Abstract
Blebs are involved in various biological processes such as cell migration, cytokinesis, and apoptosis. While the expansion of blebs is largely an intracellular pressure-driven process, the retraction of blebs is believed to be driven by RhoA activation that leads to the reassembly of the actomyosin cortex at the bleb membrane. However, it is still poorly understood how RhoA is activated at the bleb membrane. Here, we provide evidence demonstrating that myosin II–interacting guanine nucleotide exchange factor (MYOGEF) is implicated in bleb retraction via stimulating RhoA activation and the reassembly of an actomyosin network at the bleb membrane during bleb retraction. Interaction of MYOGEF with ezrin, a well-known regulator of bleb retraction, is required for MYOGEF localization to retracting blebs. Notably, knockout of MYOGEF or ezrin not only disrupts RhoA activation at the bleb membrane, but also interferes with nonmuscle myosin II localization and activation, as well as actin polymerization in retracting blebs. Importantly, MYOGEF knockout slows down bleb retraction. We propose that ezrin interacts with MYOGEF and recruits it to retracting blebs, where MYOGEF activates RhoA and promotes the reassembly of the cortical actomyosin network at the bleb membrane, thus contributing to the regulation of bleb retraction.
INTRODUCTION
Blebs are observed in various biological processes such as cell migration, cytokinesis, and apoptosis (Mills et al., 1998; Wolf et al., 2003; Fackler and Grosse, 2008; Sedzinski et al., 2011). In contrast to the mesenchymal mode of cell migration, which utilizes actin-driven membrane protrusions to allow tumor cells to migrate through the extracellular matrix, the amoeboid mode of cell migration (or bleb-based cell migration) uses intracellular pressure-driven membrane protrusions, generally termed blebs, to enable tumor cells to migrate in three-dimensional environments (Friedl and Wolf, 2003). During early embryonic development in zebrafish and Xenopus, primordial germ cells utilize bleb-based migration to migrate into the sites of developing gonads (Blaser et al., 2006; Dzementsei et al., 2013). During cytokinesis, the formation of blebs at the pole regions of dividing cells is required for the correct positioning of the cleavage furrow (Sedzinski et al., 2011). Despite this important role for membrane blebbing in various biological processes, the molecular mechanism that controls membrane blebbing is still poorly understood.
Blebs are rounded membrane protrusions on the cell surface. The formation of blebs is largely due to the broken or weakening linkage between the cell membrane and the actin cortex in a local area. In turn, intracellular pressure drives the flow of intracellular fluid into the space between the membrane and the cortex, leading to the formation of rounded membrane protrusions on the cell surface (Sheetz et al., 2006; Charras, 2008). It is generally thought that the growth and expansion of a bleb largely rely on increased intracellular pressure that results from actomyosin contraction at the cell cortex (Tinevez et al., 2009; Woolley et al., 2015b). Following bleb expansion, the cortical actomyosin network is reassembled underneath the bleb membrane and provides the driving force that leads to bleb retraction (Charras et al., 2005, 2008). Therefore, the life of a bleb generally exhibits as repeating cycles between bleb expansion and retraction (Charras et al., 2006, 2008).
The cortex is a thin actin network that is physically linked to and provides physical support for the cell membrane (Fehon et al., 2010). The ERM protein family, collectively named after three proteins (i.e., ezrin, radixin, and moesin), physically links cortical actin networks to the cell membrane (Fehon et al., 2010). In particular, it has been shown that ezrin is required for cortex reassembly and bleb retraction (Charras et al., 2006). Notably, ezrin is one of the first proteins that are recruited to the retracting bleb membrane (Charras et al., 2006). The subsequent recruitment of cytoskeleton components such as actin, actin binding proteins, and nonmuscle myosin II to the bleb membrane leads to the formation of a contractile actomyosin cortex that enables the bleb membrane to retract back to the cell body (Charras et al., 2006). Therefore, the recruitment of ezrin to the bleb membrane is believed to be critical for the subsequent reassembly of the actin cortex that leads to bleb retraction.
Rho GTPase signaling plays a crucial role in regulating the organization of the actomyosin cytoskeleton (Van Aelst and D’Souza-Schorey, 1997; Hall, 1998). In particular, the Rho GTPase protein RhoA activates its downstream effectors, such as Rho-associated protein kinase (ROCK), which in turn regulates the organization of the actomyosin cytoskeleton (Heasman and Ridley, 2008). Activation of myosin II requires the phosphorylation of myosin light chain (MLC) (Adelstein and Conti, 1975). Once activated, ROCK directly phosphorylates MLC (Amano et al., 1996). In addition, ROCK also increases MLC phosphorylation by phosphorylating and inhibiting myosin phosphatase (Kimura et al., 1996; Kawano et al., 1999; Kosako et al., 2000; Totsukawa et al., 2000). It is believed that, following bleb expansion, activation of RhoA at the bleb membrane is an early event that eventually leads to the reassembly of an actomyosin network and the retraction of blebs (Charras, 2008; Fackler and Grosse, 2008). Rho GTPase proteins are largely activated by guanine nucleotide exchange factors (GEFs) and inactivated by GTPase-activating proteins (GAPs) (Cerione and Zheng, 1996; Van Aelst and D’Souza-Schorey, 1997; Rossman et al., 2005; Zuo et al., 2014). A recent study has demonstrated that a GAP termed p190RhoGAP inactivates RhoA, thus preventing RhoA from being activated during bleb expansion. In contrast, p190RhoGAP is inactivated at the end of bleb expansion, facilitating RhoA activation and bleb retraction (Aoki et al., 2016). However, it is also possible that RhoA is activated by RhoGEFs at the bleb membrane. We have reported previously that myosin II–interacting guanine nucleotide exchange factor (MYOGEF) can activate RhoA (Wu et al., 2009). In addition, a study has also shown that ezrin can bind to PLEKHG6/MYOGEF (D’Angelo et al., 2007). In this study, we have demonstrated that ezrin–MYOGEF interactions are required for the recruitment of MYOGEF to the bleb membrane, where MYOGEF contributes to RhoA activation and bleb retraction.
RESULTS
MYOGEF is localized to the bleb membrane during bleb retraction
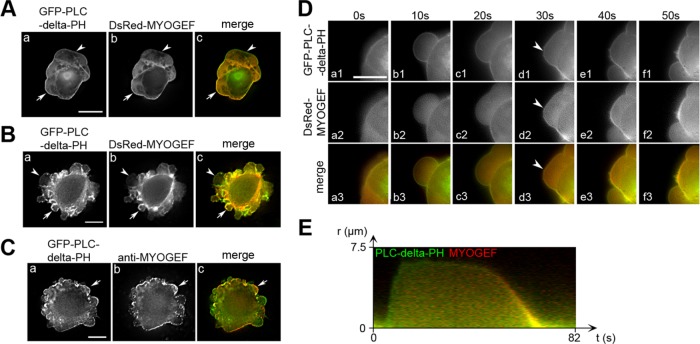
Our previous studies have shown that MYOGEF is implicated in the regulation of cell migration and cell division (Wu et al., 2006, 2009). During the course of our studies characterizing the cellular function of MYOGEF, we also noticed that GFP-tagged MYOGEF was localized to the bleb membrane in MDA-MB-231 cells. To further confirm these findings, MDA-MB-231 cells exogenously expressing DsRed-MYOGEF and GFP-PLC-delta-PH (the PH domain of phospholipase C delta) were subjected to fluorescence microscopy analyses. Our results showed that DsRed-MYOGEF was colocalized with GFP-PLC-delta-PH, a widely used membrane marker (Stauffer et al., 1998), at the bleb membrane in transfected MDA-MB-231 cells (Figure 1A, arrows in panels a–c).
FIGURE 1:
MYOGEF is localized to the bleb membrane during bleb retraction. (A, B) The localization of MYOGEF to the bleb membrane in MDA-MB-231 (A) and M2 melanoma (B) cells. Note that DsRed-MYOGEF was colocalized with the membrane marker GFP-PLC-delta-PH at the bleb membrane in some of the blebs (arrows), but not in others (arrowheads). Bar, 10 μm. (C) The localization of endogenous MYOGEF at the bleb membrane. M2 melanoma cells exogenously expressing GFP-PLC-delta-PH were subjected to immunofluorescence staining for endogenous MYOGEF with a mouse monoclonal antibody (mAb) specific for MYOGEF. Note that endogenous MYOGEF was localized to the bleb membrane (arrows). (D) Time-lapse images showing the localization of MYOGEF at the retracting bleb membrane. DsRed-MYOGEF was recruited to the bleb membrane when the bleb started retracting (arrowheads). Bar, 10 μm. (E) Kymograph showing the localization of DsRed-MYOGEF and GFP-PLC-delta-PH in a bleb cycle. Green indicates the localization of GFP-PLC-delta-PH alone to the expanding bleb membrane, while yellow indicates the colocalization of DsRed-MYOGEF and GFP-PLC-delta-PH to the retracting bleb membrane.
Filamin A (FLNA) is a filamentous actin cross-linking protein that can bind to actin filaments and maintain the branching of actin networks (Nakamura et al., 2011). M2 melanoma cells, which are deficient in FLNA, constitutively develop blebs and have been widely used to study bleb dynamics (Cunningham et al., 1992; Cunningham, 1995). Therefore, we also examined MYOGEF localization at the bleb membrane in M2 melanoma cells. Fluorescence microscopy analyses showed that DsRed-MYOGEF was also colocalized with GFP-PLC-delta-PH to the bleb membrane in transfected M2 melanoma cells (Figure 1B, arrows in panels a–c). Furthermore, like exogenously expressed DsRed-MYOGEF, endogenously expressed MYOGEF also localized to the bleb membrane (Figure 1C, arrows in panels a–c).
Next, we asked whether MYOGEF is specifically localized to retracting blebs. MDA-MB-231 cells exogenously expressing GFP-PLC-delta-PH and DsRed-MYOGEF were subjected to live-cell imaging analyses. We found that GFP-PLC-delta-PH was concentrated at the bleb membrane during bleb expansion and retraction (Figure 1D, panels a1–f1 and a3–f3). In contrast, DsRed-MYOGEF was concentrated at the bleb membrane only when blebs began to retract (Figure 1D, arrowheads in panels d2 and d3; Supplemental Video S1). Kymograph analyses also confirmed that both DsRed-MYOGEF and GFP-PLC-delta-PH were colocalized to the retracting bleb membrane. Conversely, GFP-PLC-delta-PH, but not DsRed-MYOGEF, was concentrated at the expanding bleb membrane (Figure 1E). Our results suggest that MYOGEF is specifically localized to the bleb membrane during bleb retraction. Consistent with the notion that both expanding and retracting blebs exist in a fixed blebbing cell, universal membrane makers, such as PLC-delta-PH, were localized to all blebs in a fixed cell (Figure 1, A and B, arrows and arrowheads in panels a and c), which are likely to be in either the expansion or retraction phase. In contrast, specific components for retracting blebs, such as MYOGEF, were only localized to some of the blebs (Figure 1, A and B, arrows in panels b and c), which are likely to be in the retraction phase. Similar localization patterns at the bleb membrane were also observed throughout this study for other components of retracting blebs, such as ezrin.
Video S1.
MYOGEF was localized to the bleb membrane during bleb retraction. Shown is an MDA-MB-231 cell exogenously expressing DsRed-MYOGEF and GFP-PLC-delta-PH. During bleb retraction, but not bleb expansion, DsRed-MYOGEF was localized to the bleb membrane. Images were acquired with 5s interval.
