Abstract
Lactobacillus paracasei is common in the non-starter lactic acid bacteria (LAB) community of raw milk cheeses. This species can significantly contribute to flavor formation through amino acid metabolism. In this study, the DNA and RNA of L. paracasei FAM18149 were sequenced using next-generation sequencing technologies to reconstruct the metabolism of the sulfur-containing amino acids cysteine and methionine. Twenty-three genes were found to be involved in cysteine biosynthesis, the conversion of cysteine to methionine and vice versa, the S-adenosylmethionine recycling pathway, and the transport of sulfur-containing amino acids. Additionally, six methionine-specific T-boxes and one cysteine-specific T-box were found. Five of these were located upstream of genes encoding transporter functions. RNA-seq analysis and reverse-transcription quantitative polymerase reaction assays showed that expression of genes located downstream of these T-boxes was affected by the absence of either cysteine or methionine. Remarkably, the cysK2-ctl1-cysE2 operon, which is associated with te methionine-to-cysteine conversion and is upregulated in the absence of cysteine, showed high read coverage in the 5′-untranslated region and an antisense-RNA in the 3′-untranslated region. This indicates that this operon is regulated by the combination of cis- and antisense-mediated regulation mechanisms. The results of this study may help in the selection of L. paracasei strains to control sulfuric flavor formation in cheese.
Keywords: Lactobacillus paracasei, sulfur amino acid metabolism, cysteine, methionine, RNA-seq, differential gene expression
Introduction
Lactic acid bacteria (LAB), which can ferment diverse materials such as milk, meat, and plants, are widely used in food fermentation. Lactobacillus paracasei is of particular interest as it is found in a variety of habitats, including the human body, and fermented food. It is often found in cheese at the end of ripening, and the use of this species in adjunct cultures can improve ripening and flavor development (Beresford and Williams, 2004).
Volatile sulfur compounds (VSCs) are key flavor compounds in cheese mainly derived from microbial metabolism of sulfur-containing amino acids (Landaud et al., 2008). Various strains of L. paracasei produce VSCs when incubated with amino acids in vitro (Irmler et al., 2006). The production of VSCs is probably associated with side activities of enzymes involved in cysteine and methionine biosynthesis. This hypothesis is supported by the observation that the recombinant produced cysteine synthase CysK and the C-S lyases MalY, MetC and Ctl1 of L. paracasei produce hydrogen sulfide from cysteine and/or homocysteine (Irmler et al., 2008, 2009; Bogicevic et al., 2012a). Moreover, MetC and Ctl1 release methanethiol from methionine. Consequently, these enzymes could play a role in VSC formation in cheese. However, it is not known whether these genes are expressed in a cheese environment. A deeper understanding of the regulation of these genes can be helpful for a rational selection of strains to control VSC formation in cheese.
Comparative genomics has been used to identify the regulation mechanisms of cysteine and methionine biosynthesis in Bacillales, Lactobacillales, Lactococcaceae, and Streptocococcaceae. Regulation in streptococci is predicted to involve DNA-dependent systems and transcription factors of the LysR family (Rodionov et al., 2004; Kovaleva and Gelfand, 2007). Sulfur-metabolism genes in Lactococcus lactis are also regulated by a LysR-type transcription factor (Sperandio et al., 2005). In contrast, genes involved in methionine biosynthesis and transport in lactobacilli are predicted to be regulated by T-box leader sequences, which are cis-acting RNA regulatory elements that interact with tRNAs (Rodionov et al., 2004; Gutiérrez-Preciado et al., 2009; Liu et al., 2012). The binding of the tRNA molecule results in a change in the secondary structure of the leader sequence, which determines whether downstream genes will be expressed. A similar regulatory mechanism show S-box regulons (Grundy and Henkin, 1998), which are conserved RNA motifs that bind S-adenosylmethionine (SAM). A SAM-responsive regulatory element has been identified upstream of metK, which encodes SAM synthetase, in various LAB (Fuchs et al., 2006). The S-box regulon is the key regulator of cysteine and methionine metabolism in Bacillus and Clostridium organisms (Grundy and Henkin, 1998).
A previous study has uncovered that the expression of the cysK2-ctl1-cysE2 gene cluster involved in the conversion of methionine to cysteine in L. paracasei FAM18149 was repressed in the presence of cysteine (Bogicevic et al., 2012b). This implied that cysteine may be an effector molecule that modulates gene expression. The present study extends to the overall response of L. paracasei FAM18149 to cysteine and methionine using next-generation sequencing technologies, bioinformatic analysis, and PCR-based methods. The expression of genes involved in cysteine and methionine metabolism of L. paracasei FAM18149 grown in a chemically defined medium (CDM) with cysteine as the sole sulfur source was compared to that of cells grown in CDM with methionine as the sole sulfur source.
Material and methods
Bacterial strain, media, and growth conditions
Lactobacillus paracasei FAM18149 was obtained from the Agroscope culture collection in Liebefeld (Bern, Switzerland). The strain was stored at −80°C in 10% sterile reconstituted skim milk powder and maintained at 30°C in MRS broth (De Man et al., 1960).
Pacbio sequencing, genome assembly, and annotation
Genomic DNA (gDNA) was extracted from L. paracasei FAM18149 using the EZ1 DNA Tissue Kit (Qiagen, Hombrechtikon, Switzerland) according to the manufacturer's instructions. Before extraction, the bacterial cells were first treated with 0.1 M sodium hydroxide for 15 min at room temperature and then with lysozyme (50 mg dissolved in 0.1 M Tris[hydroxymethyl]-aminomethane, 10 mM ethylenediaminetetraacetic acid, 25% [w/v] sucrose, pH 8.0) for 1 h at 37°C.
The gDNA was sheared in a Covaris g-TUBE (Covaris, Woburn, MA, USA) to obtain 20-kb fragments and the size distribution of fragmented gDNA was analyzed on a Fragment Analyzer (Advanced Analytical Technologies, Ames, IA, USA). The sheared gDNA (5 μg) was used to prepare a SMRTbell library with the PacBio SMRTbell Template Prep Kit 1 (Pacific Biosciences, Menlo Park, CA, USA) according to the manufacturer's recommendations. The resulting library was size-selected on a BluePippin system (Sage Science, Inc. Beverly, MA, USA) for molecules larger than 14 kb. The recovered library was sequenced on one SMRT cell with P6/C4 chemistry and MagBeads on a PacBio RSII system (Pacific Biosciences, Menlo Park, CA, USA) at a 240-min movie length. Sequencing yielded 101,520 reads corresponding to 1,405 Mb with a mean read length of 13,840 bases. The resulting reads were assembled using the HGAP 3 (SMRT Analysis v-2.2.0) standard procedure (Chin et al., 2013). All scaffolds were annotated using the NCBI Prokaryotic Genome Annotation Pipeline. Coding sequences (CDSs) of interest were also searched against GenBank, and, in case of putative transporters, against the Transporter Classification Database (Saier et al., 2014).
