Abstract
p62 (also known as sequestosome 1) protein, is a small regulatory protein that accumulates in autophagy-defective cells that has been demonstrated to be involved in the prognosis and survival of patients with several types of cancer. However, to the best of our knowledge, there have been no such studies for osteosarcoma (OS). In the present study, the expression of p62 in 70 OS samples was determined using immunohistochemistry and its association with various clinicopathological factors was assessed. The results demonstrated that the overexpression of p62 protein was detected in 77.1% (54/70) samples, and the expression levels were significantly associated with tumor size (P=0.001), metastasis (P=0.036), clinical staging (P=0.003) and poor prognosis (P=0.0058). Furthermore, suppression of the p62 expression by short hairpin RNA interference in F5M2 and F4 cells lines led to decreased cell proliferation, migration and invasion in vitro. These results suggested that increased expression of p62 may be involved in OS progression, and therefore the excess expression of p62 may serve as a novel prognostic biomarker for patients with OS.
Keywords: p62, immunohistochemistry, osteosarcoma, prognosis
Introduction
Osteosarcoma (OS) is the most common primary bone malignancy in children and adolescents, and accounts for ~20% of all primary bone cancer worldwide (1,2). Although there have been improvements in surgical and multi-agent chemotherapeutics, the long-term survival rate for patients with non-metastatic OS remains at ~60% (3), whereas, currently, a 5-year survival rate of 30% is observed in patients with metastatic OS (1,4). Therefore, it is essential to analyze the biological evidence at the molecular level of OS to identify novel prognostic markers and determine the underlying molecular mechanism of OS metastasis.
p62, also known as sequestosome 1, is a multifunctional scaffold protein that regulates cell proliferation, apoptosis, cell survival, inflammation and tumorigenesis through the catabolism of molecules (5,6). The p62 protein is one of the selective substrates of autophagy, localized on the membranes of autophagosomes (7). It has been identified that the accumulation of the p62 protein is variably associated with different types of tumor, including cancer of the prostate, breast, lung, colon and endometrium (8–12). An excessive expression of p62 has been identified to be a cause of poor prognosis in certain tumor types, including prostate and endometrial cancer (11,12). Furthermore, it has been demonstrated that the abnormal expression of p62 may be associated with the activation of oncogenic signaling pathways, including the mammalian target of rapamycin (mTOR), mitogen-activated protein kinase, nuclear factor κB (NF-κB), Wnt/β-catenin and potentially other signaling pathways (7,13). Therefore, it is suggested that p62 may function as an oncogene and is involved in poor prognosis in human cancer.
However, the function and clinical significance of p62 has not yet been completely elucidated in OS. In the present study, the expression of p62 was examined and its association with clinicopathological characteristics was investigated in patients with OS.
Materials and methods
Patients and tumor specimens
The present study was approved by the Ethics Committee of Xi'an HongHui Hospital (Xi'an, China). Either the patients or their guardians provided written informed consent for participation in the study. Formalin-fixed paraffin-embedded tissues were obtained from 85 patients with OS who underwent tumor resection at Xi'an HongHui Hospital between March 2007 and November 2009. Patients did not receive preoperative chemotherapy or radiotherapy. However, patients in Enneking stages I, II or III of the disease (14) received postoperative adjuvant chemotherapy [six courses of ifosfamide (2 g/m2 for 5 days/course), methotrexate (8 g/m2 for 1 day/course], and Adriamycin (50 mg/m2 for 1 day/course). All patients were followed up for ≥3 years. However, 15 patients were unreachable (17.7%), and therefore failed to complete follow-up. Consequently, the data presented were for a total of 70 patients (82.3%) with a median follow-up of 42 months (range, 2–68 months). Of the 70 patients, 22 were female and 48 were male, with a median age of 19 years (range, 8–65 years). A total of 10 osteochondroma specimens were used as normal controls, including 7 male and 3 female patients with a median age of 32 years (range, 15–54 years). All samples were evaluated for diagnosis by two experienced pathologists in Hong Hui Hospital, Xi'an Jiaotong University College of Medicine, (Xi'an, China).
