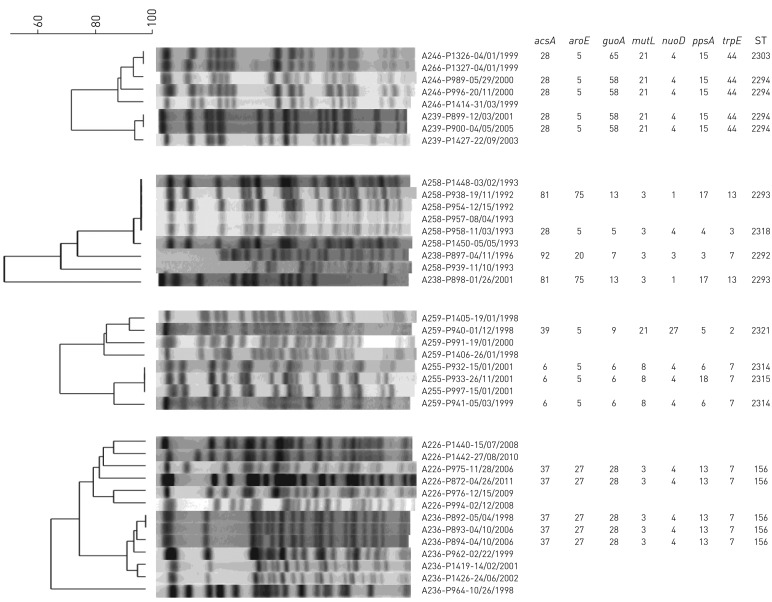
Overall, no P. aeruginosa clone within our non-CF bronchiectasis cohort was identified to be disproportionally prevalent. While four pairs of patients shared strains, this did not fit our definition for clonality (MLST types=156, 2314, 2293, 2294) (figure 3). Of these patients, one pair of patients with sequence type (ST)=156 had attended the clinic within the same year but never during the same clinic day. The remaining pairs of patients had at least ≥1 year between the collection of isolates (ST=2314, 2293, 2294). Whole-genome analysis of these strains showed that isolates from the same patient exhibited a greater degree of similar gene content, suggesting independent acquisition in each patient as opposed to transmission (figure 4).
FIGURE 3.
Cluster diagram for four patient pairs with genotyped strains sharing the same sequence typing (ST). The dendrograms were generated at 2.0% position tolerance and 1.5% optimisation using the unweighted pair-group method with arithmetic mean (UPGMA) and the Sørensen–Dice similarity coefficient. Isolates were named following the format “A(patient)-P(isolate)-(date isolated)”.
FIGURE 4.
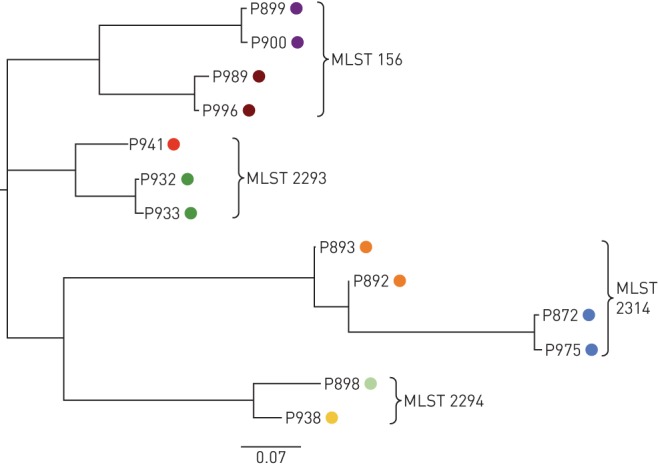
Clonal strains of Pseudomonas aeruginosa were identified from independent patient-pairs using multilocus sequence typing (MLST). Individual strains are coloured by patient. Strain relatedness was assessed using whole-genome sequencing and phylogeny generated from a matrix with the presence and absence of core and accessory genes using Roary [23].