Detection of regulatory sequences
T-box leader sequences were identified with Infernal (version 1.1rc4) (Nawrocki and Eddy, 2013), which is included in the Prokka pipeline (Seemann, 2014). To identify methionine-specific T-box leader sequences, the T-box leader sequences were aligned to methionine-specific T-boxes of the yxjH gene from Lactobacillus rhamnosus (Lebeer et al., 2007). To identify cysteine-specific T-box leader sequences, T-boxes were aligned to cysteine-specific T-box leader sequences preceding ubiG (cpe0175), cysP1 (cpe0947), cysP2 (cpe0967), and cysK (cpe1322) of Clostridium perfringens strain 13 (André et al., 2010). The Clustal Omega algorithm was used for the alignments (Sievers et al., 2011). The conserved structural elements, including AGTA box, GNTG box, F-box, T-box, and specifier codons were identified based on the annotation of the aforementioned leader sequences. The SMK box was identified by aligning the sequence described by Fuchs et al. (2006). Additional analyses were performed using RegRNA 2.0 (Chang et al., 2013).
RNA isolation, rRNA depletion, and RNA sequencing (RNA-seq)
L. paracasei FAM18149 was grown in a chemically defined medium (CDM) described for L. helveticus (Christensen and Steele, 2003), omitting either cysteine (cysteine-deficient CDM) or methionine (methionine-deficient CDM) for 25 h at 30°C. The optical density of the culture was determined at 600 nm (OD600) with a spectrophotometer (LKB Biochrom 4050 Ultrospec II). An aliquot of approximately 10 OD was collected by centrifugation (3000 g, 10 min). RNA was isolated using the TRIzol®Max™ Bacterial RNA Isolation Kit (Life Technologies, Zug, Switzerland). Instead of a conventional phase separation, Direct-zol™RNA MiniPrep centrifugation columns (Zymo Research Corp., Irvine, USA) were used according to the manufacturer's instructions. For rRNA depletion, 1 μg of total RNA was used with the RiboMinusTM Eukaryote System v2 protocol (ThermoFisher Scientific, Zug, Switzerland). The RiboMinusTM Eukaryote probes provided by the kit were replaced by bacterial probes containing two 16S rRNA and three 23S rRNA sequence-specific 5′-biotin-labeled oligonucleotides, which were provided by ThermoFisher Scientific (Chen and Duan, 2011). Barcoded cDNA libraries were prepared using the IonExpressTM RNA-Seq Barcode 1-16 Kit and the Ion Total RNA-seq v2 Kit (ThermoFisher Scientific). Two barcoded libraries were pooled and used for template preparation and enrichment with the Ion OneTouch™ 200 Template Kit v2 (ThermoFisher Scientific). Sequencing was performed on the Ion PGM™ using the Ion 318™ Chip and Ion PGM™ 200 Sequencing v2 Kits (ThermoFisher Scientific). The RNA was isolated from three biological repeats.
Differential expression analysis
Reads shorter than 20 bp were removed using Trimmomatic version 0.36 (Bolger et al., 2014). Remaining reads were mapped to the genome assembly of L. paracasei FAM18149 (GenBank: GCA_002442835.1) using Bowtie2 (version 2.2.1, default parameters) (Langmead and Salzberg, 2012). For the differential expression analysis, the reads were counted using HTSeq (Anders et al., 2014), and only the reads that aligned to a predicted CDS were included (version 0.6.1, options: -a 1 -m intersection-nonempty). Finally, the dataset was normalized (size factor normalization) and tested (Wald test) for differential gene expression using DEseq2 (version 1.6.3) (Love et al., 2014). The resulting p-values were corrected for multiple testing using the Benjamini-Hochberg approach (Benjamini and Hochberg, 1995).
Analysis of transcriptional organization
To visualize the transcriptional organization of significantly differentially expressed genes, the reads of all RNA-seq data sets were combined and aligned to the complete genome assembly of L. paracasei FAM18149 using Bowtie2 (version 2.2.1, default parameters). The read depth was determined using the Genome Analysis ToolKit (version 3.3.0) (McKenna et al., 2010) and plots for gene regions of interest were created with ggplot2 (Wickham, 2009).
Gene set enrichment analysis
The CDSs of strain FAM18149 were searched against the SWISSPROT database (March 2017) using BLAST (version 2.2.31+), and against the Pfam database (March 2017) using pfam_search (version 1.6). The Gene Ontology (GO) terms obtained by these searches were assigned to the respective CDSs. The GO enrichment analysis was performed by comparing the genes that showed an adjusted p-value of < 0.05 on the differential expression analysis to the rest of the genome as background using the elim method of topGO (Alexa and Rahnenführer, 2018). All GO terms that had a p-value of ≤ 0.01 were considered to be significantly enriched.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Lactobacillus paracasei FAM18149 that had been grown for 25 h at 30°C either in cysteine-deficient or methionine-deficient CDM was collected by centrifugation. The cells were disrupted with zirconium beads (100 μm; OPS Diagnostics, Lebanon) using an Omni Bead Ruptor (6 ms−1 for 30 s; Labforce AG, Muttenz, Switzerland) and the RNA was isolated using the NucleoSpin® RNA plus Kit (Macherey-Nagel, Oensingen, Switzerland) according to manufacturer's instructions. To remove residual DNA, RNA samples were treated with DNaseI (Zymo Research Corp., Irvine, USA) according to the manufacturer's protocol. The RNA concentration was determined using the Qubit RNA BR Assay Kit (ThermoFisher Scientific). Total RNA (260 ng) was then used for single-stranded cDNA synthesis using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, USA) according to the manufacturer's protocol.
QPCR was carried out using a MIC qPCR Cycler (Bio Molecular Systems, Upper Coomera, Australia). The PCR reactions for ctl1, cysK, and recA contained hydrolysis probes. The PCR reactions (12 μL) contained Takyon No Rox Probe MaterMix UNG (Eurogentec, Seraing, Belgium), forward primer, reverse primer, hydrolysis probe, and 2 μL of cDNA that had been diluted in water (1:25). The primer and probe concentrations for ctl1 and cysK were as described previously (Bogicevic et al., 2012a,b). For recA, 300 nM of recA_F, 300 nM of recA_R, and 100 nM of recA_FAM_probe were used. The other PCR reactions (12 μL) were based on DNA-binding dyes and contained Takyon No Rox SYBR MasterMix blue dTTP (Eurogentec), 300 nM of forward primer, 300 nM of reverse primer, and 2 μL of single-stranded cDNA that had been diluted in water (1:25). The primer sequences are listed in Table 1. The amplification conditions were 2 min at 50°C, 3 min at 95°C, followed by 40 cycles of 2 s at 95°C and 20 s at 60°C.