Materials
RPMI-1640 medium was purchased from HyClone (GE Healthcare, Logan, UT, USA). Fetal bovine serum (FBS), penicillin, streptomycin and glutamine were purchased from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Immobilon Western horseradish peroxidase chemiluminescence kit and polyvinylidene fluoride membrane were from EMD Millipore (Billerica, MA, USA). Polyclonal antibodies against p62 were purchased from Cell Signaling Technology Inc. (1:2,000; cat. no. 5114, Danvers, MA, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin and monoclonal antibodies against β-actin were purchased from Bioworld Technology, Inc. (1:5,000; cat. no. AP0060; St. Louis Park, MN, USA).
Cell culture
Human OS cell sub-lines F4 and F5M2 were established and maintained in the laboratory of Tangdu Hospital (The Fourth Military Medical University, Xi'an, China) as described previously (15). The cells were cultured in RPMI-1640 medium supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml) and glutamine (2 mM) at 37°C in a humidified incubator containing 5% CO2. The human osteoblast hFOB1.19 cell line was purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured similarly in Dulbecco's modified Eagle's medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Immunohistochemical analysis
Immunohistochemistry was performed as described in our previous study (16). The anti-p62 antibody was used at a dilution of 1:100. Images were observed at magnification, ×400 under a fluorescence microscope. The final scores of expression of p62 were calculated as described previously (16). Each specimen was categorized as follows: 0, (0); 1+, (1–3); 2+, (4–8); and 3+, (9–12). Scores of 0–3 and 4–12 were designated low and high expression, respectively.
Short hairpin RNA (shRNA) targeting p62
The lentiviruses with shRNA targeting p62 (5′-TCGAGGAAATGGGTCCACCAGGAATTCAAGA-3′) and shRNA negative control were purchased from Hanbio Biotechnology Co., Ltd. (Shanghai, China). F5M2 and F4 cells were transfected with the lentiviruses at a multiplicity of infection of 100 using TransDux transfection reagent (System Biosciences, Palo Alto, CA, USA). Stable p62-knockdown OS cells (F5M2-shp62 cells or F4-shp62 cells) were selected using 3 mg/ml puromycin (Hanbio Biotechnology Co., Ltd.) and confirmed using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting. These stable knockdown cells and controls (Green Fluorescent Protein (GFP)-F5M2 cells or GFP-F4 cells) were used for subsequent experiments following transfection for 48 h.
RT-qPCR
F5M2 and F4 cells, as well as OS adjacent healthy tissue, were harvested and lysed with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). All samples were sent to UcallM Biotech Company (Wuxi, China) for subsequent experimental procedures. The Revert Aid First-Strand RT-PCR kit (Invitrogen: Thermo Fisher Scientific, Inc.) was used to synthesize cDNA from 500 ng total RNA. Then 2 µl cDNA was amplified in 20 µl reaction mixture containing 10 µl of SYBR Green PCR mix (Invitrogen: Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The following thermocycling conditions were used: 5 min at 95°C, followed by 40 cycles of 10 sec at 95°C and 30 sec at 60°C. The p62 gene expression was calculated using the 2−ΔΔCq method (17). The primers used for amplification were as follows: Human p62 forward, 5′-TGAGGAACAGATGGAGTCGG-3′ and reverse, 5′-GAGATGTGGGTACAAGGCAG-3′; human GAPDH forward, 5′-ACCCACTCCTCCACCTTTG-3′ and reverse, 5′-CACCACCCTGTTGCTGTAG-3′. All experiments were performed in triplicate and repeated at least three times independently.
Cell counting kit-8 (CCK-8) assay
F5M2-shp62, GFP-F5M2, F4-shp62 and GFP-F4 cells were seeded into 96-well plates at a density of 2,000 cells/well. After 24 h of incubation at 37°C in 5% CO2, 10 µl/well CCK-8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to one plate, which was incubated for 2 h at 37°C and measured using a microplate reader (Thermo Fisher Scientific, Inc.) at 490 nm. Each experiment was repeated three times independently.
