Abstract
Background and Objectives:
Integrons play a major role in the transmission and accumulation of resistance factors in multidrug resistant bacteria. This study was aimed to evaluate the gene cassettes of class I integron and antimicrobial resistance in isolates of Citrobacter with multidrug resistance (MDR).
Materials and Methods:
Ninety isolates of Citrobacter spp. were collected from the largest hospital in Kermanshah, Iran. Antimicrobial resistance patterns were determined using disc diffusion method. The class I integron were detected by PCR. The integrase positive isolates were further analyzed for the presence of gene cassettes using 5′ and 3′ conserved sequences (CSs) primers and PCR products were sequenced. The data were analyzed using the chi-square test.
Results:
Of 90 Citrobacter isolates, 46 (51.1%) were multidrug resistant. Class I integron and gene cassettes were determined in 30 isolates (65.2%). Gene cassettes were found which contained genes encoded resistance to aminoglycosides and trimethoprim and a putative gene. Gene cassettes of dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1 and dfrA15-aadA2 were also found in Citrobacter isolates.
Conclusion:
Our results indicate there is a high frequency of class I integron among multi-drug resistant strains of Citrobacter isolated from clinical settings. A high frequency of class I integron associated gene cassettes, in particular dfr and aadA, present in MDR strains of Citrobacter. This data indicates an important role of integrons in the creation and transmission of MDR strains in health care centers.
Keywords: Citrobacter, Gene cassettes, Integrons, Multidrug-resistant
INTRODUCTION
Citrobacter species have been reported as nosocomial pathogens with multidrug resistance (MDR) in many countries since recent decades (1). Citrobacter species have involved in various human infections, in particular, urinary tract infections, wound infections and respiratory infections (2). The antibiotic resistance of this group of bacteria have increased and MDR isolates have frequently been reported (2). The horizontal gene transferring plays the main role in the spread of antibiotic resistance genes and subsequently the rapid emergence of antibiotic resistance among Enterobacteriaceae (3). The mobile genetic elements such as plasmids, transposons and integrons are main factor for horizontal spreading of resistance genes (3). Integrons are conserved DNA sequences, which can efficiently acquire and transfer the resistant genes among bacteria and usually located on mobile genetics elements (4). There are several different classes of integrons, each encodes a distinct integrase gene (4). Class I integron is the most common type presented in clinical isolates of the Enterobacteriaceae (5). It is capable to carry single or multiple gene cassettes, which confer resistance to various antibiotics including, aminoglycoside, β-lactams, chloramphenicol, quinolones and trimethoprim (6). Class I integron has two conserved segments; 5′-CS and 3′-CS, separated by a variable region, included the integrated gene cassettes (7). The 5′-CS encodes integrase, located next to the recombination site (att1) recognized by the integrase and the promoter (P) which controls the transcription of integrated gene cassettes (7). The 3′-CS usually includes truncated qacE (qacED1) and sul1 genes that confer resistance to quaternary ammonium compounds and sulfonamides, respectively (7). Recombination between the attl of integron and attC sites of gene cassettes leads to the insertion of gene cassettes downstream to the resident promoter mediated by integrase (7). Integrase is a member of the tyrosine site-specific recombinase family that catalyze the excision and integration of DNA fragments, including gene cassettes (7). Near two hundreds of different cassette arrays have been identified that are flanked by the 5′-CS and 3′-CS endes (6). A strong association of integrons associated gene cassettes with MDR isolates of Enterobacteriaceae has been found (8, 9). Gene cassettes encode resistance to various antimicrobial agents, including dihydrofolate reductases (dfr), chloramphenicol acetyl-transferases (cat, cml), β-lactamases (bla), aminoglycoside-modifying enzymes (aac, aad, aphA) and ADP-ribosyl transfer-ases (arr) have been frequently identified within integrons (8, 9). This study aimed to evaluate the gene cassettes of class I integron-associated antimicrobial resistance in isolates of Citrobacter with multidrug resistance (MDR).
MATERIALS AND METHODS
Bacterial isolates.
In this descriptive study, 288 different clinical samples (e.g., wound, blood, urine, stool, and other samples) from patients admitted in the largest hospital in Kermanshah were collected during 2014–2015. Using the bacteriological and API20E Kit (bio-Merieux, France) testing, 90 Citrobacter isolates were confirmed.