Table 1.
Sequences of primers and fluorogenic probes used in this study.
| Primer | Sequence (5′-3′) | Target | Source or references |
|---|---|---|---|
| metQ1_F | CAAGCCAATCTGAAGCACTTAAAG | metQ1 | This study |
| metQ1_R | ATCAGTTCTGCGTCCAAATCG | ||
| cysKI_F | CCGGCGGTTCTGTCAAAG | cysK | Bogicevic et al., 2012a |
| cysKI_R | CCCTTGTATTCGGCATCTTCA | ||
| cysKI_FAM_probe | CCGAATTGCCTTGGCCATG | ||
| metC_F | GCAGCGTCAAGGCTATTAGCA | metC | This study |
| metC_R | AGGAACGTGTTATCGACGATTGT | ||
| yckJ_F | TTGGTTCTGCGTGAAATTATCATT | yckJ | This study |
| yckJ_R | CCTTGACGAGACTGATAAAACTGTTT | ||
| metE_F | TGGCCTCAAGACTCGTGATG | metE | This study |
| metE_R | CCGCTGCCACCATGTTG | ||
| yxjH_F | ACTTGGCTTCAAAGCTGTGACA | yxjH | This study |
| yxjH_R | AACCGTTCAAGCCCCATAAA | ||
| metK_F | GCAAGCCGACTCTGGTTTG | metK | This study |
| metK_R | AAAAGGCACCACCACCATGT | ||
| metQ3_F | GATATCGGCGCAACCTACATC | metQ3 | This study |
| metQ3_R | TCGCCGTTCTTAACGTCCTT | ||
| Q-ctl1_F | GCACTGGAAAGCTTGATCGAA | ctl1 | Bogicevic et al., 2012b |
| Q-ctl1_R | ACCGAATGTCACGTGGAATTG | ||
| Q-ctl1_FAM_probe | CGGCCTTGATGACCCACGGC | ||
| glnP2_F | TACGCGTTTCCAGTGATCGG | glnP2 | This study |
| glnP2_R | TGTAACTGCCGCCGAGAAAT | ||
| glnM2_F | CAGAATTGTGCCAACCGTCG | glnM2 | This study |
| glnM2_R | CAAGGTGGCCAATTGATCGC | ||
| artM2_F | AACAGGACAAGTTCGCTGCT | artM2 | This study |
| artM2_R | GGACAGTTGCCGGGGATATG | ||
| glnH4_F | ATAAGGCGATCACCAAGGCC | glnH4 | This study |
| glnH4_R | AGGCTGCTTGGACGTGAAAT | ||
| recA_F | TTATGCGAATGGGTGCTAAGG | recA | This study |
| recA_R | CCAACACCAAGTGCATCATCA | ||
| recA_FAM _probe | CGTTTCCGTTGTCTCTAGCGGCTCACT | ||
| rpoB_F | GCTGAGCACACACGGGAAAT | rpoB | This study |
| rpoB_R | CAACTGCCACACTGGAAGCA |
All qPCR reactions were done in triplicates. Water was used as control. The crossing threshold cycle (Cq) was determined using the micPCR software v 2.4.0. The experiments were repeated three times.
Results
Reconstruction of cysteine and methionine metabolism
The assembly of the PacBio reads obtained from the sequencing of gDNA from L. paracasei FAM18149 resulted in six circular scaffolds (Supplementary Table S1). The largest scaffold, which represents the chromosome, comprised 2.7 Mbp whereas the plasmid scaffolds ranged from 28 to 84 kbp. The average read depth analysis indicated that three of the five plasmids were present in lower copy numbers.
Since the FAM18149 strain can grow in medium containing either sulfide, cysteine, or methionine as the sole sulfur source (Irmler et al., 2008; Bogicevic et al., 2016), the pathways for the conversion of cysteine to methionine and vice versa should be present. The FAM18149 genome was therefore searched for genes involved in the metabolism of these sulfur-containing amino acids. For readability purposes, the gene names are used in this report. The respective locus_tags are given in Table 2.
Table 2.
DESeq2 results for genes associated with the metabolism of sulfur-containing amino acids in L. paracasei FAM18149.
| Gene (locus_tag)a | Functionb | Induction methionine-deficient vs. cysteine-deficient CDM (log2 fold change) | Adjustedp-value | Transcriptional unit (regulatory element) |
|---|---|---|---|---|
| metQ1 (00255) | MULTISPECIES: substrate-binding protein of an ABC superfamily methionine transporter | −0.1 | 1.0 | metQ1 (T-boxmet) |
| cysE (00525) | serine O-acteyltransferasec | −1.2 | 4.1E-08 | cysE-cysK (n. d.) |
| cysK (00530) | MULTISPECIES: cysteine synthase A | −1.1 | 1.5E-09 | cysE-cysK (n. d.) |
| metC (00830) | cystathionine gamma-lyased | 1.5 | 5.7E-07 | yckJ-yckK-metC (T-boxmet) |
| yckK (00835) | MULTISPECIES: amino acid ABC transporter substrate-binding protein | 1.3 | 2.6E-04 | yckJ-yckK-metC (T-boxmet) |
| yckJ (00840) | MULTISPECIES: cysteine ABC transporter permease | 0.7 | 3.3E-01 | yckJ-yckK-metC (T-boxmet) |
| metF (00930) | MULTISPECIES: 5,10-methylenetetrahydrofolate reductase | 2.2 | 5.5E-19 | metE-metF (T-boxmet) |
| metE (00935) | MULTISPECIES: 5-methyltetrahydropteroyltriglutamate–homocysteine S-methyltransferase | 2.5 | 4.0E-36 | metE-metF (T-boxmet) |
| yxjH (01620) | MULTISPECIES: vitamin-B12 independent methionine synthase | 1.3 | 3.9E-07 | yxjH-luxS (T-boxmet) |
| luxS (01625) | MULTISPECIES: S-ribosylhomocysteine lyase / autoinducer-2 production protein LuxS | 0.4 | 7.2E-01 | yxjH-luxS (T-boxmet)e |
