Abstract
In the present study, the expression level of microRNA-146a-5p (miR-146a-5p) in breast cancer tissue and cell lines was investigated and its effects on proliferation of breast cancer cells. miR-146a-5p expression was detected by reverse transcription-quantitative polymerase chain reaction in breast cancer tissues, paraneoplastic tissue (collected by The Department of Oncology of Changhai Hospital from January 2014 to June 2015), breast cancer cell line MCF-7 and normal breast epithelial cell line MCF 10A. Bioinformatics analysis was conducted to forecast target genes of miR-146a-5p, which was further verified by fluorescent reporter gene detection. The results demonstrated the expression level of miR-146a-5p in breast cancer tissue was significantly higher, compared with paraneoplastic tissue (P<0.01), and the expression level of miR-146a-5p in MCF-7 cells was significantly higher, compared with MCF 10A cells (P<0.01). Overexpression of miR-146a-5p in MCF-7 cells can promote the proliferation, and low expression miR-146a-5p in MCF-7 can inhibit the proliferation. BRCA1 was further identified as a target gene of miR-146a-5p by bioinformatics analysis and fluorescent reporter gene detection. It was concluded that miR-146a-5p is expressed in breast cancer tissue and breast cancer cell line and may regulate the proliferation of MCF-7 via BRCA1.
Keywords: breast cancer, microRNA, epithelial cell, proliferation
Introduction
Breast cancer derived from mammary gland epithelial tissue has had an increasing morbidity rate since the 1980s (1). Breast cancer is also the most common malignant tumor in China, and accounts for 12.2% of the novel diagnosed cases and 9.6% of mortalities (2).
A microRNA (miRNA) is a small non-coding single strand RNA containing ~22 nucleotides, with a function of combining the 3′untranslated region (UTR) of target genes. With the combination of intracellular proteins to form a RNA interference silencing ribonucleoprotein complex, the target mRNA is bound to and degraded via complete complementary sequence specific binding; however, it can inhibit translation by incomplete complementary combination, to regulate the expression of gene at the post transcriptional level (3,4). At present, nearly 1,000 miRNA sequences have been cloned in vivo, including those involved in: The development of the organism (5); cell differentiation and proliferation (6); cell apoptosis (7); tumorigenesis (8); inflammation (9); and numerous other physiological and pathological processes (3). It is speculated that 1/3 of gene expression may be regulated by miRNAs (10).
Numerous studies have indicated that miRNAs serve an important part in genesis and progression of breast cancer (11–15). In the previous experimental study (16), it was determined that the expression of miR-146a-5p in breast cancer tissue was significantly higher, compared with paraneoplastic tissue.
The present study further investigated the expression of miR-146a-5p in breast cancer and its association with the proliferation of breast cancer cells to search for the target gene and understand the molecular mechanism underlying miR-146a-5p, which regulates the proliferation of breast cancer cells (17), providing a novel strategy for the prevention and treatment of breast cancer.
Materials and methods
Breast cancer tissue and paraneoplastic tissue
The surgical specimens of carcinoma tissue and paraneoplastic tissue (<3 cm from the edge of tumor specimens) were collected from 32 clinical diagnosed female patients with breast cancer from January 2014 to June 2015 in The Department of Oncology, Changhai Hospital (Shanghai, China). The median age of the patients was 49 years, and the age ranged from 27–69 years. Any patient underwent chemotherapy, immunotherapy or radiotherapy were excluded. All specimens were frozen in liquid nitrogen rapidly and stored at −80°C.
Cell culture
Human breast cancer cell line MCF-7, normal breast epithelial cells MCF 10A and human embryonic kidney cell line, 293T, were purchased from Shanghai Bioleaf Biotech Co., Ltd. (Shanghai, China). Cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% FBS in a 5% CO2 incubator at 37°C. The medium was replaced every 2–3 days. When the cells were at 70–80% confluency, the transfection experiment was carried out.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from breast cancer tissue, paraneoplastic tissue, human breast cancer cell line MCF-7 and normal breast epithelial cells MCF 10A. RT-qPCR was used to detect the expression of miR-146a-5p. Primers of miR-146a-5p and β-actin U6 were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). RT primers were of an miRNA specific stem loop structure. OligodT20 were used as RT primers for reverse transcription. The relative expression of miRNA and U6 were detected by using in SYBR Premix Ex Taq enzyme (Takara Biomedical Technology (Beijing) Co., Ltd., Beijing, China) and specific qPCR primer. The primer sequences were as follows: miR-146a-5p forward, 5′-cgagtccagttttcccagga-3′, and reverse, 5′-gtcgtatccagtgcaggg-3′; U6 forward, 5′-ctcgcttcggcagcaca-3′, and reverse, 5′-aacgcttcacgaatttgcgt-3′.
