Abstract
Cancer cells have altered metabolism and, in some cases, an increased demand for cholesterol. It is important to identify novel, rational treatments based on biology, and cellular cholesterol metabolism as a potential target for cancer is an innovative approach. Toward this end, we focused on diffuse large B-cell lymphoma (DLBCL) as a model because there is differential cholesterol biosynthesis driven by B-cell receptor (BCR) signaling in germinal center (GC) versus activated B-cell (ABC) DLBCL. To specifically target cellular cholesterol homeostasis, we employed high-density lipoprotein-like nanoparticles (HDL NP) that can generally reduce cellular cholesterol by targeting and blocking cholesterol uptake through the high-affinity HDL receptor, scavenger receptor type B-1 (SCARB1). As we previously reported, GC DLBCL are exquisitely sensitive to HDL NP as monotherapy while ABC DLBCL are less sensitive. Herein, we report that enhanced BCR signaling and resultant de novo cholesterol synthesis in ABC DLBCL drastically reduces the ability of HDL NPs to reduce cellular cholesterol and induce cell death. Therefore, we combined HDL NP with the BCR signaling inhibitor ibrutinib and the SYK inhibitor R406. By targeting both cellular cholesterol uptake and BCR-associated de novo cholesterol synthesis, we achieved cellular cholesterol reduction and induced apoptosis in otherwise resistant ABC DLBCL cell lines. These results in lymphoma demonstrate that reduction of cellular cholesterol is a powerful mechanism to induce apoptosis. Cells rich in cholesterol require HDL NP therapy to reduce uptake and molecularly targeted agents that inhibit upstream pathways that stimulate de novo cholesterol synthesis, thus, providing a new paradigm for rationally targeting cholesterol metabolism as therapy for cancer.
Keywords: Lymphoma, Cholesterol, High-density lipoproteins, Nanoparticles, B-cell receptor signaling
Introduction
Normal, healthy human cells tightly regulate cellular cholesterol in order to maintain cell membrane integrity and to stabilize membrane-anchored signaling pathways.1, 2 Evidence suggests a major role for lipids in the progression of hematological3–5 and other malignancies6–10 with specific involvement of cholesterol and cholesteryl esters, along with the high-density lipoproteins (HDL) that carry them.11–14 In the case of B-cell lymphoma, B-cell receptor (BCR) signaling, including low level, tonic signaling observed in germinal center (GC)-derived diffuse large B cell lymphoma (DLBCL) and chronic active signaling in activated B-cell (ABC) DLBCL, has been shown to directly promote cholesterol biosynthesis through intermediate kinases downstream of the BCR to maintain cell membrane integrity and BCR signaling.15 Perturbation of BCR-induced de novo cholesterol biosynthesis in malignant B cells, for example by inhibiting or depleting SYK (spleen associated tyrosine kinase), disrupts the cell membrane and inhibits BCR signaling, leading to apoptosis.15 Further delineation of this mechanism, and identification of targeted therapy that enables specific disruption of cholesterol metabolism in malignant B cells and other cholesterol-dependent cancer cells, may provide important new insights and more rational cancer treatment.16–19
The high-affinity HDL receptor, scavenger receptor type B1 (SCARB1)20 is a potential target for perturbing cellular cholesterol homeostasis. SCARB1 mediates bidirectional cholesterol flux between HDL particles and the cell membrane and provides a conduit for delivery of cholesteryl ester from the core of natural HDLs to target cells. In addition to mediating cholesterol flux, binding of natural HDL to SCARB1 activates second messenger signaling pathways, including the PI3K-AKT pro-survival pathway in multiple cell types, including breast cancer and endothelial cells.21–23 Our group previously reported that a large number of cancer cell lines, including B cell lymphoma, express SCARB1, while peripheral naïve and memory B-cells do not.19 Accordingly, SCARB1 expression in malignant cells may provide a critical opportunity to specifically and therapeutically reduce cellular cholesterol.
Synthetic, bio-inspired HDL-like nanoparticles (HDL NPs) represent a first-in-class therapy that specifically bind SCARB1 and can perturb cellular cholesterol and cholesteryl ester homeostasis, ultimately depleting cellular cholesterol and cholesteryl esters when compared with natural, cholesterol-rich HDLs.16, 18, 24, 25 HDL NPs consist of a 5nm diameter gold nanoparticle core surface-functionalized with the HDL-defining apolipoprotein A1 (apoA-I) and a phospholipid bilayer.26, 27 HDL NPs are similar to natural, mature, spherical HDL with regard to surface composition, negative surface charge, size, and shape.16, 24, 25 Functionally, HDL NPs mediate the efflux of free cholesterol from cells through SCARB1.16, 19 and, owing to the core gold nanoparticle, are not an appreciable source of cholesteryl ester.18, 19 These unique features enable HDL NPs to target SCARB1, but result in cellular cholesterol reduction when compared with natural HDLs.19 Importantly, HDL NPs induce profound cell death in GC DLBCL cell lines but are relatively ineffective against ABC DLBCL cells.19 However, a better understanding of the mechanism of action of HDL NPs and how this therapy can be rationally combined with other agents remains unexplored.1, 22, 28–30
Given the differences in BCR signaling levels and sensitivity to HDL NP, we hypothesized that ABC DLBCL cells are resistant to HDL NP-induced cholesterol depletion due to enhanced BCR-associated de novo cholesterol synthesis. Effective therapy may be realized by combining HDL NPs with agents that target BCR signaling. Our data confirm that targeted cellular cholesterol depletion is necessary to induce apoptosis in lymphoma cells, which provides a rational approach to target cholesterol metabolism in other cancer types that are cholesterol dependent.
Experimental Methods
Cell lines and authentication
The human cell lines Jurkat and SUDHL4 were obtained from ATCC and were used within 3 months of receipt and/or resuscitation. ATCC uses the Short Tandem Repeat (STR) profiling method to authenticate cell lines. The human cell lines TMD8 and HBL-1 were generously provided by Dr. Louis Staudt, whose laboratory uses the novel DNA copy number variance fingerprinting technique to verify authenticity.31 Additionally, the STR profiles for TMD8 and HBL-1 cells were analyzed by ATCC and did not match any profile in their database.
Cell culture
SUDHL4 and Jurkat cells were cultured in RPMI 1640 with L-glutamine and 25mM HEPES (Mediatech) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals), and 1% PenStrep, at 37 °C with 5% CO2 in a humidified incubator, except TMD8 and HBL-1, which were cultured with 15% FBS. The SCARB1 blocking and rabbit IgG isotype control antibodies were obtained from Novus Biologicals. The PI3K inhibitor Pilaralisib (XL-147; Selleck Chem), the AKT inhibitor GDC-0068 (Selleck Chem), the SYK inhibitor R406 (Selleck Chem) and the BTK inhibitor ibrutinib (PCI-32756; Selleck Chem) were added to cells at the same time as HDL NP treatment. The cholesterol sequesterant, methyl-β-cyclodextran (MβCD; Sigma Aldrich), was added to cells at the same time as HDL NPs, with concentrations ranging from 156.25 µM to 10 mM.
Western blot analysis
Western blot analysis was conducted as previously described.19 The SCARB1 (ab52629) and SREBP-1a (ab3259) antibodies was obtained from Abcam, and PARP (#9542) and β-actin (#4970) antibodies were obtained from Cell Signaling Technologies.