| malY (02475) | MULTISPECIES: putative C-S lyase | 0.3 | 0.7 | malY (n. a.) |
| metK (02170) | MULTISPECIES: methionine adenoysltransferase | 0.2 | 1.0 | metK (SMK box) |
| metQ2 (04115) | MULTISPECIES: MetQ/NlpA family ABC transporter substrate-binding protein | 0.7 | 1.0 | metQ2 (T-boxmet) |
| metQ3 (04120) | MULTISPECIES: MetQ/NlpA family ABC transporter substrate-binding protein | 1.2 | 5.4E-08 | metQ3-metN-metP (T-boxmet) |
| metN (04125) | MULTISPECIES: ATP-binding protein of an ABC superfamily methionine transporter | 1.1 | 9.9E-08 | metQ3-metN-metP (T-boxmet) |
| metP (04130) | MULTISPECIES: ABC transporter permease | 1.0 | 7.9E-04 | metQ3-metN-metP (T-boxmet) |
| pfs (04610) | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | −0.3 | 0.9 | pfs (n. a.) |
| glnP2 (14340) | MULTISPECIES: polar amino acid ABC transporter inner membrane subunit | −2.5 | 1.6E-19 | glnP2-glnM2-artM2-glnH4 (T-boxcys) |
| glnM2 (14345) | MULTISPECIES: amino acid ABC transporter permease | −2.5 | 3.5E-17 | glnP2-glnM2-artM2-glnH4 (T-boxcys) |
| artM2 (14350) | MULTISPECIES: polar amino acid transport system ATP-binding protein | −2.6 | 1.5E-21 | glnP2-glnM2-artM2-glnH4 (T-boxcys) |
| glnH4 (14355) | MULTISPECIES: amino acid ABC transporter substrate-binding protein | −2.9 | 1.6E-33 | glnP2-glnM2-artM2-glnH4 (T-boxcys) |
| cysE2 (14655) | serine acetyltransferase | −2.9 | 2.0E-22 | cysE2-ctl1-cysK2 (RIT, antisense RNA) |
| ctl1 (14660) | cystathionine beta and gamma-lyase | −3.2 | 8.3E-42 | cysE2-ctl1-cysK2 (RIT, antisense RNA) |
| cysK2 (14665) | MULTISPECIES: cystathionine beta-synthase | −3.1 | 5.2E-34 | cysE2-ctl1-cysK2 (RIT, antisense RNA) |
n. d, not detected; n. a, not analyzed; RIT, rho-independent terminator.
Common gene names from Bacillus subtilis and Escherichia coli are used. Gene names for transporter are based on searches of the Transporter Classification Database (Saier et al., 2014). Numbers in brackets represent the locus_tag, of which the prefix FAM18149_ was omitted.
Functions assigned based on BLAST searches.
The gene name is designated as metA in L. paracasei ATCC 334. Since the gene in fact encodes a serine acetyltransferase (Bogicevic et al., 2016), the gene name cysE is used in this study.
The recombinant-produced protein also exhibited cystathionine gamma-synthase activity in vitro (Irmler et al., 2008).
The putative promoter 5′-TTAACA-N18-TATGAT-3′, which was identified in Lactobacillus rhamnosus GG (Lebeer et al., 2007), is also located upstream of luxS in L. paracasei FAM18149, indicating a T-box-independent expression of the respective gene.
A scheme of the reconstructed pathways is illustrated in Figure 1. CysE and cysK encode the enzymes that use serine, acetyl-CoA, and sulfide to produce cysteine. MetC and malY encode enzymes that form the transsulfuration pathway that converts cysteine to homocysteine. CysK2, ctl1, and cysE2 are predicted to encode cystathionine beta-synthase, cystathionine lyase, and serine acetyltransferase, respectively, which are related to the conversion of methionine to cysteine. Based on these putative functions and the ability of recombinant Ctl1 to release sulfide from homocysteine, two pathways are proposed. In one, the sulfide produced by Ctl1 activity is used by CysE2 and CysK2 for cysteine biosynthesis. In the other, CysK2 could encode a cystathionine beta-synthase that synthesizes cystathionine from homocysteine, and Ctl1 subsequently cleaves cystathionine into cysteine and homoserine.
Figure 1.
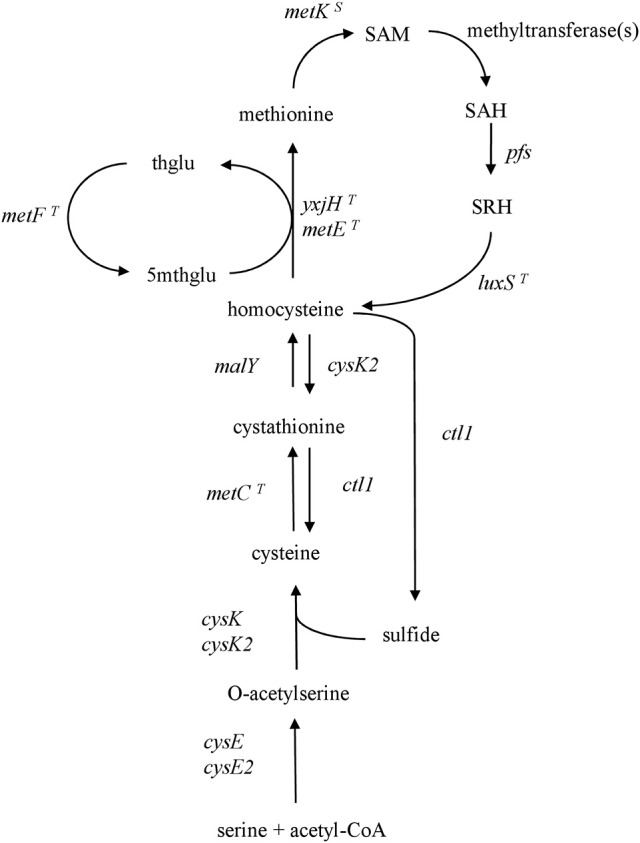
Reconstruction of cysteine and methionine metabolism in L. paracasei FAM18149. The gene names corresponding to locus-tags are presented in Table 2. A T or S in superscript indicates that the respective gene is probably regulated, respectively, by a T- or S-box regulation mechanism. thglu: tetrahydrofolate, 5mthglu: N5-methyl-tetrahydrofolate, SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SRH, S-ribosylhomocysteine.
For methionine synthesis from homocysteine, the vitamin-B12 independent methionine synthases MetE and YxjH were identified. The methylation of homocysteine to yield methionine requires 5-methyltetrahydrofolate, which is provided by MetF activity. Methionine is used by MetK for SAM synthesis, the universal methyl donor. The latter compound is regenerated to methionine via the SAM recycling pathway, which involves methyltransferases, Pfs, and LuxS.
