Abstract
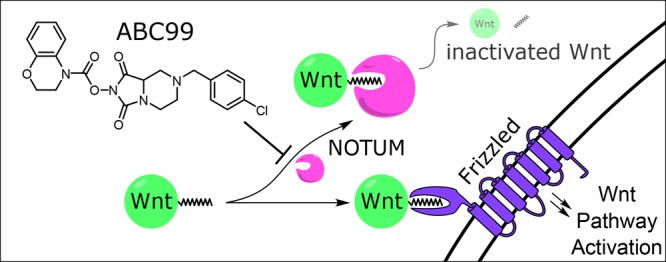
Wnt proteins are secreted morphogens that play critical roles in embryonic development and tissue remodeling in adult organisms. Aberrant Wnt signaling contributes to diseases such as cancer. Wnts are modified by an unusual O-fatty acylation event (O-linked palmitoleoylation of a conserved serine) that is required for binding to Frizzled receptors. O-Palmitoleoylation of Wnts is introduced by the porcupine (PORCN) acyltransferase and removed by the serine hydrolase NOTUM. PORCN inhibitors are under development for oncology, while NOTUM inhibitors have potential for treating degenerative diseases. Here, we describe the use of activity-based protein profiling (ABPP) to discover and advance a class of N-hydroxyhydantoin (NHH) carbamates that potently and selectively inhibit NOTUM. An optimized NHH carbamate inhibitor, ABC99, preserves Wnt-mediated cell signaling in the presence of NOTUM and was also converted into an ABPP probe for visualizing NOTUM in native biological systems.
Keywords: NOTUM, Wnt, enzyme, inhibitor, activity-based profiling, proteomics
The Wnt proteins are a family of secreted signaling molecules essential for embryogenesis, tissue homeostasis, and stem cell fate.1−3 Wnt signaling is regulated by diverse mechanisms, including inhibitory binding proteins (Dickkopf proteins4) and post-translational modifications (PTMs). Prominent among the PTMs that regulate Wnts is the O-palmitoleoylation of a conserved serine residue, which is required for Wnt trafficking, signaling, and interactions with Frizzled receptors.5 An MBOAT acyltransferase porcupine (PORCN)6 catalyzes the O-palmitoleoylation of Wnts and is a potential therapeutic target for cancers that depend on Wnt signaling for growth.7 While the role that PORCN plays in Wnt fatty acylation has been understood for more than a decade, a protein involved in deacylating Wnts was only recently identified. This protein, termed NOTUM, had long been recognized to antagonize Wnt pathways based on genetic studies,8,9 but it was not until 2015 that two groups independently elucidated the biochemical function of NOTUM as a serine hydrolase that deactivates Wnt proteins by removing the O-linked palmitoleate modification.10,11 NOTUM expression is itself regulated by Wnt signaling and may serve as a biomarker for Wnt-dependent cancers that are sensitive to PORCN inhibitors.12 NOTUM may conversely constitute a drug target for degenerative diseases that would benefit from enhanced Wnt signaling, such as osteopenia and osteoporosis.13
To date, only a handful of NOTUM inhibitors have been described, representing heteroaryl-fused thiophenes.14,15 These compounds presumably act as reversible inhibitors, but their selectivity both within and outside of the serine hydrolase family remains unknown. Past efforts by our lab and others have demonstrated that activated carbamates16,17 and ureas18 serve as a versatile source of irreversible inhibitors of serine hydrolases, especially when coupled with activity-based protein profiling (ABPP)19,20 to optimize potency and selectivity. In these past studies, however, NOTUM had not been screened by ABPP, presumably because the enzyme is secreted and has a very restricted cell and tissue distribution (http://biogps.org). Here, we show that NOTUM can be evaluated by ABPP using the conditioned media (CM) from transfected HEK293T cells (recombinant NOTUM) or SW620 cells (endogenous NOTUM). From a structurally diverse collection of activated carbamates and ureas, we discovered a set of N-hydroxyhydantoin (NHH) carbamates that act as potent and selective inhibitors of NOTUM. We show that these compounds are functional at sustaining Wnt signaling in the presence of NOTUM and can be converted into tailored ABPP probes for enhanced visualization of NOTUM in biological systems.