HDL NP synthesis and quantification
HDL NPs were synthesized and quantified as previously described.16, 18, 24, 32 Briefly, 5 nm diameter gold nanoparticles (AuNPs) are surface functionalized with apolipoprotein A-I (added at a 5 fold excess relative to [AuNP]) for one hour, followed by addition of the phospholipids 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)propionate] (PDP PE) and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) overnight (each added at 250 fold excess to the [Au NP]). The HDL NPs were purified using the KrosFlo TFF (Tangential Flow Filtration) system, with a 50 kDa cut-off PES column. To generate fluorescently labeled HDL NPs (DiI HDL NP), 1,1’-Dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine percholorate (DiI; ThermoFisher) was incorporated into the HDL NP synthesis. DiI was added at a 1µM final concentration (12.5-fold molar excess to AuNP) between addition of the PDP PE and DPPC phospholipids. Purification of DiI HDL NPs was conducted as described above for non-fluorescently HDL NPs, with special care being taken to protect the HDL NPs from light during synthesis and purification. UV/Vis spectroscopy was used to measure the concentration of the HDL NPs (ε = 9.696 × 106 M−1cm−1 for 5 nm diameter AuNPs). The size and surface charge of HDL NPs and DiI HDL NPs were determined using dynamic light scattering (DLS) and zeta potential, respectively, using a Malvern Zetasizer.
MTS assay
MTS assays were carried out as previously described19, and according to manufacturer’s instructions (CellTiter assay, Promega). Lymphoma cells were re-suspended at a concentration of 2 × 105 cells/ml, and 80 µl of cells added to each well of a 96 well plate. Cells were then treated with HDL NPs and/or various inhibitors (20 µl) for 72 hours prior to addition of the MTS reagent.
DiI HDL NP cell binding/ uptake assay
Lymphoma cells were plated at a concentration of 2.5 × 105 cells/ml and treated with 5–25 nM of fluorescently-labeled DiI HDL NPs for 24 hours. Cells were then washed twice with 1 × PBS, re-suspended in cold FACS buffer (PBS, 1% bovine serum albumin, 0.1% sodium azide) and cell associated fluorescence was obtained using the BD LSR II Fortessa flow cytometer. Data were analyzed using the FCS Express software.
Cholesterol efflux assays
Lymphoma cell cholesterol efflux assays were conducted as previously described.16, 19, 24 Lymphoma cells were labeled with 1,2 3H-cholesterol (1µCi/mL) for 24 hours in standard culture media. Post labeling, cells were spun down, re-suspended in serum free RPMI 1640, and then plated into 24 well culture plates at a concentration of 1.5 × 105 cells/well. HDL NPs and human HDL were added at a final concentration of 50 nM, with or without the addition of the SCARB1 blocking antibody (1:250), or the rabbit IgG isotype control antibody (1:250). Cells were incubated at 37°C for 4 hours. The cells were spun down and the supernatants collected, filtered and analyzed by liquid scintillation counting (countssupernatant). Total cholesterol labeling was quantified by extracting the radiolabeled cholesterol using isopropanol from lymphoma cells at the start of HDL NP and human HDL treatment (countstotal). The background cholesterol efflux in the absence of any cholesterol acceptor (e.g. hHDL or HDL NP) was subtracted from each value and the percent cholesterol efflux calculated by dividing the countssupernatant by the countstotal and multiplying by 100.
Total cholesterol quantification
Total cholesterol was quantified using the Amplex Red Cholesterol Assay (Thermo Fisher Scientific) according to manufacturer’s instructions. Briefly, lysates were prepared from SUDHL4, TMD8 and HBL-1 cells treated with PBS, HDL NPs, Ibrutinib, or HDL NP + Ibrutinib for 24 hours. Cholesterol content values were standardized to total cell number, as measured using the Countess automated cell counter (Invitrogen).
Phos-flow analysis of p-AKT and p-BTK
Cells were cultured with or without HDL NPs (50nM, 100nM), Ibrutinib (10nM) or PBS for 2 hours, then washed with FBS Stain Buffer (BD Biosciences). The cells were fixed and permeabilized with BD Phosflow Lyse/Fix buffer and BD Perm Buffer III according to manufacturer’s instructions. Following permeabilization, the cells were washed twice with Stain Buffer and then incubated for 60 minutes at room temperature with AlexaFluor 647-labeled p-AKT or p-BTK antibodies. The cells were washed twice and re-suspended in Stain Buffer for flow cytometric analysis on the BD LSR II Fortessa flow cytometer. Data were analyzed using FCS Express software.
RT-qPCR analysis
SUDHL4, TMD8 and HBL-1 cells were treated with HDL NPs (10nM, 50nM), hHDL (50nM) or PBS for 48 hours, and RNA isolated using the RNeasy Mini kit (Qiagen). RNA samples (500ng RNA per 30µl reaction) were reverse transcribed using a TaqMan Reverse Transcription kit, and qPCR for ACAT2, DHCR7, INSIG1, HMGCS1, LSS, MSMO1, MVD, SCD, p21, and SQLE run on a BioRad iCycler. Samples were standardized to β-actin, and relative expression was calculated using the ΔΔCt method. For every condition, biological triplicates were run.
Mouse tumor xenograft models
All animal work was conducted in accordance with Northwestern University’s IACUC and CCM facilities under an approved animal protocol. Flank tumors were initiated using the SUDHL4 and TMD8 cell line in SCID-beige mice (Taconic) and allowed to progress until reaching a size of approximately 50mm3. For the SUDHL4 study, mice were randomly divided into 3 treatment groups, based on their initial tumor volume: PBS, HDL NP 3 times per week or HDL NPs 5 times per week. HDL NPs (100µl of 1µM HDL NP) were injected I.V. for 21 days, at which time the mice were euthanized and blood was collected for blood chemistry analysis. For the TMD8 tumor xenograft study, mice were randomly divided into 4 treatment groups based on tumor volume at time of treatment initiation: PBS, HDL NP (1µM), Ibrutinib (3mg/kg body weight), or HDL NP (1µM) + Ibrutinib (3mg/kg body weight) and received 100µl of treatment 5 times per week for two weeks. Mice were euthanized on the day after the last dose of treatment.
Statistics
One-way ANOVAs and 2-sided student’s t-test were used to determine statistical significance, and the data were graphed using the GraphPad Prism software. Synergy calculations were performed using the Calcusyn (Biosoft) software.
Results
HDL NPs Induce Lymphoma Cell Death
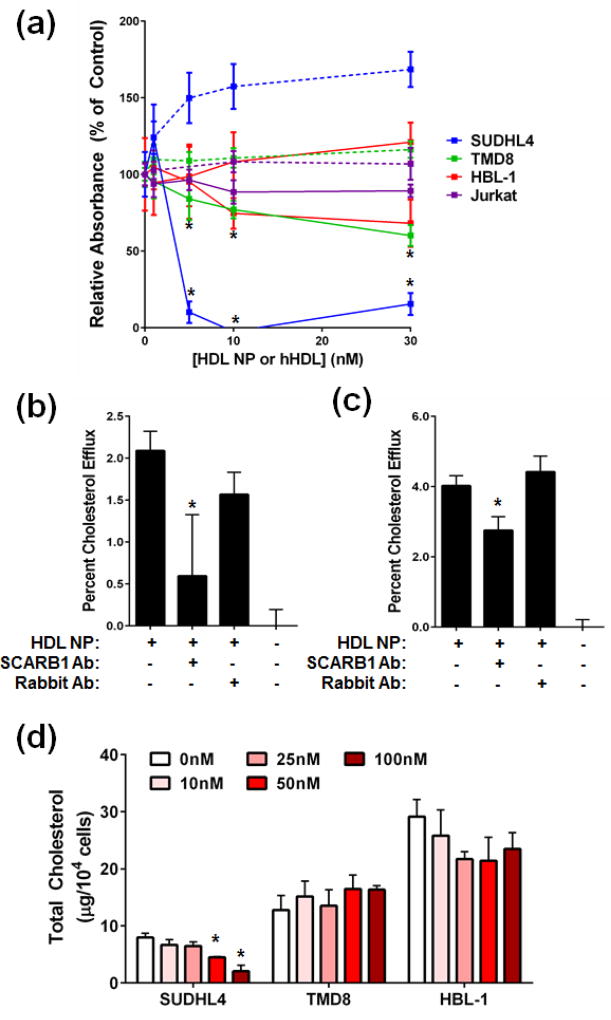
We tested the sensitivity of two commonly used ABC DLBCL cell lines, TMD8 and HBL-1, to HDL NP relative to the sensitive GC DLBCL SUDHL4 cell line. Both the GC and ABC DLBCL cell lines were positive for SCARB1 expression (Supplemental Figure S1). All B-cell lymphoma cell lines showed a dose-dependent loss of viability when treated with HDL NPs, but not when treated with human HDL (Figure 1a). The ABC DLBCL cell lines were significantly less sensitive to the HDL NP as compared with the GC DLBCL line SUDHL4 (Figure 1a). The SCARB1-negative Jurkat cell line was not sensitive to HDL NP (Figure 1a). This sensitivity correlated to HDL NP-induced PARP cleavage, with increased signal observed in the SUDHL4 cells compared with TMD8 and HBL-1 cells (Supplemental Figure S2).
