Abstract
Based on the results of a SETAC-sponsored Horizon Scanning exercise focused on advancing the adverse outcome pathway (AOP) framework, the development of guidance related to AOP network development was identified as a critical need. This not only included questions focusing directly on AOP networks, but also on related topics such as mixture toxicity assessment and the implementation of feedback loops within the AOP framework. A set of two papers has been developed to begin exploring these concepts. In the present paper (part I), derivation of AOP networks is considered in the context of how it differs from development of individual AOPs. We then propose the use of filters and layers to tailor AOP networks to suit the needs of a given research question or application. We briefly introduce a number of analytical approaches that may be used to characterize the structure of AOP networks. These analytical concepts are further described in a dedicated, complementary paper (part II). Finally, we present a number of case studies that illustrate concepts underlying development, analysis and application of AOP networks. The concepts described in this paper, and in its companion paper focused on AOP network analytics, are intended to serve as a starting point for further development of the AOP network concept, but also to catalyze AOP network development and application by the different stakeholder communities.
Keywords: Adverse outcome pathway, risk assessment, predictive toxicology, AOP network, network development, network topology
Graphical abstract
1. Introduction
Adverse outcome pathways (AOPs) are an important framework that can help support greater and more effective use of mechanistic, or pathway-based, data in risk assessment and regulatory decision-making. While the conceptual underpinnings of the AOP framework date back to at least the late 1980s (Lalone and others 2017a), AOPs have rapidly evolved from a conceptual paradigm (Ankley and others 2010) into a formalized framework for organizing biological and toxicological knowledge according to a set of principles and guidelines that are generally accepted by the scientific and regulatory communities (OECD 2013a 2015; Villeneuve and others 2014b), and for disseminating that knowledge through an internationally harmonized knowledgebase (aopwiki.org, aopkb.org). Nonetheless, further development of the framework and the tools, approaches, and concepts surrounding its application is required to fully realize its potential and acceptance by society.
In response to the recognized need to continue advancing the framework, the Society of Environmental Toxicology and Chemistry (SETAC) sponsored a global Horizon Scanning exercise to identify major outstanding topics and challenges related to the AOP framework and its application (Lalone and others 2017a). Based on a survey of the international stakeholder community, four major topics/themes that needed further development were identified: (1) enhance communication, outreach, and stakeholder engagement in the development and application of AOP knowledge, (2) enhance regulatory use and acceptance of the AOP framework and facilitate its incorporation into regulatory practices, (3) enhance the use of the framework for quantitative assessments and applications, and (4) development of approaches for deriving, interrogating, and applying networks of AOPs, which is the topic of the current paper.
As outlined by Villeneuve and others (2014b), individual AOPs are viewed as a pragmatic unit of development and evaluation. It is tractable for an individual or a research team to describe and establish, through both biological plausibility and supporting evidence, how a defined perturbation of a biological system can lead, in a causal manner, to a particular adverse outcome. It is far less tractable for that individual or team to describe all the possible adverse effects a given perturbation may cause, or conversely, all the different perturbations through which stressors may evoke a particular adverse outcome (e.g., reductions in survival, growth, reproduction, increased risk of disease). It is even more daunting to consider describing those possibilities for all the different taxa, life-stages, and sexes (where relevant) that are of interest to a stakeholder. However, at the same time it was recognized that the “one perturbation-one adverse outcome” model that an individual AOP represents is a gross oversimplification of both the complexity of biological systems and the consequences of exposures to stressors that they face. In most real world scenarios, exposures are to multiple stressors (i.e., mixtures), not just one stressor at a time. Likewise, even single stressors may induce toxicity by more than one mechanism. This may be via interaction of the chemical with multiple targets in an organism or via interaction with a single target found in multiple compartments (e.g., cell types, tissues, organs, etc.) within a complex organism. Thus, most often, AOPs cannot be considered in isolation. One needs to think about potential interactions among pathways and consider how those interactions may alter the trajectory or intensity of the effects resulting from a chemical exposure.
Recognizing this, one of the core principles of AOP development was that, in contrast to individual AOPs as pragmatic units of development, AOP networks are viewed as the most likely units of prediction (Villeneuve and others 2014b). In turn, the formalization of the AOP framework, and its implementation via a knowledgebase structure that allowed for sharing of an AOP’s modular units (key events [KEs] and key event relationships [KERs]; aopwiki.org), was conceived and designed to allow for de facto construction of more complex and comprehensive networks from individual AOPs. In this way, a more accurate representation of biological and toxicological complexity that covers more and more of the susceptible taxonomic space and biological contexts (e.g., life stage, sex, impacts in or upon different target organs) can be built up gradually over time, through the independent contributions of individuals or groups.
To date, a vision for AOP networks has just started to be realized. Following publication of principles and best practices for AOP development (Villeneuve and others 2014b; 2014c) and public release of the AOP-Wiki (aopwiki.org) in 2014, time was needed to allow for an accumulation of a sufficient number of AOPs in the AOP knowledgebase (AOP-KB) to actually begin exploring their connectivity. Likewise, technical and practical challenges in the development of sharable, modular KE and KER units in the public AOP-KB (e.g., developing naming conventions, search tools, guidance and training materials, etc.) initially hampered rapid assembly of these de facto networks. Nonetheless, over the last three years, a critical mass of AOP descriptions has started to accumulate and some of the challenges have been overcome. This has led to recent realization of some of the first examples of AOP networks (Angrish and others 2016; Angrish and others 2017; Knapen and others 2015; LaLone and others 2017b; Margiotta-Casaluci and others 2016), and along with it, opportunities to address key concepts related to the development, analysis, and application of AOP networks.
The present set of two papers begins exploring these concepts. In part I, derivation of AOP networks is considered in the context of how it differs from development of individual AOP descriptions. We then discuss the application of filters and layers to refine and enrich derived AOP networks so that they may be tailored to address specific questions of interest. Modifications to the AOP-KB that may be needed accordingly are also considered. We then briefly introduce a number of analytical and computational approaches that may be used to characterize and analyze the structure of AOP networks to derive information that can guide research and regulatory decision-making. These analytical concepts are further developed and described by Villeneuve and others (2018, part II), including the use of techniques derived from graph theory (Trudeau 2013) and network science (Lewis 2009), to analyze network topology, the identification of critical paths and the characterization of interactions among AOPs in a network (Villeneuve and others 2018, part II). Finally, we present a number of application case studies that illustrate concepts underlying development and analysis of AOP networks, and how those concepts tie in with ultimate application. While not comprehensive in scope, the intent is to provide an enhanced understanding of AOP network development, AOP network analysis (Villeneuve and others 2018, part II), their applications, and to provide perspectives on how some of the challenges identified through the Horizon Scanning exercise (Lalone and others 2017a) can be addressed.
