Abstract
In Saccharomyces cerevisiae, it is well established that Hof1, Cyk3, and Inn1 contribute to septum formation and cytokinesis. Because hof1∆ and cyk3∆ single mutants have relatively mild defects but hof1∆ cyk3∆ double mutants are nearly dead, it has been hypothesized that these proteins contribute to parallel pathways. However, there is also evidence that they interact physically. In this study, we examined this interaction and its functional significance in detail. Our data indicate that the interaction 1) is mediated by a direct binding of the Hof1 SH3 domain to a proline-rich motif in Cyk3; 2) occurs specifically at the time of cytokinesis but is independent of the (hyper)phosphorylation of both proteins that occurs at about the same time; 3) is dispensable for the normal localization of both proteins; 4) is essential for normal primary-septum formation and a normal rate of cleavage-furrow ingression; and 5) becomes critical for growth when either Inn1 or the type II myosin Myo1 (a key component of the contractile actomyosin ring) is absent. The similarity in phenotype between cyk3∆ mutants and mutants specifically lacking the Hof1–Cyk3 interaction suggests that the interaction is particularly important for Cyk3 function, but it may be important for Hof1 function as well.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, cleavage-furrow formation and cytokinesis are achieved by the coordinated actions of the septins, the contractile actomyosin ring (CAR), and the enzymes that reorganize the plasma membrane and form the septal cell wall (Weiss, 2012; Wloka and Bi, 2012; Onishi et al., 2013; Foltman et al., 2016; Meitinger and Palani, 2016). The septins are a family of GTP-binding, filament-forming proteins that localize to the bud neck throughout the cell cycle and play an essential role in cytokinesis that involves the scaffolding of other proteins but remains incompletely understood (Longtine et al., 1996; Gladfelter et al., 2001; McMurray et al., 2011a,b; Oh and Bi, 2011; Wloka and Bi, 2012; Bridges and Gladfelter, 2015). Formation of the CAR depends on Myo1 (the only type II myosin in S. cerevisiae), actin, an actin-nucleating formin, several accessory proteins, and the small GTPase Rho1 (Bi et al., 1998; Lippincott and Li, 1998a; Tolliday et al., 2002; Fang et al., 2010; Pollard and Wu, 2010; Wloka and Bi, 2012; Wloka et al., 2013; Foltman et al., 2016; Meitinger and Palani, 2016). Null mutations of MYO1 are not lethal in most strain backgrounds, despite the absence of the CAR, showing that the CAR is not essential for cytokinesis in S. cerevisiae (Bi et al., 1998; Schmidt et al., 2002; Lord et al., 2005; Ko et al., 2007; Fang et al., 2010; Wloka and Bi, 2012; Wloka et al., 2013). Indeed, some myo1∆ cells form nearly normal cleavage furrows (Schmidt et al., 2002; Fang et al., 2010; our unpublished results).
Concomitant with CAR constriction, the plasma membrane invaginates, and a chitinous primary septum (PS) is deposited by the chitin synthase Chs2 (Shaw et al., 1991; Schmidt et al., 2002; Chin et al., 2012; Devrekanli et al., 2012; Weiss, 2012; Wloka and Bi, 2012; Wloka et al., 2013; Foltman et al., 2016; Meitinger and Palani, 2016). As PS formation is completed, glucan and mannan are deposited on both sides of the PS to form the secondary septa (SS; Bowers et al., 1974; Orlean, 2012; Weiss, 2012; Onishi et al., 2013). Chitin in the PS is then digested by a chitinase secreted from the daughter cell, leading to separation of the mother and daughter cells (Kuranda and Robbins, 1991; Colman-Lerner et al., 2001; Brace et al., 2011).
The assembly and function of the CAR and PS-formation machinery also depend on a set of functionally and physically interacting proteins that includes Iqg1, Hof1, Inn1, and Cyk3. Iqg1 is the only S. cerevisiae IQGAP; it is required for the formation of both the CAR (Epp and Chant, 1997; Lippincott and Li, 1998a; Shannon and Li, 1999; Fang et al., 2010; Naylor and Morgan, 2014; Miller et al., 2015) and the PS (Lippincott and Li, 1998a; Ko et al., 2007; Onishi et al., 2013; Foltman et al., 2016). Hof1 is a member of the PCH (Pombe Cdc15 Homology) protein family (Kamei et al., 1998). Although some PCH proteins induce membrane deformation by means of their “F-BAR” domains (Chitu and Stanley, 2007; Heath and Insall, 2008; Aspenström, 2009), some others (including Hof1) may not do so (McDonald et al., 2015; Moravcevic et al., 2015; McDonald, Takizawa, et al., 2016). Hof1 also contains a C-terminal SH3 domain, by which it interacts with a proline-rich motif in Inn1 (Sanchez-Diaz et al., 2008; Jendretzki et al., 2009; Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Tonikian, Xin, Toret, et al., 2009; Meitinger et al., 2011; Devrekanli et al., 2012; Labedzka et al., 2012), and a PEST domain that appears to be responsible for its cell-cycle–specific degradation (Blondel et al., 2005). Phosphorylation of Hof1 by the mitotic-exit kinase Dbf2-Mob1 appears to regulate its interactions with septins and Myo1, which are important for septin organization and CAR constriction (Meitinger et al., 2011, 2013; Wolken et al., 2014).
Inn1 is normally essential for PS formation, but the inn1∆ defect in PS formation can be suppressed by overexpression of CYK3 (Nishihama, Schreiter, Onishi, Vallen, et al., 2009). The C2-like domain at the N-terminus of Inn1 can promote PS formation even in the absence of the rest of the protein, apparently by directly activating Chs2 (Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Devrekanli et al., 2012; Foltman et al., 2016). Cyk3 (Korinek et al., 2000) also contains an SH3 domain, by which it interacts with a proline-rich motif in Inn1 that is distinct from the Hof1-binding site (Jendretzki et al., 2009; Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Labedzka et al., 2012; Palani et al., 2012; Foltman et al., 2016), and a transglutaminase-like domain that appears to be involved in the regulation of both Chs2 and Rho1, and hence of both PS and SS formation (Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Oh et al., 2012; Onishi et al., 2013; Foltman et al., 2016). Overexpression of CYK3 also suppresses the myo1∆ and iqg1∆ growth defects without restoring the CAR (Korinek et al., 2000; Ko et al., 2007; our unpublished results), suggesting that Cyk3 acts downstream from Iqg1 to promote CAR-independent cytokinesis. Electron-microscopic analyses of septum morphology in CYK3-overexpressing cells have supported the hypotheses that Cyk3 functions in membrane invagination, PS formation, and the regulation of SS formation during cytokinesis (Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Meitinger et al., 2010; Onishi et al., 2013; our unpublished results).
Hof1 and Cyk3 have been hypothesized to function in parallel pathways because hof1 and cyk3 loss-of-function mutations, although they have only moderate growth defects on their own, are nearly lethal in combination (Korinek et al., 2000; Meitinger et al., 2011; Labedzka et al., 2012). Paradoxically, however, there is also evidence suggesting that Hof1 and Cyk3 interact physically with each other via the Hof1 SH3 domain and a proline-rich motif in Cyk3 (Tonikian, Xin, Toret, et al., 2009; Meitinger et al., 2011; Labedzka et al., 2012; Onishi et al., 2013). In this study, we have used several methods to demonstrate that the proteins indeed interact via the domains suggested previously; shown that this interaction occurs specifically at the time of cytokinesis but does not depend on the phosphorylation of both proteins that occurs at about the same time; and used point mutations that disrupt the interaction to provide evidence for its importance in the function of both proteins.
RESULTS
Physical interaction between Hof1 and Cyk3
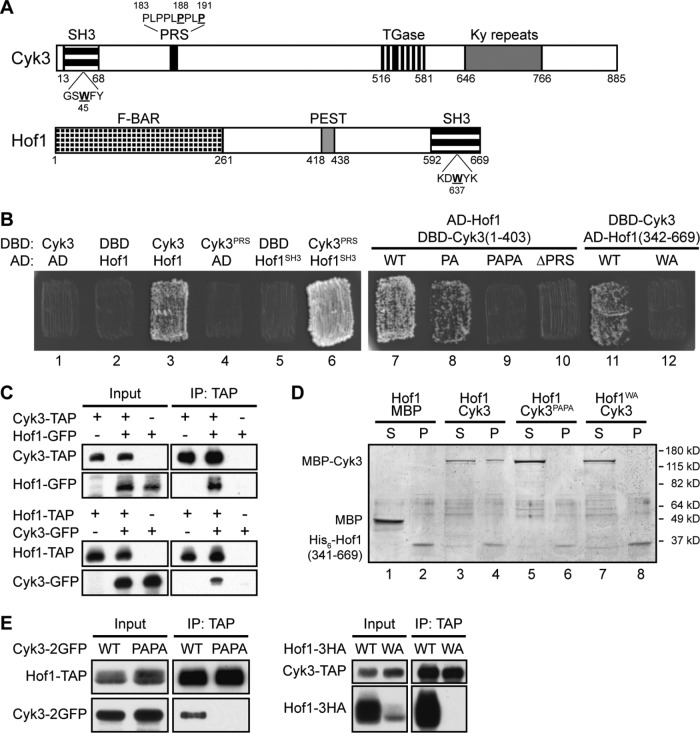
Cyk3 and Hof1 are multidomain proteins (Figure 1A) that interact with a third cytokinesis protein, Inn1, by means of the Hof1 and Cyk3 SH3 domains and distinct proline-rich sequences (PRSs) in Inn1 (see Introduction). Hof1 and Cyk3 have been hypothesized to function in parallel pathways because of the synthetic lethality between cyk3∆ and hof1∆ mutations (Korinek et al., 2000; Meitinger et al., 2011; Labedzka et al., 2012). However, in seeming conflict with this hypothesis, we and others have observed an apparent physical interaction between Hof1 and Cyk3 using yeast two-hybrid, phage-display, split-ubiquitin, and pull-down assays (Figure 1B, patches 1–3; Tonikian, Xin, Toret, et al., 2009; Meitinger et al., 2011; Labedzka et al., 2012; Onishi et al., 2013). We confirmed this interaction both by coimmunoprecipitation of the two proteins from yeast extracts (Figure 1C) and by showing coprecipitation of the two proteins in vitro after they had been expressed separately and purified from bacterial cell extracts (Figure 1D, lanes 1–4). Importantly, the in vitro coprecipitation also indicates that Hof1 and Cyk3 interact directly and not just via their mutual interactions with Inn1.
