Abstract
The formin Delphilin binds the glutamate receptor, GluRδ2, in dendritic spines of Purkinje cells. Both proteins play a role in learning. To understand how Delphilin functions in neurons, we studied the actin assembly properties of this formin. Formins have a conserved formin homology 2 domain, which nucleates and associates with the fast-growing end of actin filaments, influencing filament growth together with the formin homology 1 (FH1) domain. The strength of nucleation and elongation varies widely across formins. Additionally, most formins have conserved domains that regulate actin assembly through an intramolecular interaction. Delphilin is distinct from other formins in several ways: its expression is limited to Purkinje cells, it lacks classical autoinhibitory domains, and its FH1 domain has minimal proline-rich sequence. We found that Delphilin is an actin nucleator that does not accelerate elongation, although it binds to the barbed end of filaments. In addition, Delphilin exhibits a preference for actin isoforms, nucleating nonmuscle actin but not muscle actin, which has not been described or systematically studied in other formins. Finally, Delphilin is the first formin studied that is not regulated by intramolecular interactions. We speculate how the activity we observe is consistent with its localization in the small dendritic spines.
INTRODUCTION
Dendritic spines are postsynaptic structures essential for learning (Nimchinsky et al., 2002). Defects in spine morphology and/or number are linked to several disorders, including autism spectrum disorder, schizophrenia, and Alzheimer’s disease (Penzes et al., 2011). It is noteworthy that symptoms of the three disorders listed here present at very different ages. Thus, development and maintenance of dendritic spines are both important subjects. Spine morphology is dynamic and reflects synaptic activity levels. Spines are commonly described as mushroom-shaped structures, with a head and neck, although the morphology varies from filopodia-like to mushroom shaped (Peters and Kaiserman-Abramof, 1970; Harris et al., 1992). The head contains the so-called postsynaptic density (PSD), a collection of hundreds of proteins including receptors, signaling proteins, and cytoskeletal elements (Walikonis et al., 2000). The actin cytoskeleton is essential for formation, maintenance, and dynamic rearrangement of dendritic spines (Hlushchenko et al., 2016). It follows that many actin-binding proteins are found in spines.
Actin nucleators help build new filaments and structures in a manner related to their respective nucleation mechanisms. The Arp2/3 complex, several formins, and, perhaps, tandem monomer binding nucleators all contribute to dendritic spine morphology and dynamics. The nucleators build distinct portions of the spines and respond to different events (Hotulainen et al., 2009; Hlushchenko et al., 2016). The formin, Delphilin, is localized exclusively in neurons and primarily in cerebellar dendritic spines; however, it is not required for spine morphology and its role remains unclear (Takeuchi et al., 2008). Delphilin was first identified as a binding partner for the glutamate receptor, GluRδ2 (Miyagi et al., 2002). The GluRδ2 receptor is critical to synaptic plasticity and motor learning. The GluRδ2 receptor is also a bit of a mystery, with no known ligand or apparent electrical activity. Delphilin and GluRδ2 interact in Purkinje cells at synapses with parallel fibers (Yamashita et al., 2005). Loss of GluRδ2 results in disrupted synapse formation and loss of long-term depression (LTD), a case when synaptic connections lose efficiency (Kashiwabuchi et al., 1995). Although Delphilin is not required for spine formation, its loss does result in facilitation of LTD, consistent with its known interaction with GluRδ2 (Takeuchi et al., 2008).
Mammals have 15 formins, which represent seven of nine formin families (Higgs and Peterson, 2005; Pruyne, 2016). Delphilin is distinct from most formins in that it does not contain Diaphanous Inhibitory or Autoinhibitory Domains (DID and DAD). Formins in six of the nine formin families are regulated by intramolecular interactions between DID and DAD domains. Fmn-family formins do not have DID and DAD domains but are autoinhibited by an analogous intramolecular interaction (Bor et al., 2012). Instead of a DID domain, the three Delphilin splice variants have at least one PSD-95/disks large/ZO-1 (PDZ) domain at the N-terminus (one has two; YamPashita et al., 2005). PDZ domains are common to PSD proteins and, logically, the Delphilin-PDZ domain binds directly to the C-terminal tail of GluRδ2 (Miyagi et al., 2002). The Delphilin tail, where the DAD domain would be, is essentially absent.
It is easy to speculate that Delphilin’s actin assembly activities are essential to its function. However, Delphilin-family formins have not been extensively characterized. Therefore, to determine whether actin assembly in response to synaptic activity could be part of Delphilin’s role, we compared it with other well-studied formins, in vitro. Formins are identified based on conserved formin homology 1 and 2 (FH1 and FH2) domains, which mediate actin nucleation and processive elongation. We found that a construct containing the FH1 and FH2 domains of Delphilin assembles actin in a manner similar to the previously characterized formin, Cdc12 (Kovar et al., 2003). In the absence of profilin, new filaments only grow from their pointed ends and in the presence of profilin the barbed ends grow. However, the elongation rate is not accelerated above actin alone. Additionally, we found that, unlike other studied formins, Delphilin does not appear to be autoinhibited. Interestingly, Delphilin exhibits a strong preference for nonmuscle actin over muscle actin, which has not been described or systematically studied in other formins.
RESULTS
Delphilin is an actin assembly factor
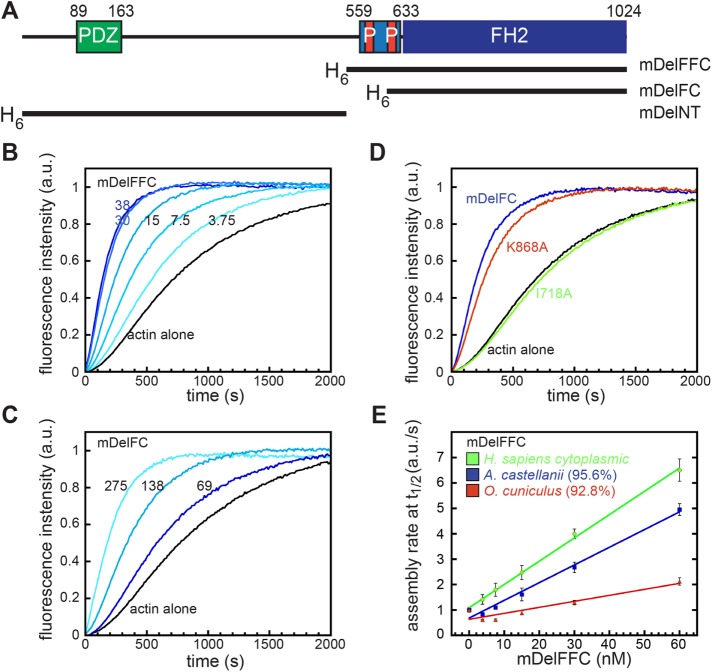
To understand the effect of Delphilin on actin assembly, we purified the C-terminal halves of both mouse and human Delphilin (mDelFFC and hDelFFC), including both the FH1 and FH2 domains through to their C-termini (Figure 1A and Supplemental Figure S1A). We tested both formins in pyrene–actin assembly assays. An addition of either DelFFC construct results in a dose-dependent increase in actin assembly (Figure 1B and Supplemental Figure S2). In this context, mDelFFC is approximately fivefold more potent than hDelFFC. The minimum domain required for actin assembly by most formins is the FH2 domain (Pruyne et al., 2002; Li and Higgs, 2003; Goode and Eck, 2007). To verify that the Delphilin FH1 domain is dispensable for actin assembly in the absence of profilin, we tested mDelFC (Figure 1A). We found that mDelFC assembles actin filaments, albeit ∼15-fold slower than mDelFFC (Figure 1C). The majority of free actin monomers within cells are bound by the actin-binding protein, profilin. Therefore, to test mDelFFC under more physiologically relevant conditions, we added profilin to the pyrene–actin assembly assays. Under these conditions, actin assembly was markedly decreased but we still observed dose-dependent stimulation of actin polymerization (Supplemental Figure S3A). These results confirm that Delphilins are capable of enhancing actin assembly, as predicted based on primary sequence.
FIGURE 1:
Delphilin is an actin assembly factor. (A) Domain structure of Delphilin. Green, PDZ domain; light blue, FH1 domain, with red proline-rich repeats; blue, FH2 domain. The numbering is based on mouse α-Delphilin (NP_579933.1). Constructs used in this paper are indicated below the diagram and shown in Supplemental Figure S1. All constructs were N-terminally His-tagged. (B) Pyrene–actin assembly assay with mDelFFC, concentrations as indicated (nM). (C) Pyrene–actin assembly assay with mDelFC, concentrations as indicated (nM). This construct is ∼15-fold weaker than mDelFFC. (D) Comparison of 150 nM wild-type mDelFC with point mutations in the same construct (I718A and K868A). (E) Comparison of actin assembly rates in pyrene assays with the indicated source of actin. Sequence similarities compared with human cytoplasmic actin are shown in parentheses. Conditions: A. castellani actin (4 μM, 10% pyrene labeled) was used in B–D. In E, the source of actin is indicated.