Transwell migration and invasion assays
The Transwell migration and invasion assays were performed as in our previous study (18). The migratory and invasive potential of cells were assessed using a Transwell (Corning Inc., Corning, NY, USA) inserts only or using inserts pre-coated with 50 µl of BD Matrigel™ Basement Membrane Matrix (1:3 dilution, BD Biosciences, Franklin Lakes, NJ, USA). The upper chambers were seeded with 1×104 cells suspended in serum-free RPMI-1640 medium, and the lower chambers were filled with complete RPMI-1640 medium with 10% FBS (0.6 ml/well). The invasion was evaluated after 16 h of incubation at 37°C. The migration was assessed after 24 h of incubation. Invasion and migration were examined under a light microscope (Nikon Eclipse 80i; Nikon Corporation, Tokyo, Japan) at a magnification, ×200 in five randomized fields. All experiments were independently repeated three times.
Western blot analysis
Cellular lysates were prepared in chilled radioimmunoprecipitation assay lysis buffer (Cell Signaling Technology, Inc.), and the protein concentrations were determined using a BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Proteins (10 µl) were loaded and separated by electrophoresis on 10% SDS-polyacrylamide gels at 110 V for 2 h. Following electrophoresis, the SDS-PAGE gels were transferred electronically to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). PVDF membranes were blocked using a solution containing 5% skimmed milk and incubated overnight at 4°C with the following antibodies: Anti-p62 and anti-β-actin (1:1,000). Following washing with Tris-buffered saline with Tween-20, the membranes were incubated for 1 h at room temperature with HRP-conjugated goat anti-rabbit immunoglobulin and monoclonal antibodies against β-actin. Reactive proteins were visualized using an Immobilon western horseradish peroxidase chemiluminescence kit (EMD Millipore, Billerica, MA, USA). Relative p62 expression levels as analyzed using ImageJ software version 1.47 (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
The statistical analyses were carried out using GraphPad Prism software (version 5.0; GraphPad Software Inc., La Jolla, CA, USA). The immunohistochemistry data were analyzed using the χ2 test. Survival data were analyzed using the Kaplan-Meier with log-rank tests estimator method. The RT-qPCR, western blot analysis, inhibitor migration assay and inhibitor invasion assay results were analyzed by one-way analysis of variance. All other data were analyzed using a two-sided Student's t-test. The data are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological features and prognostic implications of p62 expression
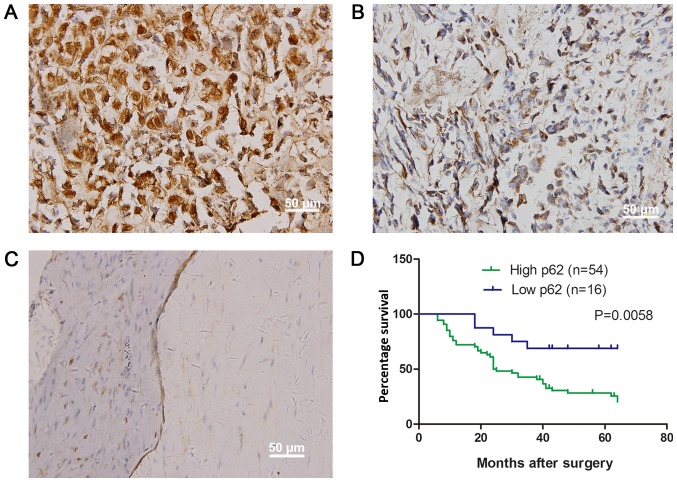
Using immunohistochemistry, it was identified that the p62 was distributed in the cytoplasm and perinuclear membrane of the OS cells (Fig. 1A and B). Compared with osteochondroma samples, the expression rate of p62 was significantly increased in OS (Table I; Fig. 1C). The increased expression of p62 protein was observed in 54/70 samples (77.14%) and was identified to be significantly associated with tumor size (P=0.001), metastasis (P=0.036) and clinical staging (P=0.003) (Table II). These patients were also significantly associated with a decreased overall survival time (P=0.0058; Fig. 1D).