Ezrin is required for MYOGEF localization to the bleb membrane
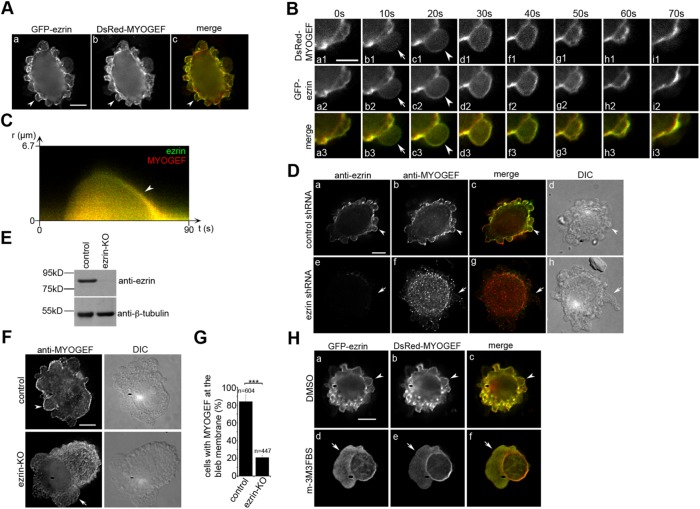
Ezrin, a member of the ERM family of proteins, plays a crucial role in organizing cortical actin networks and provides a physical linkage between the cortex and the cell membrane (Fehon et al., 2010). In particular, ezrin promotes bleb retraction by stimulating the reassembly of actomyosin networks underneath the bleb membrane during bleb retraction (Charras et al., 2006). In addition, it has been shown that the N-terminal region of ezrin interacts with the C-terminal region of PLEKHG6/MYOGEF (D’Angelo et al., 2007). Therefore, we asked whether ezrin is required for the localization of MYOGEF to the bleb membrane. To this end, we first assessed the colocalization of ezrin and MYOGEF at the bleb membrane. Fluorescence microscopy analyses showed that GFP-ezrin and DsRed-MYOGEF were colocalized at the bleb membrane in transfected M2 melanoma cells (Figure 2A, arrowheads in panels a–c). It has been shown that ezrin is specifically localized to the bleb membrane during bleb retraction (Charras et al., 2006). Our results also showed that MYOGEF was concentrated at the bleb membrane during bleb retraction (see Figure 1, D and E; Supplemental Video S1). Therefore, we went on to assess whether the colocalization of ezrin and MYOGEF occurs at the retracting bleb membrane. M2 melanoma cells exogenously expressing DsRed-MYOGEF and GFP-ezrin were subjected to time-lapse imaging analyses. We found that both GFP-ezrin and DsRed-MYOGEF were concentrated at the bleb membrane when blebs began to retract (Figure 2B, arrowheads in panels c1, c2, and c3; Supplemental Video S2). During bleb expansion, however, neither GFP-ezrin nor DsRed-MYOGEF was concentrated at the bleb membrane (Figure 2B, arrows in panels b1, b2, and b3; Supplemental Video S2). Kymograph analyses further confirmed that GFP-ezrin and DsRed-MYOGEF were colocalized to the retracting, but not the expanding, bleb membrane (Figure 2C). To assess the order of DsRed-MYOGEF and GFP-ezrin recruitment to the bleb membrane during bleb retraction, the fluorescence intensity of DsRed-MYOGEF and GFP-ezrin at the bleb membrane at different time points during a bleb cycle was quantified (Supplemental Figure S1A). We found that GFP-ezrin was recruited to the bleb membrane 7.5 ± 1.0 s earlier than DsRed-MYOGEF (Supplemental Figure S1B).
FIGURE 2:
The recruitment of MYOGEF to the bleb membrane is ezrin-dependent. (A) The colocalization of DsRed-MYOGEF and GFP-ezrin at the bleb membrane in M2 melanoma cells (arrowheads). Bar, 10 μm. (B) Time-lapse images showing that GFP-ezrin and DsRed-MYOGEF were colocalized at the bleb membrane during the retraction (arrowheads), but not the expansion (arrows), of a bleb. Bar, 10 μm. (C) Kymograph showing that DsRed-MYOGEF and GFP-ezrin were colocalized to the retracting, but not the expanding, bleb membrane. Yellow indicates the colocalization of DsRed-MYOGEF and GFP-ezrin to the bleb membrane during bleb retraction (arrowhead). (D) Ezrin knockdown impaired the localization of endogenous MYOGEF at the bleb membrane. M2 melanoma cells transfected with a control or an ezrin-specific shRNA plasmid were subjected to immunofluorescence staining for endogenous MYOGEF. Note that the colocalization of MYOGEF and ezrin at the bleb membrane was observed in control (arrowheads), but not in ezrin-knockdown (arrows) cells. Bar, 10 μm. (E) Immunoblot showing that the level of ezrin was decreased in ezrin-KO M2 melanoma cells compared to that in control cells. (F) Immunofluorescence staining showing that ezrin knockout interfered with MYOGEF localization to the bleb membrane. Bar, 10 μm. (G) Percentage of cells with endogenous MYOGEF localized at the bleb membrane in control and ezrin-KO M2 melanoma cells. Statistical significance was determined using Student’s t test. ***, p < 0.001. Data are mean ± SD. (H) Localization of GFP-ezrin and DsRed-MYOGEF at the bleb membrane in M2 melanoma cells treated with DMSO or the PLC activator m-3M3FBS. GFP-ezrin and DsRed-MYOGEF were colocalized at the bleb membrane in cells treated with DMSO (arrowheads), but not in cells treated with m-3M3FBS (arrows). Bar, 10 μm.
Video S2.
GFP-ezrin and DsRed-MYOGEF were co-localized at the bleb membrane during bleb retraction in transfected M2 melanoma cells. Images were acquired with 10s interval.
Next, we examined whether ezrin is required for the recruitment of MYOGEF to the bleb membrane. M2 melanoma cells transfected with a control or an ezrin-specific short hairpin RNA (shRNA) plasmid were analyzed for the localization of endogenous MYOGEF. The protein level of endogenous ezrin in ezrin shRNA–transfected cells was decreased as compared with that in control shRNA–transfected cells (Figure 2D, compare panel a with panel e). In addition, endogenous ezrin and MYOGEF were colocalized to the bleb membrane in control shRNA–transfected cells (Figure 2D, arrowheads in panels a–d). In contrast, MYOGEF was not concentrated at the bleb membrane in ezrin-depleted cells (Figure 2D, arrows in panels e–h). These results suggest that ezrin is required for the localization of MYOGEF to the bleb membrane.
To further confirm the role of ezrin in recruiting MYOGEF to the bleb membrane, we also used the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene editing strategy to knock out ezrin in M2 melanoma cells. Insertion/deletion (InDel) analyses confirmed that an adenine was inserted into the third exon of the ezrin gene (Supplemental Figure S2A), thus introducing a stop codon that results in premature termination of ezrin translation (Supplemental Figure S2B). Immunoblot analyses consistently showed that the protein level of ezrin decreased in ezrin-knockout (KO) M2 melanoma cells as compared with that in control M2 melanoma cells (Figure 2E). Importantly, CRISPR-mediated knockout of ezrin in M2 melanoma cells also interfered with the localization of endogenous MYOGEF to the bleb membrane (Figure 2F, compare top panels with bottom panels; Figure 2G). In addition, live-cell imaging analyses showed that ezrin knockout impaired DsRed-MYOGEF localization to the bleb membrane (Supplemental Video S3). These results clearly demonstrate an important role for ezrin in recruiting MYOGEF to the bleb membrane.
Video S3.
DsRed-MYOGEF was not localized to the bleb membrane in ezrin-KO melanoma cells co-expressing GFP-PLC-delta-PH. Images were acquired with 5s interval.
It has been demonstrated that binding of ezrin to PIP2 plays a critical role in recruiting ezrin to the cell membrane, where ezrin promotes the assembly of cortical actin networks (Barret et al., 2000; Yonemura et al., 2002). Phospholipase C (PLC) catalyzes the conversion of PIP2 to diacylglycerol (DAG) and inositol triphosphate (IP3), therefore decreasing the levels of PIP2 on the cell membrane (Hao et al., 2009). An increase in PLC activity consistently interferes with the localization of ezrin to the cell membrane (Hao et al., 2009). Therefore, we asked whether treatment of cells with an activator of PLC would interfere with the localization of ezrin or MYOGEF to the bleb membrane. M2 melanoma cells exogenously expressing GFP-ezrin and DsRed-MYOGEF were subjected to fluorescence microscopy analyses following treatment with a dimethyl sulfoxide (DMSO) vehicle or a PLC activator (m-3M3FBS). We found that both GFP-ezrin and DsRed-MYOGEF were concentrated at the bleb membrane in M2 melanoma cells treated with DMSO (Figure 2H, arrowheads in panels a–c). In contrast, neither GFP-ezrin nor DsRed-MYOGEF was concentrated at the bleb membrane in cells treated with the PLC activator m-3M3FBS (Figure 2H, arrows in panels d–f). These results further confirm that ezrin is required for the localization of MYOGEF to the bleb membrane.
The ezrin–MYOGEF interaction contributes to bleb retraction
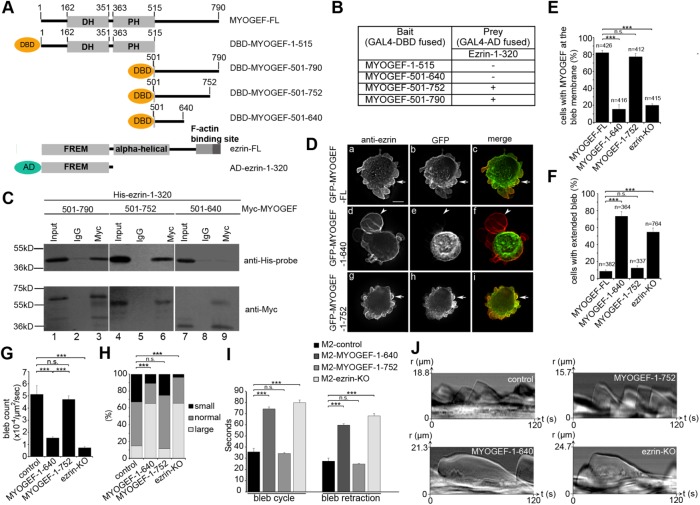
To gain insight into whether ezrin–MYOGEF interactions play a role in the localization of MYOGEF at the bleb membrane, we used yeast two-hybrid and coimmunoprecipitation assays to identify the critical region in MYOGEF that interacts with ezrin. Various truncated versions of MYOGEF were cloned into the bait vector while the N-terminal region of ezrin (amino acid residues 1–320) was cloned into the prey vector (Figure 3A). The N-terminal region of ezrin was used because a previous report has shown that it can interact with the C-terminal region of PLEKHG6/MYOGEF (D’Angelo et al., 2007). Our yeast two-hybrid assays showed that ezrin-1–320 could interact with MYOGEF-501–790 or MYOGEF-501–752, but not with MYOGEF-1–515 or MYOGEF-501–640 (Figure 3B). In addition, coimmunoprecipitation assays showed that His-ezrin-1–320 was coprecipitated with Myc-MYOGEF-501–790 or Myc-MYOGEF-501–752, but not with Myc-MYOGEF-501–640 (Figure 3C). Therefore, our results suggest that amino acid residues 640–752 in MYOGEF are required for interactions with the N-terminal region of ezrin.