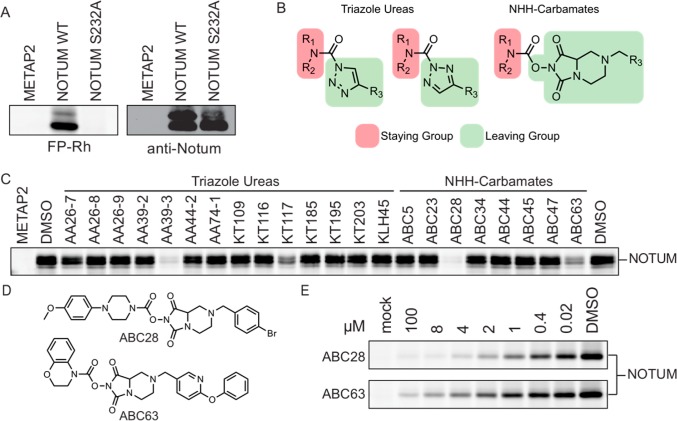
We first sought to establish whether NOTUM activity could be assayed with serine hydrolase (SH)-directed fluorophosphonate (FP) ABPP probes.21,22 Conditioned media (CM) from HEK293T cells transiently transfected with cDNAs encoding wild-type (WT) or a catalytically inactive (S232A) mutant of human NOTUM (or with a control protein METAP2) was treated with a rhodamine-tagged FP probe (FP-Rh) and analyzed by gel-based ABPP, which revealed a robustly FP-Rh-labeled ∼52 kDa band (predicted MW of full-length NOTUM is 54 kDa) in WT-NOTUM-transfected, but not S232A-NOTUM- or METAP2-transfected cell media (Figure 1A and Supplementary Figure 1). Western blotting with a commercial anti-NOTUM antibody also identified this 52 kDa protein, as well as a slightly higher MW protein (∼54 kDa) that could represent full-length unprocessed NOTUM (Figure 1A). Notably, the higher MW form of NOTUM, which matches the predicted molecular weight of the full-length protein prior to signal peptide cleavage, showed much lower relative reactivity with the FP-Rh probe, suggesting that processing may be required to fully activate NOTUM.
Figure 1.
Discovery of NOTUM inhibitors from a set of activated ureas and carbamates by competitive ABPP. (A) Gel-based ABPP and Western blot analysis of conditioned media (CM) from HEK293T cells transiently transfected with cDNAs encoding WT-NOTUM, a catalytically inactive S232A-NOTUM, or control protein (METAP2). Gel-based ABPP was performed by incubating CM proteome with the FP-Rh probe (1 μM, 30 min, room temperature). Western blotting was performed with an anti-NOTUM antibody (Sigma-Aldrich HPA023041). (B) General structures of the triazole urea18 and NHH carbamate17 classes of SH-directed inhibitors. (C) Gel-based ABPP of a collection of triazole urea and NHH carbamate compounds against the CM of WT-NOTUM-transfected HEK293T cells. CM samples were pretreated with 10 μM compound (30 min, 37 °C) followed by FP-Rh (1 μM, 30 min, room temperature), prior to SDS-PAGE analysis and in-gel fluorescence scanning. (D,E) Structures (D) and concentration-dependent inhibition profiles (E) for two NHH carbamate inhibitors of NOTUM: ABC28 and ABC63.
Having established that NOTUM activity could be visualized by ABPP, we next screened a series of serine hydrolase-directed inhibitors containing triazole urea18 and NHH carbamate17 reactive groups (10 μM, 30 min pretreatment at 37 °C) (Figure 1B). A number of compounds from both structural classes blocked FP-Rh reactivity with NOTUM (Figure 1C), and among these, we selected two NHH carbamates, ABC28 and ABC63, for follow-up studies, as we have found that this class of serine hydrolase inhibitors is generally less promiscuous than triazole ureas, while still affording the opportunity to derivatize both the staying and leaving groups to optimize potency and selectivity.