Two highly conserved residues within the FH2 domain, an isoleucine and a lysine, are essential for actin assembly activities in most formins (Xu et al., 2004; Ramabhadran et al., 2012). Although mutations in either residue abolish actin assembly in Bni1 (Xu et al., 2004), the analogous mutations have varying effects in other formins (Ramabhadran et al., 2012). We built a homology model of mDelFC based on the crystal structure of hDaam1 (Protein Data Bank [PDB] ID: 2J1D) and identified the conserved isoleucine and lysine residues, Ile718 and Lys868, respectively. We substituted alanine at these positions in mDelFC and tested the consequences in the pyrene–actin assembly assay. Actin assembly was abolished for all tested concentrations of mDelFC I718A, whereas mDelFC K868A resulted in only a mild reduction of actin assembly activity (Figure 1D). Taken together, these data suggest that Delphilin functions much like other characterized formins.
Delphilin activity is sensitive to the actin isoform
In the course of our work we noticed a difference in activity, dependent on the actin isoform present. Actin is highly conserved, often sharing >90% sequence identity across species, making this a surprising observation, although not unprecedented among actin-binding proteins (Rubenstein, 1990; De La Cruz and Pollard, 1996; Perrin and Ervasti, 2010). We tested the commonly used actin isoforms from different sources: Acanthamoeba castellanii (amoeba actin) and Oryctolagus cuniculus (rabbit skeletal muscle, α-actin), as well as Homo sapiens (cytoplasmic, 85% β-actin), reasoning that this is the actin that Delphilin is most likely to encounter in vivo. Interestingly, both mouse and human DelFFC constructs accelerated actin assembly of human cytoplasmic actin most effectively (Figure 1E [green] and Supplemental Figure S2). Amoeba actin, which shares 95.6% sequence identity to cytoplasmic actin, was assembled at ∼75% the level observed for cytoplasmic actin (Figure 1E [blue]). In contrast, Delphilin weakly assembled rabbit skeletal actin (∼20%), despite its 92.8% similarity to cytoplasmic actin (Figure 1E [red]). These data indicate that Delphilin is sensitive to actin isoform and nucleates cytoplasmic actin, the actin it would normally encounter, most effectively.
Delphilin is a nucleator
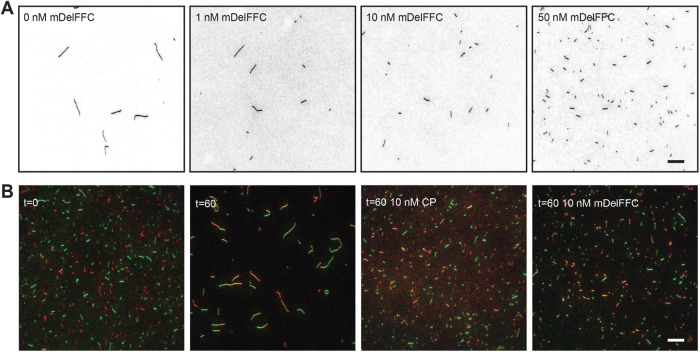
The kinetics of the pyrene–actin assembly assay reflects both nucleation and elongation of actin filaments. It is particularly important to distinguish these two activities when studying formins, which can modify each to differing degrees. Further, how Delphilin influences these two distinct processes has implications for how it functions in vivo. To elucidate between nucleation and/or elongation of actin filaments, we took advantage of total internal reflection fluorescence (TIRF) microscopy. First, we incubated 2 μM actin alone or in the presence of 1–50 nM mDelFFC for 5 min and then stabilized actin filaments with Alexa488-phalloidin. Phalloidin-stabilized filaments were adsorbed to poly-l-lysine–coated coverslips and imaged. Confirming that Delphilin is a nucleator, we observed that as concentrations of mDelFFC increase, so too do the number of actin filaments (Figure 2A). Compared to actin alone, addition of 50 nM mDelFFC results in a greater than 10-fold increase in the number of actin filaments (Figure 2A). We also noted that filaments nucleated by DelFFC were shorter than actin alone filaments (Figure 2A), reminiscent of the yeast formin Cdc12, which caps the barbed end of filaments.
FIGURE 2:
Delphilin is a nucleator. (A) Actin assembly (2 μM) was triggered by the addition of salts and the indicated concentration of mDelFFC. After 5 min, filaments were stabilized with Alexa488-phalloidin, diluted, and adsorbed to poly-l-lysine–coated coverslips. Typical fields of view are shown for a range of concentrations, as indicated. Increasing concentrations of Delphilin resulted in increasing numbers of filaments, indicating that Delphilin nucleates. The shorter filaments induced by Delphilin suggest that pointed end growth dominates under these conditions. (B) A total of 0.25 μM Alexa488- and Alexa647-phalloidin-stabilized filaments were mixed and sheared. Immediately after shearing (t = 0) filaments were short and monochromatic. After an hour (t = 60), the filament length depended on the conditions. Reannealing is readily apparent when only buffer is added. Short filaments, indistinguishable from t = 0, are seen in the presence of 10 nM capping protein or Delphilin, demonstrating that Delphilin remains bound to filament ends for long periods of time. Scale bars = 10 μm.
Because we observed short filaments in the nucleation assays, we hypothesized that Delphilin remains tightly associated with the barbed end. To test this hypothesis, we performed reannealing assays over a range of mDelFFC concentrations. We mixed and sheared two populations of actin filaments stabilized with different fluorescently labeled phalloidins in the absence and presence of mDelFFC. Capping protein was used as a positive control. We obtained images immediately after mixing actin filaments (t = 0) and an hour after incubation (t = 60; Figure 2B). Immediately after shearing, the overall length of filaments was short and filaments were single colors, revealing that no reannealing events had occurred. After an hour, filaments were significantly longer and were dual labeled indicating that reannealing of filaments occurred when no other protein was added. As expected, incubation with 10 nM capping protein prevented reannealing. mDelFFC (10 nM) produced results similar to 10 nM capping protein. These results suggest that Delphilin remains associated with barbed ends for long periods of time, similar to other formins.
Delphilin is a permissive elongation factor
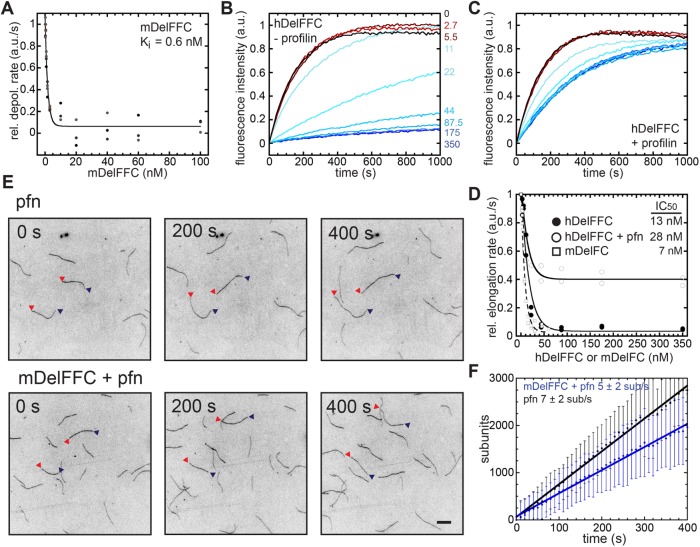
To address how Delphilin modulates the elongation rate of actin filaments, we first measured the affinity of mDelFFC for barbed ends, using a depolymerization assay. When filamentous actin was diluted to 0.1 μM in the presence of Delphilin, depolymerization was potently inhibited, reflecting tight binding (Figure 3A and Supplemental Figure S3B; Ki = 0.6 nM). We next used seeded elongation assays to bypass the nucleation step. We mixed 0–60 nM mDelFC or 0–350 nM hDelFFC and 0.5 μM G-actin with 0.25 μM actin seeds to monitor elongation. Both constructs tested (hDelFFC and mDelFC) potently inhibited barbed-end elongation (Figure 3B and Supplemental Figure S3C). hDelFFC inhibited ∼96% of elongation with an IC50 of 13 nM (Figure 3D). mDelFC bound barbed ends with a slightly higher affinity (IC50 = 7 nM) and inhibited elongation to the same degree (97%; Figure 3D). Tight barbed-end binding by Delphilin and inhibition of both elongation and depolymerization are consistent with the short filaments observed in the TIRF nucleation assays and Delphilin’s ability to inhibit reannealing (Figure 2).