FIGURE 3:
The ezrin–MYOGEF interaction is critical for bleb retraction. (A) Schematic showing different truncated versions of MYOGEF and ezrin fragments used in yeast two-hybrid assays. AD, activation domain of GAL4; DBD, DNA-binding domain of GAL4. (B, C) Yeast two-hybrid (B) and coimmunoprecipitation (C) assays indicating that amino acid residues 640–752 in MYOGEF are required for interactions with the N-terminal region of ezrin. In B, (—) indicates that yeast cells did not grow on the selective SD agar plates (SD/-Ade-His-Leu-Trp); (+) indicates that yeast cells could grow on the selective SD agar plates (SD/-Ade-His-Leu-Trp) and that X-gal filter assays were positive. (D) The ezrin-binding region (residues 640–752) in MYOGEF is required for MYOGEF localization at the bleb membrane. GFP-MYOGEF-1–640 (lacking the ezrin-binding region) was not colocalized with ezrin at the bleb membrane in transfected M2 melanoma cells (arrowheads in panels d–f), while GFP-MYOGEF-FL or GFP-MYOGEF-1–752 (containing the ezrin-binding region) was colocalized with ezrin at the bleb membrane (arrows in panels a–c and g–i). Bar, 10 μm. (E) Percentage of cells with MYOGEF at the bleb membrane was quantified. Note that lack of the ezrin-binding region or knockout of ezrin impaired the localization of MYOGEF to the bleb membrane. Statistical significance was determined using one-way ANOVA test and Tukey’s post hoc test. n.s., nonsignificant (p ≥ 0.05); ***, p < 0.001. Data are mean ± SD. (F) Percentage of cells with extended blebs was quantified in control M2 melanoma cells expressing GFP-MYOGEF-FL, GFP-MYOGEF-1–640, or GFP-MYOGEF-1–752, as well as in ezrin-KO M2 cells expressing GFP-MYOGFEF-FL. Note that extended blebs were formed in M2 melanoma cells expressing GFP-MYOGEF-1–640 and in ezrin-KO cells. Statistical significance was determined using a one-way ANOVA test and Tukey’s post hoc test. ***, p < 0.001. Data are mean ± SD. (G) Quantification of bleb number in a cell. All blebs in each cell examined were counted in a 2-min period. Three independent experiments were done and 30 cells were analyzed for each experiment. The bleb number was normalized to the cell area (μm2) and to time (s). Statistical significance was determined using one-way ANOVA and Tukey’s post hoc test. ***, p < 0.001. Data are mean ± SD. (H) Distributions of bleb size in a cell were compared using a chi-squared test. ***, p < 0.001. (I) The time required for blebs to complete a bleb cycle or bleb retraction. Statistical significance was determined using one-way ANOVA and Tukey’s post hoc test. ***, p < 0.001. Data are mean ± SD. (J) Representative kymographs demonstrating the efficiency of bleb cycling and bleb retraction. Kymographs were created from DIC images.
Next, we asked whether the ezrin-binding region (amino acid residues 640–752) is required for MYOGEF localization at the bleb membrane. M2 melanoma cells exogenously expressing GFP-MYOGEF-FL, GFP-MYOGEF-1–640, or GFP-MYOGEF-1–752 were subjected to immunofluorescence staining for ezrin (Figure 3D). It is of note that MYOGEF-FL and MYOGEF-1–752, but not MYOGEF-1–640, contain the ezrin-binding region. We found that exogenously expressed GFP-MYOGEF-FL or GFP-MYOGEF-1–752 was colocalized with endogenous ezrin at the bleb membrane in transfected M2 melanoma cells (Figure 3, D, arrows in panels a–c and g–i, and E). In contrast, exogenously expressed GFP-MYOGEF-1–640 was not colocalized with ezrin at the bleb membrane (Figure 3, D, arrowheads in panel d–f, and E). Therefore, our findings suggest that the ezrin-binding region in MYOGEF is critical not only for interactions with ezrin, but also for the localization of MYOGEF to the bleb membrane, supporting the notion that ezrin–MYOGEF interaction is required for the recruitment of MYOGEF to the bleb membrane. These results are also consistent with our observations that shRNA-mediated depletion and CRISPR-mediated knockout of ezrin disrupted the localization of MYOGEF to the bleb membrane (see Figure 2, D, F, and G).
Remarkably, M2 melanoma cells exogenously expressing GFP-MYOGEF-1–640 formed extended large blebs (Figure 3D, arrowhead in panel d; compare panel d with panels a and g; Figure 3F). However, exogenous expression of GFP-MYOGEF-FL or GFP-MYOGEF-1–752 did not alter membrane blebbing in transfected M2 melanoma cells (Figure 3D, compare panels a and g with panel d; Figure 3F). We have shown previously that the C-terminal region of MYOGEF interacts with its N-terminal region, forming an inhibitory conformation (Wu et al., 2014b). We speculated that MYOGEF-1–640 could interact with the C-terminal region of MYOGEF-FL, thus masking the ezrin-binding region and interfering with the interaction between ezrin and MYOGEF-FL. To test this possibility, we went on to examine whether MYOGEF-1–640 can bind to exogenously or endogenously expressed MYOGEF. Coimmunoprecipitation assays showed that GFP-MYOGEF-1–640 was coprecipitated with Myc-MYOGEF-FL (Supplemental Figure S3A). Myc-MYOGEF-1–640 was also coprecipitated with endogenous MYOGEF from transfected cells (Supplemental Figure S3B). These results suggest that MYOGEF-1–640 can bind to the C-terminal region of endogenous MYOGEF, interfering with the interaction of endogenous MYOGEF with ezrin (Supplemental Figure S3C). As a result, M2 melanoma cells exogenously expressing GFP-MYOGEF-1–640 formed extended large blebs (Figure 3, D and F). Importantly, these findings also suggest that ezrin–MYOGEF interactions are required for MYOGEF localization at the bleb membrane as well as for MYOGEF function in bleb retraction.
We then quantified bleb formation and blebbing dynamics in ezrin-KO M2 melanoma cells as well as in M2 melanoma cells exogenously expressing GFP-MYOGEF-1–752 or GFP-MYOGEF-1–640. The number of blebs in an individual M2 melanoma cell exogenously expressing GFP-MYOGEF-1–752 was significantly larger than that in an individual M2 melanoma cell exogenously expressing GFP-MYOGEF-1–640 (Figure 3G). In addition, the number of blebs in a control M2 melanoma cell was significantly larger than that in an individual ezrin-KO M2 melanoma cell (Figure 3G). In contrast, the percentage of large and/or extended blebs in ezrin-KO M2 melanoma cells or in M2 melanoma cells exogenously expressing GFP-MYOGEF-1–640 was significantly larger than that in control M2 melanoma cells or in M2 melanoma cells exogenously expressing GFP-MYOGEF-1–752 (Figure 3H).
A plausible reason for the presence of a higher percentage of large and/or extended blebs in ezrin-KO M2 melanoma cells or in M2 melanoma cells exogenously expressing GFP-MYOGEF-1–640 is retarded bleb retraction. To confirm this possibility, we examined whether bleb retraction is affected in ezrin-KO M2 melanoma cells or in M2 melanoma cells exogenously expressing GFP-MYOGEF-1–640. We found that knockout of ezrin or exogenous expression of GFP-MYOGEF-1–640 in M2 melanoma cells caused an increase in the cycle time and retraction time of membrane blebbing (Figure 3, I and J). Therefore, our results suggest that the ezrin–MYOGEF interaction is not only required for MYOGEF localization to the bleb membrane, but also important for bleb retraction.
The ezrin–MYOGEF interaction contributes to RhoA activation and cortex reassembly at the bleb membrane
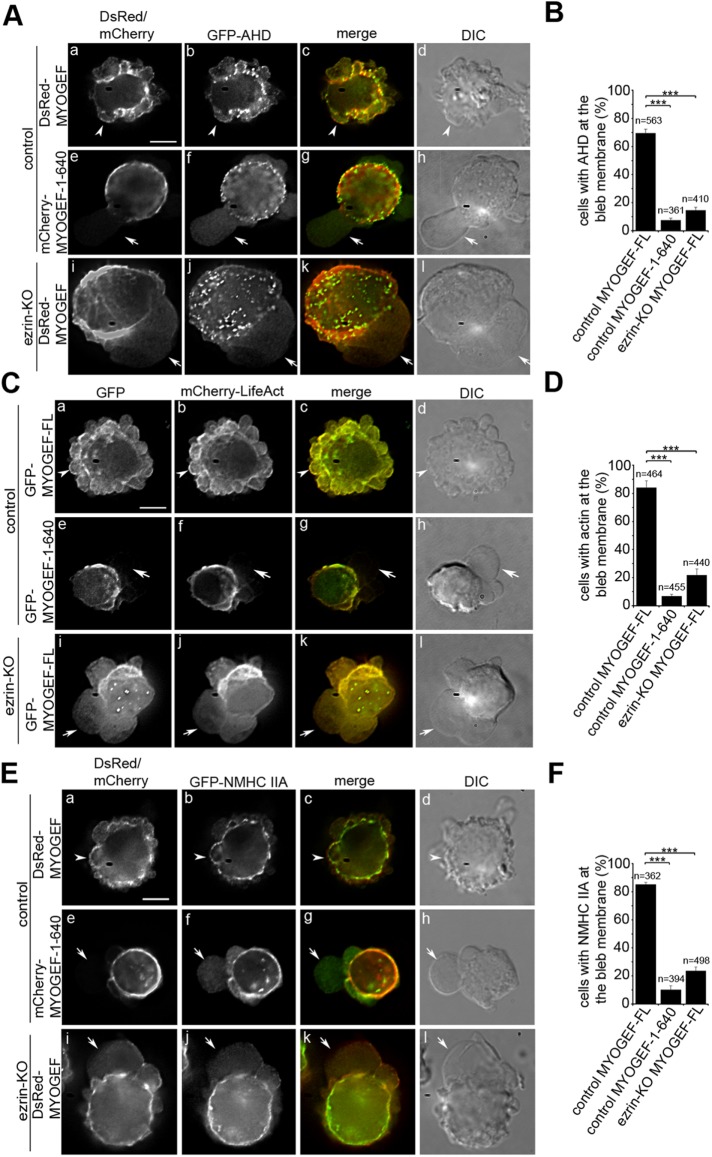
RhoA activation at the bleb membrane is believed to play a critical role in reassembling a contractile actomyosin network and promoting bleb retraction (Fackler and Grosse, 2008). Our previous studies have shown that MYOGEF can activate RhoA (Wu et al., 2009). Therefore, we asked whether MYOGEF is implicated in RhoA activation at the bleb membrane. Anillin contains an anillin homology domain (AHD) that can specifically bind to the GTP-bound form of RhoA. Therefore, GFP-AHD has been used to detect RhoA activation in transfected mammalian cells (Piekny and Glotzer, 2008; Aoki et al., 2016). We found that DsRed-MYOGEF and GFP-AHD were colocalized to the bleb membrane in transfected M2 melanoma cells (Figure 4, A, arrowheads in panels a–d, and B; Supplemental Video S4). However, in M2 melanoma cells exogenously expressing both GFP-AHD and mCherry-MYOGEF-1–640, GFP-AHD was not concentrated at the bleb membrane (Figure 4, A, arrows in panels e–h, and B; Supplemental Video S5). Furthermore, in ezrin-KO M2 melanoma cells, neither DsRed-MYOGEF nor GFP-AHD was efficiently concentrated at the bleb membrane (Figure 4, A, arrows in panels i–l, and B; Supplemental Video S6). Therefore, our results suggest that the ezrin–MYOGEF interaction and the localization of MYOGEF to the bleb membrane contribute to RhoA activation at the bleb membrane.