A comparison of the 203 non-CF bronchiectasis derived P. aeruginosa isolates revealed no known representatives from our regional and global ePA collection. While PFGE patterns between AUST-3 and five non-CF bronchiectasis isolates were observed to be 89% similar, discrepancies in three MLST alleles were found and did not meet our definition for clonally related strains. However, similarities in the PFGE profiles between 19 non-CF bronchiectasis P. aeruginosa isolates from nine patients with 31 CF-derived P. aeruginosa isolates from 14 patients were observed. Four non-CF bronchiectasis isolates were observed to share 96% similar pulsotypes with one CA-BSI derived P. aeruginosa isolate. Furthermore, five non-CF bronchiectasis isolates were found to share 89% similar PFGE profiles with one environmental isolate. MLST data from non-(non-CF bronchiectasis) isolates was not available. Interestingly, the MLST profiles of the non-CF bronchiectasis derived P. aeruginosa isolates obtained in our study included eight MLST strain types (102, 175, 291, 390, 507, 645, 847 and 1752) than had previously been identified throughout regions of Europe [26]. One strain in particular (MLST type=175) was previously isolated in Canada, the UK, Poland, Spain and France from sputum, environmental sources, bronchial lavages, urinary-tract infections and soft-tissue infections.
Comparison of PFGE and MLST as P. aeruginosa screening strategies
While all isolates underwent PFGE as the primary screening modality, the FE and the RE for each patient were characterised using MLST for 33 patients (85%). Despite repeated attempts, we were unable to fully characterise the MLST profiles of the remaining six patients. Five of these patients had only the FE fully typed by MLST and one patient had neither the RE nor the FE fully characterised. In four of these six patients, PFGE profiles were found to be the same between the FE and the RE. When confirming P. aeruginosa infection patterns, we found that the narratives obtained using PFGE were concordant with MLST in 27 out of 31 patients (87%). Differences between the MLST and PFGE findings arose where serial strains were considered clonally-related by PFGE but were assigned novel strain types due to differences at one or more MLST loci, with the FE and the RE isolates from two patients having discordant MLST profiles due to differences in three loci.
Discussion
Our longitudinal study of non-CF bronchiectasis patients has observed several distinctions in the epidemiology of P. aeruginosa infections as compared to CF. Whereas P. aeruginosa infections in CF adults are known to be relatively stable, strain displacement with unique strains was observed commonly in non-CF bronchiectasis [7, 9]. This may be more similar to early airways infections in younger CF cohorts where P. aeruginosa strain turnover appears more common until a dominant strain emerges [27]. Furthermore, when new strains were observed, they were not necessarily associated with PEx events, in a similar fashion to what has been previously observed in CF [7]. Whereas cross-infection of P. aeruginosa has been observed in CF, we did not find any evidence of cross-infection amongst non-CF bronchiectasis patients despite observing patient-pairs sharing clonal strains [8, 9]. These trends may be due to differences in other components of the respiratory microbiota, mucus physiology and potential for pathogenic exposure of non-CF bronchiectasis patients [28].
Studies surrounding the epidemiology of infection in non-CF bronchiectasis airways are recent and ongoing. De Soyza et al. [29] recently assessed P. aeruginosa obtained from 40 non-CF bronchiectasis adults between 2008 and 2011 to better understand the potential for cross-infection. In a similar fashion they noted that the majority of patients were stably infected by unique strains of P. aeruginosa with only one potential cross-infection event identified. Likewise, Mitchelmore et al. [15] observed that non-CF bronchiectasis patients commonly harboured unique strains of P. aeruginosa and, in a similar fashion to our own study, noted the occasional presence of shared isolates between patients with CF and those with non-CF bronchiectasis, which likely represent environmentally prevalent strains. Furthermore, as all patients within our study had at least two longitudinal isolates obtained over a greater duration of follow-up, we were able to observe a higher rate of emergence of new/novel strains. These mixed infections observed in our cohort have similarly been observed in recent work by Hilliam et al. [14], which may indicate co-infection of non-CF bronchiectasis patients. In their study, co-infection by multi-lineage strains of P. aeruginosa and genomic diversification was observed in seven of 24 non-CF bronchiectasis patients when multiple isolates from a single time point were assessed. The diversification and adaptation of P. aeruginosa to the lung environment was observed through the acquisition of loss-of-function mutations within virulence determinants, which has been suggested to play a role in the adaptations in both CF and non-CF bronchiectasis [30–33].
Given the ubiquity of P. aeruginosa in the natural environment and abundant data from more recent works [34] showing shared clonal but unrelated connected strains, we believe that the four patient-pairs with shared strains observed in our study are not due to cross-infection. We demonstrated through WGS that isolates differed much more between patients than within patients, suggesting independent environmental acquisition [17, 34, 35]. Our findings agree with recent evidence positing the likelihood of independent acquisition of isolates from the environment [14, 15]. However, these findings do not exclude the potential for cross-infection in non-CF bronchiectasis, as a genetically-related strain has been identified using WGS from three unrelated patients sharing the same waiting area and lung function room at a UK centre [15]. However, in our case, no environmental source of cross-infection was found. Given that the majority of patients acquiring P. aeruginosa are doing so from environmental sources, we suggest that tightening already stringent infection control standards for non-CF bronchiectasis patients in our clinic (specifically as they pertain to P. aeruginosa) is unwarranted.