2. Development of AOP Networks
A first and relevant question is: What exactly is an AOP network? An AOP network is defined as an assembly of two or more AOPs that share one or more KEs, including specialized KEs such as molecular initiating events (MIEs) and adverse outcomes (AOs, Box 1). Different AOPs diverging from a single MIE, or converging to a single AO, therefore also form AOP networks even if they do not have any other KE in common. Development of individual AOPs can be thought of as the process of (1) graphically defining a sequence of KEs that link a molecular initiating event to a defined adverse outcome, (2) describing the change in state that each KE represents and how it is measured, and (3) detailing the weight of evidence that supports inference or extrapolation from one KE to the next in the sequence based on biological plausibility, empirical support, and quantitative understanding (Villeneuve and others 2014b). AOP networks can be thought of as emerging from the description of individual AOPs, as soon as KEs are described that are shared between two or more AOPs. The description of networked KEs can either be an intentional process that is part of the strategy of an AOP developer, or the fact that certain KEs are shared among AOPs can be discovered after AOPs have been developed independently. When considering different AOP network development processes, it is therefore useful to distinguish between “network-guided AOP development” and “AOP network derivation”. While AOP network derivation is defined as a formal AOP network development process that is based on extracting and linking information that is available in the AOP-Wiki, network-guided AOP development is introduced as a rather broadly defined concept that includes many different AOP network development approaches which do not necessarily rely on database extraction procedures.
Box 1.
Coming to terms with AOP networks
| AOP network | An assembly of two or more AOPs that share one or more KEs. |
| AOP network development | Broad term referring to the description or development of AOP networks, irrespective of the strategy employed. |
| Network-guided AOP development | AOP network development strategy involving the development of at least two individual AOPs containing one or more intentionally shared KEs. |
| AOP network derivation | AOP network development by manually or programmatically extracting AOPs relevant for a given application from the AOP-Wiki. |
| AOP network analytics | Broad term referring to the analysis of AOP networks to reveal, identify or investigate specific network properties, such as topological features, critical paths, or interactions between AOPs. |
| AOP network filter | AOP network development tool to refine which KEs and KERs from a given AOP network are included in downstream applications and analysis based on specified filter criteria. |
| AOP network layer | Graphical AOP network visualization tool to overlay a given AOP network with additional data such as feedback loops to facilitate interpretation without overly complicating the underlying framework. |
| AOP network topology | The overall shape and structure of an AOP network, describing the way in which the constituent parts of the network (i.e. KEs and KERs) are interrelated or arranged. |
| Convergent topology | Topology in which KEs from two or more AOPs are directed towards a common KE or AO, representing a range of possible upstream causes. |
| Divergent topology | Topology in which two or more KERs branch off from a single MIE or KE, representing a range of possible downstream outcomes. |
| Mixed topology | Topology showing local divergent and convergent regions within the overall network, possibly featuring specific motifs such as bow tie motifs which could represent important points of biological integration. |
| Critical path | The path through an AOP network considered most significant from an investigational, biological or regulatory standpoint. A critical path does not necessarily correspond to a single AOP described in the AOP-KB. |
| Interaction between AOPs | One AOP affecting another AOP in such a way that it modulates the AO compared to the outcome that would be observed had the interaction not taken place. |
2.1. Network-guided AOP development
When AOPs are developed in the AOP-Wiki, an AOP network is created by default whenever a KE or KER description is linked to more than one AOP. This is important because it implies that AOP developers are not restricted to describing linear paths only, and can thus intentionally conceive and describe structures that are more complex than the typical “one perturbation – one outcome” unit. This process could be thought of as network-guided AOP development. The advantage of network-guided AOP development is that it is not conceptually and methodologically different from development and description of individual AOPs: the same principles, guidance, and practices in terms of description within the AOP-Wiki apply, and no additional tools are required. In order to develop an AOP network there is no need to do anything differently than one would for describing a linear AOP, other than to intentionally share KE or KER descriptions (pages) among more than one AOP, a functionality that is currently built into the AOP-Wiki.
Currently, many AOPs are being developed using this network-guided fashion (e.g., Angrish and others 2016; Cavallin and others 2017; LaLone and others 2017b; Nelson and others 2016; Stinckens and others 2016). However, it is expected that as the AOP-KB matures, AOP development will increasingly focus on filling data and knowledge gaps in the AOP-Wiki, and AOP network development has the potential to mainly become an exercise of assembling data that already exists in the AOP-KB. The process of developing AOP networks by extracting existing data from the AOP-Wiki and assembling a network based on those AOPs rather than de novo description of linked AOPs is called AOP network derivation.
2.2. AOP Network Derivation
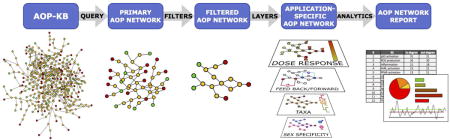
The first step of network derivation is to extract all AOPs that are relevant for a given application from the AOP-Wiki (Figure 1). The criteria that define which AOPs are relevant will vary, and will be defined by the application or stakeholder needs. Theoretically, the AOP-KB can be queried for any property of an AOP, KE, or KER that has been appropriately described and/or structurally annotated. Some examples of extraction criteria include: AOPs leading to a single AO of interest, AOPs known to be induced by a particular stressor or group of stressors, AOPs that have KEs that map to a particular data set (e.g., a collection of positive high-throughput screening assay responses observed for a particular chemical or mixture of chemicals), AOPs that have a particular species in their applicability domain, AOPs that have KEs for a particular tissue type, etc.
Figure 1.
Graphical representation of the AOP network derivation – refinement – analysis workflow. A primary AOP network is constructed by querying the AOP knowledgebase (AOP-KB). Filters are then applied to derive a filtered network containing AOPs of interest for a given application or research question. Layers can be added in a next step to add data relevant to the application. Finally, the AOP network can be analyzed to produce metrics related to the topology and other properties of the network.
Extraction can be achieved manually, for example by inspecting dedicated pages in the AOP-KB which list all the AOPs that a particular KE links to. However, manual extraction of AOP networks could rapidly become tedious as well as impractical as the AOP-KB grows. Thus, it is important to develop computational tools designed for this purpose, such as the AOP-xplorer (http://apps.cytoscape.org/search?q=aopxplorer). Using AOP-xplorer, any structured annotation field in the AOP-KB can be queried computationally in order to derive an AOP network. Once such an automated extraction process is complete, the resulting collection of AOPs could be assembled based on their topologies of shared KEs and KERs into an AOP network that is then called a primary AOP network (Figure 1). In some cases, the resulting primary network will be directly suitable for a certain application. In others, it may be desirable to refine (simplify and/or enrich) the network using a series of filters and data layer options, or to more deeply interrogate and statistically analyze the network as discussed below.