Concentration-dependent profiles revealed that ABC28 showed superior potency for NOTUM inhibition compared to ABC63 (Figure 1D and Table 1). We next confirmed that endogenously produced NOTUM could be detected by ABPP in the conditional media from SW620 cells, a human colon cancer cell line with high NOTUM expression.23 A strong FP-Rh-labeled, ∼52 kDa protein was detected in SW620 CM, and the FP-Rh reactivity of this protein was blocked by pretreatment with ABC28 (Supplementary Figure 1). Using CM from SW620 cells as a convenient source of endogenous human NOTUM, we initiated structure–activity relationship (SAR) studies for NHH carbamate inhibitors of the enzyme.
Table 1. IC50 Values for NHH Carbamate Inhibitors of NOTUM Determined by Competitive Gel-Based ABPPa.
| compd | IC50 [nM] (95% CI) |
|---|---|
| ABC28 | 677 (467–943) |
| ABC63 | 1506 (1101–2065) |
| ABC90 | 290 (221–441) |
| ABC91 | >10 μM |
| ABC92 | 200 (156–275) |
| ABC99 | 13 (9.39–17.5) |
| ABC101 | >10 μM |
Data represent average values with confidence intervals (CI) provided for three independent experiments.
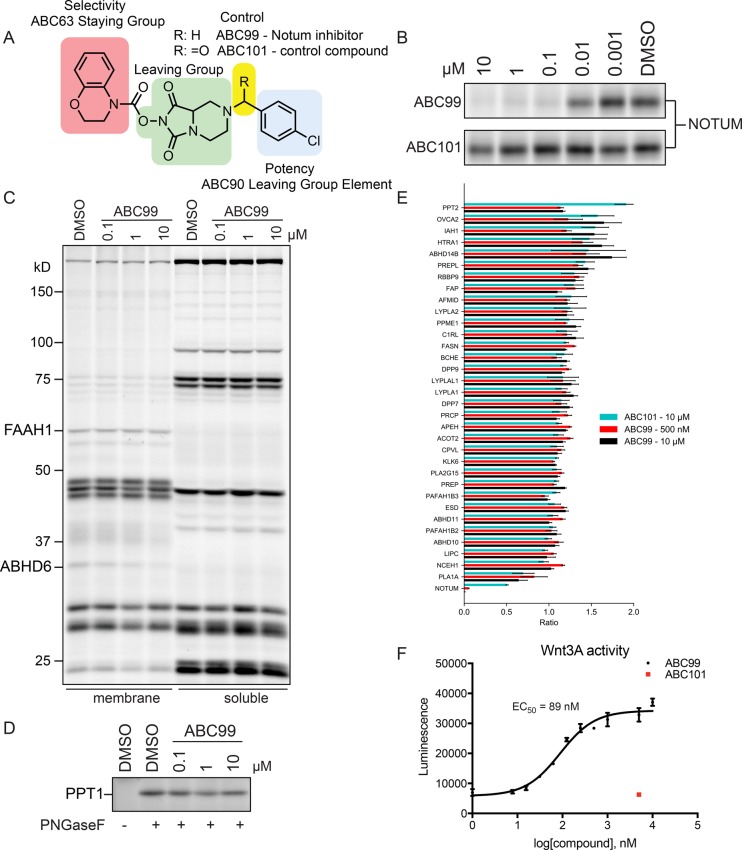
Substitution of chlorine for bromine on the distal phenyl ring of the leaving group of ABC28, combined with positional scanning of the halogen substituent (ABC90–ABC92; Supplementary Figure 2A), yielded two compounds, ABC90 (p-Cl) and ABC92 (o-Cl), that showed substantially improved potency for NOTUM inhibition (IC50 values < 300 nM; Table 1), while the m-Cl analogue (ABC91) completely lost activity (Supplementary Figure 2B). We next evaluated the selectivity of ABC90 by gel-based ABPP using soluble and membrane proteomes of SW620 cells, which express a more diverse array of serine hydrolase activities compared to the CM fraction of these cells (Supplementary Figure 2C). Among the FP-Rh-labeled proteins in SW620 soluble and membrane proteomes, only a single major site of ABC90 cross reactivity was observed; a 35 kDa protein that we suspected was ABHD6 (Supplementary Figure 2C), a common target of carbamate serine hydrolase inhibitors.17,20 We also noted, however, that the NHH carbamate scaffold represented by ABC90 was similar to that of a previously reported inhibitor of the serine hydrolase protein palmitoyl thioesterase 1 (PPT1),17 and we accordingly found that ABC90 was a potent inhibitor of PPT1, as determined using the PPT1-directed ABPP probe ABC45(17) (Supplementary Figure 2D). In contrast, the alternative NHH carbamate hit compound for NOTUM, ABC63, did not inhibit PPT1 at concentrations up to 10 μM (Supplementary Figure 2D). These data are consistent with our previous research on PPT1, where we observed that the enzyme shows reduced reactivity with NHH carbamates bearing smaller and less extended staying groups.17 We therefore reasoned that a compound, referred to hereafter as ABC99 (Figure 2A), possessing the staying group of ABC63 and leaving group of ABC90 might constitute a potent NOTUM inhibitor that does not cross-react with PPT1.