Figure 1.
Expression of p62 in osteosarcoma and osteochondroma (magnification, ×400), and prognostic implication. (A) High expression of p62 in osteosarcoma; (B) low positive expression of p62 in osteosarcoma; (C) negative expression of p62 in osteochondroma tissues; (D) overall survival curves of 70 patients with osteosarcoma according to p62 expression. Survival curves of the 54 and 16 patients with osteosarcoma that exhibited high (green) and low (blue) expression of p62.
Table I.
Expression levels of p62 in osteosarcoma and osteochondroma specimens.
| p62 expression | ||||
|---|---|---|---|---|
| Group | n | ++/+++ | −/+ | P-value |
| Osteosarcoma | 70 | 54 (77.1%) | 16 (22.9%) | <0.01 |
| Osteochondroma | 20 | 6 (30.0%) | 14 (70.0%) | |
Table II.
Association between p62 expression and clinicopathological data in patients with osteosarcoma.
| p62 expression | ||||
|---|---|---|---|---|
| Variable | n | High | Low | P-value |
| Total | 70 | 54 | 16 | |
| Sex | 0.551 | |||
| Male | 48 | 38 | 10 | |
| Female | 22 | 16 | 6 | |
| Age, years | 0.816 | |||
| ≥20 | 16 | 12 | 4 | |
| <20 | 54 | 42 | 12 | |
| Tumor size, cm2 | 0.001a | |||
| <50 | 20 | 16 | 12 | |
| ≥50 | 27 | 38 | 4 | |
| Histology | 0.355 | |||
| Osteoblastic | 31 | 25 | 6 | |
| Chondroblastic | 8 | 4 | 4 | |
| Fibroblastic | 10 | 8 | 2 | |
| Telangiectatic | 15 | 12 | 3 | |
| Mixed | 6 | 5 | 1 | |
| Metastasis | 0.036a | |||
| Yes | 29 | 26 | 3 | |
| No | 41 | 28 | 13 | |
| Clinical stage | 0.003a | |||
| I/IIA | 17 | 9 | 8 | |
| IIB | 24 | 19 | 5 | |
| III | 29 | 26 | 3 |
P<0.05.
Human OS cell lines F5M2 and F4 express p62
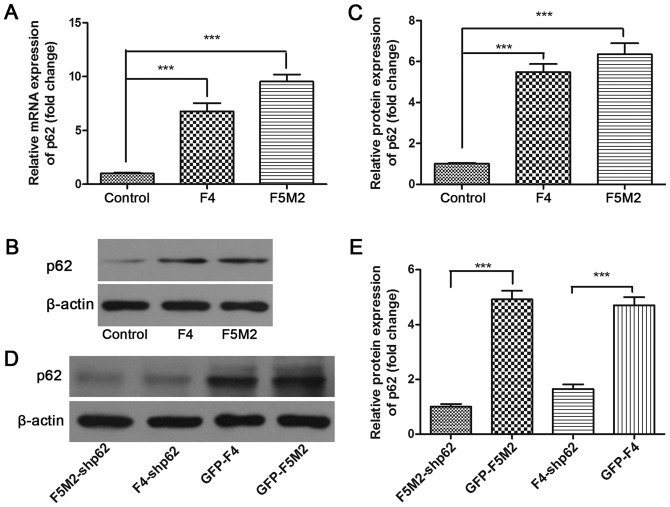
RT-qPCR results identified that the expression level of p62 mRNA was highest in the human OS cell lines F5M2 and F4 and lowest in the human osteoblastic cell line hFOB1.19 (P<0.001; Fig. 2A). The expression level of p62 protein in these cell lines was then assessed by western blotting. The results demonstrated that OS cells (F5M2 and F4) expressed increased levels of p62 protein compared with osteoblastic cells (hFOB1.19) (Fig. 2B), which was identified to be a significant difference (Fig. 2C).