FIGURE 3:
Delphilin is a permissive elongation factor. (A) Delphilin inhibits depolymerization. The initial rate of depolymerization is plotted vs. the concentration of mDelFFC added. All of the data are shown. Lines are fits to averages of the data. The Ki reported (0.6 nM) is the average of fits to three independent experiments. Representative raw data are shown in Supplemental Figure S3B. (B) Delphilin inhibits elongation. Seeded elongation assays with 0.25 μM seeds, 0.5 μM actin monomers (10% pyrene labeled), and the indicated concentration of hDelFFC. Quantification is shown in D. (C) Seeded elongation assays as in B except with 1.5 μM profilin added. (D) Initial rate of elongation vs. concentration of Delphilin. Data from two experiments each with or without profilin are shown. The concentration required to inhibit elongation 50% was similar in each case (13 vs. 28 nM). The degree of inhibition differed (96% vs. 64%), with hDelFFC inhibiting essentially all elongation at saturation and profilin partially relieving inhibition. (E) Direct observation of filament elongation by TIRF microscopy. Conditions: 2 μM actin (15% Cy3b-actin), 6 μM profilin-1 ± 10 nM mDelFFC. Scale bar = 10 μm. (F) Measurement of elongation rates from TIRF experiments in the presence or absence of mDelFFC. The elongation rates are given as mean ± SD of the slopes of all filaments from a given condition (n[−Del] = 90 and n[+Del] = 96). Data are represented as means and standard deviations at each time point. Lines fit to the time point averages (shown) give the same elongation rates. Elongation appears to be slightly slower in the presence of mDelFFC but the difference is not statistically significant.
Barbed-end elongation is often accelerated in the presence of formins and profilin. We therefore added profilin to the seeded elongation assay with hDelFFC (Figure 3C). In the presence of profilin, elongation was only ∼64% inhibited as opposed to 96%, while the affinity for the barbed end was similar (IC50 = 28 nM; Figure 3D). The decreased inhibition indicates that Delphilin allows actin assembly in the presence of profilin. We used TIRF microscopy to directly monitor single-filament elongation in the presence of mDelFFC. Under these conditions (6 μM human profilin-1 and 2 μM Mg-G-actin), the average elongation rate was 7 ± 2 sub/s (Figure 3, E and F). Addition of mDelFFC had little effect on the rate of filament elongation: 5 ± 2 sub/s (Figure 3, E and F). One could interpret the lack of change in the elongation rate as evidence that Delphilin rapidly falls off of growing barbed ends. However, the reannealing data (Figure 2B) indicate that Delphilin remains bound to barbed end for a long time, at least in the absence of elongation. We next asked whether Delphilin remains associated with barbed ends under conditions where actin is elongating. To do so, we used the pyrene assay and added both profilin and capping protein. As expected, addition of profilin and capping protein prevented spontaneous actin polymerization. Increasing concentrations of DelFFC were able to antagonize the inhibitory activity of profilin and capping protein and assemble actin filaments (Supplemental Figure S3D). We conclude that Delphilin remains associated with growing barbed ends. Thus, Delphilin “permits” but does not enhance elongation in the presence of profilin, similar to what was observed for Drosophila Daam (Barkó et al., 2010). Failure to accelerate elongation may be explained by the composition of the polyproline stretches within the FH1 domain and/or gating of the FH2 domain (see Discussion). Based on these observations, we conclude that the majority of activity observed in our original pyrene–actin assembly assays (Figure 1B) reflects nucleation by Delphilin.
Delphilin is a weak actin bundler
The C-terminal half of multiple formins bind and/or bundle actin filaments (Michelot et al., 2005; Harris et al., 2006; Quinlan et al., 2007; Schönichen et al., 2013). We first determined how tightly mDelFFC binds the sides of actin filaments. To do so, we performed high-speed cosedimentation assays, using 0.5 μM mDelFFC and various concentrations of phalloidin-stabilized actin filaments. The data reflect weak side binding with an affinity of 9 μM or higher (Figure 4A). Consistent with weak side binding, at saturation only one mDelFFC dimer was bound per 20 actin monomers. Another group reported much tighter binding (Dutta et al., 2017). When we reanalyzed their data, fitting it with a hyperbolic curve instead of a line, the Kd was ∼3 μM, which is in reasonable agreement with our findings.
FIGURE 4:
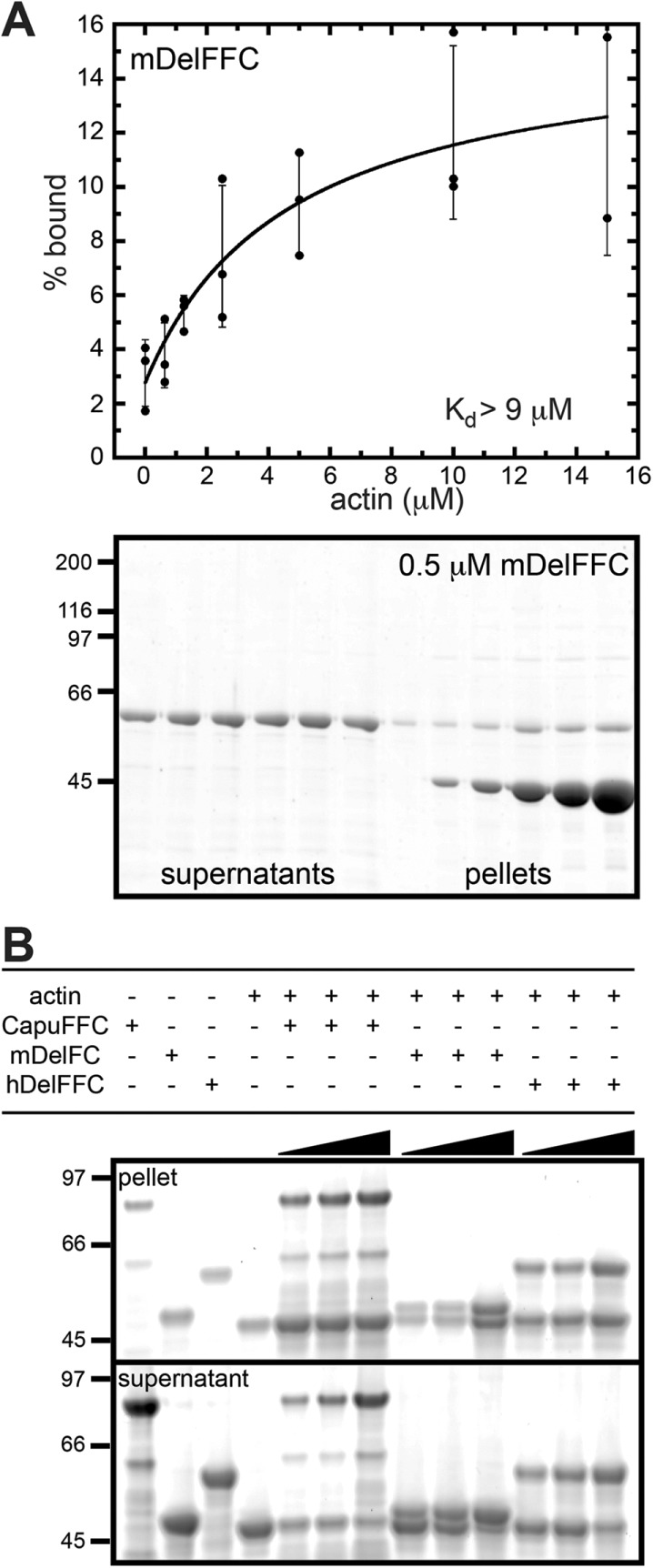
Delphilin is a weak actin bundler. (A) Using high-speed cosedimentation, we measured mDelFFC binding to actin filament sides. Various amounts of actin (0–15 μM) were mixed with 0.5 μM mDelFFC for 30 min at 25°C. Data from three independent experiments are shown. The line is a fit to averages of the data. The Kd reported (>9 μM) is the average of fits to the three independent experiments. Because the data do not reach a plateau, 9 μM is a lower limit of the Kd. At the bottom is a representative gel from a high-speed cosedimentation assay. (B) Low-speed sedimentation assays demonstrate that mDelFC and hDelFFC bundle actin filaments. Consistent with weak side binding, bundling is weak compared with CapuFFC. Various amounts of formin (1.25, 2.5, and 5 μM) were incubated with 5 μM F-actin for 30 min at 25°C. Bundles were sedimented by centrifugation at 12,000 × g for 15 min at 4°C.
Only some formins that bind filament sides also bundle actin (Harris et al., 2006). We used low-speed cosedimentation to test for bundling by mDelFC and hDelFFC (Figure 4B). Both constructs have weak bundling activity compared with the Drosophila formin Cappuccino (Capu), a known filament bundler (Rosales-Nieves et al., 2006; Quinlan et al., 2007; Vizcarra et al., 2014). Although actin is found in the low-speed pellet at all ratios of CapuFFC:F-actin (1:4, 1:2, 1:1), very little actin is in the pellet at ratios lower than 1:1 Delphilin:F-actin. At a ratio of 1:1, most of the actin is in the pellet. These data are consistent with the weak side binding we observed for mDelFFC and highly cooperative actin bundling.