2.3. Refining AOP networks using filters
The structural complexity of AOP networks will depend on various factors. Ideally, AOP network derivation tools should include ways to focus and refine the network to fit the needs of a given application and enhance the information content conveyed from the overall network diagram. For example, risk assessment of individual chemicals or mixtures might be focused on a particular effect (e.g., impaired reproduction) in a specific class of organisms. In such a scenario, one might want to remove AOPs that relate to non-reproductive endpoints, and AOPs that are relevant to other taxa. On the other hand, efforts targeting mode of action identification could benefit from examining highly branched networks encompassing many different MIEs and their associated pathways. Thus, it was conceived that one should not only be able to construct a primary network based on extraction criteria, but also to filter that network based on additional annotation terms that would allow one to focus on the pathway(s) of greatest interest.
AOP network filters are envisioned to further refine which KEs and KERs from the primary AOP network would be included in downstream applications and analysis (Figure 1). For example, the structured KE and KER domain of applicability terms selected in the AOP-KB could be used to restrict a network only to those KEs and KERs that are relevant to a given life stage, thereby simplifying the overall network. Alternatively, one might want to filter an AOP network to only those KEs measured at a defined biological level of organization, in order to select appropriate endpoints one might measure in a specific cell line or tissue. A range of different filters, based on either structured ontology terms that are part of the AOP descriptions (e.g., taxonomic applicability) or based on network metrics (e.g., how strongly connected KEs are to the network) could be envisioned. Supplementary Table S1 provides a list of possible filters that could be envisioned, including filters for taxonomic, life stage or sex applicability, network metrics, and critical paths. Each could be used to help tailor an AOP network to a given problem formulation or research question. Finally, we propose a confidence assessment filter which can be used to filter AOP networks based on various weight of evidence, biological plausibility, essentiality, etc. assessments of the constituent AOPs.
2.4. Visualizing AOP network data using layers
A simplified representation of a set of KEs and KERs (i.e., an AOP network) can easily be visualized graphically, where each unique KE is represented by a single node, and the KERs are represented by edges (Figure 1). While such a simple graphical representation can depict the general structure of an AOP network, it is not a practical means of displaying and interrogating all the complex information captured within each of its KE and KER descriptions. Additionally, one may wish to supplement a network with additional data that is external to the AOP-KB (e.g., experimental data), which can further convolute the information associated with an AOP network. To aid in the visualization and interpretation of the complex information associated AOP networks, we propose a mechanism to visually superimpose this information, as needed, as “layers” on top of an AOP network image (Figure 1). AOP network layers can be viewed as analogous to data layers employed in geographic information systems (GIS), where information relevant to interpretation or application of an AOP network could be laid over the filtered AOP network, much like for example traffic or public transportation information is laid over a city map. Ideally these layers could capture data derived from structured annotation fields within the AOP-Wiki, as well as incorporate other types of data that are not necessarily part of formal AOP descriptions.
There has been resistance to the explicit representation of additional data such as feedback loops as additional types of nodes and edges in an AOP network, as they may overly complicate network interpretation for many applications. On the other hand, for other applications, those additional levels of detail may yield insights that may allow for more accurately predicting biologically relevant outcomes. Layers add information to an AOP network without modifying or influencing the network’s overall properties, structure and topology, and are viewed as a way to address competing desires for greater information richness and detail on one hand versus clear-cut interpretive simplicity on the other hand. The consideration of feedback loops and modulating factors within AOPs and AOP networks provide a useful example of this. At present, events associated with a feedback loop may be included as KEs in the AOP in cases where a feedback response is causally linked to the adverse outcome and is measureable. In other cases however, for example when understanding of the feedback loop may aid to predict how severely a particular KE must be perturbed in order to progress further along the pathway, knowledge of the feedback loop can be included in the “quantitative understanding of the linkage” section of the relevant KER pages (Lalone and others 2017a, see Q&A 13). Therefore, feedback, feedforward or other types of signaling motifs or loops are not specifically annotated as such in AOP descriptions, and thus very difficult to identify automatically. Likewise, modulating factors that are extrinsic to the AOP network (i.e., are not driven by interactions among existing KEs found in the network) such as dietary factors, genetic susceptibility or resistance, disease states, environmental factors, etc., are currently only captured in the free-text descriptions of quantitative understanding of the KERs. While potential intrinsic modulating factors are captured de facto in the structure of the network since they arise from a shared KE or KER and, therefore, do not need explicit annotation, extrinsic modulating factors require separate descriptions and anchoring to the AOP network.
Operationally (i.e., from the perspective of further development of the AOP-KB), the implementation of certain types of layers would involve introducing additional structured annotation fields (Ives and others 2017) in the KE and KER descriptions of the AOP-KB. In the case of known modulating factors, this could for example involve introducing an optional “modulating factor” field to KER descriptions, where users could define a modulating factor and provide additional text description and supporting references. An advanced implementation of feedback loop layers could allow future KEs also affecting the feedback loop to reveal interactions between AOPs that are not necessarily evident from individual KEs. However, even at the most basic level, the ability to apply a layer that identifies those KERs for which feedback or modulating factors are known to influence response-response relationships could be very informative and signal a user to explore the additional details provided in the AOP description in order to determine whether they are relevant to the application in question. While at present, these capabilities have not been implemented as computational features of the AOP-KB, the concepts and features outlined here have been communicated to the AOP-KB development team to inform on-going software development aimed at enhancing the utility of AOP-networks.
In addition to feedback loop layers and modulating factor layers, a number of other data layers were identified which could reflect taxonomic, life stage, and sex applicability domains, genetic heterogeneity, tissue specificity and temporality, as well as quantitative response data (Supplementary Table S2). We propose that in combination, the use of filters and layers will help to achieve a network representation that is suited for the intended application and will make the AOP-KB more user-friendly and useful for other intended audiences in addition to research scientists, such as risk assessors. Importantly, by overlaying certain data types on the KERs within an AOP network, the network representation can be transformed into a mathematical construct allowing for different types of analyses to be applied (see Figure 1; Villeneuve and others 2018, part II).