Figure 2.
Generation and characterization of ABC99, a potent and selective NOTUM inhibitor. (A) Structure of ABC99 and inactive control compound ABC101. (B) Concentration-dependent inhibition of NOTUM by ABC99 and ABC101 as measured by competitive ABPP using SW620 CM. (C,D) Competitive ABPP of SW620 cells treated in situ with ABC99 (indicated concentrations, 2 h, 37 °C) followed by exposure to the general serine hydrolase probe FP-Rh (C) or the PPT1-directed probe ABC45 (D). (E) Quantitative MS-based ABPP of SW620 CM treated with ABC99 (0.5 or 10 μM, 1 h, 37 °C), ABC101 (10 μM, 1 h, 37 °C), or DMSO. Serine hydrolases were enriched with FP-biotin (4 μM, 1 h, room temperature) and streptavidin chromatography. After on-bead trypsin digestion, peptides were isotopically labeled with heavy (DMSO) or light (compound) formaldehyde, then combined and processed for MS-based analysis.24 Ratios are displayed as light/heavy; therefore, low values indicate inhibition. Data for each experimental group represent median aggregate peptide ratios across two independent experiments +/− SEM. (F) ABC99, but not ABC101, preserves Wnt-3A activity in the presence of NOTUM in a concentration-dependent manner as measured using a Super TOPflash assay. Media from Wnt-3A expressing L-cells were incubated with inhibitor treated media from SW620 cells and then added to HEK293T-STF cells, which express luciferase in response to activation of the canonical Wnt pathway. Data represent average values with confidence intervals (CI) provided for four independent experiments.
ABC99 inhibited NOTUM with an IC50 value of 13 nM (Figure 2B and Table 1) and showed excellent selectivity across the serine hydrolase family, as initially assessed by gel-based ABPP of soluble and membrane proteomes from SW620 cells (Figure 2C,D). These ABPP experiments were performed using both FP-Rh and ABC45 probes and thus confirmed that PPT1 also did not cross-react with ABC99 (Figure 2D). We next evaluated the selectivity of ABC99 (0.5 and 10 μM) by quantitative mass spectrometry (MS)-based ABPP24 in both the CM and SW620 in situ treated cells. These data confirmed the inhibition of NOTUM (Figure 2E) with virtually no cross-reactivity with the 64 additional serine hydrolases quantified by MS-ABPP (Figure 2E and Supplementary Figure 3 and Supplementary Data Set 1). A partial, concentration-dependent blockade of ABHD6 was observed, but only ∼50% of this enzyme was inhibited at concentrations of ABC99 (0.5 μM) that fully blocked NOTUM. We also noted that ABC99 more completely inhibited secreted (Figure 2E) compared to cellular (Supplementary Figure 3) NOTUM, which could reflect that the latter fraction contains a greater proportion of incompletely processed NOTUM.