FIGURE 1:
Physical interaction between Cyk3 and Hof1 and its mediation by the Cyk3 proline-rich sequence (PRS) and the Hof1 SH3 domain. (A) Domain structures of Cyk3 and Hof1. SH3, Src-homology 3 domain; PRS, proline-rich sequence; TGase, transglutaminase-like domain; Ky repeats, repeats of a motif first identified in a protein involved in kyphoscoliosis; F-BAR, FCH (Fer/CIP4 homology)-Bin-Amphiphysin-Rvs domain; PEST, putative protein-degradation signal. Amino-acid numbers are indicated, and residues mutated in Cyk3SH3, Cyk3PRS, or Hof1SH3 are underlined. (B) Two-hybrid interaction between Cyk3 and Hof1 mediated by the Cyk3PRS and Hof1SH3 domains. Patches 1–6: the Gal4 DNA-binding domain (DBD) alone and the DBD fused to either full-length Cyk3 or a segment containing its PRS (amino-acids 177–196) were tested for interaction with the Gal4 activation domain (AD) alone and the AD fused to either full-length Hof1 or its SH3 domain (amino-acids 576–669). Patches 7–10: AD–Hof1 was tested for interaction with DBD–Cyk3 (amino-acids 1–403) with a normal or mutated PRS (PA: P188A; PAPA: P188A, P191A; ∆PRS: deletion of amino-acids 183–191). Patches 11 and 12: DBD-Cyk3 was tested for interaction with AD–Hof1 (amino-acids 342–669) with a normal or mutated SH3 domain (WA: W637A). (C) Coprecipitation of Cyk3 and Hof1 from yeast extracts. Strains MWY999 (CYK3-TAP HOF1), MWY1006 (CYK3-TAP HOF1-GFP), MWY1003 (CYK3 HOF1-GFP), MWY1019 (CYK3 HOF1-TAP), MWY1025 (CYK3-GFP HOF1-TAP), and MWY1021 (CYK3-GFP HOF1) were grown to exponential phase in YM-P medium at 24°C. The TAP-tagged proteins were precipitated, and precipitates were analyzed by Western blotting as described in Materials and Methods. Protein extracts loaded in the input lanes were 1/83 of the amounts used for the IP lanes. (D) Direct interaction in vitro between Cyk3 and Hof1 and its mediation by Cyk3PRS and Hof1SH3. Bacterially expressed His6-Hof1 (amino-acids 341–669; normal or carrying the W637A mutation), MBP, and MBP-Cyk3 (normal or carrying the P188A and P191A mutations) were purified and tested for binding in vitro by precipitating His6-Hof1 and testing for the presence of MBP or MBP-Cyk3 in the supernatant (S) and pellet (P) fractions (see Materials and Methods). The experiment was performed twice with indistinguishable results. (E) Dependence of Hof1–Cyk3 coprecipitation from yeast extracts on Cyk3PRS and Hof1SH3. Strains MWY822 (CYK3-2GFP HOF1-TAP cdc15-2), MWY761 (cyk3PAPA-2GFP HOF1-TAP cdc15-2), MWY2117 (CYK3-TAP HOF1-3HA cdc15-2), and MWY2154 (CYK3-TAP hof1WA-3HA cdc15-2) were synchronized as in Figure 2A and collected 45 min after release. Coprecipitation assays were performed as in C. Protein extracts loaded in the input lanes were 1/200 of the amounts used for the IP lanes.
To localize the regions in Cyk3 and Hof1 responsible for their interaction with each other, we first extended the two-hybrid analyses. We observed a strong interaction between a short segment of Cyk3 (amino-acids 177–196) containing its PRS and a short segment of Hof1 (amino-acids 576–669) containing its SH3 domain (Figure 1B, patches 4–6). The PRS sequence in Cyk3 agrees with that predicted by Tonikian, Xin, Toret, et al. (2009) to be favored for binding by the Hof1 SH3 domain. In addition, the two-hybrid interaction was weakened or abolished by mutations that altered the Cyk3 PRS (Cyk3PA and Cyk3PAPA) or the Hof1 SH3 domain (Hof1WA; Figure 1, A and B, patches 7–12). The same mutations also effectively eliminated the coprecipitation of the two proteins from yeast cell extracts (Figure 1E) and the interaction in vitro of bacterially expressed proteins (Figure 1D, lanes 5–8). Taken together, these data indicate that the PRS of Cyk3 and the SH3 domain of Hof1 are both necessary and sufficient for a direct interaction between the two proteins. As expected, Hof1WA was also defective for interaction with Inn1 (Supplemental Figure S1, A, patches 1–4, and B; Meitinger et al., 2011).
Cytokinesis-specific association of Hof1 and Cyk3
To determine when the Hof1–Cyk3 interaction occurs during the cell cycle, we synchronized cells at mitotic exit using the cdc15-2 mutation and performed coprecipitation experiments at various times after release from the block. Although both Cyk3 and Hof1 were present throughout the period of observation (Figure 2A, input), the Hof1–Cyk3 interaction was detectable only beginning ∼40 min after the release and appeared to be maintained even as the level of Hof1 decreased (Blondel et al., 2005; Wolken et al., 2014) following cytokinesis (Figure 2A, IP: TAP). The onset of the interaction coincided approximately with several other events, including 1) the appearance of lower-mobility forms of Hof1, Cyk3, and Inn1, which in the cases of Hof1 and Inn1 have been shown previously to represent (hyper)phosphorylated species (Figure 2A; Vallen et al., 2000; Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Meitinger et al., 2010, 2011, 2013; Palani et al., 2012; Wolken et al., 2014); 2) the localization of Cyk3 to the neck in a single, central ring at the cleavage site (Figure 3, A and B; Korinek et al., 2000; Meitinger et al., 2010; Labedzka et al., 2012; Palani et al., 2012; Onishi et al., 2013); 3) the rearrangement of Hof1 from a septin-like collar at the neck into a single, central ring at the cleavage site (Figure 3B; Vallen et al., 2000; Meitinger et al., 2011, 2013; Labedzka et al., 2012; Oh et al., 2013); and 4) the initiation of CAR contraction and cleavage-furrow ingression (which typically began at ∼40 min in parallel experiments using this synchronization technique (Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Onishi et al., 2013; our unpublished data).
FIGURE 2:

Cytokinesis-specific association of Hof1 and Cyk3 and its apparent independence from the concomitant (hyper)phosphorylation. (A) Cytokinesis-specific association of Hof1 and Cyk3. Strain MWY1025 (CYK3-GFP HOF1-TAP cdc15-2) was grown to exponential phase in YM-P medium at 24°C, shifted to 37°C for 3 h to arrest the cell cycle at mitotic exit, and then released from arrest by shifting to 24°C. See Figure 3A for images of the cells from each time point. Cells were collected at the indicated times after release, and coprecipitation assays were performed as in Figure 1C. Protein extracts loaded in the input lanes were 1/40 of the amounts used for the IP lanes. (B) Phosphorylation of Cyk3 and apparent independence of Hof1–Cyk3 association from the cell-cycle–regulated (hyper)phosphorylation of both proteins. Strains MWY1025 (lanes 1–4) and MWY2117 (CYK3-TAP HOF1-3HA cdc15-2; lanes 5–8) were synchronized as in A, and cells were collected 45 min after release. Hof1-TAP or Cyk3-TAP was precipitated, and precipitates were subjected to phosphatase (PPase) treatments as indicated.