Delphilin lacks a functional tail
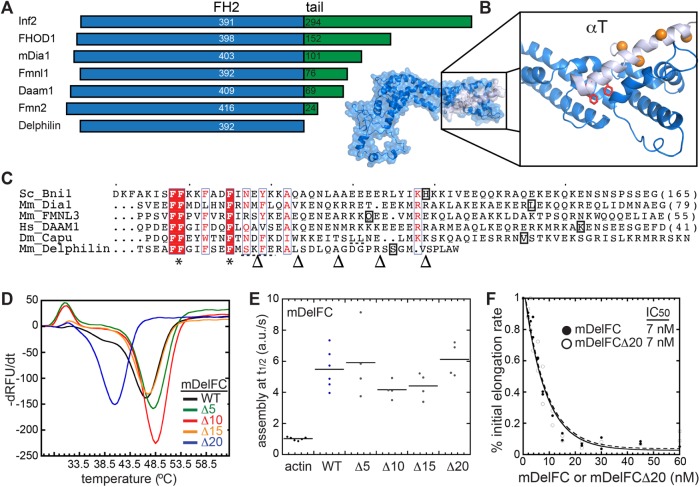
The tails of several formins, including mDia1, mDia2, Bni1, Bnr1, FRL, and Capu, are important for nucleation and processivity in addition to autoinhibition (Gould et al., 2011; Heimsath and Higgs, 2012; Vizcarra et al., 2014). Tail lengths vary a great deal among mammalian formins (Figure 5A). Based on alignments and our homology model, Delphilin does not have a C-terminal tail (Figure 5, A and B). In fact, the predicted last α-helix in the FH2 domain of Delphilin, αT, is shorter than αT helices of other formins that have been crystallized. In Delphilin, only eight residues remain beyond the predicted helix (Figure 5, B and C). To further investigate the Delphilin tail, or lack thereof, we compared mDelFC to constructs lacking 5–25 residues. mDelFCΔ25 was insoluble but the rest of the constructs were readily purified using the same protocol (Supplemental Figure S1A). We first assessed the impact of the truncations by measuring the thermal stability of the proteins. The constructs were essentially indistinguishable until 20 residues were removed, a truncation that is predicted to be well into the αT helix (Figure 5, B and C). Wild-type mDelFC has a melting temperature of 46.5°C and mDelFCΔ20 melts at 40.5°C (Figure 5D). In pyrene–actin assembly assays, mDelFC activity was not affected by truncation (Figure 5E). Even mDelFCΔ20 nucleated as potently as wild-type mDelFC. In addition, barbed-end binding was unaffected by truncation; mDelFC and mDelFCΔ20 inhibited seeded elongation to the same extent (Figure 5F). Thus, Delphilin does not have a tail that contributes to actin nucleation or end binding.
FIGURE 5:
Delphilin lacks a functional tail. (A) Comparison of tail length among the seven classes of mammalian formins. The blue FH2 domains are aligned at their C-termini. The tails are in green. Numbers indicate the length of each domain. (B) Homology model of Delphilin-FH2 based on the Daam1 crystal structure (2J1D). The C-terminus of the structure is expanded to show the short final helix (αT, light blue). Delphilin’s final eight residues are absent from the model. Conserved phenylalanines at the base of αT are shown as red sticks. Orange residues (balls) indicate where the Δ10–Δ25 truncations were made. (Δ5 is not in the structure.) (C) Alignment of αT sequences based on crystal structures (Bni1 [1UX5], mDia1 [1V9D], FMNL3 [4EAH], DAAM1 [2J1D]) and homology models (Capu [ Yoo et al., 2015], Delphilin [this work]). * indicates phenylalanines conserved in all formins, which are red in B. The boxed letters are the last residues observed in each structure. Dashed underlines indicate a predicted nonhelical insert in the middle of Delphilin and Capu αT. Arrowheads at the bottom indicate where truncations were made for experiments in D–F. (D) Thermal stability of mDelFC truncations compared with wild type (the number of residues removed is given in the legend). 1 μM mDelFC was mixed with Sypro Orange according to the manufacturers specifications and heated from 4°C to 95°C. Only mDelFCΔ20 was less stable than wild type. (E) Normalized assembly rates from pyrene assays with 4 μM actin and 150 nM mDelFC construct, as indicated. The rates did not differ significantly for any of the constructs, including mDelFCΔ20. (F) Inhibition of seeded elongation by mDelFC and mDelFCΔ20. The absence of the last 20 residues has no effect on mDelFC’s ability to inhibit barbed-end elongation (IC50 = 7 nM for both constructs). Data from two independent repeats are shown. Representative raw data are shown in Supplemental Figure S3C.
Delphilin is not autoinhibited
Autoinhibition is a hallmark of formins and occurs through conserved DID and DAD domains (Li and Higgs, 2005; Goode and Eck, 2007). Fmn-family formins lack canonical DID and DAD domains but are autoinhibited (Bor et al., 2012). Delphilin is one of two other formin families lacking these domains. As described above, Delphilin lacks a tail, which is the location of DAD domains. Nevertheless, we asked whether Delphilin can be autoinhibited. To do so, we purified the N-terminal half of α-Delphilin (α-mDelNT). Circular dichroism revealed the presence of a large amount of secondary structure, α-helix and β-sheet (>50%), suggesting that the α-mDelNT construct is folded (Figure S1C). Addition of α-mDelNT to actin alone does not affect actin polymerization (Figure 6A). Titrating α-mDelNT with mDelFFC revealed no change in the polymerization activity of mDelFFC (Figure 6A), indicating that actin assembly is not inhibited by the intramolecular interaction typical of formins. To assess whether there was an interaction between the two halves of the protein, we used size exclusion chromatography. When mixed together, α-mDelNT and mDelFC migrate independently through the column (Figure 6B).
FIGURE 6:
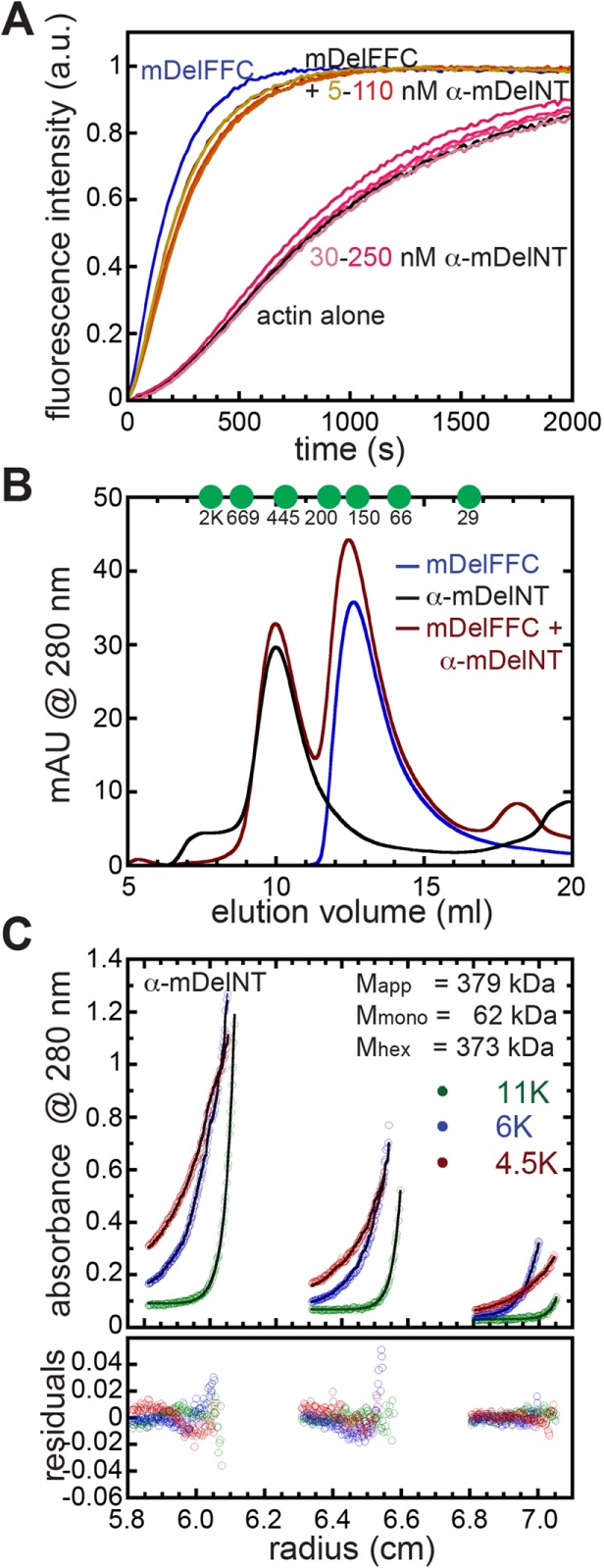
Delphilin is not autoinhibited. (A) The N-terminal half of α-Delphilin (α-mDelNT) has no effect on actin assembly whether or not mDelFFC is present. Conditions: 4 μM actin (10% pyrene labeled), 30 nM mDelFFC, and a range of α-mDelNT concentrations, as indicated. (B) Size exclusion chromatography was performed on mDelFC (blue), α-mDelNT (black), or both (red) with a Superdex S200 30/100 GL column. No differences were detected when the two proteins were mixed vs. alone. Elution peaks of standards are indicated with green circles. y-Axis is milli-absorbance units (mAU) measured at 280 nm. (C) Equilibrium sedimentation data for three concentrations of α-mDelNT at three speeds. Lines and residuals represent a global fit with a single species model. The data indicate that α-mDelNT is a hexamer. Values for the apparent mass (Mapp) and the masses predicted for α-mDelNT monomers (Mmono) and hexamers (Mhex) are given.
