Abstract
Dysregulation of microRNA (miRNA) expression has been linked to many human diseases; however, because of the challenges associated with RNA-targeted drug discovery, additional approaches are needed for probing miRNA biology. The emerging regulatory role of miRNA-binding proteins in miRNA maturation presents such an alternative strategy. Exploiting our laboratory’s click chemistry-based high-throughput screening (HTS) technology, catalytic enzyme-linked click chemistry assay or cat-ELCCA, we have designed a modular method by which to discover new chemical tools for manipulating pre-miRNA–miRNA–binding protein interactions. Using the pre-let-7d–Lin28 interaction as proof-of-concept, the results presented demonstrate how HTS using cat-ELCCA can enable the discovery of small molecules targeting RNA–protein interactions.
Keywords: MicroRNA, RNA-binding proteins, high-throughput screening, chemical probes
MicroRNAs (miRNA or miR) are small ∼21–23 nucleotide noncoding RNAs that play a crucial role in downregulating the translation of select target genes. Because an individual miRNA can target up to hundreds of mRNAs (mRNA), alterations in miRNA expression have been linked to many human diseases.1 In cancer, global downregulation of tumor suppressor miRNAs (TS-miRs) is commonly observed and has been demonstrated to be a causative feature in tumorigenesis.2−4 Loss can stem from genetic mutation or deletion, promoter methylation, or dysregulation of miRNA biogenesis.2 Of these mechanisms, alteration of global miRNA biogenesis is receiving increased attention due to recent findings demonstrating the critical role that miR-binding proteins (miR-BPs) play in the inhibition of this process, ultimately stimulating TS-miR degradation and cancer development.5−8
The canonical biogenesis of a mature miRNA derives from two intermediate hairpin loops, nuclear pri-miRNA and cytosolic pre-miRNA, and is mediated by the RNase III enzymes, Drosha and Dicer, respectively.9 Only mature miRNAs function in gene silencing.9 Importantly, the hairpin loop motif of pri- and pre-miRNAs has been found to be a critical regulatory element serving as a docking site for miR-BPs that affect maturation. The most well-characterized is the let-7–Lin28 interaction, where Lin28 protein functions as an inhibitor of let-7 maturation by binding to the hairpin loop of pri- and pre-let-7.10−12 Two isoforms of Lin28 exist in humans, Lin28A and Lin28B (collectively referred to as Lin28).12 Both interact similarly and use three RNA-binding domains to regulate let-7 maturation: an N-terminal cold shock domain and two CCHC zinc knuckle domains.13 The binding affinity (Kd) of full-length Lin28 for pre-let-7 is between 33–65 nM, and the zinc knuckle domains have been found to afford the selectivity of Lin28 for let-7 and contribute significantly to its binding affinity.14,15
The let-7 family plays an important role in cancer development and progression by downregulating cellular oncogenes including RAS and its mutant isoforms and Myc.16 Lin28 binding recruits terminal uridylyltransferases (TUTases), which polyuridylylate the 3′ terminus of pre-let-7 to inhibit Dicer processing and promote let-7 degradation.17 Loss of let-7 through this mechanism has been observed in at least 15% of all human cancers, including lung, breast, liver, esophageal, stomach, ovarian, prostate, and colon cancers, neuroblastoma, and chronic lymphocytic leukemia.3,4,12,18 Related, reduced let-7 levels have been found to correlate with poor prognosis and decreased patient survival.18 Importantly, delivery of a let-7 mimic or knockdown of Lin28 has been shown to reduce tumor growth in vivo,19−23 indicating the potential for anticancer agents targeted at restoring physiological levels of this TS-miR.
Over the past few years, our laboratory has developed high-throughput screening (HTS) technology called catalytic enzyme-linked click chemistry assay, or cat-ELCCA.24−27 Key advantages of this approach for HTS include its increased sensitivity due to catalytic signal amplification, robustness, and negligible compound interference in comparison to traditional fluorescence-based assays due to added washing steps.27,28 To date, we have applied cat-ELCCA for the discovery of pre-miRNA-selective small molecule probes29 and inhibitors of an acyltransferase30 and protein–protein interactions (PPI).31 Herein, we describe further expansion of cat-ELCCA for the discovery of inhibitors of miR–miR-BP interactions, namely, the pre-let-7–Lin28 interaction. Through these efforts, we have developed a robust screening platform for RNA–protein interactions and used this approach to discover a new chemotype capable of inhibiting a RNA-binding protein.