Antibiotic susceptibility testing.
Antimicrobial susceptibility testing for 16 antibiotics was carried out using the disk diffusion method as recommended by Clinical and Laboratory Standard Institute (CLSI) (10). The antibiotic discs were ampicillin (10μg), cefotaxime (30μg), cefpodoxime (10μg), ceftazidime (30μg), ceftriaxone (30μg), tobramycin (10μg), gentamicin (20μg), ciprofloxacin (5μg), tazobactam/ piperacillin (10μg), cefazolin (30μg), cotrimoxazole (25μg), imipenem (10μg), aztreonam (30μg), ertapenem (10 μg), meropenem (10 μg) and streptomycin (10 μg) (MAST, England). The Escherichia coli ATCC 25922 was used as a control. MDR was defined as resistance to at least one antibiotic in three or more classes of antibiotics (6).
Polymerase chain reaction-detection of class I integron.
The presence of class I integron was screened by PCR using intIF and intIR primers (SinaColon, Iran) (Table 1). Each single reaction mixture (25 μl) contained 2μl of DNA suspension, 10 pmol of each primer, 2x GoTaq Green Master Mixture (SinaColon, Iran). The PCR conditions were as follows; 94°C for 5 minutes, followed by 35 cycles at 94°C for 45 seconds, 55°C for 45 seconds, 72°C for 45 minutes and final extension at 72°C for 5 minutes.
Table 1.
Oligonucleotide primers used
Detection of the variable region of class I integrons.
PCR was performed with class I integrase positive isolates using two primers 3′CS and 5′CS (SinaColon, Iran) (Table 1) to amplify the variable region of integron. Each single reaction mixture (25μl) contained 2 μl of DNA suspension, 10 pmol of each primer, 2 × GoTaq Green Master mixtures. PCR reactions began with 5 min of primary denaturation at 94°C followed by 35 cycles of 94°C for 1 min, 58°C for 1 min and finally 72°C for 1 min. The final extension was performed at 72°C for 10 min. After electrophoresis of PCR products on 1% agarose gel (Merck Co, Germany) and staining with ethidium bromide, the gels were visualized by Gel-Documentation apparatus (Bio Rad, USA).
DNA sequence analysis.
A number of PCR products with sharp bands were cut and purified using the QIA quick PCR purification Kit (QIAGEN, Germany) followed by sequencing. The DNA sequences were performed using an ABI 3730XL DNA analyzer (Macrogen Inc., Korea). Sequences were analyzed using BLAST search (http://www.ncbi.nlm.nih.gov/BLAST).
Statistical analysis.
Data were recorded and entered into an Excel file. Statistical analyses were performed using SPSS software (Version 20). Variables were analyzed by the Chi-square test. A p-value of < 0.05 was set as the statistical significance of all analyses.
RESULTS
Clinical data.
All 90 isolates were from hospitalized patients and confirmed by API20E Kit. They included 77 (85.5%) and 13 (14.4%) C. freundii and C. koseri, respectively. The clinical samples were included urine (n=46, 51.1%), blood (n=18, 20%), stool (n=11, 12.2%), Respiratory tract secretions (n=11, 12.2%) and wound (n=4, 4.5%). The patients were 49 females (54.4%) and 41 males (45.6%) with the average age of 41.6±26-year-old.
Antibiotic susceptibility results.
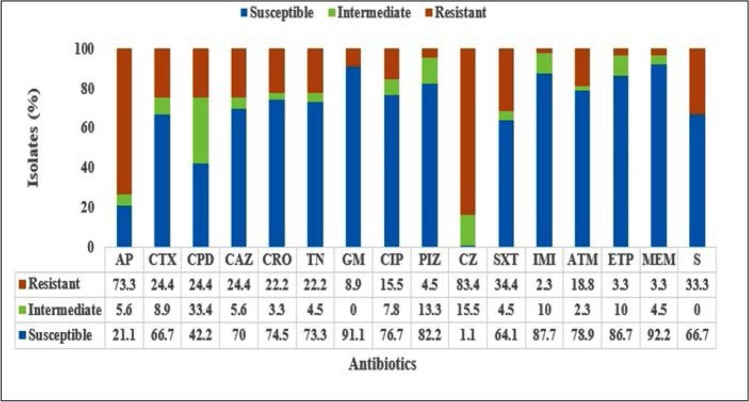
Results showed high-level of resistance to cefazolin (83.4%) and ampicillin (73.3%). Resistance rates for imipenem (2.3%), ertapenem (3.3%), meropenem (3.3%) and tazobactam/piperacillin (4.5%) was lower (Fig. 1.) Forty six isolates (51.1%) showed MDR phenotype. MDR isolates were mainly isolated from urine (43.5%), blood (21.7%), respiratory tract secretions (17.4%), stool (10.9%) and wound (6.5%).