While no clonal strains were identified in our study, we identified a strain (ST=175) with a widespread distribution within hospital centres in Canada, the UK and Central Europe [26]. As this strain is associated with multidrug resistance and severe infections, continued surveillance of ST175 will be important for its management and prevention of its transmission [36–38]. Furthermore, we did not identify any ePA amongst attendees at our regional non-CF bronchiectasis center, despite a high prevalence of ePA (∼39%) amongst CF adults attending our regional CF clinic (mere feet away but generally seen on alternate days) [9, 24]. Given that individuals with CF and non-CF bronchiectasis experience similar structural airways disease and the same predilection for lower airways infection, and given that the viability of P. aeruginosa generated through cough aerosol is independent of disease aetiology [39], we believe it is the high levels of social interconnectivity of historical CF cohorts that manifests in markedly different P. aeruginosa epidemiology in patients with non-CF bronchiectasis [12].
Studies comparing PFGE and MLST as screening techniques have inferred distinct advantages associated with each technique, with MLST suggested as offering higher predictive power with unique isolates and PFGE as being more sensitive to detecting clonality [40]. In our study, we observed several situations where longitudinal isolates of P. aeruginosa from individual patients were found to have discrepant MLST profiles, with two or more alleles differing between the RE and the FE, resulting in the designation of “distinct, novel strain-type” despite a conserved pulsotype. In these cases, while clonality was inferred using PFGE, the extended duration between testing of the isolates may allow for random mutations in the housekeeping genes over time, resulting in the designation of a new strain. Overall, we found that the use of MLST and PFGE were complimentary and that the use of both techniques may offer the highest predictive resolution of clonality.
This study has a number of limitations. Most importantly, this was a multi-decade retrospective study of prospectively collected samples. Clinical data was derived from a referral centre and therefore does not include all relevant samples nor clinical data and, given the time span of the study, there is certainly potential for effect modification by time due to changing standards of care. Whilst all samples are derived from a single centre, our clinic is similar to other referral bronchiectasis clinics and is, to our knowledge, the only clinic that prospectively collects and stores pathogens derived from both CF and non-CF bronchiectasis patients. The small sample size of our study and the slow rate of lung function decline limit the ability to identify potential relationships between patterns of infection and clinical progression. We also acknowledge the limitation of evaluating P. aeruginosa from a minimum of two time points per patient, which may not represent the evolution of P. aeruginosa infection in all cases observed. To address this, we randomly genotyped a further 63 isolates using PFGE (initially 140 isolates) from various non-CF bronchiectasis patients who experienced chronic infection or strain-displacement at various time points. Despite screening more isolates the pattern of infection for each patient remained consistent, suggesting sampling bias has not influenced our findings. While every individual morphotype that was identified (on MacConkey agar) in real-time underwent retrospective genotyping at each time point in our cohort, these results may have been insensitive to detecting multi-lineage simultaneous infections which have been detected in a recent work [27]. However, this would require that multi-lineage infections have the exact same morphotype, something that has not been previously reported. Future prospective studies should consider sampling multiple examples of each identified morphotype, something not possible in a study drawing from a biobank such as ours.
Taking advantage of a regional biobank has presented our study with a unique opportunity to follow the history of P. aeruginosa infections in non-CF bronchiectasis, in some cases over 20 years. While a majority of non-CF bronchiectasis patients experience persistent colonisation by an individual P. aeruginosa strain, a higher than expected number of individuals were observed who experienced strain displacement over the course of their disease. Clinical progression was independent of the natural history of P. aeruginosa infection and strain displacement was not linked with episodes of PEx. We did not observe the existence of any novel non-CF bronchiectasis ePA or known CF ePA in non-CF bronchiectasis. Furthermore, the sharing of strains amongst CF and non-CF bronchiectasis cases was noted in only a few instances, which we suspect represents environmentally ubiquitous isolates.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Table S1 00162-2017_tableS1 (289.6KB, pdf)
Figure S1 00162-2017_figureS1 (5MB, jpg)
Acknowledgments
We would like to thank the staff of the Calgary Adult CF Clinic, Calgary Bronchiectasis Clinic and Calgary Laboratory Services for their assistance in these matters.