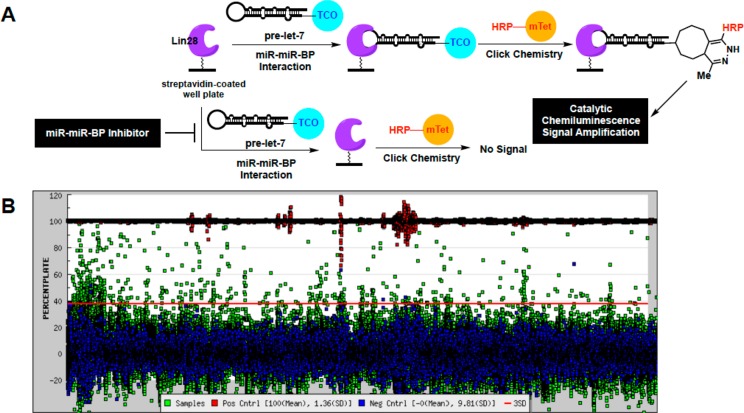
A scheme of cat-ELCCA for the pre-let-7–Lin28 interaction is shown in Figure 1A. In brief, drawing inspiration from PPI cat-ELCCA,31 murine Lin28A was first expressed as a N-terminal HaloTag fusion protein, labeled with biotin, and immobilized into the wells of a streptavidin-coated microtiter plate. Contrary to our Dicer-mediated pre-miRNA maturation cat-ELCCA,25,26,29 we chose to immobilize Lin28A, as preliminary studies revealed significantly enhanced immobilization efficiency of protein in comparison to RNA (data not shown). Following Lin28A immobilization, the wells were then incubated with pre-let-7d containing a 5′-trans-cyclooctene (TCO) click chemistry handle. Of note, this pre-let-7 isoform was chosen as its binding to Lin28 has been well-characterized.13 Importantly, binding of these modified substrates was successfully confirmed via an electrophoretic mobility shift assay (EMSA) (Figure S1A). Interaction of pre-let-7d with Lin28A was detected via click chemistry with methyltetrazine-labeled horseradish peroxidase (mTet-HRP), followed by treatment with a HRP substrate and measurement of chemiluminescence signal. As shown in Figure S1B–D, the assay exhibited a Z′ factor32 of >0.5 using automated liquid handling, was dependent on the concentration of TCO–pre-let-7d (Kd,app of 106 nM), and was amenable to competition using unlabeled prelet-7d.
Figure 1.
cat-ELCCA for the pre-let-7d–Lin28 miR–miR–BP interaction. (A) Assay scheme. (B) HTS campaign.
Encouraged by these promising results, we proceeded to HTS at the University of Michigan Center for Chemical Genomics. Our HTS assay protocol is summarized in Figure S2 and was performed using 384-well, high capacity, white streptavidin-coated well plates. In total, 127,007 small molecules were screened at 25 μM from the LOPAC (1280), Prestwick (1280), Maybridge (23 552), ChemDiv (100 000), and University of Michigan Chemistry (895) libraries. The assay performed excellently with a campaign Z′ factor of 0.5 and average plate Z′ factor of 0.71 (Figure 1B). Using primary hit criteria of ≥25% inhibition, 1468 compounds were selected for triplicate analysis, yielding a hit rate of 1.1%. Following hit confirmation, 181 molecules were selected that exhibited inhibition at ≥3SD by plate from the negative controls (12% of hits). Compounds were then analyzed in dose response; 136 molecules (75% of confirmed hits) were active with inhibitory potencies ranging from IC50 of 0.01–100 μM.33
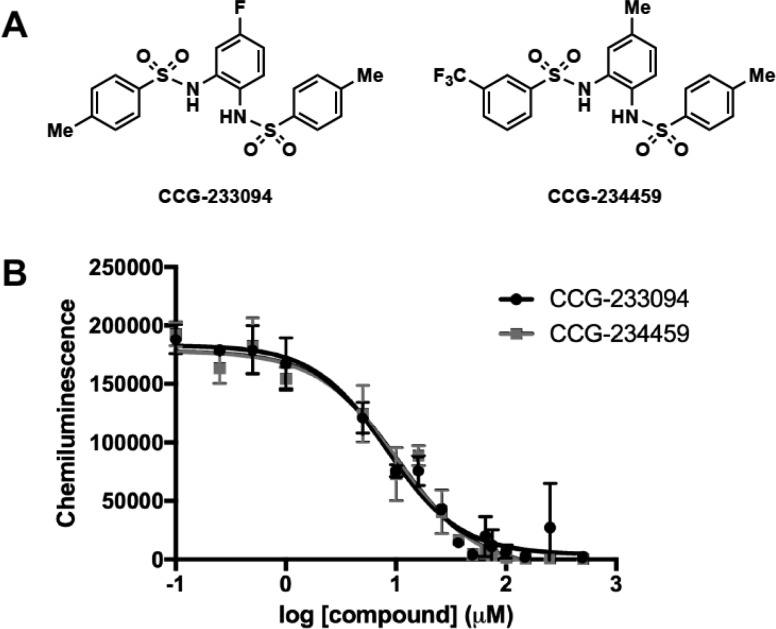