Fig. 1.
Antimicrobial susceptibility patterns of 90 Citrobacter isolates.
SXT: Trimethoprim-sulfamethoxazole, CIP: Ciprofloxacin, TN: Tobramycin, GM: Gentamicin, CRO: Ceftriaxone, CTX: Cefotaxime, CAZ: Ceftazidime, ATM: Aztreonam, AP: Ampicilin, S: Streptomycin, CZ: Cefazolin, CPD: Cefpodoxime, ETP: Ertapenem, MEM: Meropenem, PTZ: Tazobactam piperacillin, IMI: Imipeneme.
Prevalence of class 1 integrons and gene cassettes.
Of the 46 isolates with multidrug resistance, class I integron was detected in 30 (65.2%) isolates. Following PCR amplification of variable region of class 1 integrons, all isolates with class I integron contained gene cassettes with different sizes (500, 600, 700, 750, 1000, 1600 and 1700) (Table 2). The cassettes were in the form of 8 electrophoretic patterns which include three patterns with single band (750, 700, 1600 bp), two patterns with double bands (1600-1000, 1600-1700 bp), one pattern with four bands (1700-1600-1000-750 bp), one pattern with five bands (1700-1600-1000-600-500 bp) and one pattern with six bands (1700-1600-1000-750-700-500 bp) (Table 2). Sequence analysis revealed the different variants of aadA (aadA1, aadA2) and dfrA (dfrA1, dfrA12, dfrA15) gene cassettes. The sequence analysis also showed the 1700 bp fragment with 98% homology to dfrA12-orfF-aadA2 gene, the 1600 bp fragment with 100% identical to dfrA1-aadA1 gene, the 750 bp gene cassette fragment with 100% identical to dfrA15-aadA1 gene and the 1000 bp fragment was 99% homology to aadA1 gene.
Table 2.
Characterization of Class1 integrons and gene cassettes in 30 MDR isolates of integron-carrying Citrobacter.
| Isolates | Source | Hospital Ward | Antibiotic resistance profile | intI | Size of gene cassettes (bp) | Gene Cassettes |
|---|---|---|---|---|---|---|
| S1 | Urine | Surgery | AP/CTX/CPD/CAZ/CRO/CIP/CZ/ATM/S/ SXT | + | 750 | dfrA15-aadA1 |
| S75 | Urine | Surgery | AP/CIP/SXT/S | + | 750 | dfrA15-aadA1 |
| S2 | Urine | Internal | AP/CTX/CPD/CAZ/CRO/CZ/ATM/S/SXT/TN/GM | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S6 | Urine | Emergency | AP/CZ/SXT/S | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S14 | Urine | Internal ICU | AP/CTX/CPD/CAZ/CRO/TN/CIP/CZ/ATM/S/SXT | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S17 | Urine | Surgery | AP/CAZ/CZ | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S26 | Urine | Surgery | AP/CTX/CRO/CZ | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S84 | Respiratory secretions | Infectious | AP/CTX/CPD/CAZ/CRO/PTZ/CZ/SXT/S | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S86 | Blood | Pediatric | AP/CTX/CZ/S | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S94 | Stool | Infectious | AP/CTX/CPD/CAZ/ TN/ CZ | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S97 | Urine | Surgery | AP/TN/CZ | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S99 | Urine | Pediatric | AP/CZ/S | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S100 | Urine | Surgery | AP/CAZ/CZ/S | + | 1600, 1000 | dfrA1-aadA1, aadA1 |