Figure 1. HDL NPs induce B cell lymphoma cell death through modulation of cellular cholesterol.
(a) HDL NPs (solid line) potently induce cell death in the GC DLBCL cell line SUDHL4, but not the ABC DLBCL cell lines TMD8 or HBL-1. Human HDL (dotted line) does not induce lymphoma cell death. *p<0.05 v. PBS control. (b), (c) HDL NPs efflux cholesterol from SUDHL4 (b) and TMD8 (c) cells. Antibody blockade of SCARB1 reduced cholesterol efflux in both cell lines, while addition of an isotype control antibody had no effect. *p<0.05 v. No antibody. (d) Total cholesterol content, normalized to cell number, of SUDHL4, TMD8 and HBL-1 cells treated with PBS or HDL NPs. *p<0.05 v. 0nM HDL NP.
Because of differences in cell killing with HDL NPs, we tested if there was differential binding/ uptake of the HDL NPs in GC vs ABC DLBCL cells. To this end, we incubated SUDHL4 and TMD8 cells with HDL NPs labeled with the fluorescent intercalating dye DiI (DiI HDL NPs) for 24 hours prior to flow cytometric analysis. DiI HDL NPs were similar to HDL NPs in terms of their size (12.1 ± 0.3 nm diameter for HDL NP vs. 12.9 ± 0.7 nm for DiI HDL NP), surface charge (−45.9 ± 3.2 mV vs. −50.0 ± 0.2), stability and ability to induce GC DLBCL cell death (Supplemental Figure S3a, b). Interestingly, the TMD8 cells displayed enhanced DiI HDL NP fluorescence compared to SUDHL4 cells (Supplemental Figure 3c–e). These data demonstrate that the mechanism by which the HDL NPs differentially induce apoptosis in GC vs ABC DLBCL cells cannot be explained by increased HDL NP binding/ uptake.
HDL NPs Modulate Cellular Cholesterol Through SCARB1
Our previous work demonstrated that HDL NPs target and modulate cholesterol flux through SCARB1.16, 19, 24, 25 Therefore, we investigated the ability of HDL NPs to efflux cholesterol from GC and ABC DLBCL cells, with particular attention paid to SCARB1-mediated efflux. Both SUDHL4 and TMD8 cells effluxed cholesterol after exposure to HDL NPs (Figure 1b, c). Inhibition of HDL NP binding to SCARB1 by addition of a blocking antibody partially reduced cholesterol efflux to HDL NPs, which was not seen when an isotype control antibody was added (Figure 1b, c), suggesting that the nanoparticles engage with SCARB1 and modulate cholesterol flux through the receptor.
HDL NPs reduced total cholesterol in SUDHL4 cells in a dose-dependent manner (Figure 1d). The baseline cholesterol content of the ABC DLBCL cell lines TMD8 and HBL-1 was significantly higher compared with the GC-lymphoma cell lines, and was not reduced by HDL NP treatment (Figure 1d). These data correlate with the reported expanded BCR signaling observed in ABC versus GC lymphomas33 and reduced sensitivity to HDL NP-induced cell death (Figure 1a).
HDL NP Treatment Alters the Expression of Genes Involved in Cholesterol Metabolism
The transcription factor sterol response element binding protein (SREBP-1a) is activated by cellular cholesterol depletion whereupon it increases expression of cholesterol biosynthesis genes as well as INSIG1 (Insulin induced gene 1)34–37, which in combination with SCAP (SREBP cleavage-activating protein) regulates SREBP-1a activity.35, 38 Cholesterol depletion leads to uncoupling of SCAP from INSIG-1, leading to cleavage of endoplasmic reticulum-bound SREBP-1a into its free, active form, which then translocates to the nucleus and directs upregulation of cholesterol biosynthesis genes.35 HDL NP treatment increased accumulation of the active form of SREBP-1a in SUDHL4 cells (Supplemental Figure S4). In the ABC lymphoma cell lines TMD8 and HBL-1, active SREBP-1a did not change with HDL NP exposure (Supplemental Figure S4).
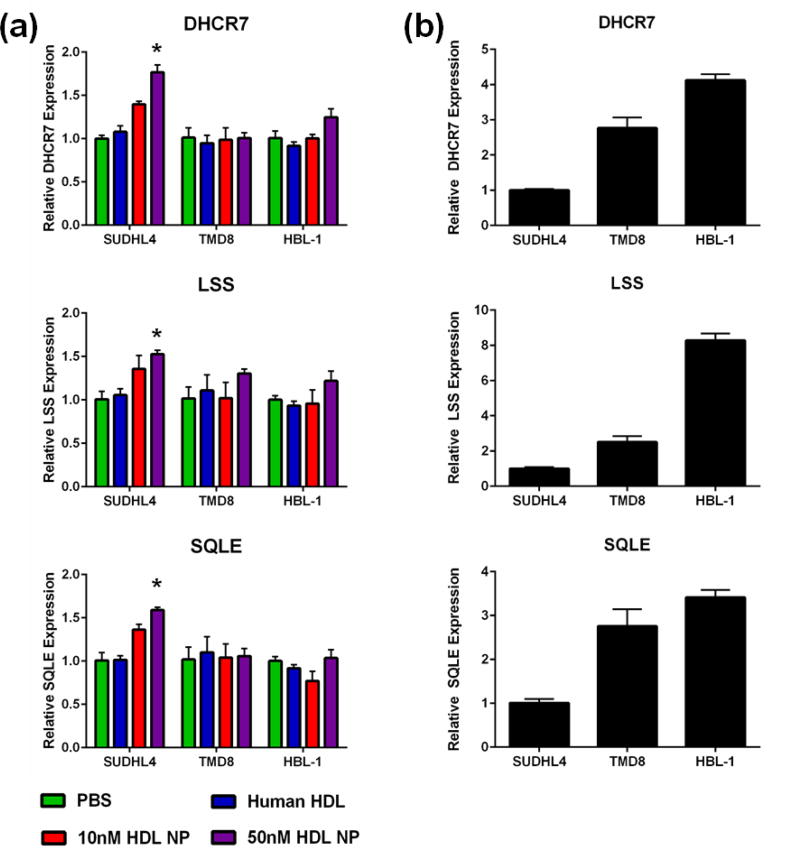
Given the ability of the HDL NPs to decrease cellular cholesterol content in the GC but not ABC DLBCL, the activation of SREBP-1a in GC but not ABC DLBCL, and the role of BCR signaling in cholesterol biosynthesis, we investigated changes in the expression of cholesterol biosynthesis genes following 48 hours of HDL NP or human HDL exposure in the SUHDL4, TMD8 and HBL-1 cell lines. Specifically, we quantified changes in expression of the cholesterol biosynthesis genes Acetyl-CoA Acetyltransferase 2 (ACAT2), Squalene Epoxidase (SQLE), Methylsterol Monooxygenase 1 (MSMO1), INSIG1, 3-Hydroxy-3-Methylglutaryl-CoA Synthase 1 (HMGCS1), 7-Dehydrocholesterol Reductase (DHCR7), Lanosterol Synthase (LSS), Stearoyl-CoA Desaturase (SCD), and Mevalonate Diphosphate Decarboxylase (MVD). These genes were selected given their role in the biosynthesis of cholesterol and their identification in previously reported studies connecting BCR signaling and cholesterol biosynthesis.15, 39 Genes were considered altered if the fold expression change was ≥1.5 or ≤-1.5, with a corresponding p < 0.05. HDL NP treatment resulted in upregulation of the cholesterol biosynthesis genes ACAT2, SQLE, MSMO1, INSIG1, HMGCS1, DHCR7, LSS, and MVD in SUDHL4 cells, with no upregulation seen when human HDL was administered (Figure 2a, Supplemental Figure S5). HDL NPs did not significantly increase ACAT2, SQLE, INSIG1, DHCR7, LSS, and MVD in the ABC lines at similar HDL NP doses. HMGCS1 and MSMO1 were elevated with 50nM HDL NP treatment in the HBL-1 but not the TMD8 cell line (Supplemental Figure S5). No significant change was observed in SCD expression in any cell line; however, expression was increased 1.473-fold with 50nM HDL NP treatment vs. PBS in SUDHL4 cells (Supplemental Figure S5). Direct comparison of baseline cholesterol biosynthesis gene expression between the GC and ABC DLBCL cell lines demonstrated higher expression levels in the ABC DLBCL cell lines relative to expression in GC DLBCL cells for all genes (Figure 2b, Supplemental Figure S6).