In the course of evaluating the SAR of NHH carbamate inhibitors, we discovered that conversion of the basic tertiary amine in ABC99 to an amide furnished a compound ABC101 that lost inhibitory activity for NOTUM (Figure 2A,B,E) but maintained inhibition of ABHD6 (Supplementary Figure 3). We therefore considered that ABC101 could serve as a useful inactive control probe for biological studies.
We next tested whether ABC99 could counteract the suppression of Wnt signaling by NOTUM in a cellular assay. Using a luciferase reporter cell line responsive to Wnt activation (HEK293T-STF),25 we found that exposure of cells to a combination of NOTUM-expressing CM (harvested from SW620 cells) preincubated with either ABC99 or the control compound ABC101 and CM from Wnt3A-producing L-cells26 resulted in the concentration-dependent preservation of Wnt signaling in the presence of ABC99, but not ABC101 (Figure 2F). The IC50 value for ABC99-mediated preservation of Wnt3A activity (Figure 2F and Table 2) was moderately higher than the IC50 value for inhibition of NOTUM measured by ABPP (Figure 2B and Table 1), which could indicate that substantially more than 50% of NOTUM needs to be inhibited for protection of Wnt-mediated cell signaling.
Table 2. EC50 Values for Preservation of Wnt Activity by ABC99 and ABC101 Determined Using a Super TOPflash Assaya.
| compound | IC50 [nM] (95% CI) |
|---|---|
| ABC99 | 89 (55–150) |
| ABC101 | >5 μM |
Data represent average values with confidence intervals (CI) provided for four independent experiments.
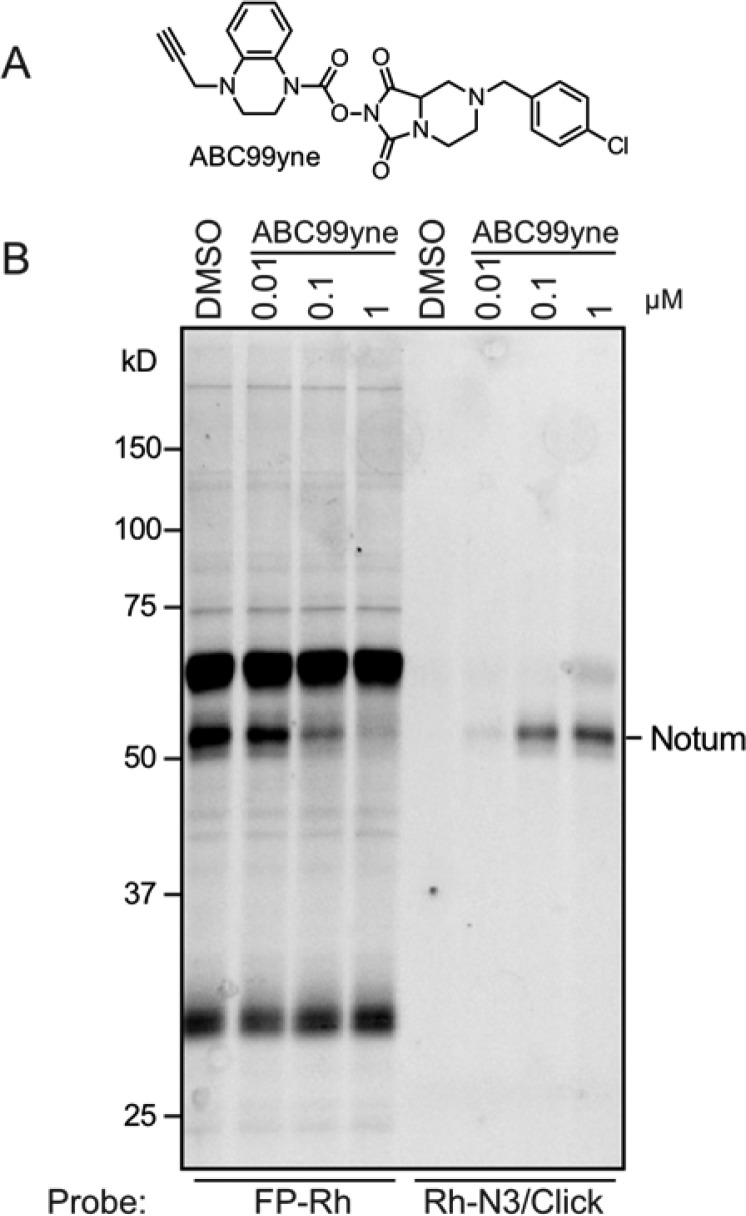
Considering that NOTUM is a secreted enzyme with highly regulated expression, including induction by Wnt signaling itself,12 it would be valuable to generate probes to specifically monitor the activity of this enzyme in native biological systems. Toward this end, we incorporated an alkyne into the staying group of the ABC99 scaffold to generate a clickable probe termed ABC99yne (Figure 3A). We found that ABC99yne blocked the FP-Rh reactivity of NOTUM in CM from SW620 cells with a sub-micromolar IC50 value, and treatment of these samples with a rhodamine-azide reporter tag27 under copper-mediated azide–alkyne cycloaddition (CuAAC, or click) chemistry conditions28,29 resulted in the highly selective labeling of NOTUM with no other cross-reactive proteins observed in the CM (Figure 3B) or cellular lysate (Supplementary Figure 4) of SW620 cells (0.1 μM ABC99yne probe).