After removing known promiscuous hits (i.e., PAINS)34 and filtering for reactivity issues and tractable medicinal chemistry scaffolds, 20 compounds were selected for repurchase (Figure S3). While 10 of the purchased compounds were found to retain activity (Figure S4), only two showed concentration-responsive activity in both cat-ELCCA (IC50 values of 8.3 and 10.3 μM (nH = −1.3 and −1.2), respectively) and EMSA: the N,N′-(1,2-phenylene)-dibenzenesulfonamide derivatives CCG-233094 and CCG-234459 (Figures 2 and S5). These compounds were also found to inhibit a cat-ELCCA of the pre-let-7d–Lin28B interaction with similar IC50 values (Figure S6). Additionally, they showed inhibition of the interaction between Lin28A and other pre-let-7 isoforms (Figure S7). Through additional analysis, the compounds were found to function through direct interaction with Lin28, as dose-responsive binding was only observed with the protein (Figure S8), not pre-let-7d (Figure S9). Importantly, these compounds showed insignificant inhibition of Dicer-mediated pre-let-7d maturation, in addition to a cat-ELCCA-based PPI assay (Figure S10), providing a promising selectivity profile for our hits.
Figure 2.
HTS hits for the pre-let-7d–Lin28A interaction. (A) Compound structures. (B) IC50 curves from 0–500 μM.
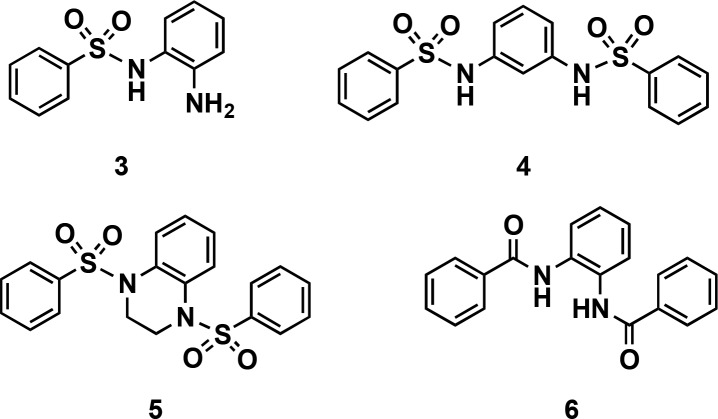
In order to further profile this scaffold, we performed initial structure–activity relationship (SAR) studies by catalogue, and 18 derivatives were available for purchase from commercial vendors. While fluoro-substituted compounds, aside from CCG-233094, could not be obtained, 11 unsubstituted derivatives were tested. As shown in Table 1, all analogues were active, showing variable inhibitory potencies of 10–62 μM. Several methyl-substituted analogues of CCG-234459 were also purchased and analyzed (Table 1). Again, each was active with IC50 values ranging between 11–28 μM. Inhibition curves for both sets of compounds are shown in Figure S11. Although none of the compounds showed enhanced potency over the parent compounds, these studies revealed that a combination of a meta-electron-withdrawing group and para-fluoro or -methyl substitution was optimal for maintaining inhibitory activity and that substituents on the center ring are not required. Additionally, two inactive molecules were identified: monosulfonamide 3 and N,N′-(1,3-phenylene)dibenzenesulfonamide 4 (Figure 3), demonstrating the necessity of the bis-sulfonamide and its 1,2-orientation for activity. This was further emphasized through synthetic derivatives 5 and 6 (Figure 3), which were also found to be inactive.