Figure 2. HDL NPs modulate cholesterol biosynthesis gene expression.
(a) HDL NPs upregulate expression of DHCR7, LSS and SQLE in SUDHL4 cells but not TMD8 or HBL-1 cells. Human HDL does not impact expression of any gene in any cell line. *p<0.05 vs. PBS control. (b) Expression of DHCR7, LSS and SQLE was higher in TMD8 and HBL-1 cells compared with SUDHL4 cells.
Interactions Between Cholesterol Depletion Agents and Targets Downstream of the B-cell Receptor
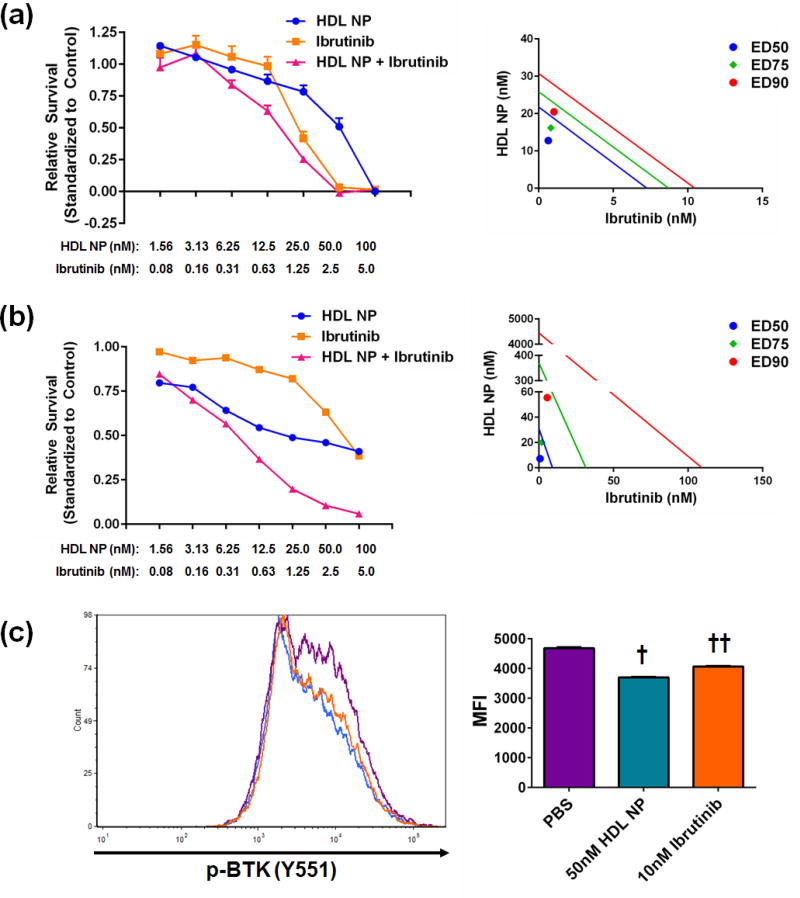
Given the ability of HDL NPs to reduce cellular cholesterol uptake and the B cell receptor’s prominent role in cholesterol biosynthesis in lymphoma cells15, we investigated the ability of both specific (HDL NP) and non-specific (methyl-β-cyclodextran, MβCD) cholesterol depletion agents to synergize with inhibitors of downstream B-cell receptor signaling, including the BTK inhibitor ibrutinib. The combination of the non-specific cholesterol sequestrant MβCD and ibrutinib synergized [combination index (CI) < 1] in TMD8 cells but not in HBL-1 cells (Supplemental Figure S7, Supplemental Table S1). Similarly, the combination of HDL NP and ibrutinib synergized in both the TMD8 and HBL-1 cells (Figure 3a, b, Supplemental Table S1).
Figure 3. HDL NPs synergize with the BTK inhibitor Ibrutinib.
A. HDL NPs synergized with the BTK inhibitor Ibrutinib in TMD8 (a) and HBL-1 (b) cells. Left- dose response curves. Right- Isobolographs depicting synergy at the ED50 (blue), ED75 (green), and ED90 levels (red). (c) Phos-flow analysis of TMD8 cells treated with HDL NPs or Ibrutinib. Cells were treated for 2 hours prior to analysis. † p<0.0001 v. all other groups by 1-way ANOVA. †† p<0.0001 v. all other groups by 1-way ANOVA. Data presented as mean ± s.e.m.
In addition to ibrutinib, we tested the ability of HDL NPs to synergize with the SYK inhibitor R406, previously shown to decrease BCR-driven cholesterol biosynthesis.15 The two agents synergized in the TMD8, but not HBL-1 cell lines (Supplemental Figure S8, Supplemental Table S1).
Beyond the BCR related kinases SYK and BTK, previous work demonstrated that engagement of SCARB1 by natural HDL activated PI3K and AKT21–23, which also play a role in both tonic and chronic active BCR signaling.33 To determine whether AKT and/or PI3K play a role in HDL NP-induced apoptosis, we investigated the interaction of clinically available small molecule inhibitors of AKT and PI3K (GDC-0068 and Pilaralisib, respectively), in combination with HDL NPs. GDC-0068 synergized with TMD8 cells, but not HBL-1 cells (Supplemental Figure S9, Supplemental Table S1), while Pilaralisib did not synergize with either cell line at any concentration (Supplemental Table S1).
To further investigate the potential involvement of AKT and BTK in HDL NP-induced lymphoma cell death, we investigated the phosphorylation status of AKT and BTK in TMD8 cells following HDL NP treatment. HDL NPs dose dependently decreased p-AKT (S473) levels in TMD8 cells (Supplemental Figure S10), while BTK phosphorylation (Y551) was decreased with 50nM HDL NP treatment in TMD8 cells similar to treatment with 10nM Ibrutinib (Figure 3c).