In addition, a search was performed for T-boxes known to be modulated by cysteine or methionine. A total of 25 T-boxes were detected, of which 24 were located on the chromosome and one on a plasmid (Supplementary Table S2). By aligning the T-box sequences to the methionine-specific T-box preceding yxjH of L. rhamnosus GG, six T-Box leader sequences with the specifier codon ATG were identified (Figure 2A). These T-boxes were upstream of CDSs, encoding ABC transporter substrate-binding proteins (locus-tags FAM18149_00255, FAM18149_00840, FAM18149_04115, and FAM18149_04120), MetE and YxjH. With regard to cysteine-specific T-boxes, the specifier codon TGC was identified in in a T-box located on a plasmid (Figure 2B). The T-box preceded a gene encoding an amino acid ABC-transporter inner-membrane subunit (locus_tag FAM18149_14340). To label the transporter encoding CDSs with gene names, the deduced amino acid sequences were analyzed using the Transporter Classification Database (Saier et al., 2014). Based on the outcome of this search the locus_tags were given gene names as indicated in Table 2. With regard to S-box elements, the SMK regulon that has previously been described for L. casei (Fuchs et al., 2006) was present in the 5′-untranslated region (UTR) of the metK gene (data not shown).
Figure 2.
Alignment of methionine-specific T-boxes (A) and cysteine-specific T-boxes (B) of L. paracasei FAM18149 with T-boxes of L. rhamnosus (LGG_yxjH) and C. perfringens (cpe0175, cpe0947, cpe0967, and cpe1322). The conserved structural motifs (AGTA box, GNTG box, specifier codon, F-box, and T-box) are highlighted in gray. Invariant nucleotide sites are marked with asteriks.
Differential expression analysis
RNA from L. paracasei FAM18149 that had been grown in the absence of either cysteine or methionine was sequenced using semiconductor sequencing technology. The number of reads obtained from repeated transcriptomes of this strain varied from 921,890 to 3,126,855 (Supplementary Table S3), of which an average of 7.0% displayed an alignment quality of < 1 (low quality). Although the rRNA depletion step was applied, an average of 43% of the reads still mapped to rRNA genes and another 9.9% were aligned to tRNAs. Of the remaining reads, an average of 12.1 and 18.0%, respectively, mapped to non-coding features and CDSs of the L. paracasei FAM18149 genome.
Differential expression analysis using the DEseq2 algorithm identified 60 significantly (adjusted p-value < 0.05) differentially expressed protein-coding loci (Supplementary Table S4). With regard to the 24 genes predicted from the genomic data to be involved in cysteine and methionine metabolism (Table 2), nine genes (cysE, cysK, glnP2, glnM2, artM2, glnH4, cysE2, clt1, and cysK2) had higher expression in the cysteine-deficient medium, while eight others (metC, yckK, metF, metE, yxjH, metQ2, metN, and metP) had higher expression in methionine-deficient medium. It is noteworthy, that metQ1 and metQ2, which are located downstream of two methionine-specific T-boxes, did not show significant differential regulation according to the DESeq2 analysis. A gene set enrichment analysis was also performed on the 60 significantly regulated genes, and nine GO terms were found that belonged to cysteine and methionine metabolism (Supplementary Table S5).
Transcriptional organization
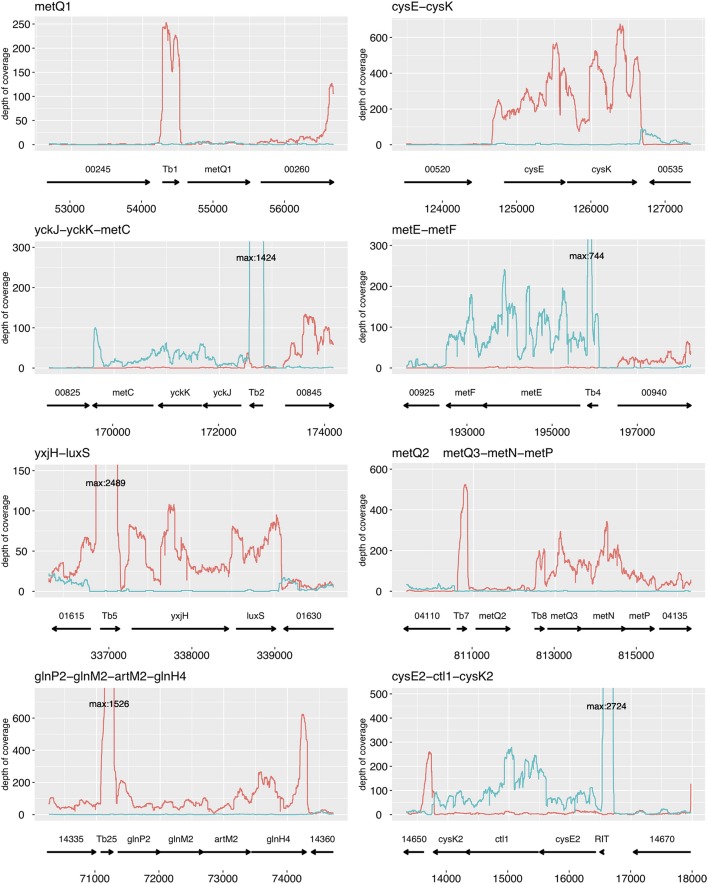
To identify operons, all six RNA-seq datasets were mapped onto the corresponding regions including the upstream and downstream loci (Figure 3). Since the strand information was retained, the reads could be assigned to the direction of transcription relative to the sense or antisense information of the loci. This process identified the cotranscription of the following genes: cysE-cysK, yckJ-yckK-metC, metE-metF, yxjH-luxS, metQ3-metN-metP, glnP2-glnM2-artM2-glnH4, and cysE2-ctl1-cysK2. Remarkably, the T-box regions displayed considerable high coverage (Figure 3). High coverage of the 5′-UTR was also oberserved for the cysK2-ctl1-cysE2 operon, and antisense-RNA was found in the 3′-UTR of this operon (Figure 3).
Figure 3.
Identification of operons that are involved in cysteine/ methionine biosynthesis and transport in L. paracasei FAM18149. The plots illustrate the sequence coverage per base obtained from six RNA-seq datasets. The orientation of the sequence reads is represented in red (forward) and blue (reverse). The arrows represent genes and T-boxes. The numbers below the arrows indicate the genomic position.
Targeted gene expression analysis using RT-qPCR
The expression of 13 genes associated with the metabolism of sulfur-containing amino acids was analyzed using RT-qPCR as an alternative method (Table 3). The recA and rpoB genes, encoding the recombinase recA and the beta subunit of the DNA-directed RNA polymerase, respectively, were used as “constitutively” expressed control genes. The selected genes confirmed the results obtained from the DESeq2 analysis except for metQ1 and cysK. MetQ1 had slightly higher expression in the methionine-deficient medium compared to the cysteine-deficient medium, while no significant differential expression was found for cysK.