RT-qPCR reaction system: 7.5 µl 2X SYBR master Mix, 0.3 µl forward Primer, 0.3 µl reverse Primer, 1.0 µl cDNA, 5.9 µl ddH2O and a total volume of 15 µl.
RT-qPCR reaction conditions: 95°C for 10 min, then 95°C 15 sec, annealing temperature (55°C) depending on the primer for 20 sec and 72°C for 30 sec, for 40 cycles, this was following with 72°C for 10 min. The 2−∆∆Cq method was used for quantification (18).
Transfection and experimental groups
The logarithmic growth phase cells were divided into blank control group, mock group, over expression group and suppression group. The blank control group had no intervention, whilst the mock, over expression and suppression group were transfected with mock (empty vector pSuper plasmid gifted from Shanghai Institutes for Biological Sciences), 100 nmol/l mimic miR-146a-5p and 100 nmol/l inhibitor miR-146a-5p (Shanghai GenePharma Co., Ltd, China), respectively, according to the Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc. CA, USA) specification. Mimic miR-146a-5p and inhibitor miR-146a-5p were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The effects of transfections were evaluated through RT-qPCR by detecting the miR-146a-5p expression level of each group.
MTT
MTT was used to detect the proliferation rate at 1 day (D1), 2 days (D2) and 3 days (D3) following transfection. The cells in the logarithmic growth phase were seeded into a 96 well plate (5×103 cells/well). A total of 20 µl 5 µg/µl MTT solution was added into each well at D1, D2 and D3 following transfection, then 150 µl dimethyl sulfoxide (Henan Tianfu Chemical Co., Ltd., Henan, China) was also added into each well following a 4-h incubation at room temperature, value the optical density of each group at detection wavelength of 490 nm. The proliferation ratio of the other groups were calculated based on comparison with the blank control group value and the experiment was repeated three times.
Bioinformatics prediction
Using TargetScan (http://www.targetscan.org/vert_71/), PicTar (http://www.pictar.org), MiRBase (http://www.mirbase.org) and BibiServ (https://bibiserv.cebitec.uni-bielefeld.de/index.html) software to predict the potential target gene of miR-146a-5p. The results were screened according to the matching condition between miRNA seed sequence and target sequence, RNA double chain free energy and miRNA target sequence conservation among different species.
Dual fluorescence reporter gene assay
The 3′UTR region of BRCA1 gene sequence that can interact with 3′UTR region of miR-146a-5p was cloned into the same region in reporter gene Luciferase of plasmid pGl3. If miR-146a-5p can act on the target gene sequence, then the translation process of report gene luciferase will be inhibited, the amount of luciferase protein with activity will be reduced, thus the catalytic substrate luminescence signal will be reduced; therefore, the inhibitory effect of miR-146a-5p on the target gene sequence can be reflected indirectly by the luminescence signal strength. Pre-miR-146a-5p was amplified with the 293T cell genome as a template, subcloned into pSuper plasmid H1 RNA promoter into multiple downstream cloning sites, and the recombinant pSuper plasmid expressing pre-miR-146a-5p was constructed. Cell lysis following transfection into 293T cells for 24 h, then the luciferase detection kit (Promega Corporation, Madison, WI, USA) was used to detect the relative expression of luciferase.
Statistical analysis
SPSS 13.0 (SPSS, Inc. Chicago, IL, USA) statistical software was applied to determine to statistical significance. Data are presented as mean ± standard deviation. Two groups were compared by Student's t-test and the comparison between multiple groups was conducted by one-way analysis of variance followed by Student-Neuman-Keuls post hoc comparisons.