FIGURE 3:
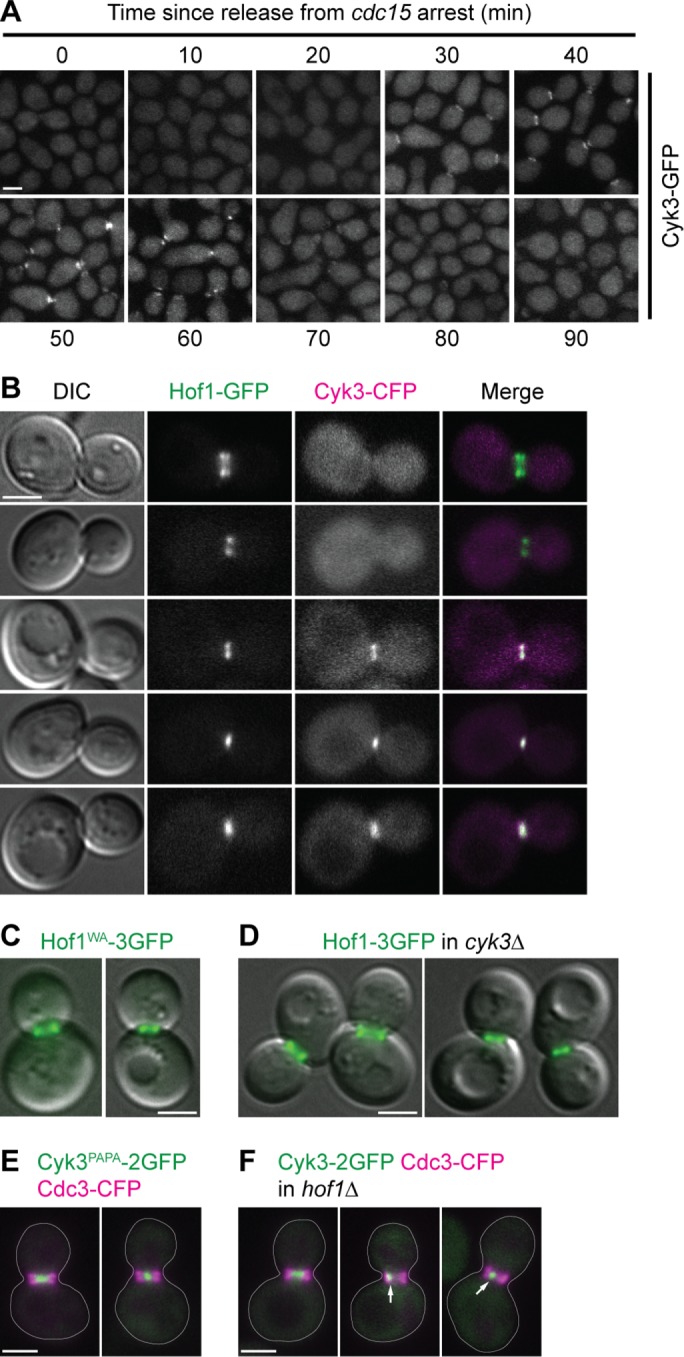
Independent localization of Hof1 and Cyk3 to the division site during cytokinesis. (A) Timing of Cyk3 localization to the cytokinesis site. Cells from the experiment in Figure 2A were examined for the localization of Cyk3–GFP. Representative fields of cells are shown. Note that these cells were from the same samples as used for the protein analyses and thus had been frozen and thawed before examination by fluorescence microscopy, presumably accounting for the dispersion of Cyk3–GFP signal (cf. panel B). (B–F) Independence of Hof1 and Cyk3 localization to the division site. Strains were grown to exponential phase in SC medium at 24°C and visualized by DIC and/or fluorescence microscopy. Each experiment was performed two or more times with indistinguishable results, and representative fields of cells are shown. (B) Localization of Hof1 to the neck before the localization of Cyk3 in wild-type cells (strain MWY1408). The top two cells are presumed to be earlier in the cell cycle than the bottom three cells. (C, D) Seemingly normal Hof1 localization in the absence of the Hof1–Cyk3 interaction (C; strain MWY1252) or in the complete absence of Cyk3 (D; strain MWY2120). Merged Hof1–3GFP fluorescence and DIC images are shown. In each panel, cells with Hof1 before (left) and after (right) rearrangement are shown. (E, F) Nearly normal Cyk3 localization in the absence of the Hof1–Cyk3 interaction (E; strain MWY2111) or in the complete absence of Hof1 (F; strain MWY2122). Merged Cyk3–2GFP (green) and Cdc3–CFP (magenta; to demarcate the entire neck region) images are shown. Cell bodies are outlined; arrows indicate asymmetric localization of Cyk3–2GFP. Scale bars (all panels), 2 µm.
Not surprisingly, protein–phosphatase experiments indicated that the slower-migrating form(s) of Cyk3, like those of Hof1 and Inn1, were phosphorylated (Figure 2B). Interestingly, in these experiments, we also observed that precipitation of Hof1-TAP pulled down both the higher- and lower-mobility forms of Cyk3, and the higher-mobility form appeared to comigrate with the species produced by phosphatase treatment (Figure 2B, lanes 1–4). Similarly, precipitation of Cyk3-TAP pulled down both higher- and lower-mobility forms of Hof1, although in this case the highest-mobility form present in the input or precipitate appeared to migrate slightly more slowly than the species produced by phosphatase treatment (Figure 2B, lanes 5–8), suggesting that a basal phosphorylation of Hof1 may be succeeded by a hyperphosphorylation at the time of cytokinesis (as suggested previously by Vallen et al., 2000). Taken together with the evidence for binding of bacterially produced Cyk3 and Hof1 (Figure 1D; Labedzka et al., 2012), these results suggest that the cell-cycle–regulated onset of association between Cyk3 and Hof1 is independent of the phosphorylation/hyperphosphorylation of the two proteins that occurs at about the same time. The hyperphosphorylation of Hof1 also does not appear to affect (positively or negatively) its association with Inn1 (Supplemental Figure S1C). Our results do not address whether the putative basal phosphorylation of Hof1 might be necessary for its association with Cyk3 and/or Inn1.
Independent localization of Hof1 and Cyk3 to the division site
The spatial and temporal coincidences noted above also suggested that the localization of Cyk3 to the neck, the rearrangement of Hof1 organization at the neck, or both might depend on the association between the two proteins. However, Cyk3-GFP became detectable at the neck 5–10 min before the association between the proteins was detectable by coprecipitation (Figures 2A and 3A), and the rearrangement of Hof1 appeared to occur even earlier: among 102 cells examined in the experiment of Figure 3B, all 36 in which Hof1 retained the collar-like arrangement showed no localized Cyk3 (e.g., Figure 3B, top row), while 10 with rearranged Hof1 still had no detectable colocalized Cyk3 (e.g., Figure 3B, second row). Moreover, both stages of Hof1 localization appeared normal either when the Hof1–Cyk3 interaction was disrupted by a hof1WA mutation (Figure 3C) or when Cyk3 was absent altogether (Figure 3D). In addition, Cyk3 localization appeared nearly normal both when the Hof1–Cyk3 interaction was disrupted by a cyk3PAPA mutation (Figure 3E) and in most cells that lacked Hof1 altogether (Figure 3F). However, in 18 of 50 hof1∆ cells examined, Cyk3-2GFP appeared as an asymmetric dot (Figure 3F, arrows). Because such abnormal Cyk3 localization was not seen in cyk3PAPA-2GFP or hof1WA cells (Figure 3E and our unpublished results) and parallels the asymmetric localizations of Myo1 (Lippincott and Li, 1998b), Inn1 (Nishihama, Schreiter, Onishi, Vallen, et al., 2009), and Chs2 (Supplemental Figure S2, arrows) that are also seen in some hof1∆ cells, we suggest that it does not directly reflect the loss of Hof1–Cyk3 interaction but rather is secondary to a more general abnormality of cleavage-furrow organization in a fraction of hof1∆ cells (see also below).
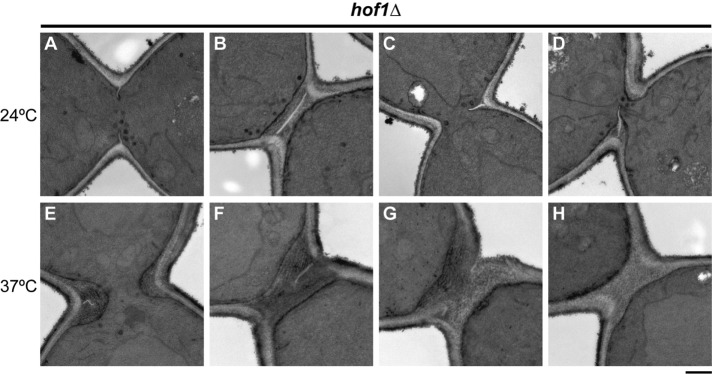
Abnormal PS formation in the absence of Hof1
As reported previously (Kamei et al., 1998; Lippincott and Li, 1998b; Vallen et al., 2000), hof1∆ cells grow well at 24°C but show obvious growth and cell-division defects at 37°C. Consistent with these observations, electron microscopy revealed approximately normal PS structures and SS structures in most of the cells examined from a culture grown at 24°C (Figure 4, A and B). However, some cells showed SS without any evident PS, and other cells showed PSs that appeared to be growing asymmetrically from one side of the neck (Figure 4, C and D). Such abnormalities were more severe in cells grown at 37°C. Of 63 cells examined, 21 showed asymmetric PSs like those seen at 24°C (Figure 4, E and F), nine showed seemingly symmetric but incomplete PSs (Figure 4G), and 33 showed SS without any evident PS (Figure 4H). These results indicate that like Inn1 (Sanchez-Diaz et al., 2008; Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Foltman et al., 2016) and Cyk3 (Onishi et al., 2013; Foltman et al., 2016), Hof1 is important for normal PS formation (as also indicated by previously published data [ Meitinger et al., 2011; Oh et al., 2013]) and appear to explain why a variety of furrow-associated proteins would show asymmetric localization in a subset of hof1∆ cells (see above).
FIGURE 4:
Abnormal PS formation in the absence of Hof1. Strain RNY370 (hof1∆) was grown to exponential phase at 24°C (A–D) or 37°C (E–H) in SC medium and examined by electron microscopy (see Materials and Methods). Representative images are shown (see text for quantitation of the phenotypes observed). Scale bar, 0.5 µm.