Table 3.
Targeted gene expression analysis using RT-qPCR.
| Gene (locus tag)a | Cysteine-deficient CDM | Methionine-deficient CDM |
|---|---|---|
| recA (02620) | 23.8 ± 0.6 | 23.6 ± 0.6 |
| rpoB (10445) | 18.3 ± 0.9 | 19.3 ± 0.9 |
| metQ1 (00255) | 25.5 ± 0.4 | 24.2 ± 0.5 |
| cysK (00530) | 22.3 ± 0.6 | 23.0 ± 0.8 |
| metC (00830) | 20.8 ± 0.4 | 18.0 ± 0.4 |
| yckJ (00840) | 24.6 ± 0.4 | 21.9 ± 0.5 |
| metE (00935) | 21.0 ± 0.4 | 17.7 ± 0.4 |
| yxjH (01620) | 22.1 ± 0.4 | 20.2 ± 0.8 |
| metK (02170) | 18.1 ± 1.1 | 18.2 ± 0.8 |
| metQ3 (04120) | 21.3 ± 0.3 | 19.8 ± 0.6 |
| glnP2 (14340) | 19.2 ± 0.3 | 22.9 ± 0.7 |
| glnM2 (14345) | 19.3 ± 0.8 | 22.4 ± 0.5 |
| artM2 (14350) | 18.7 ± 0.5 | 22.3 ± 0.5 |
| glnH4 (14355) | 17.6 ± 0.4 | 21.0 ± 0.8 |
| ctl1 (14660) | 21.4 ± 0.6 | 25.7 ± 0.6 |
The numbers represent the average of the quantification cycle value and standard deviation obtained from three independent experiments.
Numbers in brackets represent the locus_tag, of which the prefix FAM18149_ was omitted.
Discussion
Bacteria are known to synthesize the sulfur-containing amino acids cysteine and methionine from serine and homoserine, respectively. Furthermore, bacteria possess transport systems to take up amino acids from the environment. Cysteine and methionine biosynthesis has been well studied in Escherichia coli, in which the consecutive actions of serine acetyltransferase and O-acetylserine sulfhydrylase, which are respectively encoded by cysE and cysK, convert serine to cysteine (Kredich, 1996). Cysteine is then converted via cystathionine into methionine, which involves the activity of a cystathionine gamma-synthase (MetB) and a cystathionine beta-lyase (MetC). Bioinformatic analysis of the first published L. paracasei ATCC 334 (formerly Lactobacillus casei) genome indicated that this species does not possess orthologs of cysE and metB and could be auxotrophic for methionine (Makarova et al., 2006). In contrast, various L. paracasei strains including ATCC 334 grow in the absence of methionine (Irmler et al., 2008). By cloning and heterologously expressing genes that encode putative C-S lyases, it was found that the gene annotated as metC in strain ATCC 334 not only degraded but also synthesized cystathionine, depending on the in vitro conditions (Irmler et al., 2008). In addition, the gene annotated as metA in L. paracasei does not encode a homoserine O-succinyltransferase but does encode a serine acetyltransferase (Bogicevic et al., 2016). Thus, the majority of L. paracasei strains likely possess pathways for cysteine and methionine synthesis that are similar to those of E. coli. The genome analysis of strain FAM18149 also revealed the presence of all genes necessary for the SAM recycling.
Interestingly, few bacteria, including Mycobacterium tuberculosis, Klebsiella pneumoniae, Bacillus subtilis, and Clostridium acetobutylicum, possess a reverse transsulfuration pathway (Wheeler et al., 2005; Seiflein and Lawrence, 2006; Hullo et al., 2007; André et al., 2008). This pathway converts homocysteine to cysteine via the intermediate cystathionine, and requires a cystathionine beta-synthase (CBS) and a cystathionine gamma-lyase (CGL). Since L. paracasei FAM18149 can grow in cysteine-deficient medium, it is likely that it possesses the reverse transsulfuration pathway. Ctl1 could represent the cystathionine lyase needed for this pathway (Irmler et al., 2009). However, to the best of our knowledge, the required CBS activity has not yet been shown in L. paracasei. Consequently, it is proposed that as an alternative to the reverse transsulfuration pathway,the FAM18149 strain may break down homocysteine to release sulfide that is then used for cysteine biosynthesis. Figure 1 illustrates a schematic model of cysteine and methionine biosynthesis in L. paracasei FAM18149.
With regard to regulation mechanisms, six methionine-specific T-boxes and one cysteine-specific T-box were found. Rodionov et al. (2004) analyzed the unfinished genome of L. paracasei ATCC 334 and predicted methionine-specific T-boxes for metE-metF, yxjH-luxS, metQ2, and metQ3NP. Later, Gutiérrez-Preciado et al. (2009) identified a fifth methionine-specific T-box upstream of an operon designated as glnH-glnH-yjcJ in L. casei, which probably represents the yckJK-metC operon described in the present study. Five T-boxes of FAM18149 were located upstream of CDSs encoding transporter subunits. Identification of the specifier codon of T-boxes can be used to predict the substrate specificity of the downstream genes that encode transporters (Wels et al., 2008). Therefore, the genes metQ1, and metQ2 and the operon metQ3NP can be associated with methionine transport. Since the metQ3NP operon also showed homology to the metNPQ transport system of Bacillus subtilis that transports L- and D-isomers of methionine as well as an oxidation product of methionine (Hullo et al., 2004), it is assumed that the gene products are actually involved in methionine import. MetQ1 and metQ2 are predicted to encode periplasmatic subunits of transport systems. Further studies are necessary to identify the transmembrane domains (TMDs) to which these bind.
The yckJK-metC operon is of special interest. BLAST searches of GenBank and the Transporter Classification Database indicated that the gene products of yckJ and yckK form a cystine/cysteine ABC transport system. However, the genes are regulated by a methionine-specific T-box, predicting methionine as substrate. Structural analysis of various bacterial ABC transporters revealed that ABC transport systems generally consist of periplasmatic subunits that capture the substrate, subunits with TMDs, and membrane-associated subunits that possess nucleotide-binding domains (NDBs) (Wilkens, 2015). NDBs bind and hydrolyze ATP which is necessary for the transport mechanism. With regard to yckJK, metC is present instead of an NDB subunit. Based on the observation that MetC hydrolyzed O-succinyl-L-homoserine in vitro (Irmler et al., 2009), it is thought that this hydrolysis could drive the transport. Therefore, we assume that yckJK actually transports cysteine, which is subsequently used by MetC for cystathionine synthesis, the precursor for methionine biosynthesis.