Footnotes
This article has supplementary material available from openres.ersjournals.com
Author contributions: T.E. Woo was responsible for collecting strains and served as the lead on the project. T.E. Woo, J.C. Bowron, M.G. Surette, B. Waddell and J. Duong performed strain typing. R. Lim, C.H. Mody and T.E. Woo were responsible for patient data collection. T.E. Woo, M.G. Surette and R. Somayaji performed statistical analyses. The manuscript was prepared by T.E. Woo but was revised by all authors. The Calgary Non-CF Bronchiectasis Biobank is maintained by H.R. Rabin and M.D. Parkins. D.G. Storey and M.D. Parkins envisioned the project together and serve as guarantors of the work.
Conflict of interest: R. Somayaji has received research fellowship funding from Cystic Fibrosis Canada, Alberta Innovates and the Canadian Institutes of Health Research, and grants from Cystic Fibrosis Canada, the Canadian Institute of Health Research and the Royal College of Physicians and Surgeons outside the submitted work. M.D. Parkins has received research funding from Cystic Fibrosis Canada, the Canadian Institute of Health Research, the Lung Association of Alberta and the Northwest Territories, Gilead Sciences, The University of Calgary and Calgary Laboratory Services.
Support statement: This work was supported by a grant to M.D. Parkins and D.G. Storey from the Lung Association of Alberta and the Northwest Territories. T.E. Woo was supported by a Natural Sciences and Engineering Research Council and Alberta Innovates Technology Futures studentship. R. Somayaji was supported by a Cystic Fibrosis Canada fellowship. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Pasteur MC, Bilton D, Hill AT. British Thoracic Society guideline for non-CF bronchiectasis. Thorax 2010; 65: i1–i58. [DOI] [PubMed] [Google Scholar]
- 2.Brill SE, Patel AR, Singh R, et al. Lung function, symptoms and inflammation during exacerbations of non-cystic fibrosis bronchiectasis: a prospective observational cohort study. Respir Res 2015; 16: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loebinger MR, Wells AU, Hansell DM, et al. Mortality in bronchiectasis: a long-term study assessing the factors influencing survival. Eur Respir J 2009; 34: 843–849. [DOI] [PubMed] [Google Scholar]
- 4.Davies G, Wells AU, Doffman S, et al. The effect of Pseudomonas aeruginosa on pulmonary function in patients with bronchiectasis. Eur Respir J 2006; 28: 974–979. [DOI] [PubMed] [Google Scholar]
- 5.Elborn JS, Bell SC. Pulmonary exacerbations in cystic fibrosis and bronchiectasis. Thorax 2007; 62: 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang AB, Bilton D. Exacerbations in cystic fibrosis: 4·Non-cystic fibrosis bronchiectasis. Thorax 2008; 63: 269–276. [DOI] [PubMed] [Google Scholar]
- 7.Aaron SD, Ramotar K, Ferris W, et al. Adult cystic fibrosis exacerbations and new strains of Pseudomonas aeruginosa. Am J Respir Crit Care Med 2004; 169: 811–815. [DOI] [PubMed] [Google Scholar]
- 8.Williams D, Evans B, Haldenby S, et al. Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am J Respir Crit Care Med 2015; 191: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkins MD, Glezerson BA, Sibley CD, et al. Twenty-five-year outbreak of Pseudomonas aeruginosa infecting individuals with cystic fibrosis: identification of the prairie epidemic strain. J Clin Microbiol 2014; 52: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitenstein S, Walter S, Bosshammer J, et al. Direct sputum analysis of Pseudomonas aeruginosa macrorestriction fragment genotypes in patients with cystic fibrosis. Med Microbiol Immunol 1997; 186: 93–99. [DOI] [PubMed] [Google Scholar]
- 11.Lam JC, Somayaji R, Surette MG, et al. Reduction in Pseudomonas aeruginosa sputum density during a cystic fibrosis pulmonary exacerbation does not predict clinical response. BMC Infect Dis 2015; 15: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somayaji R, Waddell B, Workentine ML, et al. Infection control knowledge, beliefs and behaviours amongst cystic fibrosis patients with epidemic Pseudomonas aeruginosa. BMC Pulm Med 2015; 15: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J 2012; 40: 227–238. [DOI] [PubMed] [Google Scholar]
- 14.Hilliam Y, Moore MP, Lamont IL, et al. Pseudomonas aeruginosa adaptation and diversification in the non-cystic fibrosis bronchiectasis lung. Eur Respir J 2017; 49: 1602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchelmore PJ, Randall J, Bull MJ, et al. Molecular epidemiology of Pseudomonas aeruginosa in an unsegregated bronchiectasis cohort sharing hospital facilities with a cystic fibrosis cohort. Thorax 2017; in press [https://doi.org/10.1136/thoraxjnl-2016-209889]. [DOI] [PubMed] [Google Scholar]