Table 1. SAR by Catalogue of N,N′-(1,2-Phenylene)dibenzenesulfonamide Derivatives.
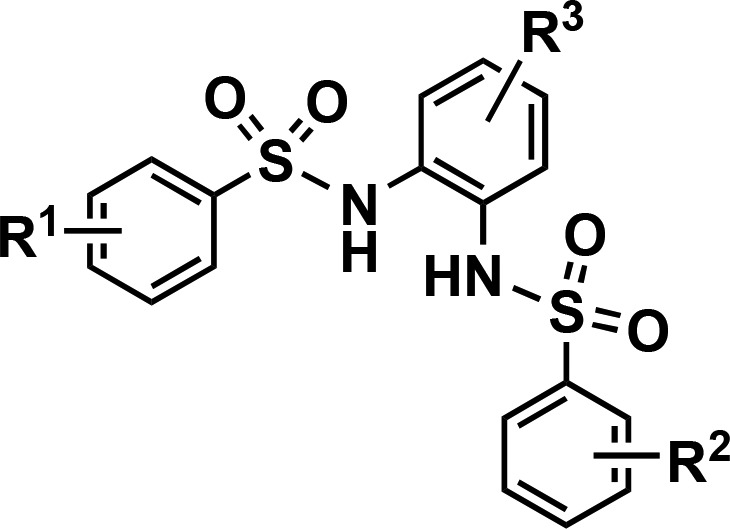
| compd | R1 | R2 | R3 | IC50 (μM) |
|---|---|---|---|---|
| 1a | H | H | H | 32.2 |
| 1b | p-F | H | H | 13.9 |
| 1c | p-F | p-F | H | 28.3 |
| 1d | p-F | p-Cl | H | 27.3 |
| 1e | p-F | p-OMe | H | 17.2 |
| 1f | p-F | m-Cl | H | 10.8 |
| 1g | p-F | m-Br | H | 16.2 |
| 1h | p-Cl | p-Cl | H | 27.5 |
| 1i | p-Br | p-Br | H | 17.5 |
| 1j | p-OMe | p-OMe | H | 16.7 |
| 1k | p-NHCOMe | p-NHCOMe | H | 62.5 |
| CCG-234459 | m-CF3 | p-Me | Me | 10.3 |
| 2a | H | H | Me | 28.8 |
| 2b | p-F | p-F | Me | 16.2 |
| 2c | p-F | p-Me | Me | 11.7 |
| 2d | p-Cl | p-Cl | Me | 28.4 |
| 2e | p-Me | p-Me | Me | 14.1 |
Figure 3.
Structures of inactive scaffolds.
To further probe the SAR of our discovered scaffold, additional derivatives were synthesized. Because the meta-chloro analogue 1f retained activity similar to that of our initial hits, we first examined if meta-substitution was tolerated on both rings. As shown in Table 2, the symmetric molecule 7a showed an improved IC50 value of 5.7 μM. Similar inhibitory activity was observed with the asymmetric methyl-substituted analogue 7b. As our screening hits contained para-methyl substituents, we next explored its combination with a meta-chloro substituent. Importantly, compounds 7c and 7d, containing unsubstituted or fluoro-substituted center rings, both showed further improvement in activity (IC50 values of 2.3 and 3.7 μM, respectively). Thus, we are hopeful that this series can be further optimized through future efforts in structure-based drug design.
Table 2. Additional SAR of N,N′-(1,2-Phenylene)dibenzenesulfonamide Derivatives.