The N-terminal halves of most formins are dimers (Li and Higgs, 2005; Rose et al., 2005; Bor et al., 2012). To determine the molecular weight of α-mDelNT, we first used size exclusion chromatography. α-mDelNT eluted as a single peak at a volume that predicts a multimer of 7 ± 1 subunits (Figure 6B). Because size exclusion chromatography is sensitive to shape as well as size, we turned to equilibrium sedimentation. Using single species modeling we calculated the molecular weight of α-mDelNT to be 379.5 kDa. The predicted molecular mass of a hexamer is 373.5 kDa, a 2% deviation from our measurement (Figure 6C). Various models, including a trimer of dimers, a dimer of trimers, and monomer-trimer-hexamer equilibria, were tested. None of these models gives better results than a single exponential fit. We note a trend in the regressions. This could reflect one or both of the following: 1) there is a contaminant that is smaller than monomeric α-mDelNT; 2) the hexamer is not homogeneous. In both cases, the data support the presence of a dominant species as opposed to variable aggregates of unfolded protein. Thus, Delphilin may be exceptional among formins in oligomer state as well.
DISCUSSION
Actin assembly by Delphilin
Although most formins slow actin elongation in the absence of profilin, only Cdc12, Drosophila Daam (dDaam), and now Delphilin completely cap barbed ends (Kovar et al., 2003; Barkó et al., 2010). In the presence of profilin, most formins accelerate elongation. This is true for Cdc12, which accelerates elongation ∼50% over actin alone (Kovar et al., 2006). In contrast, inhibition of elongation is relieved but elongation is not accelerated by Delphilin or Daam, placing them at one end of a spectrum of possible actin elongation activities (Barkó et al., 2010 and this study). Phylogenetic analysis of formins based on FH2 domains, places Delphilin closer to Diaphanous, one of the most potent elongation factors, than to Daam (Pruyne, 2016). Thus, the profilin-binding FH1 domain, and not the FH2 domain, may be largely responsible for this extreme activity.
The number and lengths of polyproline stretches and their distances from the FH2 domain all contribute to activity levels (Paul et al., 2008; Courtemanche and Pollard, 2012). For example, experiments and simulations show that higher affinity proline stretches spaced 18 residues from the FH2 domain accelerate elongation less effectively than lower affinity proline stretches with the same spacing (Courtemanche and Pollard, 2012). In fact, the closest polyproline stretch is usually a relatively weak profilin binder, presumably facilitating actin handoff to the FH2 domain (Courtemanche and Pollard, 2012). Among the mammalian formins, the number of polyproline stretches range from 2 to 19, in Delphilin and Fmn2, respectively. The linker regions, the distance between the last polyproline repeat and the beginning of the FH2 domain, vary from 10 to 47, in FHOD1 and Fmn2, respectively. The linkers of both dDaam and mDelphilin are short; 16 and 11 residues, respectively. In contrast, the Cdc12 linker is 26 residues. Even in their fully extended conformations, the short linkers of dDaam and mDelphilin are equivalent to or shorter than the diameter of an actin monomer, which could hinder handoff. Further, Delphilin has only two polyproline stretches and the longer of the two is proximal to the start of the FH2 domain, which could also hinder handoff. Thus, the Delphilin-FH1 domain is not well designed to enhance elongation, which is consistent with our measurements.
We also note the decrease of activity in mDelFC relative to mDelFFC. Nucleation activity is generally attributed to the FH2 domain. In fact, while the FH2 domain is sufficient, nucleation may be influenced by the FH1 domain, in some cases. For example, comparison of Bni1-FC and -FFC constructs reveals a large difference in nucleation strengths (Pruyne et al., 2002; Moseley et al., 2004). In contrast, the activities of Capu-FC and -FFC constructs are indistinguishable (Quinlan et al., 2007). However, the Capu-FC construct is very unstable compared with the Capu-FFC construct (our unpublished observations). It may be that the FH1 domain does not directly contribute to nucleation but instead influences FH2 structure and, therefore, activity. The contribution of the FH1 domain to nucleation is interesting in light of the fact that several formins, including Delphilin, are only active elongation factors in the presence of their FH1 domains and profilin (Kovar et al., 2003; Michelot et al., 2005; Barkó et al., 2010). It is difficult to distinguish whether the FH1 domain strictly contributes by delivering profilin-actin to the growing filament or whether the profilin-bound FH1 domains alter gating, for example. Given Delphilin’s unusual FH1 domain and lack of a tail, its FH2 domain could be an interesting base around which to further investigate how FH1 and tail domains enhance actin assembly.
Another group recently reported a biochemical study of Delphilin (Dutta et al., 2017). They described Delphilin as a barbed-end capping protein, which is consistent with our data, in that Delphilin tightly binds barbed ends and, under nucleating conditions, only pointed end growth is observed in the absence of profilin. They also report that nucleation is completely inhibited in the presence of profilin. We see strong inhibition of nucleation by profilin as well but detect activity. They observed some relief of inhibition in seeded elongation assays plus profilin but did not examine elongation directly. We believe the actin isoform used is the major source of the differences, given the very weak activity of Delphilin with rabbit skeletal muscle actin.
Actin isoform sensitivity
To date, a systematic investigation of formin sensitivity to actin isoforms has not been performed. A comparison between muscle and nonmuscle actin was performed for FMNL1, but the activities were found to be largely similar (Harris et al., 2004). Experiments are usually performed with muscle α-actin. However, most formins do not play a role in sarcomerogenesis. Thus activity differences could be functionally significant. Mammals have six actin isoforms, four of which are generally exclusive to muscle (smooth, cardiac, or skeletal). β- and γ-actin are ubiquitous and the term cytoplasmic actin refers to a mixture of these two isoforms. None of the six human isoforms differs from another by >7% and the majority of the differences are biochemically similar substitutions within the first 10 residues of the proteins. Thus, one might not expect to observe significant differences in interactions with actin-binding proteins. However, actin isoforms do not effectively compensate for one another in vivo, suggesting that their differences are mediated by more than expression patterns (Perrin and Ervasti, 2010). Indeed, several actin-binding proteins have been found to interact preferentially with cytoplasmic or muscle actin, including cofilin (De La Cruz and Pollard, 1996) and ezrin (Yao et al., 1996) among others.
We were intrigued to find such a stark difference in activity when Delphilin was combined with cytoplasmic versus muscle actin. As is the case for mammalian isoforms, conservation is high between the isoforms we tested (O. cunninculus α-actin vs. H. sapiens β-actin: 92.8% ; A. castellani vs. H. sapiens β-actin: 95.6%) and the majority of differences are at the N-terminus. To search for possible sources of actin isoform discrimination, we compared the sequences of human β-actin and rabbit α-actin. There are only 11 nonconservative amino acid substitutions between the two sequences. We examined the positions of these differences in the cocrystal structure of the Bni1 FH2 domain with rabbit α-actin (PDB ID: 1Y64). Only three of the differences are within 6 Å of the FH2 domain. One, Asn225 (rabbit α-actin numbering), is proximal to helix P within the coiled-coil region of the FH2 domain (Figure 7). The two at the N-terminus of actin, Thr5 and Thr6, are proximal to the FH2 linker. Formin linkers are flexible regions between the two “hemidimers” of the FH2 domain. They vary in length and sequence, which could be important for defining isoform specificity. Simulations also point to formin-linker/actin N-terminal interactions playing a role in actin assembly (Baker et al., 2015).