- 16.Chalmers JD, Aliberti S, Polverino E, et al. The EMBARC European Bronchiectasis Registry: protocol for an international observational study. ERJ Open Res 2016; 2: 00081–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Römling U, Kader A, Sriramulu DD, et al. Worldwide distribution of Pseudomonas aeruginosa clone C strains in the aquatic environment and cystic fibrosis patients. Environ Microbiol 2005; 7: 1029–1038. [DOI] [PubMed] [Google Scholar]
- 18.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis : criteria for bacterial strain typing. J Clin Microbiol 1995; 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curran B, Jonas D, Grundmann H. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 2004; 42: 5644–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Workentine ML, Surette MG, Bernier SP. Draft genome sequence of Burkholderia dolosa PC543 isolated from cystic fibrosis airways. Genome Announc 2014; 2: e00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015; 31: 587–589. [DOI] [PubMed] [Google Scholar]
- 22.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- 23.Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31: 3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somayaji R, Lam JC, Surette MG, et al. Long-term clinical outcomes of “prairie epidemic strain’ Pseudomonas aeruginosa infection in adults with cystic fibrosis. Thorax 2016; 72: 333–339. [DOI] [PubMed] [Google Scholar]
- 25.Lee TWR, Brownlee KG, Conway SP, et al. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J Cyst Fibros 2003; 2: 29–34. [DOI] [PubMed] [Google Scholar]
- 26.Jolley KA, Chan M-S, Maiden MCJ. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 2004; 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Don HJ, West SE, Rock MJ, et al. Pseudomonas aeruginosa in children with cystic fibrosis diagnosed through newborn screening: assessment of clinic exposures and microbial genotypes. Pediatr Pulmonol 2011; 45: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duff RM, Simmonds NJ, Davies JC, et al. A molecular comparison of microbial communities in bronchiectasis and cystic fibrosis. Eur Respir J 2013; 41: 991–993. [DOI] [PubMed] [Google Scholar]
- 29.De Soyza A, Perry A, Hall AJ, et al. Molecular epidemiological analysis suggests cross-infection with Pseudomonas aeruginosa is rare in non-cystic fibrosis bronchiectasis. Eur Respir J 2014; 43: 898–900. [DOI] [PubMed] [Google Scholar]
- 30.Winstanley C, O'Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 2016; 24: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo TE, Duong J, Jervis NM, et al. Virulence adaptations of Pseudomonas aeruginosa isolated from patients with non-cystic fibrosis bronchiectasis. Microbiology 2016; 162: 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Workentine ML, Sibley CD, Glezerson B, et al. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 2013; 8: e60225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Kosorok MR, Farrell PM, et al. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 2005; 293: 581–588. [DOI] [PubMed] [Google Scholar]
- 34.Kidd TJ, Ritchie SR, Ramsay KA, et al. Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting. PLoS One 2012; 7: e44199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heirali A, McKeon S, Purighalla S, et al. Assessment of the microbial constituents of the home environment of individuals with cystic fibrosis (CF) and their association with lower airways infections. PLoS One 2016; 11: e0148534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-Castillo M, Del Campo R, Morosini MI, et al. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J Clin Microbiol 2011; 49: 2905–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanini S, D'Arezzo S, Puro V, et al. Molecular epidemiology of a Pseudomonas aeruginosa hospital outbreak driven by a contaminated disinfectant-soap dispenser. PLoS One 2011; 6: e17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libisch B, Balogh B, Füzi M. Identification of two multidrug-resistant Pseudomonas aeruginosa clonal lineages with a countrywide distribution in Hungary. Curr Microbiol 2009; 58: 111–116. [DOI] [PubMed] [Google Scholar]
- 39.Knibbs LD, Johnson GR, Kidd TJ, et al. Viability of Pseudomonas aeruginosa in cough aerosols generated by persons with cystic fibrosis. Thorax 2014; 69: 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waters V, Zlosnik JEA, Yau YCW, et al. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 2012; 31: 3341–3350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Table S1 00162-2017_tableS1 (289.6KB, pdf)
Figure S1 00162-2017_figureS1 (5MB, jpg)