FIGURE 4:
The ezrin–MYOGEF interaction is required for RhoA activation and the reassembly of the actomyosin cortex at the bleb membrane. GFP-AHD (A, B), mCherry-LifeAct (C, D), or GFP-NMHC IIA (E, F) was colocalized with MYOGEF-FL to the bleb membrane in cells exogenously coexpressing MYOGEF-FL (arrowheads in panels Aa–Ad, Ca–Cd, and Ea–Ed), but was not localized to the bleb membrane in cells exogenously coexpressing MYOGEF-1–640 (arrows in panels Ae–Ah, Ce–-Ch, and Ee–Eh) or in ezrin-KO cells (arrows in panels Ai–Al, Ci–Cl, and Ei–El). Note that MYOGEF-FL was not localized to the bleb membrane in ezrin-KO cells (arrows in panels Ai, Ci, and Ei). Bar, 10 μm. For percentage of cells with corresponding proteins at the bleb membrane, statistical significance was determined using a one-way ANOVA test and Tukey’s post hoc test. ***, p < 0.001. Data are mean ± SD.
Video S4.
GFP-AHD was localized to the bleb membrane during bleb retraction. M2 melanoma cells expressing GFP-AHD and DsRed-MYOGEF were subjected to time-lapse microscopy analyses. Images were acquired with 2s interval.
Video S5.
GFP-AHD was not localized to the bleb membrane in M2 melanoma cells co-expressing mCherry-MYOGEF-1-640 (lack the ezrin-binding region). Images were acquired with 2s interval.
Video S6.
Ezrin-KO interfered with the localization of GFP-AHD to the bleb membrane. Shown is an ezrin-KO M2 melanoma cell expressing GFP-AHD and DsRed-MYOGEF. Neither GFP-AHD nor DsRed-MYOGEF was localized to the bleb membrane during bleb retraction. Images were acquired with 2s interval.
It is generally believed that RhoA activation at the bleb membrane stimulates the reassembly of the cortical actomyosin network, thus leading to bleb retraction (Charras et al., 2006; Fackler and Grosse, 2008). Therefore, we went on to examine actin polymerization at the bleb membrane in M2 melanoma cells exogenously expressing GFP-MYOGEF-FL or GFP-MYOGEF-1–640, as well as in ezrin-KO M2 melanoma cells. We assessed actin polymerization at the bleb membrane by examining the integration of mCherry-LifeAct into the actin filaments in transfected M2 melanoma cells. Our results showed that both GFP-MYOGEF-FL and mCherry-LifeAct were concentrated at the bleb membrane in transfected M2 melanoma cells (Figure 4, C, arrowheads in panels a–d, and D; Supplemental Video S7). However, exogenous expression of GFP-MYOGEF-1–640 disrupted the concentration of mCherry-LifeAct at the bleb membrane in transfected M2 melanoma cells (Figure 4, C, arrows in panels e–h, and D; Supplemental Video S8). More importantly, neither GFP-MYOGEF-FL nor mCherry-LifeAct was concentrated at the bleb membrane in ezrin-KO M2 melanoma cells (Figure 4, C, arrows in panels i–-l, and D; Supplemental Video S9). These results suggest that the ezrin–MYOGEF interaction may regulate membrane blebbing by promoting the reassembly of the actin cortex underneath the membrane during bleb retraction.
Video S7.
Actin cortex was reformed at the bleb membrane in M2 melanoma cells expressing GFP-MYOGEF-FL. mCherry-LifeAct was co-localized with GFP-MYOGEF-FL to the bleb membrane during bleb retraction. Images were acquired with 2s interval.
Video S8.
Exogenous expression of truncated MYOGEF lacking the ezrin-binding region interfered with actin polymerization at the bleb membrane. Actin cortex was not reformed efficiently at the bleb membrane in M2 melanoma cells exogenously expressing mCherry-LifeAct and GFP-MYOGEF-1-640. Note that mCherry-LifeAct was concentrated at the cell cortex. Images were acquired with 2s interval.
Video S9.
Reformation of actin cortex at the bleb membrane was impaired in ezrin-KO M2 melanoma cells. mCherry-LifeAct was not localized to the bleb membrane in ezrin-KO M2 cells co-expressing GFP-MYOGEF-FL. Images were acquired with 1s interval.
Next, we asked whether exogenous expression of GFP-MYOGEF-1–640 or CRISPR-mediated knockout of ezrin affects the localization of nonmuscle myosin II to the bleb membrane. Our results showed that DsRed-MYOGEF and GFP-tagged nonmuscle myosin heavy chain IIA (GFP-NMHC IIA) were colocalized to the bleb membrane (Figure 4, E, arrowheads in panels a–d, and F). In contrast, exogenous expression of mCherry-MYOGEF-1–640 disrupted the localization of GFP-NMHC IIA at the bleb membrane in transfected M2 melanoma cells (Figure 4, E, arrows in panels e–h, and F). Consistently, knockout of ezrin interfered with the localization of DsRed-MYOGEF and GFP-NMHC IIA at the bleb membrane (Figure 4, E, arrows in panels i–l, and F). It is of note that exogenous expression of MYOGEF-1–640 or knockout of ezrin led to the formation of large blebs (compare panels h and I with panel d in Figure 4, A, C, and E; also see Figure 3, D, F, and H), most likely due to defects in bleb retraction. Taken together, our results suggest that ezrin–MYOGEF interactions play a role in recruiting MYOGEF to the bleb membrane and promoting the reassembly of the actomyosin cortex at the bleb membrane.
Knockout of MYOGEF interferes with RhoA activation and cortex reassembly at the bleb membrane
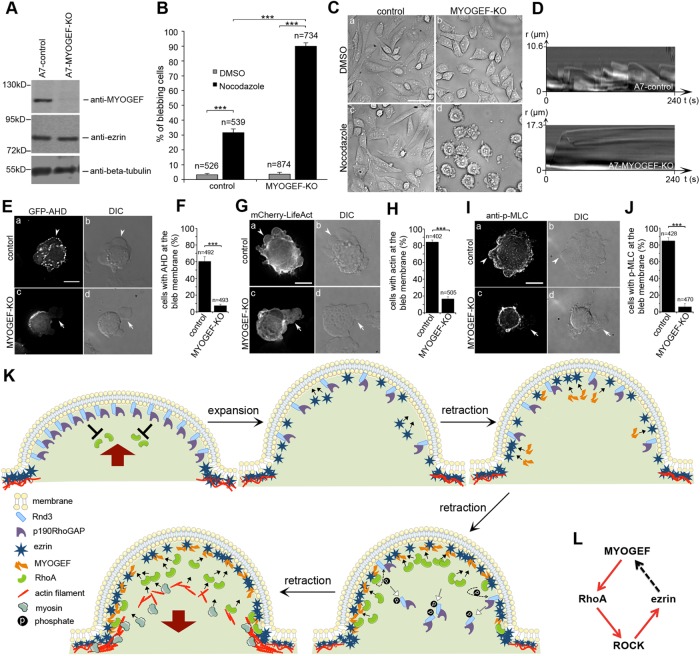
Unlike M2 melanoma cells, which lack FLNA and constantly form blebs during culture, A7 melanoma cells express FLNA and do not continually show membrane blebbing during culture (Cunningham et al., 1992; Cunningham, 1995). Therefore, we used the CRISPR/Cas9 gene editing strategy to knock out MYOGEF in A7 melanoma cells to examine the impact of MYOGEF deficiency on membrane blebbing. InDel analyses showed that an adenine insertion in the third coding exon of the MYOGEF gene introduced a stop codon that results in premature termination of MYOGEF translation (Supplemental Figure S4, A and B). Immunoblot analyses consistently showed that the expression levels of MYOGEF were decreased in MYOGEF-KO A7 melanoma cells (Figure 5A). However, we found that CRISPR-mediated knockout of MYOGEF in A7 melanoma cells did not obviously increase membrane blebbing, consistent with the notion that, like ezrin, MYOGEF is implicated in bleb retraction but not in bleb initiation and expansion.
FIGURE 5:
MYOGEF contributes to RhoA activation and cortex reassembly at the bleb membrane. (A) The protein level of MYOGEF, but not ezrin, was decreased in MYOGEF-KO A7 melanoma cells. (B) Nocodazole treatment significantly increased membrane blebbing in MYOGEF-KO A7 melanoma cells. Two-way ANOVA with Tukey’s post hoc test was used to analyze the percentage of blebbing cells. ***, p < 0.001. Data are mean ± SD. (C) Representative phase images showing membrane blebbing in control (a, c) or MYOGEF-KO (b, d) A7 melanoma cells treated with DMSO (a, b) or nocodazole (c, d). Bar, 50 μm. (D) Kymographs showing the efficiency of bleb retraction in control (top panel) or MYOGEF-KO (bottom panel) A7 melanoma cells treated with nocodazole. (E–J) GFP-AHD (E), mCherry-LifeAct (G), or phosphorylated myosin light chain (p-MLC, I) was concentrated at the bleb membrane in control (arrowheads in panels Ea, Ga, and Ia), but not in MYOGEF-KO (arrows in panels Ec, Fc, and Gc) A7 melanoma cells treated with nocodazole. Bar, 10 μm. For percentage of cells with GFP-AHD (E), mCherry-LifeAct (G), or p-MLC (I) localized to the bleb membrane, statistical significance was determined using two-tailed paired Student’s t test. ***, p < 0.001. Data are mean ± SD. (K) Schematic showing the cooperation of ezrin and MYOGEF in the regulation of bleb retraction. Red arrows indicate the direction of movement for bleb membranes. Bar-headed lines represent the inhibition of RhoA activation by p190RhoGAP. Black solid arrows in the bleb lumen indicate the recruitment of respective proteins to the bleb membrane. Black dashed arrows indicate that RhoA signaling promotes the phosphorylation of Rnd3. Open-headed arrows indicate the release of the p190RhoGAP-Rnd3 complex from the bleb membrane due to Rnd3 phosphorylation. (L) MYOGEF mediates a positive feedback loop between RhoA and ezrin at the bleb membrane. Red arrows indicate activation, and the black dash arrow indicates recruitment.
It has been shown that an increase in cortical contractility is required for bleb initiation and formation (Charras, 2008). Notably, disruption of microtubules by nocodazole treatment increases cortical contractility through stimulating RhoA activation (Waterman-Storer and Salmon, 1999). It has been shown that nocodazole treatment promotes membrane blebbing in numerous cell lines (Hagmann et al., 1999; Pletjushkina et al., 2001; Jia et al., 2006; Takesono et al., 2010). Therefore, we went on to assess the impact of nocodazole treatment on membrane blebbing in MYOGEF-KO A7 melanoma cells. We found that treatment with nocodazole significantly increased the percentage of blebbing cells in MYOGEF-KO A7 melanoma cells as compared with that in control A7 melanoma cells (Figure 5, B and C). To confirm whether CRISPR/Cas9-mediated knockout of MYOGEF interferes with bleb retraction, we performed kymograph analyses to monitor bleb dynamics in MYOGEF-KO A7 melanoma cells treated with nocodazole. Consistent with our observations that MYOGEF was specifically localized to the bleb membrane during bleb retraction (see Figure 1, D and E), retarded bleb retraction was observed in MYOGEF-KO A7 melanoma cells treated with nocodazole, but not in control A7 cells treated with nocodazole (Figure 5D; Supplemental Videos S10 and S11). Similar findings were also observed in ezrin-KO A7 melanoma cells (Supplemental Figure S5, A–C). Therefore, our results suggest that, like ezrin, MYOGEF plays a role in bleb retraction.