2.5. Analyzing AOP networks
An AOP network organizes sets of biological perturbations that may interact and influence one another in such a way that a significant understanding of the biology may be derived through examination and analysis of the structure of the network. While visual examination of the network graph is compelling, the use of techniques from graph theory (Trudeau 2013) and network science (Lewis 2009) facilitates an encompassing review of the network, especially when networks become larger and more complex. Villeneuve and others (2018, part II) address several aspects of AOP network analytics, building on the basic AOP network concepts described in this paper. They specifically focus on three key elements: (1) AOP network topology analysis, (2) critical path identification, and (3) characterization of interactions among AOPs in a network. Here, we provide a few topical examples of analytical procedures that may be applied to AOP networks to give the reader a brief introduction to some of the concepts involved. We refer to our companion paper (Villeneuve and others 2018, part II) for a complete overview and in-depth discussion.
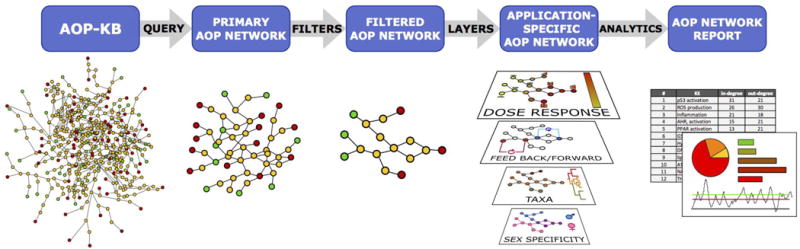
In AOP network topology analysis, a large variety of metrics can be calculated that describe the overall shape and structure of the network, or identify specific nodes in the network which may be of particular interest. For example, one of the first topological properties of interest are points of convergence and divergence within a given network (Figure 2A). In a convergent topology, AOPs are directed towards a common KE or AO, while a divergent topology involves AOPs branching off from a common MIE or KE. Conceptually, the degree of convergence or divergence of a network may affect the intensity of the AOs, and analysis of convergence/divergence of AOP networks may inform on the existence of potential additive, synergistic or antagonistic effects and interactions, or may for example be used to develop assays that would capture a broad range of MIEs, versus assays predictive of a group of related AOs, versus assays predictive of only a very specific AO. Most real-life AOP networks will likely be mixed networks (i.e. have local divergent and convergent regions within the overall network). This could lead to specific motifs, such as a node that is a local site of convergence and divergence simultaneously, a mixed structure which would create a bow tie motif (Figure 2A) and could represent important integrative biological signals. Computationally, a large number of metrics can be calculated to describe network topologies, each providing a specific view on the network, and complementary opportunities for identifying network nodes of interest. A few examples of such metrics are given in Figure 2B.
Figure 2.
Examples of AOP network analysis concepts and approaches. (A) Network topology analysis can reveal converging, diverging, or mixed patterns. A mixed pattern can take the shape of a bow tie motif. (B) Two different examples of network metrics calculated for the same hypothetical AOP network. The degree of a node (key event, KE) in the network is equal to the number of edges (key event relationships, KERs) connecting the node to the network and is one way of expressing how connected that node is to the network. The path occurrence is the number of times a node (KE) occurs in a path connecting a molecular initiation event (MIE) to an adverse outcome (AO) after evaluating all possible paths between the MIEs and AOs of the network. The path occurrence may be an indication of the relative importance of a node within the overall network.
A second and highly relevant characteristic of AOP networks is that they provide a framework for the description of the overall landscape of potential adverse outcomes resulting from particular biological perturbations. This can enable strategic identification of paths that have the greatest biological likelihood and/or relevance for risk assessment. Within an AOP network, the most significant path from an investigational or biological standpoint was termed the critical path. Here we distinguish “path” from “pathway” to recognize that the critical path may not necessarily follow an entire AOP, and may in fact emerge only through the assembly and consideration of the interactions between multiple AOPs. The interpretation of what constitutes a critical path can vary widely depending on the context and perspective of the AOP developed or end user. Critical paths may be representative of a specific research question, or of the strongest weight of evidence for certain elements of the network. They may also represent the most toxicologically relevant path which may have great importance in the application of AOP networks for risk assessment. This can in turn aid identification of endpoints or assays that can serve as useful alternatives to the direct measurement of apical adverse outcomes (OECD 2016). Also, AOP network-based critical path delineation efforts may be useful for identifying data gaps that are required to achieve a complete critical path description in scenarios where the AOP network includes poorly supported AOPs. Despite the fact that critical paths currently remain a relatively loosely defined concept and quantitative approaches (i.e., qAOP development) may be required in order to formulate a more stringent definition, Villeneuve and others (2018, part II) recognize the need for different critical path identification strategies, and distinguish among problem formulation, weight of evidence, and biologically-toxicologically defined critical paths, as well as the pure empirical identification of critical paths.
A third, and probably the most challenging, aspect of AOP network analysis is the identification and characterization of potential interactions between AOPs. AOP interactions describe how one or more components of a pathway may affect another pathway in such a way that it modulates the AO in terms of its biological properties, intensity, probability, rate, etc., compared to the outcome that would be observed had the interaction not taken place. Interactions between AOPs may be described as crosstalk between AOPs, but because the concept of crosstalk is typically associated with specific and rather stricly defined molecular processes such as signal transduction cascades, “interactions” is preferred as the descriptor. From a procedural perspective, since nodes in AOPs represent directional changes in the state of biological components (e.g., increased versus decreased testosterone concentration are two different KEs) rather than the biological components themselves (e.g., testosterone), it is recognized that tools to automatically map KEs occurring on the same components during AOP network extraction and analysis will be required before the full potential of interaction analysis is achieved. Nevertheless, interactions are anticipated to result in additive, synergistic, or antagonistic responses (Vert and Chory 2011) and their analysis may provide the opportunity to guide a more rational assessment of for example mixture toxicity (Villeneuve and others 2018, part II).
3. AOP network application: case studies
As described by Villeneuve and others (2014b), AOP networks were envisioned to be a more realistic representation of the complex biological interactions compared to single AOPs that would, for example, occur in response to exposures to chemical mixtures or single toxicants exhibiting multiple biological activities. Development and analysis of AOP networks have the potential to provide important information regarding the interactions among multiple AOPs, and represent an interface between the specific toxic outcome captured in a single AOP and modulation of those outcomes due to interactions occurring in a systems biology context. Additionally, analysis of the intersections (shared KEs and KERs) among AOPs that make up an AOP network can reveal unexpected or under-appreciated biological connections. Consequently, it is anticipated that AOP networks will ultimately be more informative than individual AOPs in a decision making context. For example, when mapping the landscape of AOPs for a particular adverse effect the network will indicate the points of convergence of different pathways, which may indicate the most promising KE for development of in vitro assays that can be tailored to capture all the pathways upstream from that KE. This approach may be very useful for informing the construction of Integrated Approaches to Testing and Assessment (IATAs) to cover the relevant biology for a wide range of potential adverse outcomes (Tollefsen and others 2014). AOP networks may also offer insights into approaches for evaluating the toxicity of mixtures to understand how a chemical acting via one AOP may be impacted by another chemical acting via another AOP in a relevant mixture.