Figure 2.
p62 expression in osteosarcoma cells and shRNA-mediated suppression. (A) p62 mRNA expression in F5M2 and F4 human osteosarcoma cells and control hFOB1.19 normal human osteoblast cells detected using the reverse transcription-quantitative polymerase chain reaction. All osteosarcoma cells expressed increased p62 mRNA levels compared with the normal osteoblasts. (B) Western blotting results demonstrating p62 protein expression in F5M2 and F4 osteosarcoma cells and control hFOB1.19 normal human osteoblast cells. (C) Relative p62 expression levels as analyzed using ImageJ software. All osteosarcoma cells expressed an increased amount of p62 protein compared with normal osteoblasts. (D) p62 protein detection by western blotting following shRNA silencing. (E) p62 relative protein expression levels analyzed using ImageJ. p62 expression was efficiently suppressed by shRNA in F5M2 and F4 cells. ***P<0.001. Data are presented as the mean ± standard deviation of three independent experiments. sh, short hairpin.
p62 knockdown decreases the proliferative capacity of F5M2 and F4 cells
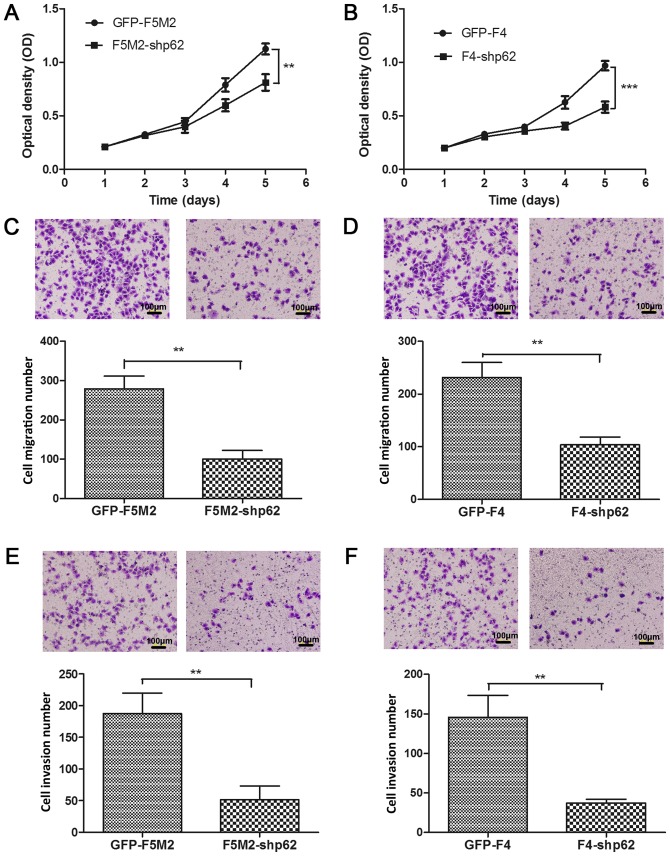
A CCK-8 cell proliferation assay was used to examine the effect of p62 inhibition on the in vitro proliferative capacity of F5M2 and F4 cells. p62 expression was stably downregulated by lentiviral-mediated shRNA. Compared with the controls (GFP-F5M2 and GFP-F4), the p62 expression was efficiently decreased in the stable p62-inhibited cells (F5M2-shp62 and F4-shp62) (Fig. 2D), which was identified to be a significant difference (Fig. 2E). Subsequently, the CCK-8 cell proliferation assay demonstrated that stable p62 knockdown decreased the proliferation of F5M2 (P<0.01; Fig. 3A) and F4 cells (P<0.001; Fig. 3B) compared with their respective controls.
Figure 3.