Video S10.
Nocodazole-induced membrane blebbing in control A7 melanoma cells. Cells were observed after treatment with nocodazole for 40 min. Images were acquired with 5s interval.
Video S11.
MYOGEF knockout interfered with the retraction of nocodazole-induced blebs in A7 melanoma cells. Nocodazole-induced blebs showed retarded retraction in MYOGEF-KO A7 melanoma cells. Images were acquired with 5s interval.
Next, we assessed whether CRISPR/Cas9-mediated knockout of MYOGEF also interferes with RhoA activation and the reassembly of the cortical actomyosin network at the bleb membrane. Control or MYOGEF-KO A7 melanoma cells exogenously expressing GFP-AHD were treated with nocodazole and then subjected to fluorescence microscopy analyses. We found that GFP-AHD was concentrated at the bleb membrane in control A7 melanoma cells treated with nocodazole (Figure 5, E, arrowheads in panels a and b, and F; Supplemental Video S12). However, GFP-AHD was not concentrated at the bleb membrane in MYOGEF-KO A7 melanoma cells treated with nocodazole (Figure 5E, arrows in panels c and d; compare panels c and d with panels a and b; Figure 5F; Supplemental Video S13). Therefore, our results suggest that knockout of MYOGEF impairs RhoA activation at the bleb membrane.
Video S12.
GFP-AHD was recruited to the bleb membrane during bleb retraction in control A7 melanoma cells treated with nocodazole. Images were acquired with 2s interval.
Video S13.
MYOGEF-KO interfered with the localization of GFP-AHD to the bleb membrane. In MYOGEF-KO A7 melanoma cells treated with nocodazole, GFP-AHD was concentrated at the cell cortex, but not recruited to the bleb membrane during bleb retraction. Images were acquired with 2s interval.
To assess whether MYOGEF knockout has an impact on actin polymerization at the bleb membrane, we examined the localization of mCherry-LifeAct at the bleb membrane in control or MYOGEF-KO A7 melanoma cells treated with nocodazole. We found that mCherry-LifeAct was concentrated at the bleb membrane in control A7 melanoma cells treated with nocodazole (Figure 5, G, arrowheads in panels a and b, and H; Supplemental Video S14). In contrast, mCherry-LifeAct was not efficiently concentrated at the bleb membrane in MYOGEF-KO A7 melanoma cells treated with nocodazole (Figure 5G, arrows in panels c and d; compare panels c and d with panels a and b; Figure 5H; Supplemental Video S15). In addition, knockout of MYOGEF impaired the localization of GFP-NMHC IIA at the bleb membrane (Supplemental Videos S16 and S17). Our results suggest that MYOGEF knockout interferes with the reassembly of the actomyosin cortex in retracting blebs.
Video S14.
Cortical actin was re-assembled at the bleb membrane in control A7 melanoma cells treated with nocodazole. mCherry-LifeAct was localized to the bleb membrane during bleb retraction. Images were acquired with 1s interval.
Video S15.
The reformation of actin cortex at the bleb membrane was inhibited in MYOGEF-KO A7 melanoma cells treated with nocodazole. mCherry-LifeAct showed defuse distribution in the bleb lumen and cell body during membrane blebbing. Images were acquired with 2s interval.
Video S16.
GFP-NMHC IIA was recruited to the bleb membrane during bleb retraction in control A7 melanoma cells treated with nocodazole. Images were acquired with 2s interval.
Video S17.
MYOGEF knockout impaired the recruitment of GFP-NMHC-IIA to the bleb membrane. GFP-NMHC IIA was not concentrated at the bleb membrane of extended blebs in MYOGEF-KO A7 melanoma cells treated with nocodazole. Images were acquired with 5s interval.
Activation of nonmuscle myosin II generally involves Thr/Ser phosphorylation of MLC (Vicente-Manzanares et al., 2009). To determine whether knockout of MYOGEF interferes with myosin II activation at the bleb membrane, the bleb membrane in MYOGEF-KO A7 melanoma cells was also examined for the presence of phosphorylated MLC. MYOGEF-KO A7 melanoma cells were treated with nocodazole and then subjected to immunofluorescence staining for phosphorylated MLC. We found that phosphorylated MLC was concentrated at the bleb membrane in control A7 melanoma cells treated with nocodazole (Figure 5I, arrowheads in panels a and b; Figure 5J). In contrast, phosphorylated MLC was not concentrated at the bleb membrane in MYOGEF-KO A7 melanoma cells treated with nocodazole (Figure 5I, arrows in panels c and d; compare panels c and d with panels a and b; Figure 5J). Our results suggest that MYOGEF stimulates the reassembly of the cortical actomyosin network at the bleb membrane via activating RhoA, thus promoting bleb retraction. Consistent with the notion that MYOGEF is recruited to the retracting bleb membrane by ezrin, we also found that the localization of DsRed-MYOGEF, GFP-AHD, mCherry-LifeAct, or phosphorylated MLC to nocodazole-induced blebs in ezrin-knockout A7 melanoma cells was severely impaired (Supplemental Figure S5, D–K).
To further confirm the role of MYOGEF in the regulation of bleb retraction, we also used the CRISPR/Cas9 system to knock out MYOGEF in M2 melanoma cells. InDel analyses showed that 13 nucleotides were deleted in the third coding exon of the MYOGEF gene (Supplemental Figure S6A). The deletion introduces a stop codon, leading to premature termination of MYOGEF translation (Supplemental Figure S6B). Immunoblot analyses confirmed that the expression levels of MYOGEF were decreased in MYOGEF-KO M2 melanoma cells (Supplemental Figure S6C). However, we found that MYOGEF knockout only slightly increased the percentage of blebbing cells (Supplemental Figure S6, D and E), consistent with the previous finding that microinjection of ezrin FERM domains into M2 melanoma cells leads to a moderate increase in membrane blebbing (Charras et al., 2006). Therefore, we treated control or MYOGEF-KO M2 melanoma cells with nocodazole and determined the impact of nocodazole treatment on membrane blebbing. We found that nocodazole treatment increased the percentage of blebbing cells in both control and MYOGEF-KO M2 melanoma cells (Supplemental Figure S6, F and G). However, we found that MYOGEF-KO, but not control, M2 melanoma cells developed extended blebs in the presence of nocodazole (Supplemental Figure S6, F and H), likely due to defects in bleb retraction. Since M2 melanoma cells lack FLNA and constantly show membrane blebbing, we did not use this cell line to analyze the impact of MYOGEF knockout on cortex reassembly at the bleb membrane during bleb retraction. Nonetheless, the formation of extended blebs in MYOGEF-KO M2 melanoma cells in the presence of nocodazole also support our conclusion that MYOGEF plays a role in regulating bleb retraction.
Overall, our results indicate that the interaction between ezrin and MYOGEF plays an important role in regulating bleb retraction. In particular, ezrin depletion or knockout interfered with the localization of MYOGEF at the bleb membrane (Figure 2). MYOGEF knockout resulted in defects in bleb retraction (Figure 5). However, a question can be raised as to whether MYOGEF knockout has an impact on ezrin localization at the bleb membrane. To address this concern, control or MYOGEF-KO M2 melanoma cells treated with DMSO or nocodazole were subjected to immunofluorescence staining for ezrin. As shown in Supplemental Figure S7, MYOGEF knockout did not affect ezrin localization at the bleb membrane in M2 melanoma cells. Notably, ezrin was still localized to the extended blebs in MYOGEF-KO M2 melanoma cells treated with nocodazole (Supplemental Figure S7A, arrowhead in panel d). Similar findings were also observed in MYOGEF-KO A7 melanoma cells (unpublished data). Therefore, our results suggest that MYOGEF knockout does not affect ezrin localization at the bleb membrane.
DISCUSSION
Previous studies have highlighted the roles of ezrin and RhoA signaling in the regulation of bleb retraction. Ezrin acts as a cortex-membrane linker protein to physically connect the newly reassembled cortex to the bleb membrane, thus promoting bleb retraction (Charras et al., 2006; Charras, 2008; Fackler and Grosse, 2008). RhoA-ROCK signaling not only is capable of activating ezrin, but also plays a critical role in promoting cortex reassembly for bleb retraction (Matsui et al., 1998; Aoki et al., 2016). Our results in this study demonstrate a critical role for ezrin in the recruitment of MYOGEF to the bleb membrane. In turn, MYOGEF contributes to RhoA activation and cortex reassembly for bleb retraction. Therefore, our findings uncover an important regulatory mechanism involving ezrin-MYOGEF-RhoA signaling for RhoA activation at the bleb membrane to promote actomyosin cortex reassembly and bleb retraction (Figure 5, K and L).
Regulation of bleb retraction by ezrin and RhoA
Membrane blebbing displays repeating cycles between bleb expansion and retraction (Charras, 2008; Fackler and Grosse, 2008). Cortical contraction and subsequent increase in intracellular pressure are the driving forces for bleb expansion, while the reassembly of actomyosin networks at the bleb membrane leads to bleb retraction (Charras, 2008; Fackler and Grosse, 2008). As a cortex–membrane linker protein, ezrin plays a pivotal role in promoting bleb retraction (Charras, 2008; Fackler and Grosse, 2008). A line of evidence also demonstrates that RhoA signaling is implicated in promoting the reassembly of actomyosin networks at the bleb membrane during bleb retraction (Charras, 2008; Fackler and Grosse, 2008). RhoA can be activated by GEFs and inactivated by GAPs (Cerione and Zheng, 1996; Van Aelst and D’Souza-Schorey, 1997; Rossman et al., 2005). Therefore, it is conceivable that either removal of a GAP from the bleb membrane or concentration of a GEF at the bleb membrane can contribute to RhoA activation at the bleb membrane. Consistent with this general concept, a recent study has demonstrated that Rnd3 and a GAP termed p190RhoGAP are concentrated in expanding blebs, where Rnd3 and p190RhoGAP cooperate to antagonize RhoA activation, thus preventing RhoA from being activated during bleb expansion (Aoki et al., 2016). When blebs continue to expand, Rnd3 and p190RhoGAP proteins lose their concentrated distribution at the bleb membrane, thus favoring RhoA activation and bleb retraction. In turn, ROCK, a downstream effector of RhoA, phosphorylates Rnd3, thus promoting the release of the Rnd3-p190RhoGAP complex from the bleb membrane (Aoki et al., 2016). Therefore, a decrease in RhoGAP activity at the bleb membrane is likely to be an important mechanism for RhoA activation during bleb retraction.