While some of the most prominent potential applications of AOP networks are noted above, there are undoubtedly other applications which may emerge. For example, AOP networks could help speed the design of new drugs or chemicals by providing early warnings of potential side effects or toxicological events that could possibly end up in adverse effects. Likewise, mapping layers of information on modulating factors onto an AOP network could help to identify vulnerable subcategories of people or wildlife whose susceptibility may be increased or decreased as a function of health status, microelement deficiencies, environmental stresses etc. These could either exacerbate the adverse effect of a chemical, or equally undesirable, undermine or counteract the effect of a drug. Given the broad range of applications, it is impractical to illustrate them all. Thus, in the context of this paper, we highlight just a few application case studies that both illustrate some of the concepts of AOP network development and analysis described above, and show how those processes can be applied to help address questions related to chemical safety assessment.
3.1. Case Study 1: AOP network for cardio-metabolic disease
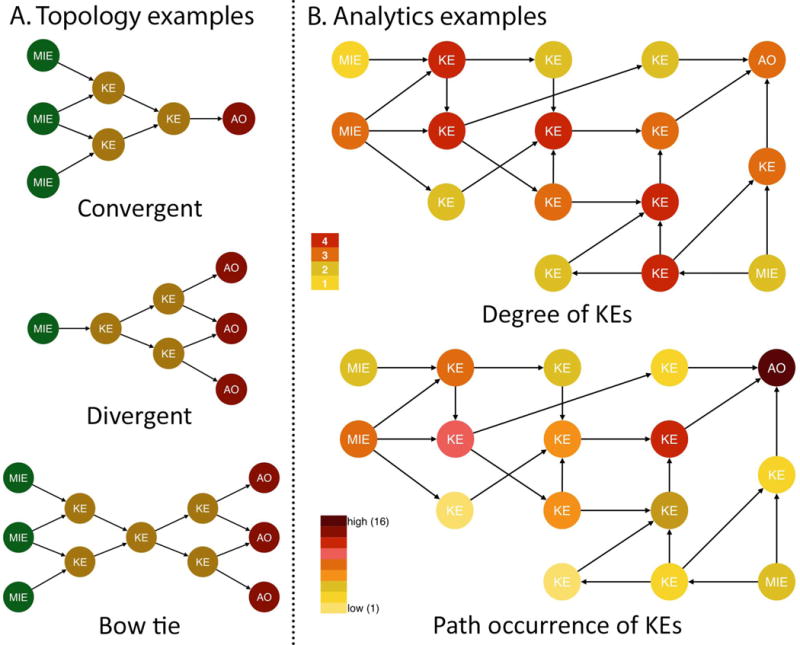
The need to develop AOP networks to effectively evaluate complex diseases was recently highlighted in the development of mechanistic toxicity tests based on an AOP network for hepatic steatosis, leveraging a large amount of publicly available mechanistic, phenotypic, and toxicological liver data (Angrish and others 2016; Angrish and others 2017; Bell and others 2016; Oki and others 2016). Steatosis, also known as fatty liver disease, is a regulatory endpoint and pathologic condition where energy metabolism is disrupted and fat accumulates in the liver. Energy homeostasis is dependent on the balance between energy intake and expenditure, a process regulated by endocrine and cellular communication between the brain, gut, and metabolic tissues such as adipose, striated (skeletal and cardiac) muscle, pancreas, and liver. At the molecular level, metabolism is coordinated by broad chemical signals, including nutrients, hormones, and environmental chemical signals that control systemic energy homeostasis by binding to cognate cell surface, cytosolic, and nuclear receptors. Chemical contact at any point along this neuro-endocrine-organ network can impact complex signal transduction, gene expression and protein activation cascades, etc. to coordinate the energy demands of a biological system. The challenge is that, because these receptors and signaling pathways crosstalk, it is difficult to adapt existing assay data (e.g., data from current ToxCast™ and Tox21 assays) to strategies predictive of a steatotic outcome. This may be because the events these assays represent are too far upstream of the AO to allow for facilitating reliable prediction of outcomes. Effectively, the interactions that occur in between are too complex to practically or reliably model.
In the steatosis AOP network this challenge was overcome by identifying a network topology converging into four KEs that were viewed as critical paths leading to steatosis (i.e., fatty acid uptake, efflux, synthesis, and oxidation, Figure 3). The assumption was that assays measuring these points of convergence would integrate the complex interplay of upstream events and translate them into KE measures or points of departure that are more proximally located relative to the AO. It is conceivable that such an approach would have the power to capture not only single chemical exposures, but also mixture effects, as long as the effects were upstream of the convergent KEs.
Figure 3.
AOP network for steatosis. The high level of crosstalk between the different receptors and associated signaling pathways complicates the use of existing high-throughput screening data as predictors of a steatotic outcome. This challenge was overcome by identifying a network topology converging into four key events (i.e., lipogenesis, and fatty acid uptake, efflux and oxidation) that were viewed as critical paths leading to steatosis. Assays measuring these points of convergence integrate the complex interplay of upstream events and translate them into measures that are more directly related to the adverse outcome. FA: fatty acid, TAG: triacylglycerol, PI3K: phosphatidylinositol-3-kinase, AKT: protein kinase B, PPAR: peroxisome proliferator-activated receptor, LXR: liver X receptor, CAR: constitutive androstane receptor, PXR: pregnane X receptor, FXR: farnesoid X receptor, RXR: retinoid X receptor.
Once the convergent KEs were identified and the corresponding assays were developed, a second step was to utilize data from those assays to predict steatotic outcomes as well as their severity. A challenge is that the compensatory actions of these 4 KEs collectively balance liver lipid levels. Consequently, progression towards a steatosis AO depends on the combination and magnitude of KEs’ change, and the interaction between all four KEs and their associated AOPs. While in some cases, only one of those four KEs may be impacted and that alone could be sufficient to elicit the adverse outcome, in most cases it is likely that more than one of the convergent KEs will be affected. This can be expected to yield different consequences than those that might be predicted based on impacts on any one of those KEs alone. For example, an exposure that increases lipid uptake may be sufficient to cause steatosis, whereas an alternative exposure that also activates lipid efflux may compensate for increased uptake and restore balance such that no AO is observed. This is a salient example of why the consideration of AOP networks has been viewed as critical to the use of the AOP framework for predictive toxicology. As such critical paths and points of convergence are identified, AOP network analyses can inform the development of complementary, biologically-based mathematical models that facilitate an alternatives-based (e.g., cell-based assays) chemical evaluation workflow.