Figure 3.
Development of ABC99yne, a clickable analogue of ABC99 for visualizing NOTUM activity in biological samples. (A) Structure of ABC99yne. (B) ABPP gel showing competitive blockade of FP-Rh labeling (left lanes) and direct labeling (right lanes) of NOTUM by ABC99yne in SW620 CM. Samples were pretreated with ABC99yne at the indicated concentrations (30 min, 37 °C) and then treated with FP-Rh (1 μM, 30 min, room temperature) or subjected to CuAAC conditions with a Rh–N3 tag prior to SDS-PAGE analysis.
We have described, to our knowledge, the first potent and selective irreversible inhibitors of the Wnt-deacylating enzyme NOTUM. The most advanced compound, ABC99, blocks NOTUM activity with low-nanomolar potency (IC50 value = 13 nM) and excellent selectivity as assessed with serine hydrolase-directed ABPP probes and by global proteomic reactivity analysis using a clickable probe analogue (ABC99yne). Over the past decade, many studies have emphasized the important role that NOTUM plays as a negative regulator of Wnt signaling in diverse biological processes, but virtually all of these efforts have relied on molecular biology (e.g., genetic) methods to perturb NOTUM function.10,11,30,31 We believe that the covalent inhibitors and related chemical probes (inactive control and clickable probes) described herein will provide valuable complementary tools to the limited set of noncovalent inhibitors reported previously14,15 to characterize the biological effects of pharmacologically inactivating NOTUM in diverse physiological and disease processes. Future studies of interest include testing the proteome-wide reactivity of ABC99 using the ABC99yne probe combined with MS-based ABPP analysis;17 directly measuring Wnt palmitoleate modification10 following exposure of biological systems to ABC99; and assessing the effect of ABC99 on Wnt signaling in vivo. Related to the last objective, we note that NOTUM appears to have a restricted tissue distribution in vivo (http://www.humanproteomemap.org/), which may provide clues as to the specific areas of Wnt biology that are most strongly influenced by NOTUM-mediated deacylation.