However, localization of a RhoGEF at the bleb membrane may also contribute to RhoA activation at the bleb membrane during bleb retraction. In this study, our results clearly demonstrate that MYOGEF is specifically recruited to the retracting bleb membrane, where MYOGEF contributes to RhoA activation and promotes actomyosin cortex reassembly. Our findings further demonstrate that ezrin is capable of recruiting MYOGEF to the retracting bleb membrane. Notably, Aoki et al. (2016) propose that the dilution of Rnd3 and p190RhoGAP at the bleb membrane during bleb expansion leads to sporadic RhoA activation that, in turn, eventually activates ezrin. However, it is less clear how RhoA activation becomes sustained during bleb retraction. Our results lead us to propose that sporadically activated ezrin recruits MYOGEF to the bleb membrane. Importantly, ezrin-mediated recruitment of MYOGEF to the bleb membrane would likely contribute to sustained RhoA activation at the bleb membrane, thus stimulating cortex reassembly and promoting bleb retraction (Figure 5K). Furthermore, it is also tempting to propose that MYOGEF mediates a positive feedback loop between ezrin and RhoA signaling (i.e., ezrin-MYOGEF-RhoA) at the bleb membrane for the regulation of cortex reassembly and bleb retraction (Figure 5L).
Redundant mechanisms in the regulation of bleb retraction
Although defects in bleb retraction were observed in ezrin-KO or MYOGEF-KO cells, those retarded blebs still eventually retract to the cell body, suggesting that multiple and/or redundant mechanisms are likely to be involved in the regulation of bleb retraction. For instance, RhoGEF KIAA0861/ARHGEF22 is localized to the bleb membrane, where the RhoGEF may contribute to RhoA activation during bleb retraction (Charras et al., 2006). Many other RhoGEFs, such as Net1 (Carr et al., 2013), LARG (Martz et al., 2013), GEF-H1 (Heasman et al., 2010), PDZ-RhoGEF (Struckhoff et al., 2013), and ECT2 (Yuce et al., 2005), are also capable of activating RhoA-ROCK signaling. However, it is still unclear whether such RhoGEFs are localized to the bleb membrane and contribute to RhoA activation at the bleb membrane. Nonetheless, our findings in this study provide a mechanism suggesting that localization of MYOGEF to the bleb membrane plays a role in controlling bleb retraction.
Activation of RhoA-ROCK signaling and subsequent increase in MLC phosphorylation are believed to be a key regulatory mechanism that promotes myosin contractility in numerous biological processes, such as mitosis, cytokinesis, cell migration, and tissue morphogenesis (Matsumura et al., 2001; Vicente-Manzanares et al., 2009). It is believed that the reassembly of a contractile actomyosin cortex at the bleb membrane during bleb retraction also involves MLC phosphorylation mediated by RhoA-ROCK signaling (Fackler and Grosse, 2008). However, other kinases, such as MLC kinase (MLCK), can also phosphorylate MLC and increase MLC phosphorylation (Totsukawa et al., 2004). Therefore, it is possible that MLC phosphorylation at the bleb membrane by MLCK may also contribute to bleb retraction. Indeed, it has been shown that MLCK and myosin II are colocalized to hypotonic stress-induced blebs, where MLCK phosphorylates MLC to promote bleb retraction (Barfod et al., 2011). However, it remains to be determined whether MLCK-mediated phosphorylation of MLC plays a role in retracting the blebs induced by cortical contraction.
Membrane blebbing is a dynamic process, with the expansion phase lasting ∼30 s and the retraction phase ∼2 min. In addition, multiple blebs in either expansion or retraction phases are likely to be present in a blebbing cell. It is conceivable that initiation and expansion of a new bleb would relieve the intracellular pressure, thus facilitating the retraction of the older blebs. Notably, findings from a recent study suggest that the contraction of the actomyosin cortex alone is not sufficient to drive bleb retraction. Instead, other factors such as bleb membrane shrinking are also likely to be involved in bleb retraction (Woolley et al., 2015a). Therefore, molecular and physical mechanisms are likely to cooperate to promote bleb retraction.
Regulation of membrane blebbing and cell migration by RhoGEFs
There are more than 70 GEFs in humans, and a subset of GEFs termed RhoGEFs are capable of increasing actomyosin contractility by activating RhoA-ROCK-myosin II signaling (Rossman et al., 2005; Zuo et al., 2014). An increase in cortical tension resulting from actomyosin contraction contributes not only to bleb formation but also the amoeboid mode of cell migration (Sahai and Marshall, 2003; Lammermann and Sixt, 2009; Tozluoglu et al., 2013; Ruprecht et al., 2015). Accordingly, RhoGEF-RhoA-ROCK signaling has been implicated in promoting membrane blebbing and/or amoeboid cell migration (Chan et al., 1996; Kitzing et al., 2007; Eitaki et al., 2012). For instance, a decrease in cortical tension resulting from disruption of a positive feedback loop between Dia1 and LARG inhibits both bleb formation and cell migration (Kitzing et al., 2007). GEF-H1, a microtubule-associated RhoGEF, becomes activated to promote actomyosin contractility when it is released from microtubules (Ren et al., 1998; Krendel et al., 2002; Meiri et al., 2012). Therefore, it is plausible that, in control A7 melanoma cells treated with the microtubule-depolymerizing agent nocodazole, GEF-H1 is released from microtubules and becomes activated to increase cortical tension through activating RhoA-ROCK-myosin II signaling, thus promoting bleb formation and expansion. In agreement with this notion, knockdown of GEF-H1 not only suppresses vincristine-induced membrane blebbing, but also inhibits cell migration (Eitaki et al., 2012). However, in MYOGEF-KO A7 melanoma cells treated with nocodazole, the absence of MYOGEF would likely compromise bleb retraction. Our results show that treatment of MYOGEF-KO A7 melanoma cells with nocodazole increases membrane blebbing (see Figure 5). Therefore, it is conceivable that RhoGEFs may contribute to bleb regulation through two different mechanisms. First, RhoGEFs such as GEF-H1 can activate RhoA-ROCK-myosin II signaling and increase the overall contractility of the cortex, therefore increasing intracellular pressure and promoting bleb formation and expansion. Second, RhoGEFs, such as MYOGEF, are localized to the bleb membrane, where they activate RhoA-ROCK-myosin II signaling and promote the reassembly of the actomyosin cortex, thus contributing to bleb retraction.
In the amoeboid mode of cell migration, blebs are formed to fill the pores of the extracellular matrix. In turn, contraction of the actomyosin cortex in the cell body leads the cell to move in the direction of the cell side with blebs that have filled the pores of the extracellular matrix (Charras and Paluch, 2008; Lammermann and Sixt, 2009; Te Boekhorst et al., 2016). Therefore, it is apparent that the regulatory machinery for cortical actomyosin contractility plays a central role in controlling the amoeboid mode of cell migration. Although it is still unclear how the reassembly of the new actomyosin cortex underneath the bleb membrane is implicated in the amoeboid mode of cell migration, our previous studies have demonstrated that MYOGEF can promote the invasion activity of MDA-MB-231 breast cancer cells (Wu et al., 2009, 2010). Investigation of whether and how MYOGEF is implicated in regulating the amoeboid mode of cell migration is currently under way.
MATERIALS AND METHODS
Plasmids
GFP-C1-PLC-delta-PH was a gift from Tobias Meyer (Stanford University Medical Center; Addgene plasmid #21179; Stauffer et al., 1998). GFP-N1-ezrin (pHJ421) was a gift from Stephen Shaw (National Cancer Institute; Addgene plasmid #20680; Hao et al., 2009). pBS/U6 ezrin siRNA was a gift from Philip Hinds (Tufts University; Addgene plasmid #8945; Yang and Hinds, 2003). pACT2.2gtwy was a gift from Guy Caldwell (University of Alabama; Addgene plasmid #11346). pGBKT7-GW was a gift from Yuhai Cui (Agriculture and Agri-Food Canada; Addgene plasmid #61703; Lu et al., 2010). pEGFP-RhoA Biosensor was a gift from Michael Glotzer (University of Chicago; Addgene plasmid #68026; Piekny and Glotzer, 2008). pDEST/LIfeAct-mCherry-N1 was a gift from Robin Shaw (Cedars-Sinai Heart Institute; Addgene plasmid #40908; Smyth et al., 2012). pCS EGFP DEST and pCS 3MT DEST were gifts from Nathan Lawson (University of Massachusetts Medical School; Addgene plasmids #13071 and #13070; Villefranc et al., 2007).
Plasmids used for in vitro translation, yeast two-hybrid assays, and bacterial/mammalian expression were generated using the pCR 8/GW/TOPO TA cloning kit and the gateway cloning system (Thermo Fisher Scientific). Detailed cloning strategies are available upon request. GFP-MYOGEF and GFP-NMHC-IIA were generated as described previously (Wei and Adelstein, 2000; Wu et al., 2006, 2014b). GFP-N1-ezrin was used as a PCR template to amplify the N-terminal region of ezrin (residues 1–320), which, in turn, was cloned into pDEST17 (Thermo Fisher Scientific) to generate His-ezrin-1–320.
Antibodies and reagents
The primary antibodies used were as follows: rabbit anti-β-tubulin (immunoblot 1:1500; Santa Cruz); mouse anti-ezrin (immunoblot 1:1000, IF 1:300; Santa Cruz); rabbit anti-ezrin (IF 1:300; Cell Signaling); rabbit anti-GFP (immunoblot 1:1000; Santa Cruz); rabbit anti-His-probe (immunoblot 1:1000; Santa Cruz); mouse anti-Myc (immunoblot 1:1500; Santa Cruz); rabbit anti-MYOGEF (immunoblot 1:100); mouse anti-MYOGEF (IF 1:100); mouse anti phosphomyosin light chain 2 (IF 1:500; Cell Signaling). Rabbit and mouse anti-MYOGEF antibodies have been described previously (Wu et al., 2006, 2014a).
Secondary antibodies include Alexa Fluor 594 Goat anti-mouse IgG (IF 1:500; Thermo Fisher Scientific), Alexa Fluor 488 Goat anti-mouse IgG (IF 1:500; Thermo Fisher Scientific), Alexa Fluor 488 Goat anti-rabbit IgG (IF 1:500; Thermo Fisher Scientific), europium-labeled anti-mouse (immunoblot 1:5000; Molecular Devices), and europium-labeled anti-rabbit (immunoblot 1:5000; Molecular Devices).
Protein A-Sepharose beads and anti-c-Myc agarose were purchased from Santa Cruz. Nocodazole and puromycin were purchased from Sigma. The phospholipase C activator m-3M3FBS was purchased from EMD Millipore.
Cell culture and transfection
MDA-MB-231 breast cancer cells (American Type Culture Collection [ATCC]) were maintained in Leibovitz’ L-15 medium with 10% fetal bovine serum (FBS) and penicillin/streptomycin at 37°C without CO2. M2 melanoma cells (a gift from Thomas Peter Stossel, Harvard Medical School) were maintained in MEM with 5% FBS and penicillin/streptomycin at 37°C, 5% CO2. A7 melanoma cells (ATCC) were maintained in MEM supplied with 5% FBS at 37°C, 5% CO2. MYOGEF-KO and ezrin-KO M2 or A7 melanoma cells were generated according to the protocol described previously (Ran et al., 2013). The resulting CRISPR cells were maintained in MEM supplied with 5% FBS and 1 μg/ml puromycin at 37°C with 5% CO2.
Plasmids were transfected into cells using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. After transfection, cells were further incubated for 18–24 h and then split onto glass coverslips for immunofluorescence analyses or onto chambered coverglasses (Thermo Fisher Scientific) for time-lapse microscopy.
Cells transiently transfected with control or ezrin-specific shRNA plasmids were incubated for 5 d prior to analyses. To induce membrane blebbing, M2 or A7 melanoma cells were treated with DMSO or 0.3 μg/ml nocodazole for 40 min. To activate phospholipase C, transfected M2 melanoma cells were treated with DMSO or 100 μM m-3M3FBS for 30 min.