3.2. Case study 2: Decreased serum thyroid hormone AOP network for alternative assay development
An example of network-guided AOP development that has led to de facto construction of an AOP network in the AOP-Wiki is centered around circulating thyroid hormone concentrations. Two major points of convergence/divergence (i.e., KEs resembling the “knot” of a bow tie motif, see section 2.4) in this multi-taxon AOP network are decreased serum thyroxine (T4) and decreased serum triiodothyronine (T3, see Figure 4A).
Figure 4.
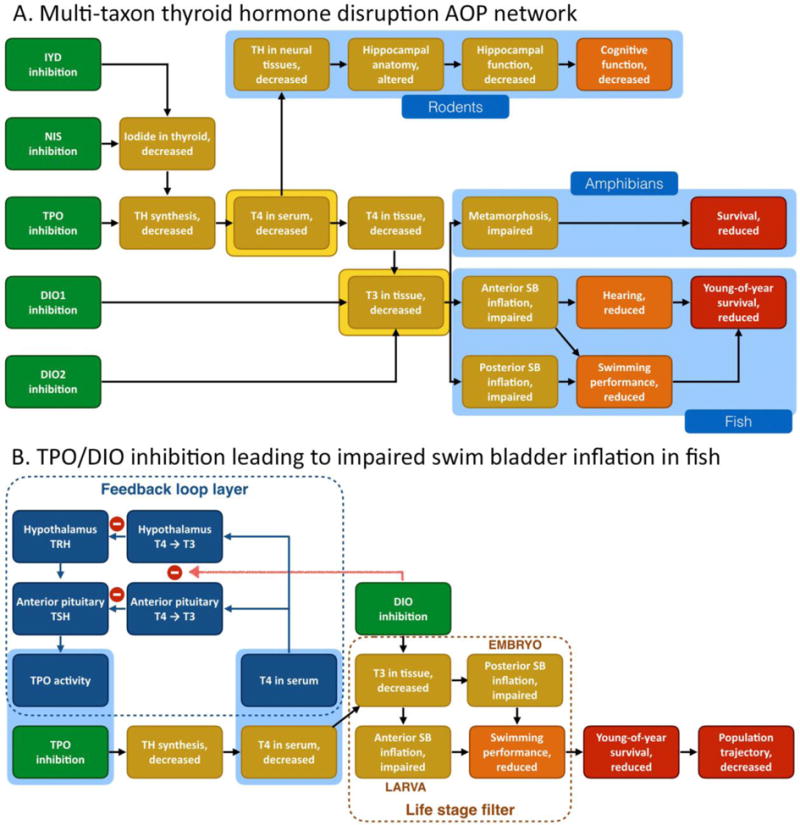
AOP networks related to disruption of the thyroid axis. (A) Multi-taxon thyroid hormone disruption AOP network including mammalian, amphibian and teleost endpoints. The blue regions illustrate how a taxonomic applicability layer may be used to add relevant data to the primary network representation. The key events highlighted in yellow indicate two major points of convergence/divergence in the network, resembling the “knot” of a bow tie motif. (B) Filtered thyroid AOP network only containing key events that are relevant to fish. The dashed brown area illustrates how additional filtering might be used to further refine the network, e.g. to only include key events that are relevant to specific life stages. The blue area illustrates the use of a layer to indicate the presence of a feedback loop acting on an AOP in the network, and the interaction between the feedback loop and one of the molecular initiating events in the network. IYD: iodotyrosine deiodinase, NIS: sodium-iodide symporter, TPO: thyroperoxidase, DIO: iodothyronine deiodinase, TH: thyroid hormone, T4: thyroxine, T3: triiodothyronine, TRH: thyrotropin-releasing hormone, TSH: thyroid stimulating hormone, thyrotropin, SB: swim bladder. Red negative sign: inhibition processes. Red arrow: DIO inhibition decreases conversion of T4 into T3, thereby inhibiting the feedback inhibition of T3 on TRH and TSH synthesis.
This thyroid hormone disruption AOP network has been employed to support the development and application of guideline toxicity tests, and subsequently alternatives to those same whole animal test guidelines. For example, the amphibian metamorphosis assay (AMA, OECD TG 231, OECD 2009) was developed for the purpose of screening chemicals for their ability to disrupt the thyroid hormone signaling axis in vertebrates. The branches in the AOP network provide the scientifically plausible and evidence-based foundation for linking the shared KE of decreased serum T4 to impaired amphibian metamorphosis as an indicator of thyroid axis disruption. Adverse neurodevelopmental outcomes in rodents build the case for the relevance of the AMA for screening thyroid disrupting chemicals that can be adverse to humans (Figure 4A). Given the time and resource-intensive nature of the AMA, there was desire to replace it with in vitro assays that could be used to screen large libraries of chemicals for their ability to disrupt the thyroid axis. Based on the AOP network, assays for thyroid peroxidase (TPO) activity, the sodium iodide symporter (NIS), and iodothyronine (DIO) and iodotyrosine (IYD) deiodinase activities were developed to assess the potential mechanisms through which chemicals could alter circulating T4 and/or tissue T3 concentrations (Figure 4A). Recognizing that not all these targets have been covered in existing high throughput screening programs (e.g., ToxCast™, Tox21), the AOP network helps inform the development of a more comprehensive screening battery for this important mode of endocrine disruption.
As part of another alternative testing development effort, a question was posed as to how the fish early life stage test (FELS test, OECD TG 210, OECD 2013b) might be replaced by more rapid and cost effective alternatives (Villeneuve and others 2014a). While a modified fish embryo test (OECD TG 236, OECD 2013c) was proposed as an alternative that could cover much of the toxicological space encompassed by the FELS test, it was recognized that certain developmental events occurring after hatch, during larval to juvenile transition, could be missed. One example was swim bladder inflation, which in common laboratory model cyprinids like zebrafish and fathead minnow occurs in two stages: inflation of the posterior chamber shortly after hatch, followed by inflation of the anterior chamber several days to weeks later (Cavallin and others 2017; Nelson and others 2016; Stinckens and others 2016; Villeneuve and others 2014a). While a range of biological perturbations may disrupt this event, decreases in circulating T4 and/or deiodination of T4 to T3 have been defined, through development of an AOP network, as a means through which chemicals could impact swim bladder inflation in fish, a KE which has been linked to reduced young of year survival (Czesny and others 2005; Woolley and Qin 2010). The AOP network focused on swim bladder inflation in fish was subsequently integrated with the broader amphibian/mammalian AOP network described above, resulting in a multi-taxon thyroid AOP network (see Figure 4). Consequently, the same battery of in vitro assays that can plausibly screen for thyroid disrupting chemicals in amphibian and mammalian models, could also cover toxicological space that might be missed if a fish embryo test were employed as the only alternative to a FELS test.