Functional significance of the Hof1–Cyk3 interaction
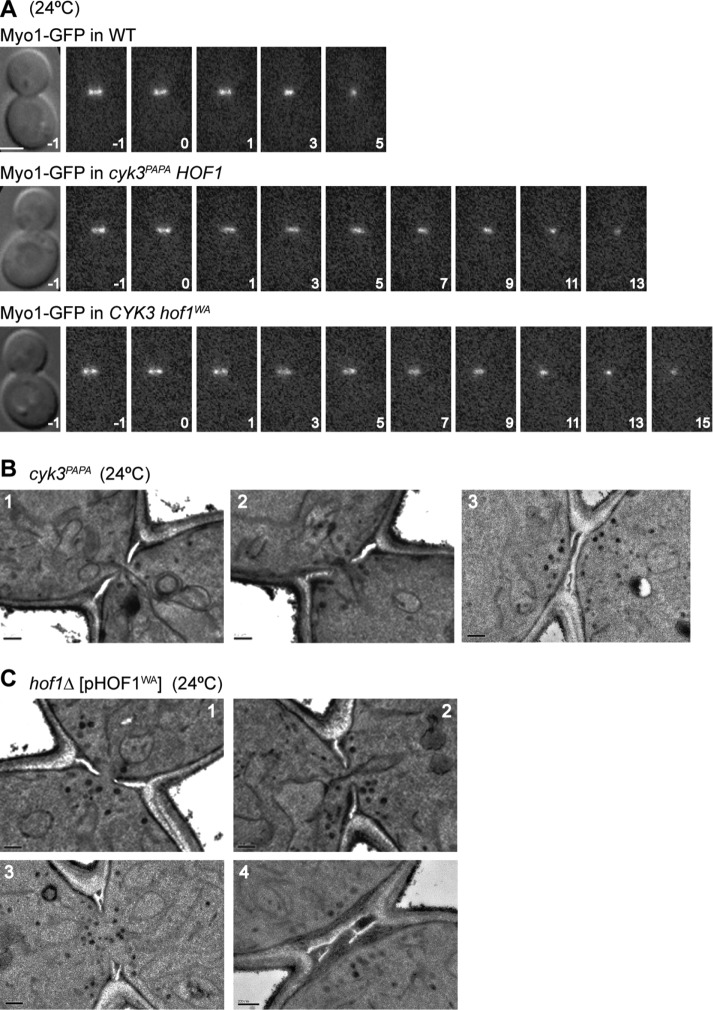
Cells expressing Cyk3PAPA instead of Cyk3 or Hof1WA instead of Hof1 showed no obvious growth defect at temperatures from 24 to 37°C (our unpublished results). However, because the association between Cyk3 and Hof1 begins approximately coincident with the onset of CAR constriction, cleavage-furrow ingression, and PS formation (see above), it seemed possible that the Hof1–Cyk3 interaction is important for these processes. Indeed, time-lapse microscopy revealed that CAR constriction and furrow ingression in cyk3PAPA and hof1WA cells proceeded at about half the speed seen in wild-type cells (Figure 5A). In addition, electron microscopy revealed abnormal septal structures in 38 of 47 cyk3PAPA cells and 39 of 45 hof1WA cells. In most of the cells with abnormal septa, PS and SS appeared to be forming simultaneously (Figure 5, B, panels 1 and 2, and C, panels 1 and 2), as seen also in cyk3∆ cells (Onishi et al., 2013), suggesting that PS formation is delayed or slowed, and/or that SS formation is not properly suppressed during PS formation, in the absence of the Hof1–Cyk3 interaction. The remaining cells showed either asymmetric PS formation, as in hof1∆ cells (see Figure 4), or branched or multiple PSs (Figure 5, B, panel 3, and C, panels 3 and 4). These effects may not be due solely to the loss of the Hof1–Cyk3 interaction, because Hof1WA is known to be defective in at least one other relevant interaction (i.e., that with Inn1: Supplemental Figure S1, A and B; Meitinger et al., 2011), and Cyk3PAPA may conceivably also lack some other significant interaction (given the slightly weaker growth of a cyk3PAPA hof1∆ strain relative to that of a CYK3 hof1∆ strain: Figure 6A, rows 1 and 2).
FIGURE 5:
Importance of the Hof1–Cyk3 interaction for cleavage-furrow ingression and the formation of normal septa. (A) Slow constriction of the CAR in the absence of the Hof1–Cyk3 interaction. Constriction of the Myo1–GFP ring was observed by time-lapse microscopy in wild-type (MWY1327), cyk3PAPA (MWY1315), and hof1WA (MWY1317) strains. All strains were grown to exponential phase and examined in SC medium at 24°C. Times are indicated in minutes; the onset of constriction is set at 0. Mean ± SD constriction times for 10 cells of each genotype were 6.6 ± 0.8 min (wild type), 13.4 ± 2.7 min (cyk3PAPA), and 11.4 ± 2.4 min (hof1WA). Scale bar, 2 µm. (B, C) Abnormal septal structures in the absence of the Hof1–Cyk3 interaction. Strains (B) MWY732 (cyk3PAPA) and (C) MWY864 (hof1∆ [pHOF1WA]) were grown to exponential phase at 24°C in SC (MWY732) or SC–Leu (MWY864) medium and examined by electron microscopy. Representative images are shown (see text). Scale bars, 0.2 µm.
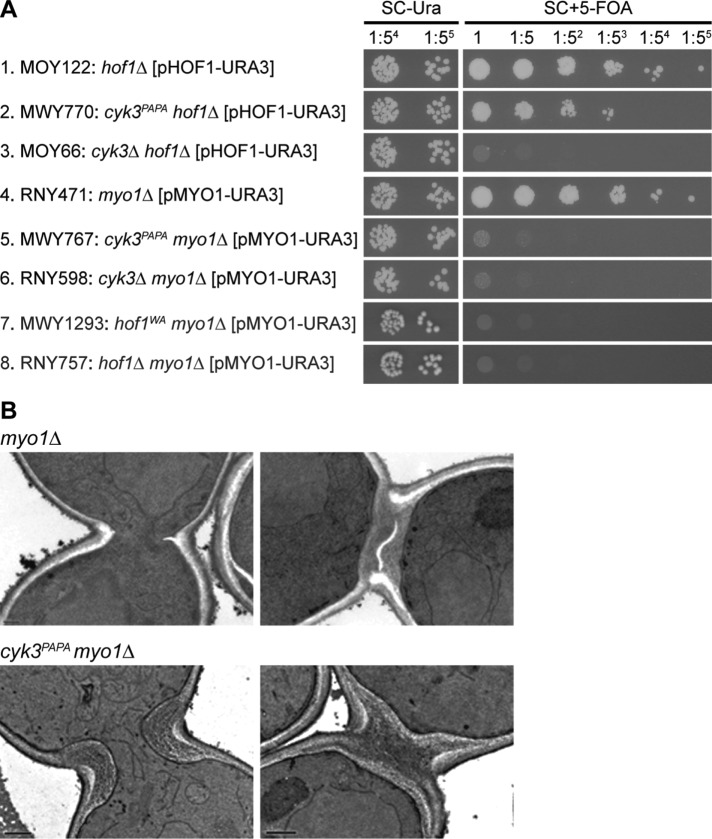
FIGURE 6:
Importance of the Hof1–Cyk3 interaction for growth (A) and PS formation (B) in myo1∆ cells. (A) The indicated strains were grown overnight on SC–Ura plates at 24°C, suspended in SC–Ura medium, spotted onto both SC–Ura and SC± 5-FOA plates (4-µl aliquots of 5×-serial-dilution series; 1× = 105 cells in 4 µl), and incubated at 24°C for 4 d. The experiment was performed twice with indistinguishable results. (B) Strains RNY471 (myo1∆) and MWY767 (cyk3PAPA myo1∆) were grown to exponential phase at 24°C in SC medium and examined by electron microscopy; both strains had previously been grown on an SC + 5-FOA plate at 24°C for 3 d to eliminate the URA3 MYO1 plasmid. Representative images are shown (see text). Scale bars, 0.2 µm.
The importance of the Hof1–Cyk3 interaction was even more evident in cells lacking the CAR because of a myo1∆ mutation. A cyk3PAPA hof1∆ strain grew only slightly less well than a CYK3 hof1∆ strain and much better than a cyk3∆ hof1∆ strain (Figure 6A, rows 1–3), indicating that Cyk3PAPA retains much of its function despite its inability to interact with Hof1. In contrast, a cyk3PAPA myo1∆ strain grew much worse than a CYK3 myo1∆ strain and nearly as badly as a cyk3∆ myo1∆ strain (Figure 6A, rows 4–6), indicating that the Hof1–Cyk3 interaction becomes critical when Myo1 (and thus the CAR) is absent. Similarly, a hof1WA myo1∆ strain was inviable like the hof1∆ myo1∆ double mutant (Figure 6A, rows 4, 7, and 8), although this result could reflect the loss of Hof1–Inn1 interaction, as well as of Hof1–Cyk3 interaction, in the Hof1WA strain (Supplemental Figure S1, A and B; Meitinger et al., 2011).
Electron microscopy has shown that >80% of myo1∆ cells form a readily observable PS, although these PSs are typically abnormal in structure and/or orientation (Figure 6B, top panels; Fang et al., 2010; our unpublished observations). In contrast, no PSs were observed in 50 cyk3PAPA myo1∆ cells examined (Figure 6B, bottom panels). Taken together, the data indicate that the Hof1–Cyk3 interaction is important for PS formation even in otherwise wild-type cells and becomes critical for PS formation in the absence of Myo1 and the CAR.