Protein expression and purification
A plasmid encoding His-ezrin-1–320 was transfected into BL21 bacterial cells. Isopropyl-beta-d-1-thiogalactoside (IPTG; Teknova) was used to induce protein expression. Cell pellets were resuspended in 50 mM sodium phosphate (pH 8.0) and 300 mM NaCl and homogenized by sonication. His-ezrin-1–320 polypeptides were then purified using a nickel–nitrilotriacetic acid (Ni-NTA) agarose (QIAGEN) column, followed by elution with 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, and 250 mM imidazole. His-ezrin-1–320 polypeptides were dialyzed against 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl. GFP-MYOGEF-1–640, Myc-MYOGEF-FL, Myc-MYOGEF-501–790, Myc-MYOGEF-501–752, and Myc-MYOGEF-501–640 were produced using the TNT Quick Coupled in vitro transcription/translation system (Promega) according to the manufacturer’s instructions.
Immunoprecipitation assays
Purified His-ezrin-1–320 polypeptides were mixed with in vitro translated Myc-MYOGEF-501–790, Myc-MYOGEF-501–752, or Myc-MYOGEF-501–640 in BC100 buffer. Cell lysates were also collected from M2 melanoma cells exogenously expressing Myc-MYOGEF-1–640. Protein mixtures or cell lysates were then incubated with anti-c-Myc agarose beads for 12 h at 4°C. Agarose beads were precipitated using Pierce microcentrifuge spin columns (Thermo Fisher Scientific) and washed three times in BC100 buffer. Proteins bound to agarose beads were eluted with 2X SDS sample buffer, heated at 95°C for 5 min, and subjected to SDS–PAGE, followed by immunoblot analyses.
Immunoblot
To examine the levels of protein expression, cells were lysed with BC100 buffer supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitor mix, followed by 10-min incubation on ice. After centrifugation, the supernatant of lysates was mixed with the SDS gel loading buffer and incubated at 95°C for 5 min. Samples from cell lysates or coimmunoprecipitation assays were separated on 8%, 10%, or 12% SDS–PAGE gels. Proteins on the gel were transferred to an Immobilon-P transfer membrane (EMD Millipore) using a Trans-Blot semidry electrophoretic transfer cell (Bio-Rad). The membrane was then blocked in ScanLater 1X blocking buffer (Molecular Devices) for 1 h at 22.5°C and incubated with primary antibodies for 12 h at 4°C. After being washed three times in 1X TBS supplemented with 0.05% Tween 20 (1X TBST), the blot was incubated with europium-labeled anti-rabbit or anti-mouse ScanLater secondary antibodies (Molecular Devices), followed by washing in 1X TBST. SpectraMax i3 with a ScanLater Western blot cartridge was used to scan the immunoblot according to the manufacturer’s instructions (Molecular Devices).
Yeast two-hybrid assays
Ezrin-1–320 was inserted into the prey vector pACT2.2-gtwy to generate pACT2.2-gtwy-ezrin-1–320, in which ezrin-1–320 was fused in frame with GAL4 activation domain (AD). MYOGEF fragments were subcloned into the bait vector pGBKT7-GW to generate pGBKT7-GW-MYOGEF fragments, in which MYOGEF fragments were fused in frame with GAL4 DNA-binding domain (DBD). AH109 yeast cells transformed with prey or bait plasmids alone were plated on SD selective agar plates lacking leucine or tryptophan, respectively. AH109 yeast cells cotransformed with prey and bait plasmids were plated on SD selective agar plates lacking adenine, histidine, leucine, and tryptophan (SD/-Ade-His-Leu-Trp). X-gal filter assays were conducted according to the instructions of the manufacturer (Clontech).
Fluorescence microscopy
Transfected cells were fixed in 4% paraformaldehyde for 15 min at 22.5°C. To analyze endogenous MYOGEF, cells were fixed in 10% ice-cold trichloroacetic acid (TCA) for 30 min. For immunofluorescence (IF) staining, fixed cells were blocked in 1% bovine serum albumin (BSA) for 1 h at 22.5°C, followed by incubation with primary antibodies for 12 h at 4°C. Cells were washed three times in 1X PBS and incubated with secondary antibodies for 1 h at 22.5°C. Coverslips were washed three times in 1X PBS, dried, and mounted using a Prolong Antifade kit (Thermo Fisher Scientific). Images were taken using the Nikon TiE Perfect Focus digital fluorescence imaging system (Morrell Instrument Company) with an Andor Zyla sCMOS 2560 × 2160 camera. Raw images were processed by deconvolution.
Time-lapse microscopy
Cells were plated on chambered coverglasses (Thermo Fisher Scientific). MDA-MB-231 cells were incubated for 5 h at 37°C without CO2 to allow cells to attach to the surface before analyses. Control and ezrin-KO M2 melanoma cells were incubated for 3 h at 37°C, 5% CO2, prior to time-lapse microscopy. Control and MYOGEF-KO A7 melanoma cells were incubated on chambered coverglasses for 16 h at 37°C, 5% CO2, prior to drug treatment. For single-cell time-lapse experiments, cells were maintained in L-15 medium supplied with 5% FBS in an environmental control unit (In Vivo Scientific) at 37°C without CO2. Images were acquired using the Nikon TiE Perfect Focus digital fluorescence imaging system (Morrell Instrument Company) with the 100X objective. Differential interference contrast (DIC) images were taken at a 500-ms interval for kymographs. Fluorescence images were taken for four min at a 1-s interval for multichannel imaging. Kymographs were generated using the ND processing measurement tool from the Nikon TiE microscope system (Morrell Instrument Company).
Genomic editing by CRISPR
MYOGEF or ezrin knockout cell lines were generated using the CRISPR/Cas9 system. Single guide RNAs (sgRNAs) for MYOGEF/PLEKHG6 and ezrin have been designed and validated previously (Sanjana et al., 2014). The sgRNA sequences are as follows: MYOGEF-sgRNA (5′-CTCCCTCCAGGACTTCTCGA-3′, complementary strand) and ezrin-sgRNA (5′-CAATGTCCGAGTTACCACCA-3′). sgRNA sequences were inserted into the lentiviral vector pRSGC1-U6-sg-CMV-Cas9-2A-Puro (Cellecta) using a PCR/ligation strategy. Briefly, sgRNA sequences were inserted into the lentiviral vector by PCR. The PCR products were then treated with the restriction enzyme Dpn1 to remove the PCR template. Amplified DNAs (i.e., the lentiviral vectors containing the respective sgRNA sequences) were phosphorylated with T4 polynucleotide kinase and circularized with T4 DNA ligase. Lentiviruses containing the sequences of interest were produced in HEK293T cells by cotransfecting lentiviral CRISPR plasmids into cells with psPAX2 and pMD2.G, followed by incubation for 72 h at 37°C, 5% CO2. M2 and A7 melanoma cells were infected with lentivirus for 96–120 h prior to puromycin selection.
Stable cells carrying Cas9 and sgRNAs were collected 2–3 wk after antibiotic selection (1 μg/ml puromycin). The protein levels of MYOGEF and ezrin were analyzed by immunoblot. To validate the genomic deletion or insertion in the gene of interest, genomic DNA was extracted from cells using QIAamp DNA mini and blood mini kits (QIAGEN). The region flanking the Cas9 cutting site was amplified by PCR. Amplified DNA fragments were subcloned into T-vectors for sequencing. After the presence of InDels was confirmed, infected cells were diluted to isolate single cell clones. DNA sequences were aligned using A-plasmid-editor (ApE) software. Protein alignments were performed using pairwise sequence alignment (www.ebi.ac.uk/Tools/psa/).
Statistical analysis
To quantify the percentage of cells with corresponding proteins at the bleb membrane in control, ezrin-KO, and MYOGEF-KO cells, transfected blebbing cells were fixed with 4% paraformaldehyde and observed using a 100X objective. Antibodies specific for MYOGEF and p-MLC were used to detect the localization of endogenous MYOGEF or p-MLC under the microscope.
To quantify the percentage of cells that produce extended blebs (area > 40 μm2), M2 melanoma cells expressing GFP-MYOGEF-FL, GFP-MYOGEF-1–640, or GFP-MYOGEF-1–752 as well as in ezrin-KO M2 cells expressing GFP-MYOGEF-FL were fixed and analyzed under the microscope. Statistical significance was determined using one-way analysis of variance (ANOVA) and Tukey’s post hoc test.
To quantify the bleb number in a cell, all blebs produced in a cell within a 2-min period were counted. The bleb number was then normalized to the cell area (μm2) and to time (s). Thirty cells were examined for each treatment per cell type. Bleb number was compared using one-way ANOVA and Tukey’s post hoc test.
To assess the distribution of bleb size in a cell, blebs were categorized as small blebs (area < 10 μm2), normal blebs (area = 10–40 μm2), and large blebs (area > 40 μm2). All blebs in a cell during a 4-min period were measured at the end of bleb expansion. Thirty cells were quantified for each group of cells. Distributions of bleb size were compared using a chi-squared test.
To assess the efficiency of a bleb in completing a bleb cycle and bleb retraction, the time required for the cycle and retraction was quantified for all blebs in a cell during a 4-min period. Thirty cells were analyzed in an individual experiment. Differences in bleb cycle or bleb retraction were compared using one-way ANOVA and Tukey’s post hoc test.
To quantify blebbing cells, fixed cells with or without blebs were counted under the microscope using the 20X objective. The percentages of blebbing cells in control or MYOGEF-KO A7 melanoma cells with or without drug treatment were compared using two-way ANOVA and Tukey’s post hoc test.
The fluorescence intensity of GFP-ezrin and DsRed-MYOGEF at the bleb membrane was quantified using the ADAPT tool and normalized to the respective cortical intensity before bleb formation (Barry et al., 2015). Images were taken for 4 min at a 1-s interval. Five blebs were quantified in each cell, and five cells were analyzed in each experiment. The starting points for GFP-ezrin or DsRed-MYOGEF to be recruited to the bleb membrane were determined by detecting the lowest normalized fluorescence intensity followed by subsequent increasing signals of the respective proteins to the bleb membrane.
Each experiment was repeated three times. Values of p < 0.05 were deemed statistically significant.
Supplementary Material
Acknowledgments
We thank Thomas P. Stossel (Harvard Medical School) for providing the M2 melanoma cell line and Robert S. Adelstein (National Heart, Lung, and Blood Institute) and Mary Anne Conti (National Heart, Lung, and Blood Institute) for critical reading and comments on the manuscript. This work was supported by National Institutes of Health Grant R15GM097702 to Q.W.
Abbreviations used:
- AHD
anillin homology domain
- FLNA
filamin A
- MYOGEF
myosin II–interacting guanine nucleotide exchange factor
- NMHC IIA
nonmuscle myosin II-A heavy chain
- PLC-delta-PH
the PH domain of phospholipase C delta
- PLEKHG6
pleckstrin homology and RhoGEF domain containing G6
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0579) on January 10, 2018.