Finally, given the ability of HDL NPs to reduce BTK phosphorylation, and thus activation, we investigated whether HDL NPs would synergize with MβCD. Synergy between MβCD and the HDL NPs was observed in TMD8 and HBL-1 cells (Supplemental Figure S11, Supplemental Table S1), suggesting that the HDL NPs may play a larger role than simply blocking cholesterol influx. Taken together, these data suggest that HDL NPs act both to deplete cellular cholesterol and disrupt membrane-linked receptor signaling critical for lymphoma cell survival.
HDL NP Synergize with Ibrutinib to Reduce Cellular Cholesterol
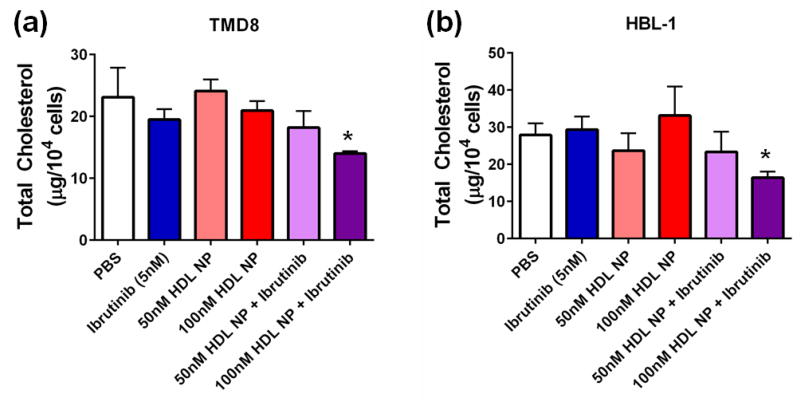
As demonstrated above, the ABC lymphoma cell lines are more resistant to HDL NP-mediated reduction in cellular cholesterol. Based on synergy data between the HDL NP and ibrutinib, we hypothesized that the combination would reduce the total cholesterol content of TMD8 and HBL-1 cells. Administration of either drug alone did not alter the cholesterol content of the cells; however, the combination of HDL NPs and Ibrutinib (5nM) significantly reduced total cellular cholesterol in both the TMD8 and HBL-1 cell lines (Figure 4a, b).
Figure 4. HDL NPs synergize with Ibrutinib to reduce total cellular cholesterol in ABC DLBCL cells.
Total cholesterol of TMD8 (a) and HBL-1 (b) cells treated with Ibrutinib (5nM) in combination with HDL NPs. N=4 for each condition. *p=0.0432 v. PBS control by 2-sided t-test for TMD8 and p=0.0319 v. PBS control by 2-sided t-test for HBL-1. Data presented as mean ± s.e.m.
HDL NP Inhibits Tumor Growth in GC DLBCL Xenografts and Synergizes with Ibrutinib in ABC DLBCL Xenografts
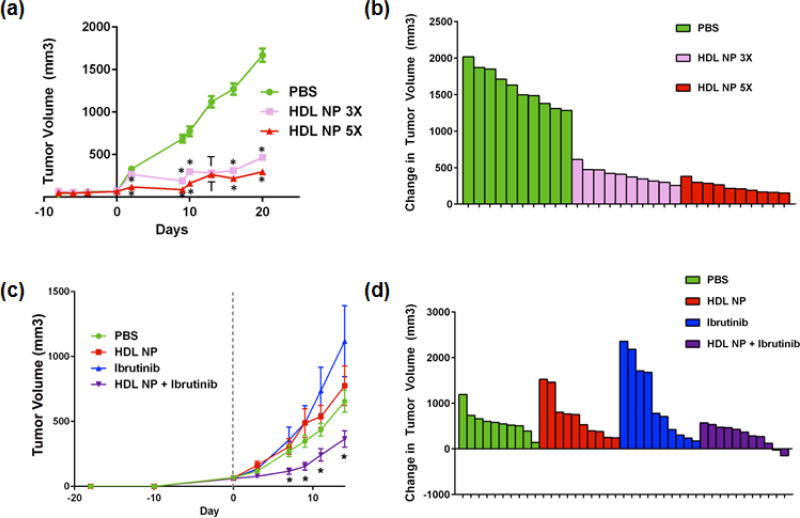
We first investigated the in vivo efficacy of HDL NPs against the GC DLBCL cell line, SUDHL4. SUDHL4 xenografts were initiated in SCID-beige mice and then treated with 100µl of 1µM HDL NPs either 3 or 5 times per week (Figure 5a, b). Both dosing schedules resulted in significantly decreased tumor volumes relative to PBS control and were well tolerated by the mice, with no observed adverse effects. Blood chemistries demonstrated no major change in kidney or liver function, and hematopoietic function (hemoglobin, white count and platelet count) remained normal (Supplemental Table S2). Based on these results, we selected the 5 times per week dosing for the subsequent ABC DLBCL tumor xenograft study.
Figure 5. In vivo efficacy of HDL NPs as monotherapy and in combination with ibrutinib.
(a) SUDHL4 tumor xenografts were initiated in SCID-beige mice, and treated either 3 times or 5 times per week with 100µl of 1µM HDL NPs, injected via the tail vein. N=10 for each treatment group. *p<0.0001 vs. all other treatment groups by 1-way ANOVA; † p<0.0001 vs. PBS control only by 1-way ANOVA. (b) Waterfall plot of the change in tumor volume from Day 0 (start of treatment) to Day 21. (c) TMD8 tumor xenografts were initiated in SCID-beige mice, and treated 5 times per week with PBS, 100µl of 1µM HDL NPs, 3mg/kg ibrutinib, or a combination of HDL NPs and ibrutinib for two weeks. N=10 for PBS, HDL NP alone, and ibrutinib alone. N=11 for HDL NP + ibrutinib. *p<0.05 v. all other treatment groups by 1-way ANOVA. (d) Waterfall plot of the change in tumor volume from Day 0 (start of treatment) to Day 14.
To further investigate potential synergy between HDL NPs and ibrutinib, a recently described second line therapy for ABC DLBCL40, and to investigate whether HDL NPs can function as a monotherapy against ABC DLBCL in an in vivo setting, TMD8 tumor xenografts were initiated in SCID-beige mice, which were then treated with HDL NPs (100µl of 1µM HDL NPs, i.v.), ibrutinib (3mg/kg body weight, i.p.), HDL NP + ibrutinib or vehicle (PBS) control 5 times per week for 14 days. The dose of HDL NPs was selected based on our previous tumor xenograft models, while the ibrutinib dosing was based on previously reported studies.41 HDL NPs and ibrutinib alone were ineffective (Figure 5c, d) while their combination significantly reduced tumor volumes compared with vehicle control and ibrutinib and HDL NP alone (Figure 5c, d).
Discussion
In this study, we present a rationale for targeting cellular cholesterol as a therapeutic intervention in DLBCL. Our data demonstrate that HDL NPs deplete cellular cholesterol levels in GC DLBCL cells, which in turn activates cholesterol biosynthesis pathways. However, HDL NP-mediated cholesterol depletion eventually results in apoptosis both in vitro and in vivo. In ABC DLBCL, HDL NPs alone are insufficient to result in cholesterol depletion and lymphoma cell death, due to enhanced BCR-driven cholesterol biosynthesis and the corresponding higher baseline cellular cholesterol content. To overcome this, combination therapy of small molecule inhibitors of BCR signaling with the HDL NPs ultimately reduces cellular cholesterol levels, resulting in lymphoma cell death, both in vitro and in an in vivo tumor xenograft model. Correspondingly, the combination of non-specific cholesterol sequestrants with BCR signaling inhibitors also synergizes, leading to enhanced ABC DLBCL cell death. These results demonstrate the feasibility of targeted cholesterol depletion as a therapeutic intervention for lymphoma.