| S8 | Urine | Surgery | AP/CTX/CPD/CAZ/CRO/CIP/CZ/ATM | + | 700 | Hypothetical protein |
| S10 | Urine | Emergency | AP/CTX/CAZ/CPD/CRO/CZ/SXT/S | + | 1600 | dfrA1-aadA1 |
| S32 | Stool | Internal | AP/CPD/CZ | + | 1600 | dfrA1-aadA1 |
| S42 | Respiratory secretions | Surgery | AP/CTX/CAZ/CPD/CRO/TN/GM/IMI/CZ/ S | + | 1600 | dfrA1-aadA1 |
| S47 | Respiratory secretions | Infectious | AP/CTX/CAZ/CPD/CRO/TN/GM/IMI/CZ/SXT/S | + | 1600 | dfrA1-aadA1 |
| S60 | Urine | Surgery ICU | CRO/CZ/SXT/S | + | 1600 | dfrA1-aadA1 |
| S67 | Wound | Surgery | AP/CTX/CRO/CIP/PTZ/IMI/ATM/MEM/SXT/S | + | 1600 | dfrA1-aadA1 |
| S46 | Respiratory secretions | NICU | AP/CZ/SXT/S | + | 1700, 1600, 1000, 750 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, dfrA15-aadA1 |
| S92 | Stool | Infectious | AP/TN/CZ/SXT/S | + | 1700, 1600, 1000, 750 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, dfrA15-aadA1 |
| S54 | Urine | Surgery | AP/CZ/SXT/SXT/S | + | 1700, 1600, 1000, 600, 500 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, Hypothetical protein |
| S88 | Blood | Infectious | AP/TN/CZ/SXT/S | + | 1700, 1600, 1000, 600, 500 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, Hypothetical protein |
| S91 | Blood | Surgery | AP/CTX/CPD/TN/CZ/SXT/S | + | 1700, 1600, 1000, 600, 500 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, Hypothetical protein |
| S73 | Urine | Infectious | AP/CRO/CZ/SXT/S | + | 1700, 1600 | dfrA12-orfF-aadA2, dfrA1-aadA1 |
| S85 | Urine | Surgery | AP/CPD/TN/CZ/SXT | + | 1700, 1600 | dfrA12-orfF-aadA2, dfrA1-aadA1 |
| S79 | Stool | Surgery | AP/CPD/CZSXT/S/SXT | + | 1700, 1600, 1000, 750, 700, 500 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, dfrA15-aadA1, Hypothetical protein |
| S80 | Urine | Surgery | AP/CPD/CAZ/CIP/CZ/SXT/S | + | 1700, 1600, 1000, 750, 700, 500 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, dfrA15-aadA1, Hypothetical protein |
| S89 | Blood | Pediatric | AP/CPD/CZ/S | + | 1700, 1600, 1000, 750, 700, 500 | dfrA12-orfF-aadA2, dfrA1-aadA1, aadA1, dfrA15-aadA1, Hypothetical protein |
SXT: Trimethoprim-sulfamethoxazole, CIP: Ciprofloxacin, TN: Tobramycin, GM: Gentamicin, CRO: Ceftriaxone, CTX: Cefotaxime, CAZ: Ceftazidime, ATM: Aztreonam, AP: Ampicilin, S: Streptomycin, CZ: Cefazolin, CPD: Cefpodoxime.
Gene cassettes of dfrA1-aadA1, aadA1, dfrA12-orfF-aadA2 and dfrA15-aadA2 were found in 26 (56.5%), 20 (43.5%), 10 (21.7%) and 7 (15.2%) isolates, respectively. The 1000 and 1600 bp fragments, which contained dfrA1-aadA1, aadA1, were the most frequents. The nucleotide sequences of gene cassettes reported in this study have been submitted to GenBank under accession numbers MF589545 for aadA and MF589546 and MF589547 for dfrA. The relationship between class I integron with resistance to 16 antibiotics was statistically analyzed (Table 3). Isolates contained class I integron showed significantly higher resistance to ciprofloxacin (p= 0.002), streptomycin (p=0.004) and Cotrimoxazole (p=0.041).
Table 3.