REFERENCES
- Adelstein RS, Conti MA. (1975). Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature , 597-598. [DOI] [PubMed] [Google Scholar]
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. (1996). Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J Biol Chem , 20246-20249. [DOI] [PubMed] [Google Scholar]
- Aoki K, Maeda F, Nagasako T, Mochizuki Y, Uchida S, Ikenouchi J. (2016). A RhoA and Rnd3 cycle regulates actin reassembly during membrane blebbing. Proc Natl Acad Sci USA , E1863-E1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfod ET, Moore AL, Van de Graaf BG, Lidofsky SD. (2011). Myosin light chain kinase and Src control membrane dynamics in volume recovery from cell swelling. Mol Biol Cell , 634-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret C, Roy C, Montcourrier P, Mangeat P, Niggli V. (2000). Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP(2)) binding site in the NH(2)-terminal domain of ezrin correlates with its altered cellular distribution. J Cell Biol , 1067-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry DJ, Durkin CH, Abella JV, Way M. (2015). Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J Cell Biol , 163-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, Kawakami K, Solnica-Krezel L, Heisenberg CP, Raz E. (2006). Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell , 613-627. [DOI] [PubMed] [Google Scholar]
- Carr HS, Zuo Y, Oh W, Frost JA. (2013). Regulation of focal adhesion kinase activation, breast cancer cell motility, and amoeboid invasion by the RhoA guanine nucleotide exchange factor Net1. Mol Cell Biol , 2773-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. (1996). The Dbl family of oncogenes. Curr Opin Cell Biol , 216-222. [DOI] [PubMed] [Google Scholar]
- Chan AM, Takai S, Yamada K, Miki T. (1996). Isolation of a novel oncogene, NET1, from neuroepithelioma cells by expression cDNA cloning. Oncogene , 1259-1266. [PubMed] [Google Scholar]
- Charras G, Paluch E. (2008). Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol , 730-736. [DOI] [PubMed] [Google Scholar]
- Charras GT. (2008). A short history of blebbing. J Microsc , 466-478. [DOI] [PubMed] [Google Scholar]
- Charras GT, Coughlin M, Mitchison TJ, Mahadevan L. (2008). Life and times of a cellular bleb. Biophys J , 1836-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras GT, Hu CK, Coughlin M, Mitchison TJ. (2006). Reassembly of contractile actin cortex in cell blebs. J Cell Biol , 477-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. (2005). Non-equilibration of hydrostatic pressure in blebbing cells. Nature , 365-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CC. (1995). Actin polymerization and intracellular solvent flow in cell surface blebbing. J Cell Biol , 1589-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. (1992). Actin-binding protein requirement for cortical stability and efficient locomotion. Science , 325-327. [DOI] [PubMed] [Google Scholar]
- D’Angelo R, Aresta S, Blangy A, Del Maestro L, Louvard D, Arpin M. (2007). Interaction of ezrin with the novel guanine nucleotide exchange factor PLEKHG6 promotes RhoG-dependent apical cytoskeleton rearrangements in epithelial cells. Mol Biol Cell , 4780-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzementsei A, Schneider D, Janshoff A, Pieler T. (2013). Migratory and adhesive properties of Xenopus laevis primordial germ cells in vitro. Biol Open , 1279-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitaki M, Yamamori T, Meike S, Yasui H, Inanami O. (2012). Vincristine enhances amoeboid-like motility via GEF-H1/RhoA/ROCK/Myosin light chain signaling in MKN45 cells. BMC Cancer , 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler OT, Grosse R. (2008). Cell motility through plasma membrane blebbing. J Cell Biol , 879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. (2010). Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol , 276-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K. (2003). Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer , 362-374. [DOI] [PubMed] [Google Scholar]
- Hagmann J, Burger MM, Dagan D. (1999). Regulation of plasma membrane blebbing by the cytoskeleton. J Cell Biochem , 488-499. [PubMed] [Google Scholar]
- Hall A. (1998). Rho GTPases and the actin cytoskeleton. Science , 509-514. [DOI] [PubMed] [Google Scholar]
- Hao JJ, Liu Y, Kruhlak M, Debell KE, Rellahan BL, Shaw S. (2009). Phospholipase C-mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J Cell Biol , 451-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman SJ, Carlin LM, Cox S, Ng T, Ridley AJ. (2010). Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J Cell Biol , 553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. (2008). Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol , 690-701. [DOI] [PubMed] [Google Scholar]
- Jia Z, Vadnais J, Lu ML, Noel J, Nabi IR. (2006). Rho/ROCK-dependent pseudopodial protrusion and cellular blebbing are regulated by p38 MAPK in tumour cells exhibiting autocrine c-Met activation. Biol Cell , 337-351. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Fukata Y, Oshiro N, Amano M, Nakamura T, Ito M, Matsumura F, Inagaki M, Kaibuchi K. (1999). Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J Cell Biol , 1023-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. (1996). Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science , 245-248. [DOI] [PubMed] [Google Scholar]
- Kitzing TM, Sahadevan AS, Brandt DT, Knieling H, Hannemann S, Fackler OT, Grosshans J, Grosse R. (2007). Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev , 1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosako H, Yoshida T, Matsumura F, Ishizaki T, Narumiya S, Inagaki M. (2000). Rho-kinase/ROCK is involved in cytokinesis through the phosphorylation of myosin light chain and not ezrin/radixin/moesin proteins at the cleavage furrow. Oncogene , 6059-6064. [DOI] [PubMed] [Google Scholar]
- Krendel M, Zenke FT, Bokoch GM. (2002). Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat Cell Biol , 294-301. [DOI] [PubMed] [Google Scholar]
- Lammermann T, Sixt M. (2009). Mechanical modes of “amoeboid” cell migration. Curr Opin Cell Biol , 636-644. [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J , 259-270. [DOI] [PubMed] [Google Scholar]
- Martz MK, Grabocka E, Beeharry N, Yen TJ, Wedegaertner PB. (2013). Leukemia-associated RhoGEF (LARG) is a novel RhoGEF in cytokinesis and required for the proper completion of abscission. Mol Biol Cell , 2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. (1998). Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol , 647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F, Totsukawa G, Yamakita Y, Yamashiro S. (2001). Role of myosin light chain phosphorylation in the regulation of cytokinesis. Cell Struct Funct , 639-644. [DOI] [PubMed] [Google Scholar]
- Meiri D, Marshall CB, Greeve MA, Kim B, Balan M, Suarez F, Bakal C, Wu C, Larose J, Fine N, et al. (2012). Mechanistic insight into the microtubule and actin cytoskeleton coupling through dynein-dependent RhoGEF inhibition. Mol Cell , 642-655. [DOI] [PubMed] [Google Scholar]
- Mills JC, Stone NL, Erhardt J, Pittman RN. (1998). Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J Cell Biol , 627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. (2011). The filamins: organizers of cell structure and function. Cell Adh Migr , 160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny AJ, Glotzer M. (2008). Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol , 30-36. [DOI] [PubMed] [Google Scholar]
- Pletjushkina OJ, Rajfur Z, Pomorski P, Oliver TN, Vasiliev JM, Jacobson KA. (2001). Induction of cortical oscillations in spreading cells by depolymerization of microtubules. Cell Motil Cytoskeleton , 235-244. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc , 2281-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Li R, Zheng Y, Busch H. (1998). Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem , 34954-34960. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. (2005). GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol , 167-180. [DOI] [PubMed] [Google Scholar]
- Ruprecht V, Wieser S, Callan-Jones A, Smutny M, Morita H, Sako K, Barone V, Ritsch-Marte M, Sixt M, Voituriez R, Heisenberg CP. (2015). Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell , 673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ. (2003). Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol , 711-719. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods , 783-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Biro M, Oswald A, Tinevez JY, Salbreux G, Paluch E. (2011). Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature , 462-466. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Sable JE, Dobereiner HG. (2006). Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu Rev Biophys Biomol Struct , 417-434. [DOI] [PubMed] [Google Scholar]
- Smyth JW, Vogan JM, Buch PJ, Zhang SS, Fong TS, Hong TT, Shaw RM. (2012). Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma membrane. Circ Res , 978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. (1998). Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol , 343-346. [DOI] [PubMed] [Google Scholar]
- Struckhoff AP, Rana MK, Kher SS, Burow ME, Hagan JL, Del Valle L, Worthylake RA. (2013). PDZ-RhoGEF is essential for CXCR4-driven breast tumor cell motility through spatial regulation of RhoA. J Cell Sci , 4514-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takesono A, Heasman SJ, Wojciak-Stothard B, Garg R, Ridley AJ. (2010). Microtubules regulate migratory polarity through Rho/ROCK signaling in T cells. PLoS One , e8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Boekhorst V, Preziosi L, Friedl P. (2016). Plasticity of cell migration in vivo and in silico. Annu Rev Cell Dev Biol , 491-526. [DOI] [PubMed] [Google Scholar]
- Tinevez JY, Schulze U, Salbreux G, Roensch J, Joanny JF, Paluch E. (2009). Role of cortical tension in bleb growth. Proc Natl Acad Sci USA , 18581-18586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Wu Y, Sasaki Y, Hartshorne DJ, Yamakita Y, Yamashiro S, Matsumura F. (2004). Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J Cell Biol , 427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. (2000). Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol , 797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoglu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. (2013). Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat Cell Biol , 751-762. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. (1997). Rho GTPases and signaling networks. Genes Dev , 2295-2322. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol , 778-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn , 3077-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon E. (1999). Positive feedback interactions between microtubule and actin dynamics during cell motility. Curr Opin Cell Biol , 61-67. [DOI] [PubMed] [Google Scholar]
- Wei Q, Adelstein RS. (2000). Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol Biol Cell , 3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. (2003). Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol , 267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley TE, Gaffney EA, Goriely A. (2015a). Membrane shrinkage and cortex remodelling are predicted to work in harmony to retract blebs. R Soc Open Sci , 150184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley TE, Gaffney EA, Oliver JM, Waters SL, Baker RE, Goriely A. (2015b). Global contraction or local growth, bleb shape depends on more than just cell structure. J Theor Biol , 83-97. [DOI] [PubMed] [Google Scholar]
- Wu D, Asiedu M, Adelstein RS, Wei Q. (2006). A novel guanine nucleotide exchange factor MyoGEF is required for cytokinesis. Cell Cycle , 1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Asiedu M, Matsumura F, Wei Q. (2014a). Phosphorylation of myosin II-interacting guanine nucleotide exchange factor (MyoGEF) at threonine 544 by aurora B kinase promotes the binding of polo-like kinase 1 to MyoGEF. J Biol Chem , 7142-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Asiedu M, Wei Q. (2009). Myosin-interacting guanine exchange factor (MyoGEF) regulates the invasion activity of MDA-MB-231 breast cancer cells through activation of RhoA and RhoC. Oncogene , 2219-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Haruta A, Wei Q. (2010). GIPC1 interacts with MyoGEF and promotes MDA-MB-231 breast cancer cell invasion. J Biol Chem , 28643-28650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Jiao M, Zu S, Sollecito CC, Jimenez-Cowell K, Mold AJ, Kennedy RM, Wei Q. (2014b). Intramolecular interactions between the Dbl homology (DH) domain and the carboxyl-terminal region of myosin II-interacting guanine nucleotide exchange factor (MyoGEF) act as an autoinhibitory mechanism for the regulation of MyoGEF functions. J Biol Chem , 34033-34048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Hinds PW. (2003). Increased ezrin expression and activation by CDK5 coincident with acquisition of the senescent phenotype. Mol Cell , 1163-1176. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Matsui T, Tsukita S, Tsukita S. (2002). Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J Cell Sci , 2569-2580. [DOI] [PubMed] [Google Scholar]
- Yuce O, Piekny A, Glotzer M. (2005). An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol , 571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Oh W, Frost JA. (2014). Controlling the switches: Rho GTPase regulation during animal cell mitosis. Cell Signal , 2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.