Acknowledgments
This work was supported by the NIH (DA033760, CA193994) and the Skaggs Institute for Chemical Biology.
Glossary
ABBREVIATIONS
- ABPP
activity-based protein profiling
- CM
conditioned media
- FP
fluorophosphonate
- NHH
N-hydroxyhydantoin
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00191.
Author Present Address
† Department of Chemistry, University of Washington, Seattle, WA 98195, USA.
Author Contributions
‡ R.S. and A.B.C. contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): B.F.C. is a cofounder and scientific advisor to Abide Therapeutics, a biotechnology company interested in developing SH inhibitors as therapeutics.
Supplementary Material
References
- Gordon M. D.; Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 2006, 281 (32), 22429–33. 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- Logan C. Y.; Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Zeng Y. A.; Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 2010, 6 (6), 568–77. 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 2006, 25 (57), 7469–81. 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- Janda C. Y.; Garcia K. C. Wnt acylation and its functional implication in Wnt signalling regulation. Biochem. Soc. Trans. 2015, 43 (2), 211–6. 10.1042/BST20140249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr P.; Hausmann G.; Basler K. WNT secretion and signalling in human disease. Trends Mol. Med. 2012, 18 (8), 483–93. 10.1016/j.molmed.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Ho S. Y.; Keller T. H. The use of porcupine inhibitors to target Wnt-driven cancers. Bioorg. Med. Chem. Lett. 2015, 25 (23), 5472–6. 10.1016/j.bmcl.2015.10.032. [DOI] [PubMed] [Google Scholar]
- Gerlitz O.; Basler K. Wingful, an extracellular feedback inhibitor of Wingless. Genes Dev. 2002, 16 (9), 1055–9. 10.1101/gad.991802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giráldez A. J.; Copley R. R.; Cohen S. M. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev. Cell 2002, 2 (5), 667–676. 10.1016/S1534-5807(02)00180-6. [DOI] [PubMed] [Google Scholar]
- Kakugawa S.; Langton P. F.; Zebisch M.; Howell S.; Chang T. H.; Liu Y.; Feizi T.; Bineva G.; O’Reilly N.; Snijders A. P.; Jones E. Y.; Vincent J. P. Notum deacylates Wnt proteins to suppress signalling activity. Nature 2015, 519 (7542), 187–192. 10.1038/nature14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Cheong S. M.; Amado N. G.; Reis A. H.; MacDonald B. T.; Zebisch M.; Jones E. Y.; Abreu J. G.; He X. Notum is required for neural and head induction via Wnt deacylation, oxidation, and inactivation. Dev. Cell 2015, 32 (6), 719–30. 10.1016/j.devcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B.; Ke Z.; Lei Z. D.; Oliver F. A.; Oshima M.; Lee M. A.; Rozen S.; Virshup D. M. NOTUM is a potential pharmacodynamic biomarker of Wnt pathway inhibition. Oncotarget 2016, 7 (11), 12386–92. 10.18632/oncotarget.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. G.; Fotsch C.; Babij P. Emerging targets in osteoporosis disease modification. J. Med. Chem. 2010, 53 (11), 4332–53. 10.1021/jm9018756. [DOI] [PubMed] [Google Scholar]
- Han Q.; Pabba P. K.; Barbosa J.; Mabon R.; Healy J. P.; Gardyan M. W.; Terranova K. M.; Brommage R.; Thompson A. Y.; Schmidt J. M.; Wilson A. G.; Xu X.; Tarver J. E. Jr.; Carson K. G. 4H-Thieno[3,2-c]chromene based inhibitors of Notum Pectinacetylesterase. Bioorg. Med. Chem. Lett. 2016, 26 (4), 1184–7. 10.1016/j.bmcl.2016.01.038. [DOI] [PubMed] [Google Scholar]
- Tarver J. E. Jr.; Pabba P. K.; Barbosa J.; Han Q.; Gardyan M. W.; Brommage R.; Thompson A. Y.; Schmidt J. M.; Wilson A. G. E.; He W.; Lombardo V. K.; Carson K. G. Stimulation of cortical bone formation with thienopyrimidine based inhibitors of Notum Pectinacetylesterase. Bioorg. Med. Chem. Lett. 2016, 26 (6), 1525–1528. 10.1016/j.bmcl.2016.02.021. [DOI] [PubMed] [Google Scholar]