Suppressing p62 expression inhibited proliferation, migration, and invasion. (A) CCK-8 assay for the GFP-F5M2 and F5M2-shp62 cells. The proliferation rate of the F5M2-shp62 cells was significantly decreased compared with that of the GFP-F5M2 cells. (B) CCK-8 assay for the GFP-F4 and F4-shp62 cells. The proliferation rate of the F4-shp62 cells was significantly decreased compared with that of the GFP-F4 cells. (C) Migration assay with GFP-F5M2 and F5M2-shp62 cells. The migratory capacity of the F5M2-shp62 cells was significantly decreased compared with that of the GFP-F5M2 cells. (D) Migration assay with GFP-F4 and F4-shp62 cells. The migratory capacity of the F4-shp62 cells was significantly decreased compared with that of the GFP-F4 cells. (E) Invasion assay with GFP-F5M2 and F5M2-shp62 cells. The invasion rate of the F5M2-shp62 cells was significantly decreased compared with that of the GFP-F5M2 cells. (F) Invasion assay with GFP-F4 and F4-shp62 cells. The invasion rate of the F4-shp62 cells was significantly decreased compared with that of the GFP-F4 cells (vs. F4-shp62). Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01; ***P<0.001. CCK-8, Cell Counting kit-8; sh, short hairpin.
p62 knockdown decreases the migration and invasion of F5M2 and F4 cells
The migration assay results demonstrated that p62 inhibition significantly decreased the migratory ability of F5M2 cells (P<0.01; Fig. 3C) and F4 cells (P<0.01; Fig. 3D) when compared with the corresponding control groups. The invasion assay results revealed that p62 inhibition significantly decreased the invasive ability of F5M2 (P<0.01; Fig. 3E) and F4 cells (P<0.01; Fig. 3F).
Discussion
The results of the present study demonstrated that the increased expression of p62 protein was significantly associated with tumor size, metastasis and clinical staging in human OS, and may be used as a prognostic factor. However, patients with increased expression of p62 protein were identified to be significantly associated with decreased overall survival times compared with those with low p62 expression. To the best of our knowledge, this present study is the first to investigate the clinical significance of p62 expression in OS.
The abnormal expression of p62 detected in a variety of tumors, including breast, prostate, lung, liver and oral cancer, serves an active function in tumorigenesis and metastasis (12,19,20). To determine the p62 protein levels in prostate cancer, Burdelski et al (11) used immunohistochemistry to assess 7,822 prostate cancers and found that p62 stained in 73% (5,716/7,822). In addition, the protein was markedly associated with high Gleason grade, positive nodal status, advanced pathological tumor stage and positive resection margin (12). Therefore, the accumulation of p62 may be considered as a predictor of the detrimental prognostic behavior of prostate cancer (12). The expression of p62 is substantially weak in normal gastric tissue samples, whereas increased cytoplasmic and nuclear levels are detected in gastric cancer (21). The immunohistochemical analysis of the present study indicated increased expression p62 protein in 77.14% (54/70) of the analyzed OS samples.
Previous studies have demonstrated that the autophagy system constantly degraded the p62 protein (22,23), whereas other studies indicated that p62 expression is regulated at the transcriptional level (24,25). However, the specific function of p62 in tumors remains unclear, and it has been demonstrated that increased p62 mRNA expression is associated with increased p62 protein expression in the oral squamous cell carcinoma (19). In the present study, it was demonstrated that the p62 mRNA and protein levels were upregulated in human F5M2 and F4 OS cells, suggesting that the protein regulation occurred at the transcriptional and translational levels.