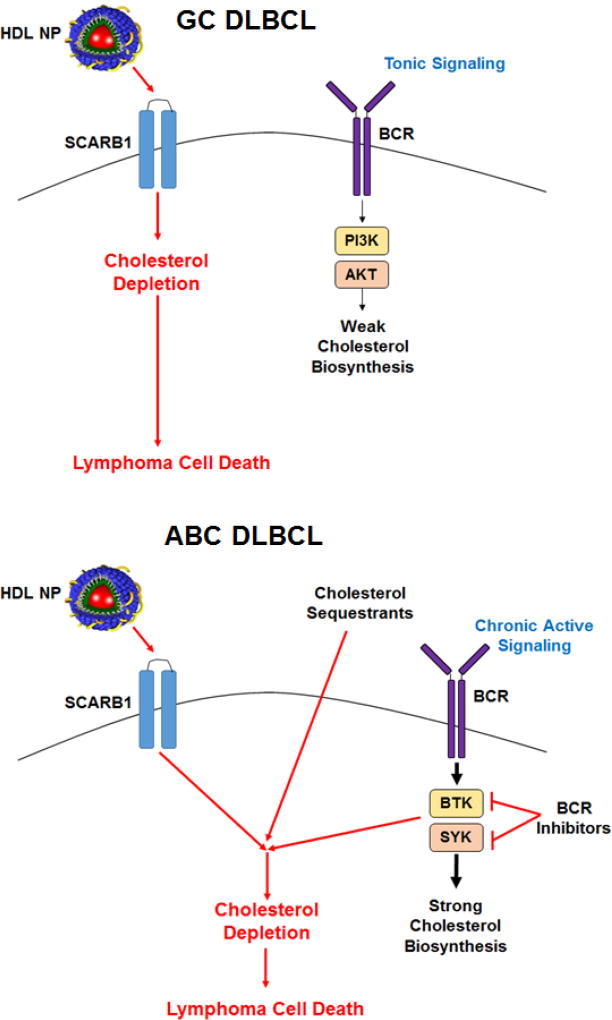
The proposed mechanism of HDL NP-induced cell death is presented in Figure 6. In GC DLBCLs, tonic BCR signaling results in a requirement for cellular cholesterol such that HDL NP potently induces cholesterol depletion and activation of the transcription factor SREBP-1a, a master regulator of cholesterol biosynthesis. This, in turn, drives a cellular response to increase de novo cholesterol biosynthesis that, ultimately, cannot overcome HDL NP-mediated cholesterol depletion and the cell undergoes apoptosis. In ABC DLBCL, our data suggest that more active BCR signaling results in a baseline increased need for cholesterol that must be met with increased synthesis and SREBP-1a activity, leading to higher baseline expression of cholesterol biosynthesis genes and correspondingly higher baseline cholesterol content. As such, a two pronged approach, 1) limitation of cholesterol and cholesteryl ester uptake from HDL and 2) impairment of de novo cholesterol biosynthesis, is necessary to appreciably cholesterol-starve ABC DLBCL cells and induce cell death. Clinical outcomes with ABC DLBCL are typically worse than in GC DLBCL42, and this novel combination approach may become a new paradigm for treatment. Of note, other HDL-based therapeutics, such as statin-loaded reconstituted HDLs, have been developed to decrease cellular cholesterol content by inhibiting de novo cholesterol synthesis.43 Such materials should be tested for their inherent and drug-loaded cellular cholesterol depletion properties in the context of lymphoma.
Figure 6. Targeted cholesterol depletion and cell death induction in GC (top) and ABC DLBCL (bottom) mediated by HDL NPs.
Taken together, these molecular and functional studies clearly demonstrate the potential of targeted disruption of cellular cholesterol homeostasis in lymphoma, and provide a framework for further studies and novel therapeutic combinations, not only in lymphoma but other cholesterol-dependent malignancies.
Beyond lymphoma, recent work in prostate cancer shows that depletion of cholesteryl ester accumulation, which occurred as a result of PI3K/AKT activation, reduced cancer cell proliferation, impaired cancer invasion capability and suppressed tumor xenograft growth44, thus providing further rationale for targeting cholesterol metabolism in cancer. Additionally, the high affinity HDL receptor SCARB1 was reported to be over expressed in a wide range of malignancies, including breast cancer45, pancreatic cancer46, renal cell carcinoma47, chronic lymphocytic leukemia48, and prostate cancer9, and has been identified as an independent prognostic factor associated with worse outcomes in breast cancer.45 The HDL NPs are a unique cholesterol depletion agent that, due to their composition, specifically target SCARB117–19, 48, 49, making them an ideal component of a cholesterol depleting therapeutic regimen. Whether or not combination therapy is required, in lymphoma or other cancers, may be answered by measurement of the level of cholesterol, cholesteryl ester, or the level of critical de novo cholesterol synthesis targets (e.g. LSS or DHCR7), in addition to quantification of the expression level of SCARB1. Based on our data in ABC DLBCL, a new paradigm may emerge that depends upon optimal cholesterol depletion, where the combination of drugs that best reduces cholesterol biosynthesis and uptake of cholesterol from lipoproteins optimizes treatment outcomes.
Supplementary Material
Acknowledgments
This work was supported by the Northwestern University Flow Cytometry Core Facility and a Cancer Center Support Grant (NCI CA060553), as well as the Northwestern University Immunobiology Center Flow Cytometry Core Facility. The authors would also like to acknowledge the Robert H. Lurie Comprehensive Cancer Center of Northwestern University Pathology Core Facility for their assistance in processing and staining of patient tissue samples. Gene microarray analysis was performed by the Genomics Core at Northwestern University. J.S.R. thanks the National Institutes of Health/National Heart, Lung, and Blood Institute for support from the Vascular Surgery Scientist Training Program grant (T32HL094293). C.S.T. thanks the Howard Hughes Medical Institute (HHMI) for a Physician Scientist Early Career Award, the Department of Defense/Air Force Office of Scientific Research (FA95501310192), and the National Institutes of Health/National Cancer Institute (U54CA151880 and R01CA167041). We thank Dr. Louis Stoudt for providing the TMD8 and HBL-1 cell lines, and for valuable suggestions.
Funding Sources
J.S.R. is supported by NIH T32HL094293. R.L. is supported by NIH CA073507. L.I.G. is supported by Cancer Center Support Grant (NCI CA060553), the Brookstone Research Fund, and the Lymphoma Research Fund. C.S.T. is supported by NIH R01CA167041 and NHLBI R01HL116577.
ABBREVIATIONS
- HDL
high-density lipoprotein
- HDL NP
high-density lipoprotein-like nanoparticle
- BCR
B-cell receptor
- DLBCL
diffuse large B cell lymphoma
- GC
germinal center
- ABC
activated B-cell
- BTK
Bruton’s tyrosine kinase
- SYK
spleen-associated tyrosine kinase
- SCARB1
scavenger receptor type B1
- apoA-I
apolipoprotein A-I
- MβCD
methyl-β-cyclodextran
- ACAT2
acetyl-CoA acetyltransferase 2
- SQLE
squalene epoxidase
- MSMO1
methylsterol monooxygenase 1
- INSIG1
insulin-induced gene 1
- HMGCS1
3-hydroxy-3-methylglutaryl-CoA synthase 1
- DHCR7
7-dehydrocholesterol reductase
- LSS
lanosterol synthase
- SCD
stearoyl-CoA desaturase
- MVD
mevalonate diphosphate decarboxylase
- hHDL
human HDL
- SREBP-1a
sterol response element binding protein-1a
- STR
short tandem repeat
- RPMI
Roswell Park Memorial Institute
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- PARP
poly (ADP-ribose) polymerase
- FBS
fetal bovine serum
- PDP PE
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)propionate]
- DPPC
1,2-dipalmitoyl-sn-3-glycero-phosphocholine
- TFF
tangential flow filtration
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- PBS
phosphate buffered saline
- RBC
red blood cell
- PLT
platelet
- HGB
hemoglobin
- HCT
hematocrit
- LDL
low density lipoprotein
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BUN
blood urea nitrogen
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supplemental Figure S1 (SCARB1 western blot), S2 (PARP western blot), S3 (DiI HDL NP characterization and internalization by DLBCL cells), S4 (SREBP-1a western blot), S5 (RT-qPCR data), S6 (RT-qPCR data), S7 (MβCD: Ibrutinib synergy graphs), S8 (R406: HDL NP synergy graphs), S9 (GDC-0068: HDL NP synergy graphs), S10 (Phos-flow for p-AKT), S11 (MβCD: HDL NP synergy graphs); Supplemental Table S1 (CI synergy values), S2 (blood chemistry values).