Because Inn1 interacts with both Cyk3 and Hof1 and is also implicated in the control of PS formation (Sanchez-Diaz et al., 2008; Jendretzki et al., 2009; Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Devrekanli et al., 2012; Labedzka et al., 2012; Palani et al., 2012; Foltman et al., 2016), we explored the interplay among these three proteins by investigating the role of the Hof1–Cyk3 interaction in Inn1-deficient cells. As shown previously (Nishihama, Schreiter, Onishi, Vallen, et al., 2009), overexpression of wild-type Cyk3 can suppress both the growth and PS-formation defects of inn1∆ cells (Figure 7, A, rows 1 and 2, and B, left panel). In striking contrast, overexpression of Cyk3PAPA could suppress neither defect (Figure 7, A, row 3, and B, right two panels). Similarly, overexpression of wild-type Cyk3 in a hof1WA inn1∆ strain also produced little or no suppression of the inn1∆ growth defect (Figure 7A, rows 5–7). Thus, the Hof1–Cyk3 interaction becomes critical for PS formation in the absence of either Myo1 or Inn1.
FIGURE 7:

Importance of the Hof1–Cyk3 interaction in the absence of Inn1, and roles of the Cyk3 SH3 domain. (A, B) Inability of Cyk3 overexpression to suppress inn1∆ in the absence of the Hof1–Cyk3 interaction or of a normal Cyk3 SH3 domain. (A) Lack of suppression of the inn1∆ growth defect. Strains LY1310 (inn1∆ [pUG36-INN1]; rows 1–4) and MWY1296 (inn1∆ hof1WA [pUG36-INN1]; rows 5–7) were transformed with LEU2-marked, high-copy plasmids carrying no insert (pRS425, 2µ vector), CYK3 (pRS425-CYK3), or cyk3 mutants (pRS425GW-CYK3PAPA and pRS425GW-CYK3WA), or with pRS315GW-INN1 (CEN INN1). Transformants were grown, spotted, and examined as described in Figure 6A except that SC–Ura-Leu and SC–Leu + 5-FOA plates were used. (B) Lack of suppression of the inn1∆ defect in PS formation. Strains MWY938 (inn1∆ [pUG36-INN1] [pRS425-CYK3]) and MWY934 (inn1∆ [pUG36-INN1] [pRS425GW-CYK3PAPA]) were grown on SC–Leu + 5-FOA plates at 24°C for 3 d to eliminate the URA3 INN1 plasmid and then grown to exponential phase at 24°C in SC–Leu medium and examined by electron microscopy. Cells shown are representative of more than 50 cells examined for MWY938 (see also Nishihama, Schreiter, Onishi, Vallen, et al., 2009) and of 50 cells examined for MWY934; of the latter, only two cells had recognizable PS-like structures. Scale bar, 0.5 µm. (C) Loss of Cyk3 localization in the absence of both Hof1–Cyk3 interaction and a functional Cyk3 SH3 domain. (Top panel) Merged GFP (green) and CFP (magenta) images are shown for strains MWY1436 (left; cyk3WA-2GFP [YCp111-CDC3-CFP]) and MWY1438 (right; cyk3WA,PAPA-2GFP [YCp111-CDC3-CFP]). Representative cells are shown; cell bodies are outlined. Scale bar, 2 µm. Bottom panel, protein levels of wild-type and mutant Cyk3. Strains MWY815 (CYK3-2GFP), MWY1414 (cyk3WA-2GFP), MWY673 (cyk3PAPA-2GFP), and MWY1418 (cyk3WA,PAPA-2GFP) were grown to exponential phase in YM-P medium at 24°C, and protein extracts were analyzed by Western blotting (see Materials and Methods).
Because Cyk3 is able to localize to the division site in the absence of either Inn1 (Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Palani et al., 2012) or Hof1 (Figure 3F), we asked whether interaction with both Inn1 and Hof1 might contribute to Cyk3 localization. To test this, we constructed a W45A mutation in the SH3 domain of Cyk3, which was expected to eliminate the interaction of Cyk3 with Inn1 (Jendretzki et al., 2009; Nishihama, Schreiter, Onishi, Vallen, et al., 2009; Labedzka et al., 2012) and indeed appeared to do so (Supplemental Figure S1A, patches 5–8). By itself, this mutation did not detectably affect the localization of Cyk3 to the division site (Figure 7C, top left two panels), but Cyk3 carrying both the WA and PAPA mutations failed to localize (Figure 7C, top right two panels), although the protein appeared to be present at normal levels in cells (Figure 7C, bottom panel). Thus, it appears that interaction with either Hof1 or Inn1 (and/or perhaps with another binding partner[s] of the Cyk3 SH3 or PRS domain—see below) is necessary and sufficient for Cyk3 localization.
Evidence for a Cyk3 binding partner(s) other than Hof1 and Inn1
In the course of the above studies, we unexpectedly observed that overexpression of Cyk3WA was also unable to suppress the growth defect of an inn1∆ mutant (Figure 7A, row 4). In the absence of Inn1, a mutation whose only effect was to disrupt the interaction with Inn1 should be irrelevant. Thus, it appears that the SH3 domain of Cyk3 must also interact with at least one other protein and that this interaction also contributes to Cyk3 function.
DISCUSSION
Since the original descriptions of Hof1 (Kamei et al., 1998), Cyk3 (Korinek et al., 2000), and Inn1 (Sanchez-Diaz et al., 2008; Nishihama, Schreiter, Onishi, Vallen, et al., 2009), much has been learned about the regulation of these proteins and their function in promoting PS formation and regulating SS formation (see Introduction). However, a remaining puzzle has been whether Hof1 and Cyk3 function in parallel pathways, as suggested by the synthetic lethality of hof1∆ and cyk3∆ mutations (Korinek et al., 2000; Meitinger et al., 2011; Labedzka et al., 2012), or as components of a single functional complex, as suggested both by the evidence for physical interaction between them (Tonikian, Xin, Toret, et al., 2009; Meitinger et al., 2011; Labedzka et al., 2012; Onishi et al., 2013) and by their isolation in association with Inn1, Iqg1, Myo1, and Chs2 (Foltman et al., 2016). In this study, we focused on clarifying the nature and functional significance of the putative Hof1–Cyk3 physical interaction.
First, we used yeast two-hybrid assays, coimmunoprecipitation from yeast extracts, and coprecipitation in vitro of bacterially synthesized proteins to provide strong confirmation that the interaction indeed occurs and is direct. Moreover, we used all three assays to show that the interaction is disrupted by point mutations (hof1W637A, cyk3P188A, cyk3P191A) in either the SH3 domain of Hof1 or the proline-rich motif of Cyk3, thus confirming the suggestion from previous work that the interaction involves these domains of the proteins.
Second, we used synchronized cultures and coimmunoprecipitation to show that the interaction occurs specifically at the time of cytokinesis, even though both proteins are readily detectable in cell extracts for a considerable time before this. Moreover, although the onset of the interaction coincides approximately with the onset of hyperphosphorylation of Hof1 and of phosphorylation of Cyk3, the interaction does not appear to depend on these phosphorylation events, given 1) that pull downs of either protein contain both less-phosphorylated and more-phosphorylated forms of the other and 2) that the interaction is also observed in vitro with bacterially synthesized proteins. However, until more detailed determinations of binding affinities have been performed, this interpretation must remain tentative.
Third, we found that Hof1 can localize normally to the cytokinesis site in the absence of its normal interaction with Cyk3, and vice versa. However, Cyk3 could not localize normally if its interactions with both Hof1 and Inn1 were disrupted by point mutations in both its proline-rich and SH3 domains (cyk3W45A,P188A,P191A). Meitinger et al. (2010) also reported normal localization of Cyk3 to the neck in hof1∆ cells, whereas Labedzka et al. (2012) reported that Cyk3 localization was largely lost in hof1∆ cells. This discrepancy might possibly be explained if the interaction of Cyk3 with Inn1 were less robust in the strain background used by Labedzka et al. (2012).
Fourth, we found that the Hof1–Cyk3 interaction is important for normal PS formation and a normal rate of cleavage-furrow ingression. As the PS and SS formed by cyk3P188A,P191A and hof1W637A cells at 24°C appeared more similar to those formed at that temperature by cyk3∆ cells than those formed by hof1∆ cells, the results suggest that the Hof1–Cyk3 interaction may be particularly important for Cyk3 function, although it seems likely to be important for Hof1 function as well. In any case, it appears that the Hof1–Cyk3 interaction becomes critical when either Inn1 or Myo1 is absent, given 1) the lethality or near lethality of cyk3P188A,P191A myo1∆ and hof1W637A myo1∆ cells, 2) the complete loss of PS formation in a cyk3P188A,P191A myo1∆ strain, and 3) the inability of high-copy cyk3P188A,P191A to suppress the inn1∆ growth and PS-formation defects and the inability of high-copy CYK3 to suppress inn1∆ in a hof1W637A background.
In summary, it seems clear that Hof1 and Cyk3 interact directly by means of their respective SH3 and proline-rich domains and that this interaction is important for their function. The interaction occurs specifically at the time of cytokinesis and is triggered by a mechanism that remains unclear. Taking all the available data together, the most plausible model is that Hof1 and Cyk3 do not function in parallel pathways but rather as interacting components of a common complex that also contains Inn1 and (for at least part of its lifetime) also Iqg1 and Myo1. The stoichiometry of complex components is likely to matter, perhaps explaining the observation by Jendretzki et al. (2009) that overexpression of Cyk3 can rescue the defects due to overexpression of Hof1. The complex may also contain other proteins, as suggested by the evidence that the Cyk3 SH3 domain has an interaction partner(s) other than Inn1 (see above; Tonikian, Xin, Toret, et al., 2009; Labedzka et al., 2012; our unpublished results). The complex functions both to promote PS formation, through activation of Chs2 (a role that Inn1 and Cyk3 both seem able to fill), and to regulate SS formation. Complex formation and function appear able to withstand single insults (e.g., the loss of Hof1, Cyk3, or their direct interaction) but not multiple ones (e.g., the loss of two core components of the complex). Despite the recent progress in understanding the composition and function of this complex, more work will be needed to fully understand its function and, in particular, how this function relates to that of the CAR during cleavage-furrow formation.