Relatitionship of class I integron and antibiotic resistance among 46 MDR Citrobacter isolates.
| Antibiotics | Integron-positive isolates | Integron-negative isolates | p value | ||||
|---|---|---|---|---|---|---|---|
| R (%) | S (%) | I (%) | R (%) | S (%) | I (%) | ||
| Aztreonam | 15 (32.6) | 14 (30.4) | 1 (2.2) | 4 (8.7) | 11 (23.9) | 1 (2.2) | 0.257 |
| Imipenem | 2 (4.3) | 24 (52.2) | 4 (8.7) | 0 (0) | 13 (28.3) | 3 (6.5) | 0.53 |
| Cefazolin | 26 (56.2) | 0 (0) | 4 (8.7) | 16 (34.8) | 0 (0) | 0 (0) | 0.126 |
| Ceftazidime | 14 (30.4) | 12 (26) | 4 (8.7) | 8 (17.4) | 8 (17.4) | 0 (0) | 0.302 |
| Cefpodoxime | 15 (32.6) | 10 (21.7) | 5 (10.9) | 6 (13) | 7 (15.2) | 3 (6.5) | 0.708 |
| Tazobactam-Piperacillin | 6 (13) | 17 (36.9) | 7 (15.2) | 0 (0) | 12 (26) | 4 (8.7) | 0.152 |
| Gentamicin | 6 (13) | 24 (52.2) | 0 (0) | 2 (4.3) | 14 (30.4) | 0 (0) | 0.523 |
| Tobramycin | 16 (34.8) | 13 (28.3) | 1 (2.2) | 6 (13) | 9 (19.5) | 1 (2.2) | 0.573 |
| Meropenem | 3 (6.5) | 25 (54.3) | 2 (4.3) | 0 (0) | 14 (30.4) | 2 (4.3) | 0.362 |
| Ciprofloxacin | 15 (32.6) | 13 (28.3) | 2 (4.3) | 0 (0) | 15 (32.6) | 1 (2.2) | 0.002* |
| Ertapenem | 2 (4.3) | 22 (47.8) | 6 (13) | 1 (2.2) | 13 (28.3) | 2 (4.3) | 0.808 |
| Cotrimoxazole | 22 (47.8) | 6 (13) | 2 (4.3) | 6 (13) | 9 (19.5) | 1 (2.2) | 0.041* |
| Cefotaxime | 18 (39.1) | 9 (19.5) | 3 (6.5) | 6 (13) | 9 (19.5) | 1 (2.2) | 0.221 |
| Ampicillin | 29 (63) | 0 (0) | 1 (2.2) | 15 (32.6) | 1 (2.2) | 0 (0) | 0.299 |
| Ceftriaxone | 18 (39.1) | 10 (21.7) | 2 (4.3) | 4 (8.7) | 10 (21.7) | 2 (4.3) | 0.077 |
| Streptomycin | 23 (50) | 7 (15.2) | 0 (0) | 5 (10.9) | 11 (23.9) | 0 (0) | 0.004* |
DISCUSSION
Recent research shows an increase of Citrobacter isolates among urinary tract infection agents with high antibiotic resistance in developed countries (12–14). Our results of antibiotic susceptibility testing showed the highest resistance of Citrobacter isolates to cefazolin and ampicillin and highest sensitivity to carbapenems, tazobactam and gentamicin. These findings are consistent with the results of previous studies (15–17). A high percentage of Citrobacter isolates in Kermanshah with multi-drug resistance indicates the dissemination of antibiotic resistance genes in this opportunistic pathogen (7). On the other hand, the accumulation of resistance genes within integrons contributes to the spread of MDR strains among Enterobacteriaceae isolates (18). Class I integron is widely distributed among multidrug resistance of Enterobacteriaceae isolates (19). Studies in Malaysia and Egypt have reported the rate of class I integron in isolates of Citrobacter and Enterobacteriaceae with 50% and 51%, respectively (5, 20). The above results are compatible with our results for the frequency of class I integron among Citrobacter isolates in Kermanshah.