Author Contributions
J.S.R., L.I.G. and C.S.T. designed the experiments and analyzed the data presented here. J.S.R. synthesized the HDL NPs, conducted the cell death assays on all lymphoma cell lines, the HDL NP binding/ uptake fluorescent assay, all cholesterol efflux and total cholesterol quantification assays, synergy cell death assays and calculations, phos-flow experiments, RT-qPCR assays, western blot assays for SREBP-1a, SCARB1 and PARP, and assisted S.M., K.M.M. and O.C. on the TMD8 tumor xenograft study. K.M.M. and C.S.T. conducted the SUDHL4 tumor xenograft study. J.S.R., L.I.G. and C.S.T. wrote the manuscript. All authors reviewed and approved the manuscript prior to submission.
References
- 1.Silvente-Poirot S, Poirot M. Cholesterol metabolism and cancer: the good, the bad and the ugly. Current opinion in pharmacology. 2012;12(6):673–6. doi: 10.1016/j.coph.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290(5497):1721–6. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- 3.Dessi S, Batetta B, Pani A, Spano O, Sanna F, Putzolu M, Bonatesta R, Piras S, Pani P. Role of cholesterol synthesis and esterification in the growth of CEM and MOLT4 lymphoblastic cells. Biochem J. 1997;321(Pt 3):603–8. doi: 10.1042/bj3210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goncalves RP, Rodrigues DG, Maranhao RC. Uptake of high density lipoprotein (HDL) cholesteryl esters by human acute leukemia cells. Leuk Res. 2005;29(8):955–9. doi: 10.1016/j.leukres.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Mulas MF, Abete C, Pulisci D, Pani A, Massidda B, Dessi S, Mandas A. Cholesterol esters as growth regulators of lymphocytic leukaemia cells. Cell Prolif. 2011;44(4):360–71. doi: 10.1111/j.1365-2184.2011.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Gonzalo-Calvo D, Lopez-Vilaro L, Nasarre L, Perez-Olabarria M, Vazquez T, Escuin D, Badimon L, Barnadas A, Lerma E, Llorente-Cortes V. Intratumor cholesteryl ester accumulation is associated with human breast cancer proliferation and aggressive potential: a molecular and clinicopathological study. Bmc Cancer. 2015;15 doi: 10.1186/s12885-015-1469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drabkin HA, Gemmill RM. Cholesterol and the development of clear-cell renal carcinoma. Current opinion in pharmacology. 2012;12(6):742–50. doi: 10.1016/j.coph.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Gu D, Lee SS, Song B, Bandyopadhyay S, Chen S, Konieczny SF, Ratliff TL, Liu X, Xie J, Cheng JX. Abrogating cholesterol esterification suppresses growth and metastasis of pancreatic cancer. Oncogene. 2016;35(50):6378–6388. doi: 10.1038/onc.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Ren SC, Piao HL, Wang FB, Yin PY, Xu CL, Lu X, Ye GZ, Shao YP, Yan M, Zhao XJ, Sun YH, Xu GW. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Scientific reports. 2016;6 doi: 10.1038/srep20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue SH, Li JJ, Lee SY, Lee HJ, Shao T, Song B, Cheng L, Masterson TA, Liu XQ, Ratliff TL, Cheng JX. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3K/AKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014;19(3):393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz PM, Mo H, McConathy WJ, Sabnis N, Lacko AG. The role of cholesterol metabolism and cholesterol transport in carcinogenesis: a review of scientific findings, relevant to future cancer therapeutics. Frontiers in pharmacology. 2013;4:119. doi: 10.3389/fphar.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cvetkovic Z, Cvetkovic B, Petrovic M, Ranic M, Debeljak-Martarcic J, Vucic V, Glibetic M. Lipid profile as a prognostic factor in cancer patients. Journal of B.U.ON. : official journal of the Balkan Union of Oncology. 2009;14(3):501–6. [PubMed] [Google Scholar]
- 13.Kuliszkiewicz-Janus M, Malecki R, Mohamed AS. Lipid changes occuring in the course of hematological cancers. Cell Mol Biol Lett. 2008;13(3):465–474. doi: 10.2478/s11658-008-0014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mooberry LK, Sabnis NA, Panchoo M, Nagarajan B, Lacko AG. Targeting the SR-B1 Receptor as a Gateway for Cancer Therapy and Imaging. Frontiers in pharmacology. 2016;7:466. doi: 10.3389/fphar.2016.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Monti S, Juszczynski P, Ouyang J, Chapuy B, Neuberg D, Doench JG, Bogusz AM, Habermann TM, Dogan A, Witzig TE, Kutok JL, Rodig SJ, Golub T, Shipp MA. SYK Inhibition Modulates Distinct PI3K/AKT- Dependent Survival Pathways and Cholesterol Biosynthesis in Diffuse Large B Cell Lymphomas. Cancer cell. 2013;23(6):826–38. doi: 10.1016/j.ccr.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luthi AJ, Lyssenko NN, Quach D, McMahon KM, Millar JS, Vickers KC, Rader DJ, Phillips MC, Mirkin CA, Thaxton CS. Robust passive and active efflux of cellular cholesterol to a designer functional mimic of high density lipoprotein. Journal of lipid research. 2015;56(5):972–85. doi: 10.1194/jlr.M054635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon KM, Thaxton CS. High-density lipoproteins for the systemic delivery of short interfering RNA. Expert opinion on drug delivery. 2014;11(2):231–47. doi: 10.1517/17425247.2014.866089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plebanek MP, Mutharasan RK, Volpert O, Matov A, Gatlin JC, Thaxton CS. Nanoparticle Targeting and Cholesterol Flux Through Scavenger Receptor Type B-1 Inhibits Cellular Exosome Uptake. Scientific reports. 2015;5:15724. doi: 10.1038/srep15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Damiano MG, Zhang H, Tripathy S, Luthi AJ, Rink JS, Ugolkov AV, Singh AT, Dave SS, Gordon LI, Thaxton CS. Biomimetic, synthetic HDL nanostructures for lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(7):2511–6. doi: 10.1073/pnas.1213657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271(5248):518–20. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 21.Assanasen C, Mineo C, Seetharam D, Yuhanna IS, Marcel YL, Connelly MA, Williams DL, de la Llera-Moya M, Shaul PW, Silver DL. Cholesterol binding, efflux, and a PDZ-interacting domain of scavenger receptor-BI mediate HDL-initiated signaling. The Journal of clinical investigation. 2005;115(4):969–77. doi: 10.1172/JCI200523858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danilo C, Gutierrez-Pajares JL, Mainieri MA, Mercier I, Lisanti MP, Frank PG. Scavenger receptor class B type I regulates cellular cholesterol metabolism and cell signaling associated with breast cancer development. Breast cancer research : BCR. 2013;15(5):R87. doi: 10.1186/bcr3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saddar S, Mineo C, Shaul PW. Signaling by the high-affinity HDL receptor scavenger receptor B type I. Arterioscler Thromb Vasc Biol. 2010;30(2):144–50. doi: 10.1161/ATVBAHA.109.196170. [DOI] [PubMed] [Google Scholar]