MATERIALS AND METHODS
Growth conditions, strains, and genetic methods
Standard yeast culture media (Guthrie and Fink, 1991) were used except where noted; cells were grown at 24°C on YM-P rich liquid medium (Lillie and Pringle, 1980), YPD rich solid medium, or liquid or solid synthetic-complete (SC) medium lacking appropriate nutrients as needed to select plasmids or transformants. Dextrose (2%) was used as carbon source. 5-Fluoroorotic acid (5-FOA; 1 mg/ml; Research Products International) was used to select for the loss of URA3 plasmids or chromosomal URA3, and the antibiotic Geneticin (G418; Lonza) was used to select for kanR cells.
Except for PJ69-4A, the yeast strains used (Table 1) are all in the S288C genetic background (Mortimer and Johnston, 1986) and were derived specifically from YEF473 (Bi and Pringle, 1996). Genes were deleted or tagged at their C-termini using the PCR method (Baudin et al., 1993) and either one of the template plasmids described by Longtine et al. (1998) or pFA6a-TAP-His3MX6 (provided by P. Walter, University of California, San Francisco). Oligonucleotide primers were purchased from Integrated DNA Technologies, and nonstandard primers are described in Supplemental Table 1. To construct strains containing mutant and/or tagged versions of CYK3 or HOF1 in place of the normal chromosomal genes, we used pCR8GW-based plasmids containing the desired versions of the genes (see below and Table 2). For CYK3, the plasmids were digested with MfeI, which releases a fragment containing the CYK3 coding region plus ∼300 base pairs of upstream and ∼300 base pairs of downstream sequence. The digests were transformed into cyk3∆::URA3:kanMX6 strain RNY2150, and transformants were collected on SC+5-FOA plates to screen for loss of the URA3 marker and then replicated onto SC+G418 plates to ensure that the kanR marker had been lost simultaneously. For HOF1, the plasmids were digested with BamHI and NsiI, which releases a fragment containing the HOF1 coding region plus ∼430 base pairs of upstream and ∼470 base pairs of downstream sequence; the digests were then transformed into hof1∆::URA3:kanMX6 strain RNY2240 and screened as just described. In some cases, the introduced chromosomal genes were tagged with TRP1 or His3MX6 in a subsequent step using the PCR method in order to provide a convenient marker for later crosses. Other steps in strain constructions involved conventional genetic crosses and plasmid transformations (Guthrie and Fink, 1991).
TABLE 1:
Saccharomyces cerevisiae strains used in this study.
| Strain | Genotypea | Source |
|---|---|---|
| PJ69-4A | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4∆ gal80∆ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al., 1996 |
| YEF473A | MATa his3-∆200 leu2-∆1 lys2-801 trp1-∆63 ura3-52 | Bi and Pringle, 1996 |
| YEF473B | MATα his3-∆200 leu2-∆1 lys2-801 trp1-∆63 ura3-52 | Bi and Pringle, 1996 |
| LY1310 | as YEF473A except inn1∆::kanMX6 [pUG36-INN1] | Nishihama, Schreiter, Onishi, Vallen, et al., 2009 |
| MOY66 | as YEF473B except cyk3∆::TRP1 hof1∆::kanMX6 [pRS316GW-HOF1] | Onishi et al., 2013 |
| MOY122 | as YEF473B except hof1∆::TRP1 [pRS316GW-HOF1] | This study |
| RNY370 | as YEF473A except hof1∆::TRP1 | This study |
| RNY471 | as YEF473B except myo1∆::kanMX6 [pRS316-MYO1] | This study |
| RNY598 | as YEF473A except myo1∆::kanMX6 cyk3∆::TRP1 [pRS316-MYO1] | This study |
| RNY757 | as YEF473A except myo1∆::kanMX6 hof1∆::TRP1 [pRS316-MYO1] | This study |
| RNY2150 | as YEF473A except cyk3∆::URA3:kanMX6 | Onishi et al., 2013 |
| RNY2240 | as YEF473B except hof1∆::URA3:kanMX6 | This studyb |
| RNY2720 | as YEF473A except CHS2-GFP:TRP1 hof1∆::TRP1 [YCp111-CDC3-CFP] | This study |
| MWY636 | as YEF473A except cyk3P188A,P191A | See text |
| MWY673 | as YEF473A except cyk3P188A,P191A-2GFP | See text |
| MWY732 | as YEF473A except cyk3P188A,P191A:TRP1 | See text |
| MWY761 | as YEF473B except cdc15-2 HOF1-TAP:His3MX6 cyk3P188A,P191A-2GFP | This study |
| MWY767 | as YEF473A except cyk3P188A,P191A:TRP1 myo1∆::kanMX6 [pRS316-MYO1] | This study |
| MWY770 | as YEF473A except cyk3P188A,P191A:TRP1 hof1∆::TRP1 [pRS316GW-HOF1] | This study |
| MWY815 | as YEF473A except CYK3-2GFP | See text |
| MWY822 | as YEF473B except cdc15-2 HOF1-TAP:His3MX6 CYK3-2GFP | This study |
| MWY864 | as YEF473B except hof1∆::TRP1 [pRS315GW-HOF1W637A-3GFP] | See text |
| MWY934 | as YEF473A except inn1∆::kanMX6 [pUG36-INN1] [pRS425GW-CYK3P188A,P191A] | This study |
| MWY938 | as YEF473A except inn1∆::kanMX6 [pUG36-INN1] [pRS425-CYK3] | This study |
| MWY999 | as YEF473A except cdc15-2 CYK3-TAP:His3MX6 | This study |
| MWY1003 | as YEF473A except cdc15-2 HOF1-GFP:kanMX6 | This study |
| MWY1006 | as YEF473A except cdc15-2 CYK3-TAP:His3MX6 HOF1-GFP:kanMX6 | This study |
| MWY1019 | as YEF473A except cdc15-2 HOF1-TAP:His3MX6 | This study |
| MWY1021 | as YEF473A except cdc15-2 CYK3-GFP:kanMX6P | This study |
| MWY1025 | as YEF473A except cdc15-2 HOF1-TAP:His3MX6 CYK3-GFP:kanMX6 | This study |
| MWY1252 | as YEF473B except hof1W637A-3GFP | See text |
| MWY1256 | as YEF473B except hof1W637A | See text |
| MWY1259 | as YEF473B except hof1W637A-His3MX6 | See text |
| MWY1293 | as YEF473A except hof1W637A:His3MX6 myo1∆::kanMX6 [pRS316-MYO1] | This study |
| MWY1296 | as YEF473A except hof1W637A:His3MX6 inn1∆::kanMX6 [pUG36-INN1] | This study |
| MWY1315 | as YEF473B except cyk3P188A,P191A:TRP1 MYO1-GFP:kanMX6 | This study |
| MWY1317 | as YEF473B except hof1W637A:His3MX6 MYO1-GFP:kanMX6 | This study |
| MWY1327 | as YEF473B except MYO1-GFP:kanMX6 | This study |
| MWY1408 | as YEF473A except CYK3-CFP:kanMX6 HOF1-GFP:kanMX6 | This study |
| MWY1414 | as YEF473A except cyk3W45A-2GFP | See text |
| MWY1418 | as YEF473A except cyk3W45A,P188A,P191A-2GFP | See text |
| MWY1436 | as YEF473A except cyk3W45A-2GFP [YCp111-CDC3-CFP] | This study |
| MWY1438 | as YEF473A except cyk3W45A,P188A,P191A-2GFP [YCp111-CDC3-CFP] | This study |
| MWY2111 | as YEF473A except cyk3P188A,P191A-2GFP [YCp111-CDC3-CFP] | This study |
| MWY2117 | as YEF473A except cdc15-2 CYK3-TAP:His3MX6 HOF1-3HA:TRP1 | This study |
| MWY2119 | as YEF473A except cdc15-2 INN1-TAP:His3MX6 HOF1-3HA:TRP1 | This study |
| MWY2120 | as YEF473B except cyk3∆::TRP1 HOF1-3GFP | See text |
| MWY2122 | as YEF473B except hof1∆::TRP1 CYK3-2GFP [YCp111-CDC3-CFP] | This study |
| MWY2150 | as YEF473A except cdc15-2 INN1-TAP:His3MX6 hof1W637A-3HA:TRP1 | This study |
| MWY2154 | as YEF473A except cdc15-2 CYK3-TAP:His3MX6 hof1W637A-3HA:TRP1 | This study |
aPlasmids are indicated in square brackets and are described in Table 2.
bConstructed in the same way as RNY2150.