FIGURE 7:
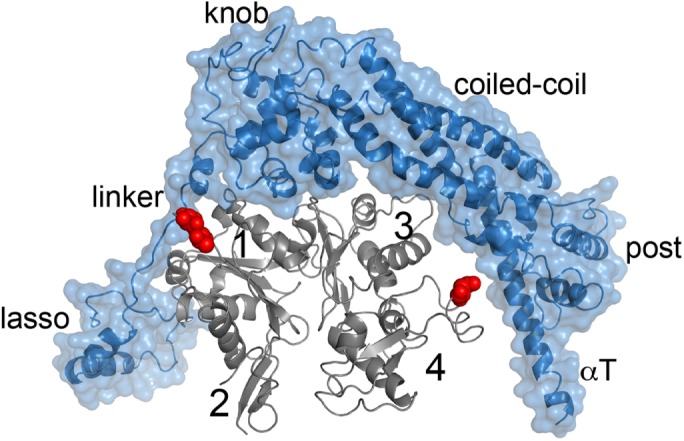
Actin sequence variation near the FH2/actin interface. The Bni1 FH2 domain (blue)/ Oryctolagus cunniculus α-actin (gray) cocrystal structure (1Y64) provides a model for how actin sequence variation may determine formin interactions. Of the 11 nonconservative mutations between human β-actin and O. cunninculus α-actin, three residues (Thr5, Thr6, and Asn225) are within 6 Å of the FH2 domain. They are highlighted with red spheres. The N-terminal Thr residues are positioned to make van der Waals contacts with the FH2 linker region.
We also found that human Fhod-1 is sensitive to actin isoform, nucleating A. castellani actin more potently than rabbit muscle actin, similar to Delphilin (unpublished data). Fhod1 is found in nonmuscle cells and costameres but not sarcomeres of muscle cells (Al Haj et al., 2015; Antoku et al., 2015). The second human Fhod-family isoform, Fhod3, is found in sarcomeres. Drosophila have one Fhod gene with multiple splice variants that all contain the same FH1 and FH2 domains, and Drosophila Fhods are found both in sarcomeres and nonmuscle structures (Lammel et al., 2014; Shwartz et al., 2016). Logically, Drosophila Fhod nucleates both A. castellani and muscle actin potently. A comparison of Drosophila Fhod with Fhod1 and Fhod3 reveals that the linkers are less well conserved than the rest of the FH2 domains, supporting the idea that specificity may be conferred by this domain. Intriguingly, α-actin is expressed in motoneurons and distinct roles for α-, β-, and γ-actin have been reported (Moradi et al., 2017). Thus, there may be a link between isoform specificity and function for some formins, including Fhod and Delphilin.
While revising this paper, a report was published suggesting that mDia2 specifically nucleates β-actin over γ-actin (Chen et al., 2017), further reinforcing the need to consider actin isoform when studying formin activity.
Regulation of Delphilin
Unlike all other characterized formins, Delphilin does not appear to be autoinhibited. The absence of an appreciable tail-domain C-terminal to the FH2 domain eliminates the possibility of canonical DID/DAD autoinhibition (Li and Higgs, 2005; Goode and Eck, 2007). However, an intramolecular interaction involving the FH2 domain could also inhibit actin assembly. We detected no evidence of such an interaction.
The N-terminal halves of mDia1, Capu, and most other formins are dimers (Otomo et al., 2005; Rose et al., 2005; Bor et al., 2012). In contrast, the N-terminus of Delphilin forms a hexamer, further distinguishing it from canonical DID/DAD regulation. Multimerization is reported for other PDZ domain–containing proteins. Specifically, PSD-95 has been shown to multimerize, clustering other proteins at membranes (Kim et al., 1995). Two N-terminal cysteines (Cys-3 and Cys-5) in the PDZ domain of PSD-95 are required for multimerization (Hsueh et al., 1997). There are two cysteines in the N-terminal portion of Delphilin-PDZ (Cys-3 and Cys-29) suggesting that the multimerization may be mediated by a mechanism similar to that of PSD-95. Although our data, circular dichroism, SEC, and equilibrium sedimentation all provide evidence that α-mDelNT is folded, we cannot exclude the possibility that the purified α-mDelNT construct is partially unfolded in a manner that impacts autoinhibitory interactions. However, it is interesting to consider the possibility that that the oligomeric state of the N-terminus may prevent an interaction with the C-terminal domain. In this case, Delphilin could be autoinhibited and activation would lead to oligomerization. If so, activation could be stimulated or stabilized by interaction of the N-terminal PDZ domains with GluRδ2.
We note that three splice variants of Delphilin have been reported: α-, β-, and L-Delphilin (Yamashita et al., 2005; Matsuda et al., 2006). They vary in their N-terminal halves but not their C-terminal halves, meaning that none has a C-terminal “tail.” The α variant is the shortest transcript and can be palmitoylated at Cys-3 (Matsuda et al., 2006). The first five residues of α-Delphilin are spliced out and replaced either with 12 residues (β-Delphilin) or 184 residues including a second PDZ domain (L-Delphilin). Given the small difference between α- and β-Delphilin, we do not expect this variant to be autoinhibited. We cannot rule out intramolecular interactions in the case of L-Delphilin. Immunofluorescence images in cultured neurons indicate that the N-termini are important for localization. α-Delphilin is specifically localized in dendritic spines, whereas β-Delphilin is relatively diffuse in both dendritic spines and the shafts of spines (Yamashita et al., 2005; Matsuda et al., 2006). In contrast, L-Delphilin forms clusters that are predominantly within the neuron shafts (Matsuda et al., 2006). Coordination of localization and activity has been reported for other formins (Seth et al., 2006; Ramalingam et al., 2010). Whether this is the case for Delphilin remains to be determined. In the absence of autoinhibition, one can only speculate about regulation. Two likely possibilities are interactions with an unknown partner and/or posttranslational modification. Although Delphilin is a weak elongation factor, it is a relatively strong nucleator, making it unlikely that its activity goes unchecked.
Two new families of formins were recently identified: Multiple Wing Hairs–related Formin (MWHF) and Pleckstrin Homology domain–containing Formins (PHCF; Pruyne, 2016). Formins in the MWHF family have conserved DID and DAD domains and are likely to be autoinhibited. Formins in the PHCF family do not have these domains. Instead, they have Pleckstrin homology (PH) domains. Most have more than one PH domain and some have C-terminal PH domains in addition to or instead of N-terminal PH domains. It is interesting to note that PDZ and PH domains both play roles in coordinating proteins at the membrane. Perhaps the Delphilin and PHCF families of formins evolved, function, and/or are regulated in similar ways, although they are not closely related based on FH2 sequences.
Role in dendritic spines
How the actin cytoskeleton is regulated within dendritic spines affects their structure and function (Hlushchenko et al., 2016; Lei et al., 2016). We set out to determine the biochemical characteristics of the formin, Delphilin, because it is the only formin specifically expressed in dendritic spines. mDia2, Daam1, and the Arp2/3 complex are all implicated in distinct stages of spine formation, including filopodia formation and head expansion (Salomon et al., 2008; Hotulainen et al., 2009). Interestingly, Delphilin is not required for spine formation (Takeuchi et al., 2008). Instead, it binds to the C-terminus of GluRδ2, a receptor crucial to LTD (Miyagi et al., 2002). Other PDZ domain proteins bind the same site of GluRδ2 and have been implicated in transduction of signals leading to LTD (Kohda et al., 2007). In contrast, induction of LTD is enhanced in the absence of Delphilin, suggesting that it plays an inhibitory role in this process (Takeuchi et al., 2008). Competition for GluRδ2-binding is an obvious possibility. At this time, we can only speculate about a mechanism and the significance of nucleation by Delphilin. For example, Delphilin could build a structure that stabilizes its interaction with GluRδ2, limiting the downstream response to activation of this receptor. Based on the work presented here, the knockout mouse already made by others, and tools available to perform “rescue” experiments in cerebellar slices, it is now possible to directly test whether nucleation activity is essential (Kohda et al., 2007; Takeuchi et al., 2008).
Finally, we consider the other formin known to be a poor elongation factor, dDaam (Barkó et al., 2010). It is implicated in a range of roles, including tracheal tube formation, growth cone filopodia formation, sarcomerogenesis, axonal growth, and even noncanonical Wnt signaling (Sato et al., 2006; Salomon et al., 2008; Barkó et al., 2010; Higashi et al., 2010; Li et al., 2011). dDaam could be specialized to build small structures needed in a wide number of cells and processes. Alternatively, elongation by formins like dDaam and Delphilin may be enhanced by additional binding partners in a manner similar to that recently described for mDia1 and CLIP-170 (Henty-Ridilla et al., 2016).
MATERIALS AND METHODS
DNA constructs
mDelFFC (aa 559–1024; NP_579933.1), mDelFC (aa 626–1024), and α-mDelNT (aa 1–558) were generated by PCR amplification of a mDelphilin-FL template that was kindly provided by S. Kawamoto (Chiba University). These constructs were subcloned into a modified pET-15b+ plasmid with a BamHI site immediately following the N-terminal His tag. mDelFC point mutations, I718A and K868A, as well as the mDelFC tail truncations were introduced by QuikChange Site Directed Mutagenesis (Stratagene; Santa Clara, CA). hDelFFC (aa: 744–1211; NP_01138590) was assembled from synthetic DNA, which was codon optimized for Escherichia coli expression (IDT), and cloned into pET-15b+.