The presence of a cysteine-specific T-boxs upstream of the glnP2-glnM2-artM2-glnH4 operon indicates that the gene products also form a cysteine ABC transport system. Sequence analysis of the CDSs encoded by this operon showed a relationship to glutamine/glutamate transporters. As the operon was more highly expressed in the absence of cysteine, it may indeed be involved in the transport of cysteine or of the precursors needed for cysteine biosynthesis. This transport system was also found on a plasmid in L. paracasei ATCC 334 (GenBank acc. no.CP000424) but not in other L. paracasei genomes in the GenBank database. Since cheese is a cysteine-poor environment, the presence of a cysteine transport system apparently does not provide an evolutionary advantage for this bacterium, which could explain why this transport system is not widely distributed in L. paracasei strains, especially those of dairy origin.
Analysis of the RNA-seq data enabled identification of the genes that formed transcriptional units (Figure 3). This made it possible to confirm that the cysE-cysK and cysK2-ctl1-cysE2 gene clusters were transcribed as polycistronic RNAs, which had been previously demonstrated by PCR methods (Bogicevic et al., 2012a,b). Furthermore, the RNA-seq and the gene set enrichment analysis clearly showed that the expression of many genes involved in cysteine and methionine biosynthesis and transport respond to the availability of cysteine and methionine. With regard to the SAM recycling pathway, the expression levels for pfs and luxS did not show significant differences between cells grown in cysteine-deficient or methonine-deficient medium. In the closely related species L. rhamnosus, monocistronic, and polycistronic RNAs of luxS were identified (Lebeer et al., 2007). This may explain why this gene seems to be unaffected by cysteine or methionine, although it forms an operon with yxjH, which is controlled by a methionine-specific T-box.
RNA-seq analysis showed that the expression of genes located downstream of methionine- and cysteine-specific T-boxes, except for metQ1 and metQ2, were affected by the absence of either cysteine or methionine. RT-qPCR analysis confirmed this observation. Additionally, RT-qPCR showed that the expression of metQ1 was lower in the absence of methionine. The extremely low read coverage for this gene in the RNA-seq data set was likely the reason it was not determined by the DESeq2 algorithm to be significantly expressed.
Remarkably, all T-box leader sequences displayed extremely high read coverage (Figure 3). There are probably two main reasons for this finding. First, the T-boxes mediate the premature termination of transcription yielding more transcript sequence reads for the 5′-UTR than for the downstream located CDS regions. Second, RNase III, which was used in this study for RNA fragmentation, preferentially cleaves double-stranded RNA. T-boxes that form complex secondary structures (Vitreschak et al., 2008) are therefore probably prone to RNase III activity, which could subsequently lead to enrichment of T-boxes in the cDNA libraries.
The 5′-UTR of the cysE-cysK and cysK2-ctl1-cysE2 operons did not contain T-box leader sequences. The DESeq2 analysis showed a small difference of expression for the cysE-cysK operon between the cysteine- and methionine-deficient media. In-depth analysis of the nucleotide sequence of the 5′-UTR did not provide hints of any regulation mechanisms (data not shown). When cysK expression was studied using RT-qPCR analysis, no significant difference was observed between the cysteine- and methionine-deficient media-grown cells. Therefore, it is assumed that the cysE-cysK operon was constitutively expressed in the present study.
The high read coverage in the 5′-UTR of the cysK2-ctl1-cysE2 operon indicates a mechanism of regulation similar to that of S-box and T-box leader sequences. However, these regulons could not be identified. Liu et al. (2012) identified the palindromic motif AAAGGGCGCGAA-N(11−18)-TTCGCGCCTTTT as a potential regulatory motif upstream of genes encoding cystathionine synthase and cystathionine lyase in lactobacilli. In strain FAM18149, the similar motif AAGGGCGCGAAA-N16-TTTCGCGCCCTT can be found upstream of the cysK2-ctl1-cysE2 operon. When the 5′-UTR was analyzed using RegRNA 2.0, a 60-bp rho-independent terminator region that covered the aforementioned motif was identified. The sequence of this region can base-pair with itself and form a stem-loop structure. This stem-loop structure could cause the RNA polymerase to stall or stop the transcription. The high read coverage present in the 5′-UTR of the cysK2-ctl1-cysE2 operon supports the hypothesis that expression is regulated by premature termination of transcription. The occurrence of antisense RNA in the 3′-UTR of this operon could be an additional regulatory element. The combination of a cis- and antisense-mediated regulation was recently detected in Clostridium acetobutylicum (André et al., 2008). Interestingly, in this species it regulates the ubiG operon, which is also involved in methionine-to-cysteine conversion.
The present study showed that the genes involved in the metabolism of sulfur-containing amino acids in L. paracasei are regulated mainly at the transcriptional level. Methionine-specific T-box regulons regulate methionine biosynthesis and uptake, but the molecular mechanism of regulation for the expression of the cysK2-ctl1-cysE2 operon involved in the methionine-to-cysteine conversion is yet to be discovered. The RNA-seq analysis in this study was a valuable tool for studying cis-mediated regulation, verifying operon structure, and identifying active and inactive genes. Combined with the enzymatic characterization, RNA-seq helps to reveal metabolic pathways used by bacteria species in cheese providing a clearer understanding of the biochemistry of cheese.