TABLE 2:
Plasmids used in this study.
| Plasmid | Descriptiona | Source |
| pGBDU | 2µ, URA3, GAL4-DBD | James et al., 1996 |
| pGAD | 2µ, LEU2, GAL4-AD | James et al., 1996 |
| pRS316-MYO1 | CEN, URA3, MYO1 | Ko et al., 2007 |
| YCp111-CDC3-CFP | CEN, LEU2, CDC3-CFP | Nishihama, Schreiter, Onishi, Vallen, et al., 2009 |
| pUG36-INN1 | CEN, URA3, pMET25-yEGFP-INN1 | Nishihama, Schreiter, Onishi, Vallen, et al., 2009 |
| pCR8GW-INN1 | INN1 | See text |
| pRS315GW-INN1 | CEN, LEU2, INN1 | This studyb |
| pCR8GW-HOF1 | HOF1 | See text |
| pCR8GW-HOF1-3GFP | HOF1-3GFP | See text |
| pRS316GW-HOF1 | CEN, URA3, HOF1 | This studyc |
| pRS315GW-HOF1W637A-3GFP | CEN, LEU2, hof1W637A-3GFP | This studyd |
| pRS425-CYK3 | 2µ, LEU2, CYK3 | Ko et al., 2007 |
| pCR8GW-CYK3 | CYK3 | See text |
| pCR8GW-CYK3-2GFP | CYK3-2GFP | See text |
| pRS425GW-CYK3P188A,P191A | 2µ, LEU2, CYK3 P188A,P191A | This studye |
| pRS425GW-CYK3W45A | 2µ, LEU2, CYK3W45A | This studye |
| pMALC2-CYK3 | malE-CYK3 | See text |
| pMALC2-CYK3P188A,P191A | malE-CYK3P188A,P191A | This studyf |
| pCOLADuet-His6-HOF1341–669 | His6-HOF1 (codons 341–669) | Nishihama, Schreiter, Onishi, Vallen, et al., 2009 |
| pCOLADuet-His6-HOF1341–669(W637A) | His6-HOF1 (codons 341–669) with W637A mutation | This studyf |
aCEN indicates low-copy-number plasmids; 2µ indicates high-copy-number plasmids.
bConstructed by Gateway recombination between pCR8GW-INN1 and pRS315-attR (see text).
cConstructed by Gateway recombination between pCR8GW-HOF1 and pRS316-attR (see text).
dConstructed by Gateway recombination between pRS315-attR (see text) and pCR8GW-HOF1-3GFP into which the indicated mutation had been introduced (see text).
eConstructed by Gateway recombination between pRS425-attR (see text) and pCR8GW-CYK3 into which the indicated mutations had been introduced (see text).
fSite-directed mutagenesis was performed (see text) on pMALC2-CYK3 and pCOLADuet-His6-HOF1341–669, respectively.
Plasmid constructions
The plasmids used in this study are listed in Table 2 and/or described below. To construct plasmids pRS315-attR, pRS316-attR, and pRS425-attR, a fragment containing attR1-ccdB-attR2 was PCR amplified with primers attR1+SacI and attR2+XhoI using the Gateway vector-conversion system cassette A (Invitrogen) as a template, digested with SacI and XhoI, and cloned into SacI/XhoI–digested pRS315 (Sikorski and Hieter, 1989), pRS316 (Sikorski and Hieter, 1989), or pRS425 (Christianson et al., 1992). To construct plasmids pCR8GW-INN1, pCR8GW-HOF1, and pCR8GW-CYK3, genomic fragments containing the genes (positions relative to the start codons: INN1, −939 to +1689 base pairs; HOF1, −1000 to +2510; CYK3, −900 to +3248) were amplified by PCR and cloned into the pCR8/GW/TOPO/TA vector (Invitrogen) following the manufacturer’s instructions. To construct plasmids pCR8GW-HOF1-3GFP and pCR8GW-CYK3-2GFP, a 9–base pair sequence (GGCGGCCGC) containing a NotI site was introduced immediately before the stop codons of the HOF1 and CYK3 ORFs in plasmids pCR8GW-HOF1 and pCR8GW-CYK3 using the QuickChange II Site-Directed Mutagenesis kit (Stratagene) and primers CYK3-C-NotI-QCF, CYK3-C-NotI-QCR, HOF1-C-NotI-QCF, and HOF1-C-NotI-QCR. NotI fragments (2.2 and 1.5 kb) containing 3 × GFP or 2 × GFP (our unpublished results) were then inserted at the new NotI sites to generate the desired plasmids. Mutant versions of HOF1 and CYK3 (used both for plasmid constructions and for replacement of the wild-type chromosomal genes as described above) were generated in the pCR8GW-based plasmids by site-directed mutagenesis using the QuickChange kit and the primers described in Supplemental Table 1.
Plasmids for in vitro protein-binding assays were constructed as follows. A DNA fragment encoding full-length CYK3 was PCR amplified using primers CYK3_1004F+RI and CYK3_3658R+Pst, digested with EcoRI and PstI, and cloned into pMAL-c2 (New England Biolabs) to create pMALC2-CYK3. Plasmids pMALC2-CYK3 and pCOLADuet-His6-HOF1341–669 (Table 2) were subjected to site-directed mutagenesis using the QuickChange kit to generate mutations in the Cyk3 proline-rich sequence and the Hof1 SH3 domain, which were confirmed by sequencing.
Two-hybrid analyses
The two-hybrid vectors were the DNA-binding–domain (DBD) plasmid pGBDU and the activation domain (AD) plasmid pGAD (Table 2). Both full-length CYK3 and HOF1 and fragments of the genes were PCR amplified and cloned into pGBDU and pGAD. Plasmids pGBDU-CYK31–403 and pGAD-HOF1342–669 were subjected to site-directed mutagenesis using the QuickChange kit to generate mutations in the Cyk3 proline-rich sequence and the Hof1 SH3 domain, which were confirmed by sequencing. For two-hybrid analyses, strain PJ69-4A was transformed with various DBD and AD plasmids. Transformants were first selected on SC–Ura-Leu plates and then streaked or spotted onto SC–Ura-Leu-His plates and incubated at 30°C for 3 d to detect interactions. All experiments were performed two or more times with indistinguishable results.
Immunoprecipitation
Cells were collected by centrifugation, and the pellets were frozen immediately in liquid nitrogen. Extracts were prepared by several rounds of vortexing with glass beads on ice in NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1% Nonidet P-40, 150 mM NaCl, 2 mM EDTA) supplemented with 50 mM NaF, 0.1 mM Na3VO4, 1 mM dithiothreitol (DTT), and “complete protease-inhibitor cocktail” (Roche #11697498001) and centrifuged at 13,000 × g for 10 min. To precipitate TAP-tagged proteins, 25 mg of protein extract were incubated with 25 µl Dynabeads Pan Mouse IgG (Invitrogen #11041) at 4°C for 2 h, washed five times with NP-40 buffer containing the supplements indicated above, and eluted with SDS sample buffer. Samples were analyzed by SDS–PAGE and Western blotting using a mouse anti-GFP antibody (Roche #11814460001) and a horseradish peroxidase–conjugated rabbit anti-mouse-immunoglobulin-G antibody (MP Biomedicals #0861204) to detect GFP-fusion proteins, a horseradish peroxidase–conjugated rat anti-3HA antibody (Roche #12013819001) to detect 3HA-fusion proteins, and peroxidase–anti-peroxidase soluble complex (Sigma-Aldrich #P1291) to detect TAP-tagged proteins. All experiments were performed two or more times with indistinguishable results.
Analysis of protein phosphorylation
Protein extracts were prepared, incubated with Dynabeads, and washed as described above; the Dynabeads were then separated into four aliquots. SDS sample buffer was added immediately to one aliquot as a control. The other three aliquots were washed twice with lambda–protein-phosphatase buffer (New England Biolabs #P0753S) and incubated at 30°C for 30 min in 50 µl of the same buffer with or without lambda protein phosphatase and phosphatase inhibitors (50 mM NaF and 1 mM Na3VO4). Reactions were terminated by adding SDS sample buffer, and samples were analyzed by SDS–PAGE and Western blotting as described above. All experiments were performed two or more times with indistinguishable results.
In vitro protein-binding assays
To purify wild-type and mutant His6-Hof1341–669, Escherichia coli strain BL21 (Invitrogen) was transformed with pCOLADuet-His6-Hof1341–669 or pCOLADuet-His6-Hof1341–669(W637A), grown to exponential phase at 37°C, and induced with 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) for 3 h at 24°C. Cells were collected by centrifugation, and the pellets were frozen immediately in liquid nitrogen. Subsequently, the pellets were suspended in Ni-NTA lysis buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 20 mM imidazole, 10 mM 2-mercaptoethanol, and 0.1% Triton X-100) containing 1 mM phenylmethylsulfonyl fluoride, sonicated twice for 10 s at low power, placed on ice for 15 min, and centrifuged at 12,000 × g for 15 min. The supernatants were then mixed with Ni-NTA agarose beads (Qiagen #30210) that were preequilibrated with Ni-NTA lysis buffer. After 90 min of rocking at 4°C, the beads were collected by centrifugation, washed four times with Ni-NTA lysis buffer, and stored in aliquots at −80°C.
To purify wild-type and mutant MBP-Cyk3, E. coli BL21 was transformed with pMALC2-CYK3 or pMALC2-CYK3P188A,P191A, grown to exponential phase at 37°C, and induced with 1 mM IPTG for 3 h at 24°C. Protein extracts were prepared as described above except that lysis was in STE buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol, and 0.1% Triton X-100) containing the complete protease-inhibitor cocktail. The supernatants obtained after centrifugation (12,000 × g for 15 min) were mixed with amylose resin (New England Biolabs #E8021S) that had been preequilibrated with STE buffer containing the protease-inhibitor cocktail (see above), and rocked for 1 h at 4°C. The beads were collected by centrifugation, washed four times with STE buffer, and eluted three times with elution buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 100 mM maltose, 1 mM DTT). Eluates were separated into aliquots and stored at −80°C.