Protein expression and purification
All Delphilin constructs were expressed in Rosetta-I or Rosetta-II (DE3) pLysS-competent cells. Cells were grown in Terrific Broth at 37°C until they reached an O.D. 600 nm of 0.7–1.0; the temperature was then lowered to 18°C for ∼1 h. Cells were induced with 0.25 mM isopropyl-β-d-thiogalactoside and harvested after 14–17 h. Cell pellets were then flash frozen in liquid nitrogen and stored at −80°C.
All C-terminal constructs were purified and stored using published protocols, with the exception that mDelFFC was stored in 10 mM Tris, pH 8, 50 mM KCl, 1 mM dithiothreitol (DTT), and 50% glycerol (Vizcarra et al., 2011). Briefly, protein was purified on a Talon column followed by a monoQ column. α-mDelNT cell pellets were resuspended in lysis buffer (50 mM Tris, pH 7, 150 mM KCl, 1 mM DTT) supplemented with 2 mM phenylmethanesulfonyl fluoride (PMSF) and 1 μg/ml DNaseI. All following steps were carried out on ice or at 4°C. Resuspended cells were lysed with a microfluidizer (Microfluidics, Newton, MA) and clarified lysate was run over a Talon column (GE Healthcare). α-mDelNT was eluted from resin with 200 mM imidazole and spin concentrated in an Amicon 10-kDa–molecular weight cut off centrifugal filter unit and gel filtered on a Superdex 200 10/300 GL column (GE Healthcare) in 10 mM Tris, pH 8, 50 mM KCl, and 1 mM DTT. Fractions of α-mDelNT were pooled based on SDS–PAGE analysis and then dialyzed into 10 mM Tris, pH 8, 50 mM KCl, 1 mM DTT, and 50% glycerol. Aliquots were flash frozen in liquid nitrogen and stored at −80°C. The resulting samples were >95% pure (Supplemental Figure 1A).
Protein concentrations of C-terminal constructs are reported in terms of dimer concentration. α-mDelNT concentration is reported as a monomer. Protein concentrations were determined by analyzing five serial dilutions of the sample and a standard by SDS−PAGE. The gels were stained with SyproRed (Invitrogen) and imaged on a Pharos FX Plus molecular imager with Quantity One software (Bio-Rad).
A. castellani actin purification and labeling with Oregon green 488 and Cy3b was performed according to previously published protocols (MacLean-Fletcher and Pollard, 1980; Bor et al., 2012). Rabbit skeletal actin was a generous gift from the Reisler laboratory (University of California, Los Angeles). Cytoplasmic actin purified from human platelets (85% beta, 15% gamma) was purchased from Cytoskeleton (APHL99-A). Unless otherwise indicated, assays were performed with Acanthamoeba actin.
Pyrene–actin polymerization assays
Pyrene–actin assembly assays were performed as described in Zalevsky et al. (2001). Briefly, 4 μM A. castellani actin (10% pyrene labeled) was incubated for 2 min at 25°C with ME buffer (final concentration, 200 μM ethylene glycol tetraacetic acid [EGTA] and 50 μM MgCl2) to convert Ca-G–actin to Mg-G–actin. Polymerization was initiated by adding KMEH polymerization buffer (final concentration, 10 mM Na-HEPES, pH 7.0, 1 mM EGTA, 50 mM KCl, 1 mM MgCl2) to the Mg-G–actin. Additional components, such as mDelFFC, hDelFFC, m-αDelNT, mDelFC, and its associated mutants were combined in the polymerization buffer before addition to Mg-G–actin. For experiments that included profilin, a 3:1 M ratio of profilin was added to actin before ME. In these and all pyrene–actin-based assays we used profilin from Schizosaccharomyces pombe because it is not sensitive to labeled versus unlabeled actin. In control experiments, we did not detect a difference in nucleation when profilin from either S. pombe or H. sapiens was used. In all other assays we used H. sapiens profilin-1. For seeded elongation assays 5 μM actin was polymerized overnight. A. castellani actin (0.5 μM; 10% pyrene labeled) and F-actin seeds (0.25 μM) were used. Delphilin constructs were added to either actin alone or 1.5 μM profilin plus 0.5 μM actin.
Pyrene actin fluorescence was monitored using a TECAN F200 with  .
.
TIRF microscopy assays
Coverslips were PEGylated by two different methods: 1) incubation with poly-l-lysine PEG or 2) treatment with 3-aminopropyltrethoxysilane followed by PEG-NHS (3% biotin-PEG-NHS) as described (Bor et al., 2012). No difference in rate was observed between the two immobilization methods. Flow cells were blocked for 2 min with Pluronic F-127 (Sigma) and 50 μg/ml κ-casein in phosphate-buffered saline and then washed with TIRF buffer (final concentration, 10 mM Na-HEPES, pH 7.0, 1 mM EGTA, 50 mM KCl, 1 mM MgCl2). Experiments were performed with 2 μM Mg-G–actin (15% Cy3b labeled), 6 μM human profilin-1, 5 nM phalloidin-stabilized actin seeds, and 10 nM mDelFFC in TIRF buffer, supplemented with 0.5% methylcellulose, 50 mM DTT, 0.2 mM ATP, 50 μg/ml catalase, 50 μg/ml κ-casein, 250 μg/ml glucose oxidase, and 20 mM glucose. Images were collected every 10 s on a DMI6000 TIRF microscope (Leica). Data were analyzed using the JFilament plug-in (Smith et al., 2010) to Fiji (Schindelin et al., 2012).
Actin filament cosedimentation assays
Side binding.
Actin (25 μM) was polymerized in 1 × KMEH for 1 h at room temperature before adding a 1:1 M ratio of phalloidin. mDelFFC was precleared by centrifugation at 117,000 × g for 20 min at 4°C. mDelFFC (0.5 μM) was incubated for 30 min at 25°C with actin (0, 0.625, 1.25, 2.5, 5, 10, and 15 μM). The polymerized filaments were transferred using cut pipette tips to avoid shearing. Samples were centrifuged at 89,000 × g for 20 min at 4°C. The supernatants and pellets were analyzed by SDS–PAGE. The gels were stained with SyproRed and imaged using a Pharos FX Plus molecular imager with Quantity One software (Bio-Rad).
Bundling.
Each formin (1.25, 2.5, and 5 μM) was incubated for 30 min at 25°C with 5 μM (rabbit skeletal muscle) actin. The Drosophila formin CapuFFC was used as a positive control for bundling. Samples were centrifuged at 12,000 × g for 15 min at 4°C. Supernatants and pellets were separated, and pellet fractions were concentrated fourfold for SDS–PAGE analysis. Protein bands were stained with SyproRed and imaged on a Typhoon FLA 7000 (GE).
Circular dichroism
α-mDelNT was dialyzed into 10 mM Tris, 50 mM KCl, and 1 mM DTT (pH 8). Circular dichroism (CD) spectra were measured on a J-715 spectropolarimeter (Jasco, Tokyo, Japan) by averaging two wavelength scans from 195 to 280 nm. Data were analyzed using Jasco and Sofsec1 software packages, which compare the acquired data to a database of spectra from proteins with known secondary structure (Sreerama and Woody, 1993).
hDelFFC was brought to 1 μM concentration with 10 mM NaCl, 50 mM sodium phosphate, and 1 mM DTT (pH 7). CD spectra were collected on a 62DS spectropolarimeter (Aviv Biomedical). For thermal denaturation, temperature was increased in increments of 1°C and held at each temperature for 1 min.
Analytical ultracentrifugation
Sedimentation equilibrium analytical ultracentrifugation was performed on α-mDelNT dialyzed in 10 mM Tris, 50 mM KCl, and 0.5 mM TCEP (pH 8.0). Three different concentrations of protein (4.8, 2.5, and 1.1 μM) were spun at 4°C in an Optima XL-A Analytical Ultracentrifugation system (Beckman Coulter, Brea, CA) at three speeds: 4500, 6000, and 11,000 rpm. Scans between 24 and 28 h were collected and analyzed.
Supplementary Material
Acknowledgments
We thank Susumu Kawamoto (Chiba University) for the original mouse Delphilin constructs and the Reisler laboratory for rabbit skeletal muscle. Thanks also to the Reisler, Quinlan, and Vizcarra laboratories for helpful discussions throughout the project. This work was supported by the generous support of Barnard College, the Sally Chapman Fund, and the Sherman Fairchild Foundation (C.L.V. and G.C.N.), and a National Institute of General Medical Sciences grant (No. R01 GM096133) to M.E.Q.
Abbreviations used:
- DAD
Diaphanous autoinhibitory domain
- DID
Diaphanous inhibitory domain
- Dlg1
Drosophila disk large tumor suppressor
- FH1
formin homology 1
- FH2
formin homology 2
- LTD
long-term depression
- PDZ
postsynaptic density protein (PSD95)
- PSD
postsynaptic density
- ZO-1
zonula occludens-1 protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0363) on December 27, 2017.