In order to investigate the function of p62 in tumor cell proliferation, migration and invasion, stable p62 knockdown cell lines were established using specific shRNA. Using a CCK-8 assay, p62 knockdown was determined to suppress the proliferation of F5M2 and F4 OS cells. Furthermore, the migration and invasion assays demonstrated that p62 knockdown decreased the migration and invasion of F5M2 and F4 cells in vitro. Thus, increased p62 expression increased the proliferation, migration and invasion of OS cells in vitro. Previous studies have also established that the p62 protein functions as a hub for various signal transduction pathways for cell survival and cell death (20). The overexpression of p62 has been identified to promote tumor cell survival via impairment of the NF-κB signaling pathway (26). The NRF2-Keap1 pathway consisted of the transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), and the cytoplasmic contact protein Keap1. NRF2 is a central regulator of the cellular antioxidant stress reaction. Furthermore, NRF2 is able to regulate the expression of several detoxification enzymes and a series of antioxidative protein genes, thereby serving a major function in antitumor mechanisms (6). Under normal conditions, NRF2 was degraded by the ubiquitin-proteasome pathway through interaction with Keap1. When exposed to the pro-electronic reagent, reactive oxygen species and nitric oxide, Keap1 is activated, leading to NRF2 heterodimerization and translocation to the nucleus (6). Subsequently, the nuclear NRF2 is able to induce a series of transcription cell protective genes (6). The p62 protein is able to directly interact with Keap1 of NRF2, and the excessive expression of p62 is able to continuously stimulate NRF2, which is hypothesized to lead to tumor progression (27). Aberrant mitosis often leads to tumor occurrence (28). The active association of cyclin-dependent kinase (CDKs) with cell cycle proteins is the key to an orderly cell cycle (28). The lack of cell cycle proteins or the presence of CDK inhibitor leads to cell cycle arrest and cell death (29). The study of Linares et al (30) elucidated the expression of p62 protein in different cells at various stages of the cell cycle, and identified that the protein had been phosphorylated in the early stage of mitosis. Following inhibition of the activity of CDKs, the p62 protein was phosphorylated at Thr269 and Ser272, suggesting that it may be involved in the mitotic division of cells (30). In addition, it has been demonstrated that p62 is able to activate the mTOR signaling pathway to regulate cell growth and autophagy (31).
There were several limitations to the present study. First, the study was primarily limited by the number of patient samples. Secondly, despite osteochondroma as the control group, the study lacked normal tissue as the control group. Finally, the regulatory mechanism of p62 in OS was not elucidated. Further studies with larger patient populations involving normal tissue, mechanism of p62 in OS, and animal experiment are essential to assess p62 more accurately as a therapeutic target for OS.
In summary, the results of the present study demonstrated that p62 was upregulated in human OS. Furthermore, the overexpression of p62 protein is associated with poor survival rates in patients with OS. Additionally, p62 may be involved in osteosarcoma progression; however, further studies are a prerequisite for understanding the significance of increased expression of p62 in order to develop specific and comprehensive treatments.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Project of Science and Technology Department of Shaanxi Province (grant nos. 2015SF116, 2015SF110 and 2013K14-02-12).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HX, ML and KZ analyzed and interpreted the patient data. LS and BH performed the immunohistochemical analysis, YL, QW and YZ performed RT-qPCR, western blot, CCK-8 assay and Transwell and were major contributors in writing the manuscript. CR and ND performed cell transfection. HL and CZ performed statistical analysis. ZL and TM designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Xi'an HongHui Hospital (Xi'an, China). Either the patients or their guardians provided written informed consent for participation in the study.