To test for protein binding in vitro, 5 µg of His6-Hof1341–669 or His6-Hof1341–669(W637A) (on the Ni-NTA beads) were mixed with 5 µg of MBP, MBP-Cyk3, or MBP-Cyk3P188A,P191A and rocked for 1 h at 4°C. The beads were washed two times with Ni-NTA lysis buffer, collected by brief centrifugation, and resuspended in 40 µl SDS sample buffer; they were then analyzed by SDS–PAGE and Coomassie staining.
Light and electron microscopy
To visualize proteins tagged with fluorescent markers, cells were grown to exponential phase and examined using a Nikon Eclipse 600-FN microscope equipped with an Apochromat 100×/1.40 NA oil-immersion objective and an ORCA-2 cooled charge-coupled-device camera (Hamamatsu Photonics). Both fluorescence and differential-interference-contrast (DIC) images were acquired and processed with Metamorph version 7.0 software (Molecular Devices).
Electron microscopy was performed essentially as described by Nishihama, Schreiter, Onishi, Vallen, et al. (2009): exponential-phase cells were collected by filtration, fixed with glutaraldehyde and potassium permanganate, embedded in LR white resin (Sigma-Aldrich), and stained with uranyl acetate and lead citrate. Images were acquired and processed using a JEOL (Tokyo, Japan) TEM1230 transmission electron microscope equipped with a Gatan Orius SC1000 cooled charge-coupled-device camera. For each experiment, three grids were prepared and examined, and representative cells are shown.
Supplementary Material
Acknowledgments
We thank past and present members of our laboratory for their support and helpful discussions and J. Mulholland and J. Perrino (Stanford University Cell Sciences Imaging Facility) for assistance with electron microscopy. This work was supported in part by National Institutes of Health Grant no. 31006, National Science Foundation EAGER Grant no. 1548533, National Institutes of Health Shared Instrumentation Grant no. 1S10RR02678001 (to J.M.), and the Stanford University Department of Genetics.
Abbreviations used:
- CAR
contractile actomyosin ring
- PRS
proline-rich sequence
- PS
primary septum
- SS
secondary septa.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-04-0227) on January 10, 2018.
REFERENCES
Boldface names denote co–first authors.
- Aspenström P. (2009). Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol , 1-31. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res , 3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. (1998). Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol , 1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Pringle JR. (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol , 5264-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. (2005). Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J , 1440-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers B, Levin G, Cabib E. (1974). Effect of polyoxin D on chitin synthesis and septum formation in Saccharomyces cerevisiae. J Bacteriol , 564-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace J, Hsu J, Weiss EL. (2011). Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol Cell Biol , 721-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges AA, Gladfelter AS. (2015). Septin form and function at the cell cortex. J Biol Chem , 17173-17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CF, Bennett AM, Ma WK, Hall MC, Yeong FM. (2012). Dependence of Chs2 ER export on dephosphorylation by cytoplasmic Cdc14 ensures that septum formation follows mitosis. Mol Biol Cell , 45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V, Stanley ER. (2007). Pombe Cdc15 homology (PCH) proteins: coordinators of membrane-cytoskeletal interactions. Trends Cell Biol , 145-156. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene , 119-122. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R. (2001). Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell , 739-750. [DOI] [PubMed] [Google Scholar]
- Devrekanli A, Foltman M, Roncero C, Sanchez-Diaz A, Labib K. (2012). Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J Cell Sci , 5453-5466. [DOI] [PubMed] [Google Scholar]
- Epp JA, Chant J. (1997). An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol , 921-929. [DOI] [PubMed] [Google Scholar]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. (2010). Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol , 1333-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M, Molist I, Arcones I, Sacristan C, Filali-Mouncef Y, Roncero C, Sanchez-Diaz A. (2016). Ingression progression complexes control extracellular matrix remodelling during cytokinesis in budding yeast. PLoS Genet , e1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. (2001). The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol , 681-689. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. (1991). Guide to yeast genetics and molecular biology. Methods Enzymol , 933. [PubMed] [Google Scholar]
- Heath RJ, Insall RH. (2008). F-BAR domains: multifunctional regulators of membrane curvature. J Cell Sci , 1951-1954. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics , 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendretzki A, Ciklic I, Rodicio R, Schmitz HP, Heinisch JJ. (2009). Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol Genet Genomics , 437-451. [DOI] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. (1998). Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem , 28341-28345. [DOI] [PubMed] [Google Scholar]
- Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, Pringle JR. (2007). Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol Biol Cell , 5139-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Bi E, Epp JA, Wang L, Ho J, Chant J. (2000). Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol , 947-950. [DOI] [PubMed] [Google Scholar]
- Kuranda MJ, Robbins PW. (1991). Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem , 19758-19767. [PubMed] [Google Scholar]
- Labedzka K, Tian C, Nussbaumer U, Timmermann S, Walther P, Müller J, Johnsson N. (2012). Sho1p connects the plasma membrane with proteins of the cytokinesis network through multiple isomeric interaction states. J Cell Sci , 4103-4113. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. (1980). Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol , 1384-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. (1998a). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol , 355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. (1998b). Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol , 1947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. (1996). The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol , 106-119. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953-961. [DOI] [PubMed] [Google Scholar]
- Lord M, Laves E, Pollard TD. (2005). Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol Biol Cell , 5346-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Takizawa Y, Feoktistova A, Xu P, Ohi MD, Vander Kooi CW, Gould KL. (2016). The tubulation activity of a fission yeast F-BAR protein is dispensable for its function in cytokinesis. Cell Rep , 534-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. (2015). Oligomerization but not membrane bending underlies the function of certain F-BAR proteins in cell motility and cytokinesis. Dev Cell , 725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Bertin A, Garcia G, 3rd, Lam L, Nogales E, Thorner J. (2011a). Septin filament formation is essential in budding yeast. Dev Cell , 540-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Stefan CJ, Wemmer M, Odorizzi G, Emr SD, Thorner J. (2011b). Genetic interactions with mutations affecting septin assembly reveal ESCRT functions in budding yeast cytokinesis. Biol Chem , 699-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. (2011). Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev , 875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Palani S. (2016). Actomyosin ring driven cytokinesis in budding yeast. Semin Cell Dev Biol , 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Palani S, Hub B, Pereira G. (2013). Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol Biol Cell , 1290-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Petrova B, Lombardi IM, Bertazzi DT, Hub B, Zentgraf H, Pereira G. (2010). Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci , 1851-1861. [DOI] [PubMed] [Google Scholar]
- Miller DP, Hall H, Chaparian R, Mara M, Mueller A, Hall MC, Shannon KB. (2015). Dephosphorylation of Iqg1 by Cdc14 regulates cytokinesis in budding yeast. Mol Biol Cell , 2913-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Alvarado D, Schmitz KR, Kenniston JA, Mendrola JM, Ferguson KM, Lemmon MA. (2015). Comparison of Saccharomyces cerevisiae F-BAR domain structures reveals a conserved inositol phosphate binding site. Structure , 352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. (1986). Genealogy of principal strains of the yeast genetic stock center. Genetics , 35-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor SG, Morgan DO. (2014). Cdk1-dependent phosphorylation of Iqg1 governs actomyosin ring assembly prior to cytokinesis. J Cell Sci , 1128-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. (2009). Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J Cell Biol , 995-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Bi E. (2011). Septin structure and function in yeast and beyond. Trends Cell Biol , 141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Chang KJ, Orlean P, Wloka C, Deshaies R, Bi E. (2012). Mitotic exit kinase Dbf2 directly phosphorylates chitin synthase Chs2 to regulate cytokinesis in budding yeast. Mol Biol Cell , 2445-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. (2013). Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell , 1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Ko N, Nishihama R, Pringle JR. (2013). Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol , 311-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P. (2012). Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics , 775-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S, Meitinger F, Boehm ME, Lehmann WD, Pereira G. (2012). Cdc14-dependent dephosphorylation of Inn1 contributes to Inn1-Cyk3 complex formation. J Cell Sci , 3091-3096. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu JQ. (2010). Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol , 149-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A, Marchesi V, Murray S, Jones R, Pereira G, Edmondson R, Allen T, Labib K. (2008). Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat Cell Biol , 395-406. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. (2002). In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci , 293-302. [DOI] [PubMed] [Google Scholar]
- Shannon KB, Li R. (1999). The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell , 283-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Durán A, Cabib E. (1991). The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol , 111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics , 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday N, VerPlank L, Li R. (2002). Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol , 1864-1870. [DOI] [PubMed] [Google Scholar]
- Tonikian R, Xin X, Toret CP, Gfeller D, Landgraf C, Panni S, Paoluzi S, Castagnoli L, Currell B, Seshagiri S, et al. (2009). Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol , e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. (2000). Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell , 593-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL. (2012). Mitotic exit and separation of mother and daughter cells. Genetics , 1165-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. (2012). Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) , 710-726. [DOI] [PubMed] [Google Scholar]
- Wloka C, Vallen EA, Thé L, Fang X, Oh Y, Bi E. (2013). Immobile myosin-II plays a scaffolding role during cytokinesis in budding yeast. J Cell Biol , 271-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolken DM, McInnes J, Pon LA. (2014). Aim44p regulates phosphorylation of Hof1p to promote contractile ring closure during cytokinesis in budding yeast. Mol Biol Cell , 753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.