Author contributions
SI and HB conceived and designed the study. SI and TB performed the RNA-seq experiments. DW, SI, and RB performed the bioinformatic analysis. HB and CW carried out the RT-qPCR analysis. SI, HB, DW, RB, and CW contributed to data interpretation. DW and SI wrote the manuscript. All authors approved the work for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Keith Harshman, Emmanuel Beaudoing, and Mélanie Dupasquier from the Genomic Technologies Facility of the University of Lausanne for performing the PacBio RSII sequencing.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01261/full#supplementary-material
References
- Alexa A., Rahnenführer J. (2018). Gene Set Enrichment Analysis With topGO. Available online at: https://bioconductor.org/packages/devel/bioc/vignettes/topGO/inst/doc/topGO.pdf
- Anders S., Pyl P. T., Huber W. (2014). HTSeq - a Python framework to work with high-throughput sequencing databio. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André G., Even S., Putzer H., Burguière P., Croux C., Danchin A., et al. (2008). S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 36, 5955–5969. 10.1093/nar/gkn601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André G., Haudecoeur E., Monot M., Ohtani K., Shimizu T., Dupuy B., et al. (2010). Global regulation of gene expression in response to cysteine availability in Clostridium perfringens. BMC Microbiol. 10:234. 10.1186/1471-2180-10-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. [Google Scholar]
- Beresford T., Williams A. (2004). The microbiology of cheese ripening, in Cheese: Chemistry, Physics and Microbiology: General Aspects, eds Fox P. F., McSweeney P. L. H., Cogan T. M., Guinee T. P. (Oxford: Elsevier Ltd.), 287–317. [Google Scholar]
- Bogicevic B., Berthoud H., Portmann R., Bavan T., Meile L., Irmler S. (2016). Cysteine biosynthesis in Lactobacillus casei: identification and characterization of a serine acetyltransferase. FEMS Microbiol. Lett. 363:fnw012. 10.1093/femsle/fnw012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogicevic B., Berthoud H., Portmann R., Meile L., Irmler S. (2012a). CysK from Lactobacillus casei encodes a protein with O-acetylserine sulfhydrylase and cysteine desulfurization activity. Appl. Microbiol. Biotechnol. 94, 1209–1220. 10.1007/s00253-011-3677-5 [DOI] [PubMed] [Google Scholar]
- Bogicevic B., Irmler S., Portmann R., Meile L., Berthoud H. (2012b). Characterization of the cysK2-ctl1-cysE2 gene cluster involved in sulfur metabolism in Lactobacillus casei. Int. J. Food Microbiol. 152, 211–219. 10.1016/j.ijfoodmicro.2011.06.015 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2144–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. H., Huang H. Y., Hsu J. B., Weng S. L., Horng J. T., Huang H. D. (2013). An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics 2013, 14(Suppl. 2):S4. 10.1186/1471-2105-14-S2-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Duan X. (2011). Ribosomal RNA depletion for massively parallel bacterial RNA-sequencing applications. Methods Mol. Biol. 733, 93–103. 10.1007/978-1-61779-089-8_7 [DOI] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Christensen J. E., Steele J. L. (2003). Impaired growth rates in milk of Lactobacillus helveticus peptidase mutants can be overcome by use of amino acid supplements. J. Bacteriol. 185, 3297–3306. 10.1128/JB.185.11.3297-3306.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Man J. C., Rogosa M., Sharpe M. E. (1960). A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23, 130–135. 10.1111/j.1365-2672.1960.tb00188.x [DOI] [Google Scholar]
- Fuchs R. T., Grundy F. J., Henkin T. M. (2006). The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 13, 226–233. 10.1038/nsmb1059 [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. (1998). The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol. Microbiol. 30, 737–749. 10.1046/j.1365-2958.1998.01105.x [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A., Henkin T. M., Grundy F. J., Yanofsky C., Merino E. (2009). Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. 73, 36–61. 10.1128/MMBR.00026-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullo M. F., Auger S., Dass E., Danchin A., Martin-Verstraete I. (2004). The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, D- and L-methionine. Res. Microbiol. 155, 80–86. 10.1016/j.resmic.2003.11.008 [DOI] [PubMed] [Google Scholar]
- Hullo M. F., Auger S., Soutourina O., Barzu O., Yvon M., Danchin A., et al. (2007). Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J. Bacteriol. 189, 187–197. 10.1128/JB.01273-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler S., Heusler M. L., Raboud S., Schlichtherle-Cerny H., Casey M. G., Eugster-Meier E. (2006). Rapid volatile metabolite profiling of Lactobacillus casei strains: selection of flavour producing cultures. Aust. J. Dairy Technol. 61, 123–127. [Google Scholar]
- Irmler S., Raboud S., Beisert B., Rauhut D., Berthoud H. (2008). Cloning and characterization of two Lactobacillus casei genes encoding a cystathionine lyase. Appl. Environ. Microbiol. 74, 99–106. 10.1128/AEM.00745-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler S., Schäfer H., Beisert B., Rauhut D., Berthoud H. (2009). Identification and characterization of a strain-dependent cystathionine β/γ-lyase in Lactobacillus casei potentially involved in cysteine biosynthesis. FEMS Microbiol. Lett. 295, 67–76. 10.1111/j.1574-6968.2009.01580.x [DOI] [PubMed] [Google Scholar]
- Kovaleva G. Y., Gelfand M. S. (2007). Transcriptional regulation of the methionine and cysteine transport and metabolism in streptococci. FEMS Microbiol. Lett. 276, 207–215. 10.1111/j.1574-6968.2007.00934.x [DOI] [PubMed] [Google Scholar]
- Kredich N. M. (1996). Biosynthesis of cysteine in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, eds Neidhardt F. C., Curtiss R., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. (Washington, DC: ASM Press; ), 514–527. [Google Scholar]
- Landaud S., Helinck S., Bonnarme P. (2008). Formation of volatile sulfur compounds and metabolism of methionine and other sulfur compounds in fermented food. Appl. Microbiol. Biotechnol. 77, 1191–1205. 10.1007/s00253-007-1288-y [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S., De Keersmaecker S. C., Verhoeven T. L., Fadda A. A., Marchal K., Vanderleyden J. (2007). Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189, 860–871. 10.1128/JB.01394-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Prakash C., Nauta A., Siezen R. J., Francke C. (2012). Computational analysis of cysteine and methionine metabolism and its regulation in dairy starter and related bacteria. J. Bacteriol. 194, 3522–3533. 10.1128/JB.06816-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K., Slesarev A., Wolf Y., Sorokin A., Mirkin B., Koonin E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616. 10.1073/pnas.0607117103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P., Eddy S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. (2004). Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 32, 3340–3353. 10.1093/nar/gkh659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Reddy V. S., Tamang D. G., Västermark Å. (2014). The transporter classification database. Nucleic Acids Res. 42, D251–D258. 10.1093/nar/gkt1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Seiflein T. A., Lawrence J. G. (2006). Two transsulfurylation pathways in Klebsiella pneumoniae. J. Bacteriol. 188, 5762–5774. 10.1128/JB.00347-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gigson T. J., Karplus K., Weizhong L., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Sys. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio B., Polard P., Ehrlich D. S., Renault P., Guédon E. (2005). Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187, 3762–3778. 10.1128/JB.187.11.3762-3778.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak A. G., Mironov A. A., Lyubetsky V. A., Gelfand M. S. (2008). Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14, 717–735. 10.1261/rna.819308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wels M., Groot Kormelink T., Kleerebezem M., Siezen R. J., Francke C. (2008). An in silico analysis of T-box regulated genes and T-box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genomics 9:330. 10.1186/1471-2164-9-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler P. R., Coldham N. G., Keating L., Gordon S. V., Wooff E. E., Parish T., et al. (2005). Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J. Biol. Chem. 280, 8069–8078. 10.1074/jbc.M412540200 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer. [Google Scholar]
- Wilkens S. (2015). Structure and mechanism of ABC transporters. F1000Prime Rep. 7:14. 10.12703/P7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.