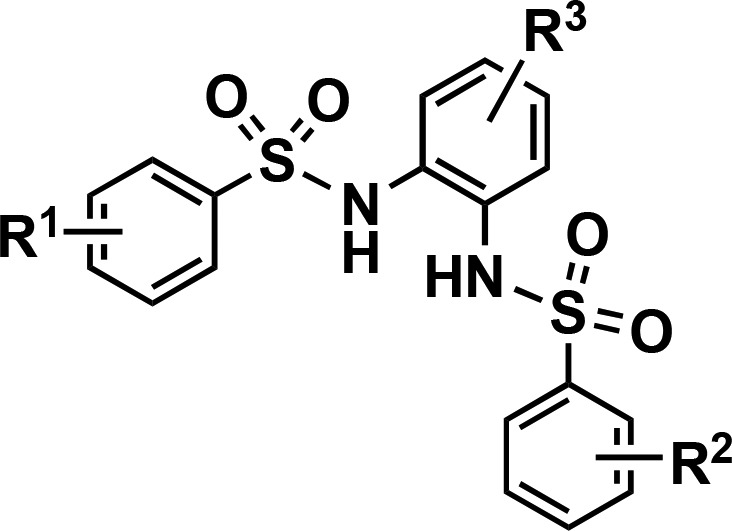
| compd | R1 | R2 | R3 | IC50 (μM) |
|---|---|---|---|---|
| 7a | m-Cl | m-Cl | H | 5.7 |
| 7b | m-Cl | m-Cl | Me | 4.3 |
| 7c | m-Cl, p-Me | m-Cl, p-Me | H | 2.3 |
| 7d | m-Cl, p-Me | m-Cl, p-Me | F | 3.7 |
In conclusion, using the adaptability of cat-ELCCA, we have developed new assay technology for analyzing RNA–protein interactions. Using this approach, we subsequently performed HTS and identified a new class of inhibitor for the pre-let-7–Lin28 interaction.35−37 As the important roles that RNA-binding proteins play in regulating RNA biology are emerging,38,39 there is a need for methods by which to discover small molecule chemical probes and drug leads for these targets. Similar to PPIs, RNA–protein interaction assays have relied on traditional methods such as fluorescence polarization,40 FRET,35,36 or AlphaScreen.41 Although useful for high-throughput experimentation, these assays can be limited by size requirements and the need for structural information for assay design. Because cat-ELCCA is amenable to HTS, utilizes a simple N- or C-terminus protein modification strategy, and does not require structural information, it should be an enabling tool for these future research endeavors. While there are many benefits to this new technology, limitations include the need for washing steps, which can hinder throughput and restrain its use to interactions with binding affinities ≤1.0 μM, similar to ELISA. Efforts are currently underway to engineer cat-ELCCA as a homogeneous chemiluminescence-based assay.
Acknowledgments
We thank Martha Larsen and Steve Vander Roest for assistance with the HTS. We also thank Glory Velazquez for assistance in preparing Lin28B.
Glossary
ABBREVIATIONS
- miRNA or miR
microRNA
- miR-BP
microRNA-binding protein
- HTS
high-throughput screening
- cat-ELCCA
catalytic enzyme-linked click chemistry assay
- TCO
trans-cyclooctene
- mTet
methyltetrazine
- HRP
horseradish peroxidase
- nH
Hill slope
- EMSA
electrophoretic mobility shift assay
- SD
standard deviation
- SAR
structure–activity relationship
Biography
Amanda Garner received her Ph.D. in Chemistry from the University of Pittsburgh working under the supervision of Prof. Kazunori Koide and completed NIH-funded postdoctoral studies in the laboratory of Prof. Kim Janda at The Scripps Research Institute. She began her independent career in 2013 in the Department of Medicinal Chemistry at the University of Michigan. Her laboratory uses chemical biology, medicinal chemistry, and molecular and cellular biology approaches to investigate the high-risk/high-reward areas of targeting microRNAs and RNA–protein and protein–protein interactions for probe and drug discovery.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00126.
General synthetic, protein expression/purification, and bioconjugation methods, protocol for pre-let-7–Lin28 cat-ELCCA, HTS details, supplemental figures, 95% confidence intervals for measured IC50 values, and compound characterization and purity spectra (PDF)
Author Contributions
The manuscript was written through contributions of D.A.L., T.K., and A.L.G. All authors have given approval to the final version of the manuscript.
This work was supported through a Catalyst Award from the Dr. Ralph and Marian Falk Medical Research Trust.