Class I integron has been found to carry resistance to several antimicrobial agents in bacteria. For instance, cassettes for resistance to fluoroquinolones, β-lactams, aminoglycosides, trimethoprim and chloramphenicol have been identified (5). According to research data, most of the resistant genes for aminoglycosides (aad, aac) are transmitted by class I integrin (21). The results of our study also suggest a statistically significant association between the presence of class I integron and resistance to streptomycin. In our study, a significant correlation between the presence of class I integron and resistance to ciprofloxacin was also noted, which is consistent with other studies (21, 22). Although the mutation in topoisomerase genes is the main mechanism of resistance to fluoroquinolones, recently proteins have been identified encoded by integrons and carried on plasmids which increases the bacterial permeability for quinolones (23, 24).
Gene cassettes with different sizes carried by class I integron in Citrobacter isolates are consistent with the results of other studies on Enterobacteriaceae family (25, 26). Similar to other studies, our results indicate that integrons can carry several cassettes simultaneously and contribute to the emergence of MDR strains (27, 28). In our study, the phenotypic resistance to the certain antibiotics was observed in isolates carried the corresponding gene cassettes. For instance, there is a significant association between the presence of dehydrofolate reductase and aminoglycoside aden-yltransferase cassettes with phenotypic resistance to trimethoprim-sulfamethoxazole and streptomycin, respectively. This indicates the expression of integron genes and their role in the phenotype of bacteria.
Our results show eight different patterns of class I integron gene cassettes. The DNA analysis of gene cassettes indicates several antibiotic resistance gene cassettes. Two variants of aminoglycoside adenyl transfers (aadA1/aadA2) were detected which encode aminoglycoside 3′-9-adenylyltransferases and confer the resistance to streptomycin and spectinomycin (29). Sequence analysis also revealed three variants of dfrA (dfrA1/dfrA12/dfrA15), which encode the dihydrofolate reductase gene, confer resistance to trimethoprim (29). The horizontal transmission of resistance genes between bacteria can occur and expand the gene cassettes (30). As indicated by our results and also supported by other studies, the dfr cassette is mostly associated with the aadA gene cassette (30). These observations suggest this combination of gene cassettes can reflect their co-transmission and stable integration (31). In the present study, the dfrA1-aadA1 gene cassettes showed a high prevalence in Class I integron, which is consistent with the results of other studies on Enterobacteriaceae isolates (32, 33). According to the previous research, Class I integron contained the aadA1 or dfrA1-aadA1 cassettes are commonly found in E. coli isolates in Europe (34–36). Similarly, these gene cassettes have been reported in Asian countries (37, 38). In some studies in Iran, the 5-arr, aacA4-orfD, aadA5-dfrA17, dfrA1, aadA1-dfrA1 and aadA2-dfrA12-orfF cassettes have been reported as the most common cassettes in Klebsiella pneumoniae and E. coli isolates (29, 39–40). It seems that the aadA and dfrA gene cassettes in Iran are also prevalent
In conclusion, our results indicate a high prevalence of MDR among Citrobacter isolates in Kermanshah. A high frequency of class I integron and the associated gene cassettes, in particular dfr and aadA, present in MDR strains of Citrobacter isolated from hospitalized patients. They may play an important role in the creation and transmission of MDR strains. Statistical analysis indicates the association of integration class I and MDR isolates in this opportunistic pathogen, which needs continues surveillance in health care centers.