REFERENCES
- Al Haj A, Mazur AJ, Radaszkiewicz K, Radaszkiewicz T, Makowiecka A, Stopschinski BE, Schönichen A, Geyer M, Mannherz HG. (2015). Distribution of formins in cardiac muscle: FHOD1 is a component of intercalated discs and costameres. Eur J Cell Biol , 101-113. [DOI] [PubMed] [Google Scholar]
- Antoku S, Zhu R, Kutscheidt S, Fackler OT, Gundersen GG. (2015). Reinforcing the LINC complex connection to actin filaments: the role of FHOD1 in TAN line formation and nuclear movement. Cell Cycle Georget Tex , 2200-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Courtemanche N, Parton DL, McCullagh M, Pollard TD, Voth GA. (2015). Electrostatic interactions between the Bni1p Formin FH2 domain and actin influence actin filament nucleation. Struct Lond Engl 1993 , 68-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkó S, Bugyi B, Carlier M-F, Gombos R, Matusek T, Mihály J, Nyitrai M. (2010). Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. J Biol Chem , 13154-13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, Vizcarra CL, Phillips ML, Quinlan ME. (2012). Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol Biol Cell , 3801-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Arora PD, McCulloch CA, Wilde A. (2017). Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat Commun , 1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. (2012). Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem , 7812-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Pollard TD. (1996). Kinetics and thermodynamics of phalloidin binding to actin filaments from three divergent species. Biochemistry (Mosc.) , 14054-14061. [DOI] [PubMed] [Google Scholar]
- Dutta P, Das S, Maiti S. (2017). Non diaphanous formin delphilin acts as a barbed end capping protein. Exp Cell Res , 163-169. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. (2007). Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem , 593-627. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. (2011). The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol , 384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. (1992). Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci , 2685-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. (2004). The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem , 20076-20087. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. (2006). Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem , 14383-14392. [DOI] [PubMed] [Google Scholar]
- Heimsath EG, Higgs HN. (2012). The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem , 3087-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Rankova A, Eskin JA, Kenny K, Goode BL. (2016). Accelerated actin filament polymerization from microtubule plus ends. Science , 1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Ikeda T, Murakami T, Shirakawa R, Kawato M, Okawa K, Furuse M, Kimura T, Kita T, Horiuchi H. (2010). Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase. J Biol Chem , 16231-16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. (2005). Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell , 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlushchenko I, Koskinen M, Hotulainen P. (2016). Dendritic spine actin dynamics in neuronal maturation and synaptic plasticity. Cytoskeleton (Hoboken) , 435-441. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P. (2009). Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol , 323-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Kim E, Sheng M. (1997). Disulfide-linked head-to-head multimerization in the mechanism of ion channel clustering by PSD-95. Neuron , 803-814. [DOI] [PubMed] [Google Scholar]
- Kashiwabuchi N, Ikeda K, Araki K, Hirano T, Shibuki K, Takayama C, Inoue Y, Kutsuwada T, Yagi T, Kang Y. (1995). Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell , 245-252. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. (1995). Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature , 85-88. [DOI] [PubMed] [Google Scholar]
- Kohda K, Kakegawa W, Matsuda S, Nakagami R, Kakiya N, Yuzaki M. (2007). The extreme C-terminus of GluRdelta2 is essential for induction of long-term depression in cerebellar slices. Eur J Neurosci , 1357-1362. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell , 423-435. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol , 875-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel U, Bechtold M, Risse B, Berh D, Fleige A, Bunse I, Jiang X, Klämbt C, Bogdan S. (2014). The Drosophila FHOD1-like formin Knittrig acts through Rok to promote stress fiber formation and directed macrophage migration during the cellular immune response. Development , 1366-1380. [DOI] [PubMed] [Google Scholar]
- Lei W, Omotade OF, Myers KR, Zheng JQ. (2016). Actin cytoskeleton in dendritic spine development and plasticity. Curr Opin Neurobiol , 86-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Hallett MA, Zhu W, Rubart M, Liu Y, Yang Z, Chen H, Haneline LS, Chan RJ, Schwartz RJ, et al. (2011). Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development , 303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. (2003). The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol , 1335-1340. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. (2005). Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem , 6986-6992. [DOI] [PubMed] [Google Scholar]
- MacLean-Fletcher S, Pollard TD. (1980). Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem Biophys Res Commun , 18-27. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Matsuda S, Gladding CM, Yuzaki M. (2006). Characterization of the delta2 glutamate receptor-binding protein delphilin: splicing variants with differential palmitoylation and an additional PDZ domain. J Biol Chem , 25577-25587. [DOI] [PubMed] [Google Scholar]
- Michelot A, Guérin C, Huang S, Ingouff M, Richard S, Rodiuc N, Staiger CJ, Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell , 2296-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi Y, Yamashita T, Fukaya M, Sonoda T, Okuno T, Yamada K, Watanabe M, Nagashima Y, Aoki I, Okuda K, et al. (2002). Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci , 803-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi M, Sivadasan R, Saal L, Lüningschrör P, Dombert B, Rathod RJ, Dieterich DC, Blum R, Sendtner M. (2017). Differential roles of α-, β-, and γ-actin in axon growth and collateral branch formation in motoneurons. J Cell Biol , 793-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. (2004). A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell , 896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K. (2002). Structure and function of dendritic spines. Annu Rev Physiol , 313-353. [DOI] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. (2005). Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell , 273-281. [DOI] [PubMed] [Google Scholar]
- Paul AS, Paul A, Pollard TD, Pollard T. (2008). The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol , 9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen J-E, Woolfrey KM. (2011). Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci , 285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, Ervasti JM. (2010). The actin gene family: function follows isoform. Cytoskeleton , 630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Kaiserman-Abramof IR. (1970). The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat , 321-355. [DOI] [PubMed] [Google Scholar]
- Pruyne D. (2016). Revisiting the phylogeny of the animal formins: two new subtypes, relationships with multiple wing hairs proteins, and a lost human formin. PLoS One , e0164067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science , 612-615. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. (2007). Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol , 117-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramabhadran V, Gurel PS, Higgs HN. (2012). Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J Biol Chem , 34234-34245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam N, Zhao H, Breitsprecher D, Lappalainen P, Faix J, Schleicher M. (2010). Phospholipids regulate localization and activity of mDia1 formin. Eur J Cell Biol , 723-732. [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. (2006). Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol , 367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. (2005). Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature , 513-518. [DOI] [PubMed] [Google Scholar]
- Rubenstein PA. (1990). The functional importance of multiple actin isoforms. Bioessays , 309-315. [DOI] [PubMed] [Google Scholar]
- Salomon SN, Haber M, Murai KK, Dunn RJ. (2008). Localization of the Diaphanous-related formin Daam1 to neuronal dendrites. Neurosci Lett , 62-67. [DOI] [PubMed] [Google Scholar]
- Sato A, Khadka DK, Liu W, Bharti R, Runnels LW, Dawid IB, Habas R. (2006). Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development , 4219-4231. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönichen A, Mannherz HG, Behrmann E, Mazur AJ, Kühn S, Silván U, Schoenenberger C-A, Fackler OT, Raunser S, Dehmelt L, Geyer M. (2013). FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J Cell Sci , 1891-1901. [DOI] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK. (2006). Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol , 701-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz A, Dhanyasi N, Schejter ED, Shilo B-Z. (2016). The Drosophila formin Fhos is a primary mediator of sarcomeric thin-filament array assembly. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MB, Li H, Shen T, Huang X, Yusuf E, Vavylonis D. (2010). Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton (Hoboken) , 693-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. (1993). A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem , 32-44. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Ohtsuki G, Yoshida T, Fukaya M, Wainai T, Yamashita M, Yamazaki Y, Mori H, Sakimura K, Kawamoto S, et al. (2008). Enhancement of both long-term depression induction and optokinetic response adaptation in mice lacking delphilin. PLoS One , e2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Bor B, Quinlan ME. (2014). The role of formin tails in actin nucleation, processive elongation, and filament bundling. J Biol Chem , 30602-30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. (2011). Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci USA , 11884-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW, Mercer JA, Kennedy MB. (2000). Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci , 4069-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell , 711-723. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Miyagi Y, Ono M, Ito H, Watanabe K, Sonoda T, Tsuzuki K, Ozawa S, Aoki I, Okuda K, et al. (2005). Identification and characterization of a novel Delphilin variant with an alternative N-terminus. Brain Res Mol Brain Res , 83-94. [DOI] [PubMed] [Google Scholar]
- Yao X, Cheng L, Forte JG. (1996). Biochemical characterization of ezrin-actin interaction. J Biol Chem , 7224-7229. [DOI] [PubMed] [Google Scholar]
- Yoo H, Roth-Johnson EA, Bor B, Quinlan ME. (2015). Drosophila Cappuccino alleles provide insight into formin mechanism and role in oogenesis. Mol Biol Cell , 1875-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, Mullins RD. (2001). Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem , 3468-3475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.