- 24.Luthi AJ, Zhang H, Kim D, Giljohann DA, Mirkin CA, Thaxton CS. Tailoring of biomimetic high-density lipoprotein nanostructures changes cholesterol binding and efflux. ACS nano. 2012;6(1):276–85. doi: 10.1021/nn2035457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thaxton CS, Daniel WL, Giljohann DA, Thomas AD, Mirkin CA. Templated spherical high density lipoprotein nanoparticles. J Am Chem Soc. 2009;131(4):1384–5. doi: 10.1021/ja808856z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W, Kewalramani S, Hujsak K, Zhang H, Bedzyk MJ, Dravid VP, Thaxton CS. Mesophase in a thiolate-containing diacyl phospholipid self-assembled monolayer. Langmuir : the ACS journal of surfaces and colloids. 2015;31(10):3232–41. doi: 10.1021/la504822q. [DOI] [PubMed] [Google Scholar]
- 27.Sun WQ, Wu WQ, McMahon KM, Rink JS, Thaxton CS. Mosaic Interdigitated Structure in Nanoparticle-Templated Phospholipid Bilayer Supports Partial Lipidation of Apolipoprotein A-I. Part Part Syst Char. 2016;33(6):300–305. doi: 10.1002/ppsc.201600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danilo C, Frank PG. Cholesterol and breast cancer development. Current opinion in pharmacology. 2012;12(6):677–82. doi: 10.1016/j.coph.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez-Pajares JL, Ben Hassen C, Chevalier S, Frank PG. SR-BI: Linking Cholesterol and Lipoprotein Metabolism with Breast and Prostate Cancer. Frontiers in pharmacology. 2016;7:338. doi: 10.3389/fphar.2016.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Ren S, Piao HL, Wang F, Yin P, Xu C, Lu X, Ye G, Shao Y, Yan M, Zhao X, Sun Y, Xu G. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Scientific reports. 2016;6:20984. doi: 10.1038/srep20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Queiroz KC, Tio RA, Zeebregts CJ, Bijlsma MF, Zijlstra F, Badlou B, de Vries M, Ferreira CV, Spek CA, Peppelenbosch MP, Rezaee F. Human plasma very low density lipoprotein carries Indian hedgehog. Journal of proteome research. 2010;9(11):6052–9. doi: 10.1021/pr100403q. [DOI] [PubMed] [Google Scholar]
- 32.McMahon KM, Mutharasan RK, Tripathy S, Veliceasa D, Bobeica M, Shumaker DK, Luthi AJ, Helfand BT, Ardehali H, Mirkin CA, Volpert O, Thaxton CS. Biomimetic high density lipoprotein nanoparticles for nucleic acid delivery. Nano Lett. 2011;11(3):1208–14. doi: 10.1021/nl1041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nature reviews. Drug discovery. 2013;12(3):229–43. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon KM, Foit L, Angeloni NL, Giles FJ, Gordon LI, Thaxton CS. Synthetic high-density lipoprotein-like nanoparticles as cancer therapy. Cancer treatment and research. 2015;166:129–50. doi: 10.1007/978-3-319-16555-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao WM, Murao K, Imachi H, Yu X, Abe H, Yamauchi A, Niimi M, Miyauchi A, Wong NC, Ishida T. A mutant high-density lipoprotein receptor inhibits proliferation of human breast cancer cells. Cancer Res. 2004;64(4):1515–21. doi: 10.1158/0008-5472.can-03-0675. [DOI] [PubMed] [Google Scholar]
- 37.Phoenix TN, Patmore DM, Boop S, Boulos N, Jacus MO, Patel YT, Roussel MF, Finkelstein D, Goumnerova L, Perreault S, Wadhwa E, Cho YJ, Stewart CF, Gilbertson RJ. Medulloblastoma Genotype Dictates Blood Brain Barrier Phenotype. Cancer Cell. 2016;29(4):508–22. doi: 10.1016/j.ccell.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncology. 2017;19(1):139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X, Tang JJ, Peng C, Wang Y, Fu L, Qiu ZP, Xiong Y, Yang LF, Cui HW, He XL, Yin L, Qi W, Wong CC, Zhao Y, Li BL, Qiu WW, Song BL. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Molecular cell. 2017;66(1):154–162 e10. doi: 10.1016/j.molcel.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, Lih CJ, Williams PM, Shaffer AL, Gerecitano J, de Vos S, Goy A, Kenkre VP, Barr PM, Blum KA, Shustov A, Advani R, Fowler NH, Vose JM, Elstrom RL, Habermann TM, Barrientos JC, McGreivy J, Fardis M, Chang BY, Clow F, Munneke B, Moussa D, Beaupre DM, Staudt LM. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–6. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Shaffer AL, 3rd, Emre NC, Ceribelli M, Zhang M, Wright G, Xiao W, Powell J, Platig J, Kohlhammer H, Young RM, Zhao H, Yang Y, Xu W, Buggy JJ, Balasubramanian S, Mathews LA, Shinn P, Guha R, Ferrer M, Thomas C, Waldmann TA, Staudt LM. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer cell. 2012;21(6):723–37. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JG, Sabet H, Tran T, Yu X, Powell JI, Yang LM, Marti GE, Moore T, Hudson J, Lu LS, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 43.Duivenvoorden R, Tang J, Cormode DP, Mieszawska AJ, Izquierdo-Garcia D, Ozcan C, Otten MJ, Zaidi N, Lobatto ME, van Rijs SM, Priem B, Kuan EL, Martel C, Hewing B, Sager H, Nahrendorf M, Randolph GJ, Stroes ES, Fuster V, Fisher EA, Fayad ZA, Mulder WJ. A statin-loaded reconstituted high-density lipoprotein nanoparticle inhibits atherosclerotic plaque inflammation. Nature communications. 2014;5:3065. doi: 10.1038/ncomms4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, Cheng L, Masterson TA, Liu X, Ratliff TL, Cheng JX. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19(3):393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan B, Wu C, Wang X, Wang D, Liu H, Guo L, Li XA, Han J, Feng H. High scavenger receptor class B type I expression is related to tumor aggressiveness and poor prognosis in breast cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-4141-4. [DOI] [PubMed] [Google Scholar]
- 46.McMahon KM, Scielzo C, Angeloni NL, Deiss-Yehiely E, Scarfo L, Ranghetti P, Ma S, Kaplan J, Barbaglio F, Gordon LI, Giles FJ, Thaxton CS, Ghia P. Synthetic high-density lipoproteins as targeted monotherapy for chronic lymphocytic leukemia. Oncotarget. 2017;8(7):11219–11227. doi: 10.18632/oncotarget.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, Salic A. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell. 2016;166(5):1176–1187 e14. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hentschel A, Zahedi RP, Ahrends R. Protein lipid modifications--More than just a greasy ballast. Proteomics. 2016;16(5):759–82. doi: 10.1002/pmic.201500353. [DOI] [PubMed] [Google Scholar]
- 49.Tripathy S, Vinokour E, McMahon KM, Volpert OV, Thaxton CS. High Density Lipoprotein Nanoparticles Deliver RNAi to Endothelial Cells to Inhibit Angiogenesis. Particle & particle systems characterization : measurement and description of particle properties and behavior in powders and other disperse systems. 2014;31(11):1141–1150. doi: 10.1002/ppsc.201400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.