REFERENCES
- 1.Nayar R, Shukla I, Sultan A. Epidemiology, prevalence and identification of Citrobacter species in clinical specimens in a tertiary care hospital in India. IJSRP 2014;4(4):1–6. [Google Scholar]
- 2.Akya A, Jafari S, Ahmadi K, Elahi A. Frequency of blaCTX-M, blaTEM and blaSHV genes in Citrobacters isolated from Imam Reza Hospital in Kermanshah. J Mazandaran Univ Med Sci 2015; 25(127): 65–73. [Google Scholar]
- 3.White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 2001;45:2658–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azam H, Mikaili Ghezeljeha S, Shavandi M. Prevalence of class 1 and 2 integrons among the multidrug resistant uropathogenic strains of Escherichia coli. Asian Biomed 2015; 9:49–54. [Google Scholar]
- 5.Malek MM, Amer FA, Allam AA, El-Sokkary RH, Gheith T, Arafa MA. Occurrence of classes I and II integrons in Enterobacteriaceae collected from Zagazig University Hospitals, Egypt. Front Microbiol 2015;6:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoramrooz SS, Sharifi A, Yazdanpanah M, Malek Hosseini SA, Emaneini M, Gharibpour F, et al. High frequency of class 1 integrons in Escherichia coli isolated from patients with urinary tract infections in Yasuj, Iran. Iran Red Crescent Med J 2016;18(1):e26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guangchao Y, Yanmei L, Xiaochen L, Xihong Z, Yanyan L. Role of integrons in antimicrobial resistance: a review. Afr J Microbiol Res 2013;7(15):1301–1310. [Google Scholar]
- 8.El-Sokkary MMA, Abdelmegeed ES. Characterisation of class 1 integron among Escherichia coli isolated from Mansoura University Hospitals in Egypt. AiM 2015;5:269–277. [Google Scholar]
- 9.Guo X, Xia R, Han N, Xu H. Genetic diversity analyses of class 1 integrons and their associated antimicrobial resistance genes in Enterobacteriaceae strains recovered from aquatic habitats in China. Lett Appl Microbiol 2011;52:667–675. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement M100-S22. Wayne, PA, USA: CLSI; 2012. [Google Scholar]
- 11.Mirnejad R, Mostofi S, Masjedian F. Antibiotic resistance and carriage class 1 and 2 integrons in clinical isolates of Acinetobacter baumannii from Tehran, Iran. Asian Pac J Trop Biomed 2013;3(2):140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basavaraj CM, Jyothi P. Antibiotic sensivity pattern of Citrobacter spp. Isolated from patients wirh urinary tract infections in tertiary care hospital in south india. Int J Pharm Pharm Sci 2015;7(1): 252–254. [Google Scholar]
- 13.Metri BC, Jyothi P, Peerapur BV. Antibiotic resistance in Citrobacter spp. isolated from urinary tract infection. Urol Ann 2013;5(4):312–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leski TA, Taitt CR, Bangura U, Stockelman MG, Ansumana R, Cooper WH, et al. High prevalence of multidrug resistant Enterobacteriaceae isolated from out-patient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infect Dis 2016;16:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansouri S, Abbasi S. Prevalence of multiple drug resistant clinical isolates of extended-spectrum β-Lactamase producing Enterobacteriaceae in southeast Iran. IJMS 2010; 35:101–107. [Google Scholar]
- 16.Hareendranath G, Dominic RMS, Saralaya V. Clinical microbiological study of Citrobacter isolates from various clinical specimens and detection of β-lactamase production. J Int Med Dent 2015;2:36–46. [Google Scholar]
- 17.Shrestha A, Manandhar S, Pokharel P, Panthi P, Chaudhary DK. Prevalence of extended spectrum beta-Lactamase (ESBL) producing multidrug resistance gram-negative isolates causing urinary tract infection. EC Microbiol 2016;4:749–755. [Google Scholar]
- 18.Ahangarzadeh Rezaee M, Sheikhalizadeh V, Hasani A. Detection of integrons among multi-drug resistant (MDR) Escherichia coli strains isolated from clinical specimens in northern west of Iran. Braz J Microbiol 2011; 42:1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu HS, Lee JC, Kang HY, Ro DW, Chung JY, Jeong YS, et al. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J Clin Microbiol 2003;41:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim N, Wajidi MF, Yusof MY, Tay ST. The integron prevalence of extended-spectrum beta-lactamase producing enterobacterial isolates in a Malaysian teaching hospital. Trop Biomed 2011;28:668–671. [PubMed] [Google Scholar]