Consent for publication
The patient or their guardian provided written informed consent for the publication of any associated data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the surveillance, epidemiology, and end results (SEER) program database. Cancer Epidemiol. 2015;39:593–599. doi: 10.1016/j.canep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Maugg D, Rothenaigner I, Schorpp K, Potukuchi HK, Korsching E, Baumhoer D, Hadian K, Smida J, Nathrath M. New small molecules targeting apoptosis and cell viability in osteosarcoma. PLoS One. 2015;10:e0129058. doi: 10.1371/journal.pone.0129058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest. 2001;19:292–315. doi: 10.1081/CNV-100102557. [DOI] [PubMed] [Google Scholar]
- 5.Manley S, Williams JA, Ding WX. Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med (Maywood) 2013;238:525–538. doi: 10.1177/1535370213489446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue D, Suzuki T, Mitsuishi Y, Miki Y, Suzuki S, Sugawara S, Watanabe M, Sakurada A, Endo C, Uruno A, et al. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci. 2012;103:760–766. doi: 10.1111/j.1349-7006.2012.02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo RZ, Yuan ZY, Li M, Xi SY, Fu J, He J. Accumulation of p62 is associated with poor prognosis in patients with triple-negative breast cancer. Onco Targets Ther. 2013;6:883–888. doi: 10.2147/OTT.S46222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inui T, Chano T, Takikita-Suzuki M, Nishikawa M, Yamamoto G, Okabe H. Association of p62/SQSTM1 excess and oral carcinogenesis. PLoS One. 2013;8:e74398. doi: 10.1371/journal.pone.0074398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdelski C, Reiswich V, Hube-Magg C, Kluth M, Minner S, Koop C, Graefen M, Heinzer H, Tsourlakis MC, Wittmer C, et al. Cytoplasmic accumulation of sequestosome 1 (p62) is a predictor of biochemical recurrence, rapid tumor cell proliferation, and genomic instability in prostate cancer. Clin Cancer Res. 2015;21:3471–3479. doi: 10.1158/1078-0432.CCR-14-0620. [DOI] [PubMed] [Google Scholar]
- 12.Iwadate R, Inoue J, Tsuda H, Takano M, Furuya K, Hirasawa A, Aoki D, Inazawa J. High expression of p62 protein is associated with poor prognosis and aggressive phenotypes in endometrial cancer. Am J Pathol. 2015;185:2523–2533. doi: 10.1016/j.ajpath.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Puissant A, Fenouille N, Auberger P. When autophagy meets cancer through p62/SQSTM1. Am J Cancer Res. 2012;2:397–413. [PMC free article] [PubMed] [Google Scholar]
- 14.Jawad MU, Scully SP. In brief: Classifications in brief: Enneking classification: Benign and malignant tumors of the musculoskeletal system. Clin Orthop Relat Res. 2010;468:2000–2002. doi: 10.1007/s11999-010-1315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Yang TT, Wang W, Sun HH, Ma BA, Li CX, Ma Q, Yu Z, Fan QY. Establishment and characterization of human osteosarcoma cell lines with different pulmonary metastatic potentials. Cytotechnology. 2009;61:37–44. doi: 10.1007/s10616-009-9239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Guan GF, Chen J, Hu B, Sun C, Ma Q, Wen YH, Qiu XC, Zhou Y. Aberrant CXCR4 and β-catenin expression in osteosarcoma correlates with patient survival. Oncol Lett. 2015;10:2123–2129. doi: 10.3892/ol.2015.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Ma Q, Liu T, Guan G, Zhang K, Chen J, Jia N, Yan S, Chen G, Liu S, et al. Interleukin-6 suppression reduces tumour self-seeding by circulating tumour cells in a human osteosarcoma nude mouse model. Oncotarget. 2016;7:446–458. doi: 10.18632/oncotarget.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JL, Chen FF, Lung J, Lo CH, Lee FH, Lu YC, Hung CH. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br J Cancer. 2014;111:944–954. doi: 10.1038/bjc.2014.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Yang Z, Dong J. P62: An emerging oncotarget for osteolytic metastasis. J Bone Oncol. 2016;5:30–37. doi: 10.1016/j.jbo.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed A, Ayman A, Deniece J, Wang T, Kovach C, Siddiqui MT, Cohen C. P62/Ubiquitin IHC expression correlated with clinicopathologic parameters and outcome in gastrointestinal carcinomas. Front Oncol. 2015;5:70. doi: 10.3389/fonc.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, Reischl M, Carnio S, Labeit D, Sandri M, et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 2014;10:123–136. doi: 10.4161/auto.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. P62/SQSTM1 at the interface of aging, autophagy, and disease. Age (Dordr) 2014;36:9626. doi: 10.1007/s11357-014-9626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirai H, Nakatsuru Y. Evaluating chemical CDK inhibitors as cell death inducers. Methods Mol Biol. 2016;1336:167–178. doi: 10.1007/978-1-4939-2926-9_14. [DOI] [PubMed] [Google Scholar]
- 30.Linares JF, Amanchy R, Greis K, Diaz-Meco MT, Moscat J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol Cell Biol. 2011;31:105–117. doi: 10.1128/MCB.05371-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.