From a network development perspective, the thyroid AOP network demonstrates how some of the proposed filters and layers might be applied (Figure 4B). For example, application of a life-stage filter would show that the AOP mediated via inhibition of the TPO enzyme is only relevant to larval fish. If the exposure was during the embryo stage only and the focus was inflation of the posterior chamber, DIO enzyme inhibition would represent the critical path in the network. Alternatively, if the exposure occurred or was sustained until after hatch, both TPO and DIO inhibition would be inferred to be contributing to reduced anterior swim bladder inflation, suggesting that the outcome may be more severe than that triggered by a chemical exhibiting only one of the two bioactivities. Further, invoking the feedback loop layer in the AOP network visualization could unveil additional detail relevant to predicting the interactive effect of these two AOPs, since the MIE of DIO inhibition also impacts the negative feedback loop mechanism itself. Adding the quantitative properties of this feedback mechanism to the response-response relationship of the KER linking decreased T4 levels to reduced anterior swim bladder inflation might provide for a more accurate prediction of the joint effect of the two AOPs than the basic AOP network alone would provide.
3.3. Additional case studies
Two additional, fully described case studies are given in the Supplementary Materials to provide further examples to the interested reader illustrating AOP network development and application in more advanced scenarios. The first case study illustrates the application of AOP networks to support the assessment of complex mixtures. A water sample extract of a metropolitan wastewater treatment plant was tested using a number of ToxCast™ assays to evaluate the ability of the sample to activate different nuclear receptors and transcription factor promoter-regulated reporter sequences. Assay activity was mapped to MIEs described in the AOP-Wiki and the resulting AOP network was filtered to focus on KEs that were directly relevant to the observed bioactivities. The resulting set of AOP networks was further filtered to exclude AOPs that did not terminate at AOs that would be considered relevant to ecological risk assessment. Focusing on the remaining AOPs, known potential hazards to aquatic vertebrate wildlife associated with this mixture could be identified. The second case study provides an example of how an AOP network approach was used to explore the polypharmacological profile of the pharmaceutical beclomethasone dipropionate (BDP) using the fathead minnow. Thanks to its ability to modulate the glucocorticoid receptor BDP is used to treat chronic inflammatory conditions, but the drug also has the ability to modulate the androgen and progesterone receptors. Data generated during drug development were used to identify the cascades of KEs likely to be triggered and this information was organized within an AOP network. Chronic in vivo exposures to BDP were then carried out to generate a quantitative AOP network which provided evidence that the polypharmacology profile of the BDP was indeed critically important to interpret and accurately predict the toxicological profile of the drug (Margiotta-Casaluci and others 2016).
4. Summary and conclusions
Based on the results of a SETAC-sponsored Horizon Scanning exercise focused on advancing the AOP framework, the development of guidance and best practices related to AOP network derivation and application was identified as a critical need. This not only included questions and concerns focusing directly on AOP networks, but also on different related topics such as mixture toxicity assessment, the implementation and graphical representation of feedback loops within the AOP framework, the characterization of interactions among pathways, the ability to include information on extrinsic modulating factors, etc. While the concept of constructing networks has always been deliberately, but possibly rather implicitly, built into the AOP framework (Villeneuve and others 2014b; 2014c), the number of available AOPs has only recently reached a level sufficient to begin developing AOP networks. Recognizing different needs and strategies for developing AOP networks, we distinguish between network-guided AOP development and AOP network derivation based on the AOP-KB. We then propose the use of filters and layers to simplify visualization and interpretation of AOP networks, and to tailor them to suit the needs of a given research question or application. AOP networks can subsequently be analyzed in a variety of ways to extract useful information, including topological analyses, critical path identification and characterization of interactions among AOPs within a network. The concepts described in this paper, and in its companion paper focused on AOP network analytics, are intended to serve as a starting point for further development of the AOP network concept and of the AOP-KB to increase its capabilities for managing and analyzing AOP networks, but also to catalyze AOP network development and application by the different stakeholder communities. Along with other manuscripts produced as a result of the April 2017 SETAC Pellston workshop on Advancing the AOP Framework (Lalone and others 2017a), we hope to serve the ongoing development of the AOP framework in general as a critical concept to support 21st century approaches to toxicological research and regulation.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Society of Environmental Toxicology and Chemistry (SETAC) North America staff, in particular Greg Schiefer, Nikki Mayo, and Tamar Schlekat who provided support to the workshop co-chairs, steering committee, and workshop participants before, during, and after the Pellston workshop. We appreciate the funding support from the Society of Environmental Toxicology and Chemistry, United States Environmental Protection Agency, American Cleaning Institute, Cefic-LRI (European Chemical Industry Council Long-Range Research Initiative), Chevron-Environmental, ECETOC (European Center for Ecotoxicology and Toxicology of Chemicals), European Commission Joint Research Centre, European Crop Protection, ExxonMobil, Humane Society International, The Humane Society of the United States, Human Toxicology Project Consortium, Syngenta, and Unilever. In addition, we thank the groups from academia, industry, and government who supported participants’ travel. The authors thank the workshop co-chairs, Dr. Carlie LaLone and Dr. Markus Hecker for their coordination, organization, and guidance of the workshop. We acknowledge the other workshop participants for their stimulating discussions and feedback, and the respondents to the Horizon Scanning effort for the charge questions and themes that informed our discussion. Contents of this paper represent the personal opinions of the authors and neither constitute, nor necessarily reflect the policies or viewpoints of their employers or institutes.