- 21.Reyes A, Bello H, Dominguez M, Mella S, Zemelman R, Gonzalez G. Prevalence and types of class 1 integrons in aminoglycoside-resistant Enterobacteriaceae from several Chilean hospitals. J Antimicrob Chemother 2003;51:317–321. [DOI] [PubMed] [Google Scholar]
- 22.Huan W, Fan XL. Relationship between class 1 integron and resistance in gram negative bacteria. J Dalian Med Univ 2007;29:584–586. [Google Scholar]
- 23.Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet 1998; 351:797–799. [DOI] [PubMed] [Google Scholar]
- 24.Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb Drug Resist 1995; 1:195–202. [DOI] [PubMed] [Google Scholar]
- 25.Mahluji Z, Firoozeh F, Khorshidi A, Zibaei M. The frequency of class 1 integrons in multidrug resistant Klebsiella pneumoniae isolated from clinical samples using polymerase chain reaction assay. SJKU 2016;21:68–78. [Google Scholar]
- 26.Mehdipour Moghaddam MJ, Mirbagheri AA, Salehi Z, Habibzade SM. Prevalence of class 1 integrons and extended spectrum beta Lactamases among multi-drug resistant Escherichia coli isolates from north of Iran. Iran Biomed J 2015;19:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima AM, de Melo ME, Alves LC, Brayner FA, Lopes AC. Investigation of class 1 integrons in Klebsiella pneumoniae clinical and microbiota isolates belonging to different phylogenetic groups in Recife, State of Pernambuco. Rev Soc Bras Med Trop 2014;47:165–169. [DOI] [PubMed] [Google Scholar]
- 28.Chang CY, Fang YT, Tsai SM, Chang LL, Yu WL. Characterization of class 1 integrons and gene cassettes in clinical isolates of Klebsiella pneumoniae from Taiwan. Diagn Microbiol Infect Dis 2009;65:214–216. [DOI] [PubMed] [Google Scholar]
- 29.Shahcheraghi F, Rahmati Ghezelgeh F, Nobari S, Torabi E, Mousavi SF, Aslani MM, et al. Identification and characterization of class 1 integrons among atypical enteropathogenic Escherichia coli isolated from children under 5 years of age. Iran J Microbiol 2014;6:156–162. [PMC free article] [PubMed] [Google Scholar]
- 30.Yan H, Li L, Zong M, Alam MJ, Shinoda S, Shi L. Occurrence and characteristics of class 1 and 2 integrons in clinical bacterial isolates from patients in south china. J Health Sci 2010;56:442–450. [Google Scholar]
- 31.Chang LL, Chang TM, Chang CY. Variable gene cassette patterns of class 1 integron-associated drug-resistant Escherichia coli in Taiwan. Kaohsiung J Med Sci 2007;23:273–280. [DOI] [PubMed] [Google Scholar]
- 32.Sung JY, Oh JE. Distribution and characterization of integrons in Enterobacteriaceae isolates from chickens in Korea. J Microbiol Biotechnol 2014;24:1008–1013. [DOI] [PubMed] [Google Scholar]
- 33.Sunde M. Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J Antimicrob Chemother 2005;56:1019–1024. [DOI] [PubMed] [Google Scholar]
- 34.Van Essen-Zandbergen A, Smith H, Veldman K, Mevius D. Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in the Netherlands. J Antimicrob Chemother 2007;59:746–750. [DOI] [PubMed] [Google Scholar]
- 35.Cocchi S, Grasselli E, Gutacker M, Benagli C, Convert M, Piffaretti JC. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol Med Microbiol 2007;50:126–132. [DOI] [PubMed] [Google Scholar]
- 36.Moura A, Henriques I, Ribeiro R, Correia A. Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J Antimicrob Chemother 2007;60:1243–1250. [DOI] [PubMed] [Google Scholar]
- 37.Wang GQ, Wu CM, Du XD, Shen ZQ, Song LH, Chen X, et al. Characterization of integrons-mediated antimicrobial resistance among Escherichia coli strains isolated from bovine mastitis. Vet Microbiol 2008;127(1–2):73–78. [DOI] [PubMed] [Google Scholar]
- 38.Waturangi DE, Suwanto A, Schwarz S, Erdelen W. Identification of class 1 integrons-associated gene cassettes in Escherichia coli isolated from Varanus spp. in Indonesia. J Antimicrob Chemother 2003;51:175–177. [DOI] [PubMed] [Google Scholar]
- 39.Salimizand H, Shahcheraghi F, Kalantar E, Badmasti F, Mousavi SF. Molecular characterization of class 1 integrons and gene cassettes in multidrug resistant (MDR) Klebsiella spp. isolated from hospitalized and outpatients in Iran, 2009. Iran J Microbiol 2013;5:48–55. [PMC free article] [PubMed] [Google Scholar]
- 40.Masoumian N, Haghi F. Analysis of integrons and associated gene cassettes in clinical isolates of Escherichia coli. J Zanjan Univ Med Sci 2015;23:74–82. [Google Scholar]