The authors declare no competing financial interest.
Supplementary Material
References
- Li Z.; Rana T. M. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discovery 2014, 13, 622–638. 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- Garzon R.; Marcucci G.; Croce C. M. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discovery 2010, 9, 775–789. 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H.; Fabbri M.; Calin G. A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discovery 2013, 12, 847–865. 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R.; Slack F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discovery 2017, 16, 203–221. 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- van Kouwenhove M.; Kedde M.; Agami R. MicroRNA regulation by RNA-binding proteins and its implications in cancer. Nat. Rev. Cancer 2011, 11, 644–656. 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- Thomson J. M.; Newman M. A.; Parker J. S.; Morin-Kensicki E. M.; Wright T.; Hammond S. M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006, 20, 2202–2207. 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. S.; Lu J.; Mercer K. L.; Golub T. R.; Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007, 39, 673–677. 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lee E. J.; Baek M.; Gusev Y.; Brackett D. J.; Nuovo G. J.; Schmittgen T. D. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 2008, 14, 35–42. 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M.; Kim V. N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Viswanathan S. R.; Daley G. Q.; Gregory R. I. Selective blockade of microRNA processing by Lin28. Science 2008, 320, 97–100. 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. A.; Thomson J. M.; Hammond S. M. Lin-28 interaction with the let-7 precursor loop mediates regulated microRNA processing. RNA 2008, 14, 1539–1549. 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Gregory R. I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.; Chen C.; Gregory R. I.; Chou J. J.; Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin F. E.; Gebert L. F. R.; Towbin H.; Brunschweiger A.; Hall J.; Allain F. H.-T. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2012, 19, 84–89. 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- Wang L.; Nam Y.; Lee A. K.; Yu C.; Roth K.; Chen C.; Ransey E. M.; Sliz P. LIN28 zinc knuckle domain is required and sufficient to induce let-7 oligouridylation. Cell Rep. 2017, 17, 2664–2675. 10.1016/j.celrep.2017.02.044. [DOI] [PubMed] [Google Scholar]
- Roush S.; Slack F. J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Heo I.; Joo C.; Cho J.; Ha M.; Han J.; Kim V. N. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 2008, 32, 276–284. 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Viswanathan S. R.; Powers J. T.; Einhorn W.; Hoshida Y.; Ng T. L.; Toffanin S.; O’Sullivan M.; Lu J.; Phillips L. A.; Lockhart V. L.; Shah S. P.; Tanwar P. S.; Mermel C. H.; Beroukhim R.; Azam M.; Teixeira J.; Meyerson M.; Hughes T. P.; Llovet J. M.; Radich J.; Mullighan C. G.; Golub T. R.; Sorensen P. H.; Daley G. Q. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848. 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A.; Trang P.; Wiggins J. F.; Patrawala L.; Cheng A.; Ford L.; Weidhaas J. B.; Brown D.; Bader A. G.; Slack F. J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008, 7, 759–764. 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Kumar M. S.; Erkeland S. J.; Pester R. E.; Chen C. Y.; Ebert M. S.; Sharp P. A.; Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 3903–3908. 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. H.; Robinton D. A.; Seligson M. T.; Wu L.; Li L.; Rakheja D.; Comerford S. A.; Ramezani S.; Sun X.; Parikh M. S.; Yang E. H.; Powers J. T.; Shinoda G.; Shah S. P.; Hammer R. E.; Daley G. Q.; Zhu H. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014, 26, 248–261. 10.1016/j.ccr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar J. J.; Domingo-Fernandez R.; Ebus M. E.; Lindner S.; Koster J.; Drabek K.; Mestdagh P.; van Sluis P.; Valentijn L. J.; van Nes J.; Broekmans M.; Haneveld F.; Volckmann R.; Bray I.; Heukamp L.; Sprussel A.; Thor T.; Kieckbusch K.; Klein-Hitpass L.; Fischer M.; Vandesompele J.; Schramm A.; van Noesel M. M.; Varesio L.; Speleman F.; Eggert A.; Stallings R. L.; Caron H. N.; Versteeg R.; Schulte J. H. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- Hamano R.; Miyata H.; Yamasaki M.; Sugimura K.; Tanaka K.; Kurokawa Y.; Nakajima K.; Takiguchi S.; Fujiwara Y.; Mori M.; Doki Y. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients with oesophagus cancer. Br. J. Cancer 2012, 106, 1415–1423. 10.1038/bjc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. L.; Janda K. D. cat-ELCCA: a robust method to monitor the fatty acid acyltransferase activity of ghrelin O-acyltransferase(GOAT). Angew. Chem., Int. Ed. 2010, 49, 9630–9634. 10.1002/anie.201003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz D. A.; Song J. M.; Garner A. L. High-throughput platform assay technology for the discovery of pre-microRNA-selective small molecule probes. Bioconjugate Chem. 2015, 26, 19–23. 10.1021/bc500544v. [DOI] [PubMed] [Google Scholar]
- Lorenz D. A.; Garner A. L. A click chemistry-based microRNA maturation assay optimized for high-throughput screening. Chem. Commun. 2016, 52, 8267–8270. 10.1039/C6CC02894B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. L. cat-ELCCA: catalyzing drug discovery through click chemistry. Chem. Commun. 2018, 10.1039/C8CC02332H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz D. A.; Garner A. L. Approaches for the discovery of small molecule ligands targeting microRNAs. Top. Med. Chem. 2017, 27, 79–110. 10.1007/7355_2017_3. [DOI] [Google Scholar]
- Lorenz D. A.; Vander Roest S.; Larsen M. J.; Garner A. L. Development and implementation of an HTS-compatible assay for the discovery of selective small-molecule ligands for pre-microRNAs. SLAS Disc. 2018, 23, 47–54. 10.1177/2472555217717944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. L.; Janda K. D. A small molecule antagonist of ghrelin O-acyltransferase(GOAT). Chem. Commun. 2011, 47, 7512–7514. 10.1039/c1cc11817j. [DOI] [PubMed] [Google Scholar]
- Song J. M.; Menon A.; Mitchell D. C.; Johnson O. T.; Garner A. L. High-throughput chemical probing of full-length protein-protein interactions. ACS Comb. Sci. 2017, 19, 763–769. 10.1021/acscombsci.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-H.; Chung T. D. Y.; Oldenburg K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screening 1999, 4, 67–73. 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Of note, the compound displaying a 10 nM IC50 value from our HTS contained a tetrazine that was found to react with our TCO-labeled prelet-7d (Figure S3).
- Baell J. B.; Holloway G. A. New substructure filters for removal of pan assay interference compounds(PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Roos M.; Pradere U.; Ngondo R. P.; Behera A.; Allegrini S.; Civenni G.; Zagalak J. A.; Marchand J.-R.; Menzi M.; Towbin H.; Scheuermann J.; Neri D.; Caflisch A.; Catapano C. V.; Claudo C.; Hall J. A small-molecule inhibitor of Lin28. ACS Chem. Biol. 2016, 11, 2773–2781. 10.1021/acschembio.6b00232. [DOI] [PubMed] [Google Scholar]
- Lim D.; Byun W. G.; Koo J. Y.; Park H.; Park S. B. Discovery of a small-molecule inhibitor of protein-microRNA interaction using binding assay with a site-specifically labeled Lin28. J. Am. Chem. Soc. 2016, 138, 13630–13638. 10.1021/jacs.6b06965. [DOI] [PubMed] [Google Scholar]
- Lightfoot H. L.; Miska E. A.; Balasubramanian S. Identification of small molecule inhibitors of the Lin28-mediated blockage of pre-let-7g processing. Org. Biomol. Chem. 2016, 14, 10208–10216. 10.1039/C6OB01945E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S.; Hafner M.; Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M. W.; Castello A.; Schwarzl T.; Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327. 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- Wu X.; Lan L.; Wilson D. M.; Marquez R. T.; Tsao W.-C.; Gao P.; Roy A.; Turner B. A.; McDonald P.; Tunge J. A.; Rogers S. A.; Dixon D. A.; Aube J.; Xu L. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem. Biol. 2015, 10, 1476–1484. 10.1021/cb500851u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills N. L.; Shelat A. A.; Guy R. K. Assay optimization and screening of RNA-protein interactions by AlphaScreen. J. Biomol. Screening 2007, 12, 946–955. 10.1177/1087057107306128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.