References
- Angrish MM, Kaiser JP, McQueen CA, Chorley BN. Tipping the Balance: Hepatotoxicity and the 4 Apical Key Events of Hepatic Steatosis. Toxicol Sci. 2016;150(2):261–268. doi: 10.1093/toxsci/kfw018. [DOI] [PubMed] [Google Scholar]
- Angrish MM, McQueen CA, Cohen-Hubal E, Rooney JP, Bruno M, Ge Y, Chorley BN. Mechanistic Toxicity Tests Based on an Adverse Outcome Pathway Network for Hepatic Steatosis. Toxicol Sci. 2017;159(1):159–169. doi: 10.1093/toxsci/kfx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29(3):730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Bell SM, Angrish MM, Wood CE, Edwards SW. Integrating Publicly Available Data to Generate Computationally Predicted Adverse Outcome Pathways for Fatty Liver. Toxicol Sci. 2016;150(2):510–20. doi: 10.1093/toxsci/kfw017. [DOI] [PubMed] [Google Scholar]
- Cavallin JE, Ankley GT, Blackwell BR, Blanksma CA, Fay KA, Jensen KM, Kahl MD, Knapen D, Kosian PA, Poole ST, Randolph EC, Schroeder AL, Vergauwen L4, Villeneuve DL. Impaired swim bladder inflation in early life stage fathead minnows exposed to a deiodinase inhibitor, iopanoic acid. Environ Toxicol Chem. 2017;36(11):2942–2952. doi: 10.1002/etc.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesny SJ, Graeb BDS, Dettmers JM. Ecological consequences of swim bladder noninflation for larval yellow perch. Trans Am Fish Soc. 2005;134(4):1011–1020. [Google Scholar]
- Ives C, Campia I, Wang RL, Wittwehr C, Edwards SW. Creating a structured AOP knowledgebase via ontology-based annotations. Applied in vitro toxicology. 2017;3(4):298–311. doi: 10.1089/aivt.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D, Vergauwen L, Villeneuve DL, Ankley GT. The potential of AOP networks for reproductive and developmental toxicity assay development. Reprod Toxicol. 2015;56:52–55. doi: 10.1016/j.reprotox.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Lalone CA, Ankley GT, Belanger SE, Embry MR, Hodges G, Knapen D, Munn S, Perkins EJ, Rudd MA, Villeneuve DL, Whelan M, Willett C, Zhang X, Hecker M. Advancing the adverse outcome pathway framework - an international horizon scanning approach. Environ Toxicol Chem. 2017a;36(6):1411–1421. doi: 10.1002/etc.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Wu-Smart J, Milsk RY, Sappington K, Garber KV, Housenger J, Ankley GT. Weight of evidence evaluation of a network of adverse outcome pathways linking activation of the nicotinic acetylcholine receptor in honey bees to colony death. Sci Total Environ. 2017b;584:751–775. doi: 10.1016/j.scitotenv.2017.01.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TG. Network Science: Theory and Applications. Wiley; 2009. [Google Scholar]
- Margiotta-Casaluci L, Owen SF, Huerta B, Rodriguez-Mozaz S, Kugathas S, Barcelo D, Rand-Weaver M, Sumpter JP. Internal exposure dynamics drive the Adverse Outcome Pathways of synthetic glucocorticoids in fish. Sci Rep 6. 2016;6:21978. doi: 10.1038/srep21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KR, Schroeder AL, Ankley GT, Blackwell BR, Blanksma C, Degitz SJ, Flynn KM, Jensen KM, Johnson RD, Kahl MD, Knapen D, Kosian PA, Milsk RY, Randolph EC, Saari T, Stinckens E, Vergauwen L, Villeneuve DL. Impaired anterior swim bladder inflation following exposure to the thyroid peroxidase inhibitor 2-mercaptobenzothiazole part I: Fathead minnow. Aquat Toxicol. 2016;173:192–203. doi: 10.1016/j.aquatox.2015.12.024. [DOI] [PubMed] [Google Scholar]
- OECD. OECD guidelines for the testing of chemicals. No 231: Amphibian Metamorphosis Assay 2009 [Google Scholar]
- OECD. (Series on Testing and Assessment No. 184).Guidance on developing and assessing adverse outcome pathways. 2013a [Google Scholar]
- OECD. OECD guidelines for the testing of chemicals. No 210: Fish, Early-life Stage Toxicity Test 2013b [Google Scholar]
- OECD. OECD guidelines for the testing of chemicals. No 236: Fish Embryo Acute Toxicity (FET) Test 2013c [Google Scholar]
- OECD. Users’ handbook supplement to the guidance document for developing and assessing AOPs. [ http://dx.doi.org/10.1787/5jlv1m9d1g32-en]
- OECD. (Series on Testing & Assessment No. 260).Guidance document for the use of adverse outcome pathways in developing integrated approaches to testing and assessment (IATA) 2016 [Google Scholar]
- Oki NO, Nelms MD, Bell SM, Mortensen HM, Edwards SW. Accelerating Adverse Outcome Pathway Development Using Publicly Available Data Sources. Curr Environ Health Rep. 2016;3(1):53–63. doi: 10.1007/s40572-016-0079-y. [DOI] [PubMed] [Google Scholar]
- Stinckens E, Vergauwen L, Schroeder AL, Maho W, Blackwell BR, Witters H, Blust R, Ankley GT, Covaci A, Villeneuve DL, Knapen D. Impaired anterior swim bladder inflation following exposure to the thyroid peroxidase inhibitor 2-mercaptobenzothiazole part II: Zebrafish. Aquat Toxicol. 2016;173:204–217. doi: 10.1016/j.aquatox.2015.12.023. [DOI] [PubMed] [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, Edwards SW, de Knecht J, Crofton K, Garcia-Reyero N, Hartung T, Worth A, Patlewicz G. Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA) Regul Toxicol Pharmacol. 2014;70(3):629–640. doi: 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Trudeau RJ. Introduction to Graph Theory. Dover Publications; 2013. [Google Scholar]
- Vert G, Chory J. Crosstalk in Cellular Signaling: Background Noise or the Real Thing? Dev Cell. 2011;21(6):985–91. doi: 10.1016/j.devcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve D, Volz DC, Embry MR, Ankley GT, Belanger SE, Leonard M, Schirmer K, Tanguay R, Truong L, Wehmas L. Investigating alternatives to the fish early-life stage test: a strategy for discovering and annotating adverse outcome pathways for early fish development. Environ Toxicol Chem. 2014a;33(1):158–169. doi: 10.1002/etc.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Angrish MM, Fortin MC, Katsiadaki I, Leonard M, Margiotta-Casaluci L, Munn S, O’Brien JM, Pollesch NL, Smith LC, Zhang X, Knapen D. Adverse Outcome Pathway Networks II: Network Analytics. Environ Toxicol Chem. 2018 doi: 10.1002/etc.4124. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, Whelan M. Adverse Outcome Pathway (AOP) Development I: Strategies and Principles. Toxicol Sci. 2014b;142(2):312–320. doi: 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, Whelan M. Adverse Outcome Pathway Development II: Best Practices. Toxicol Sci. 2014c;142(2):321–330. doi: 10.1093/toxsci/kfu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley LD, Qin JG. Swimbladder inflation and its implication to the culture of marine finfish larvae. Rev Aquacult. 2010;2(4):181–190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