- Niphakis M. J.; Cognetta A. B. 3rd; Chang J. W.; Buczynski M. W.; Parsons L. H.; Byrne F.; Burston J. J.; Chapman V.; Cravatt B. F. Evaluation of NHS carbamates as a potent and selective class of endocannabinoid hydrolase inhibitors. ACS Chem. Neurosci. 2013, 4 (9), 1322–32. 10.1021/cn400116z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognetta A. B. 3rd; Niphakis M. J.; Lee H. C.; Martini M. L.; Hulce J. J.; Cravatt B. F. Selective N-Hydroxyhydantoin Carbamate Inhibitors of Mammalian Serine Hydrolases. Chem. Biol. 2015, 22 (7), 928–37. 10.1016/j.chembiol.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibekian A.; Martin B. R.; Wang C.; Hsu K. L.; Bachovchin D. A.; Niessen S.; Hoover H.; Cravatt B. F. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 2011, 7 (7), 469–78. 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin D. A.; Cravatt B. F. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discovery 2012, 11 (1), 52–68. 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin D. A.; Ji T.; Li W.; Simon G. M.; Blankman J. L.; Adibekian A.; Hoover H.; Niessen S.; Cravatt B. F. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (49), 20941–6. 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Patricelli M. P.; Cravatt B. F. Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (26), 14694–9. 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli M. P.; Giang D. K.; Stamp L. M.; Burbaum J. J. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 2001, 1 (9), 1067–71. . [DOI] [PubMed] [Google Scholar]
- Scherf U.; Ross D. T.; Waltham M.; Smith L. H.; Lee J. K.; Tanabe L.; Kohn K. W.; Reinhold W. C.; Myers T. G.; Andrews D. T.; Scudiero D. a.; Eisen M. B.; Sausville E. a.; Pvommier Y.; Botstein D.; Brown P. O.; Weinstein J. N. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000, 24 (3), 236–44. 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- Inloes J. M.; Hsu K. L.; Dix M. M.; Viader A.; Masuda K.; Takei T.; Wood M. R.; Cravatt B. F. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (41), 14924–9. 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q.; Wang Y.; Dabdoub A.; Smallwood P. M.; Williams J.; Woods C.; Kelley M. W.; Jiang L.; Tasman W.; Zhang K.; Nathans J. Vascular development in the retina and inner ear: Control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 2004, 116 (6), 883–895. 10.1016/S0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- Willert K.; Brown J. D.; Danenberg E.; Duncan A. W.; Weissman I. L.; Reya T.; Yates J. R.; Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 2003, 423 (6938), 448–452. 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- Speers A. E.; Cravatt B. F. Chemical strategies for activity-based proteomics. ChemBioChem 2004, 5 (1), 41–7. 10.1002/cbic.200300721. [DOI] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ″ligation″ of azides and terminal alkynes. Angew. Chem., Int. Ed. 2002, 41 (14), 2596–9. . [DOI] [PubMed] [Google Scholar]
- Tornoe C. W.; Christensen C.; Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67 (9), 3057–64. 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Gerhardt B.; Leesman L.; Burra K.; Snowball J.; Rosenzweig R.; Guzman N.; Ambalavanan M.; Sinner D. Notum attenuates Wnt/beta-catenin signaling to promote tracheal cartilage patterning. Dev. Biol. 2018, 436 (1), 14–27. 10.1016/j.ydbio.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P.; Reddien P. W. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 2011, 332 (6031), 852–5. 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.