Results
Expression of miR-146a-5p in breast cancer and breast cancer cell lines
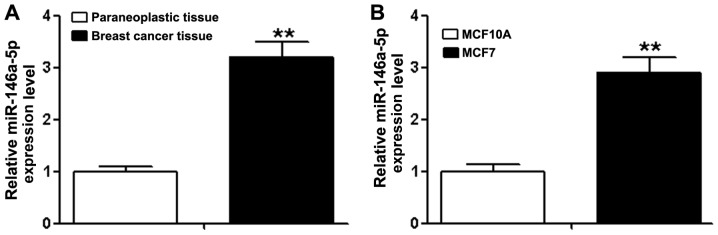
The results of RT-qPCR demonstrated that the expression of miR-146a-5p in breast cancer tissue was 3.2±0.3 times more, compared with paraneoplastic tissue (Fig. 1A, P<0.01), the expression in MCF-7 cell lines was 2.9±0.3 times more, compared with MCF 10A cells (Fig. 1B, P<0.01).
Figure 1.
Expression of miR-146a-5p in breast cancer tissue and breast cancer cell line. (A) The expression of miR-146a-5p in breast cancer tissues was significantly higher, compared with the paraneoplastic tissue. **P<0.01. (B) The expression of miR-146a-5p in breast cancer cell line MCF-7 was significantly higher, compared with the normal breast epithelial cells MCF 10A. **P<0.01. miR-146a-5p, microRNA-146a-5p.
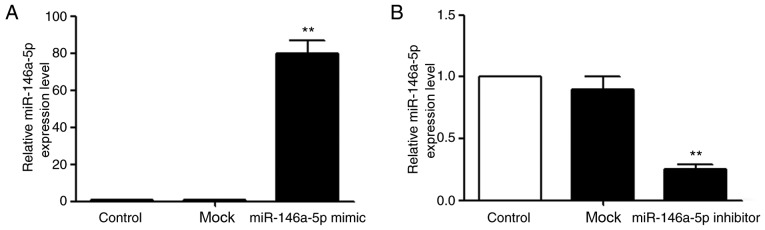
miR-146a-5p mimic and inhibitor transfection into MCF-7 cells
According to the miR mimic and inhibitor transfection introduction, the transfection concentration was 50 nM. The results indicated that the expression of miR-146a-5p in MCF-7 cells transfected with miR-146a-5p mimic was 80±7 times more, compared with original expression (Fig. 2A, P<0.01), whilst cells transfected with miR-146a-5p inhibitor decreased to about 0.25±0.04 times compared with original expression (Fig. 2B, P<0.01). These results indicated that the transfections were successful.
Figure 2.
Mimic miR-146a-5p and inhibitor miR-146a-5p transfected MCF-7 cells. (A) Transfected with mimic miR-146a-5p, the expression of miR-146a-5p in MCF-7 cells was significantly upregulated, compared with blank control group. **P<0.01. (B) Transfected with inhibitor miR-146a-5p, the expression of miR-146a-5p in MCF-7 cells was significantly downregulated, compared with the blank control group. **P<0.01. miR-146a-5p, microRNA-146a-5p.
Effect of miR-146a-5p on the proliferation of breast cancer cell line MCF-7
The proliferation (OD 490) of MCF-7 cells increased gradually following D3. The proliferation of the overexpression group significantly increased following transfection of the miR-146a-5p mimic at D2 (0.51±0.03, P<0.05) and D3 (0.73±0.04, P<0.01), whilst the mock group cells at D2 and D3 (D2, 0.42±0.03; D3, 0.54±0.05) had no significant difference (Fig. 3) with the blank control group (D2, 0.40±0.04; D3, 0.56±0.05). By contrast, the proliferation of suppression group following the transfection of miR-146a-5p inhibitor was significantly decreased, compared with the blank control group at D2 (0.30±0.03, P<0.05) and D3 (0.41±0.04, P<0.01). These results indicated that high expression of miR-146a-5p in MCF-7 cells promoted proliferation, and low expression of miR-146a-5p inhibited proliferation significantly (Fig. 3).
Figure 3.
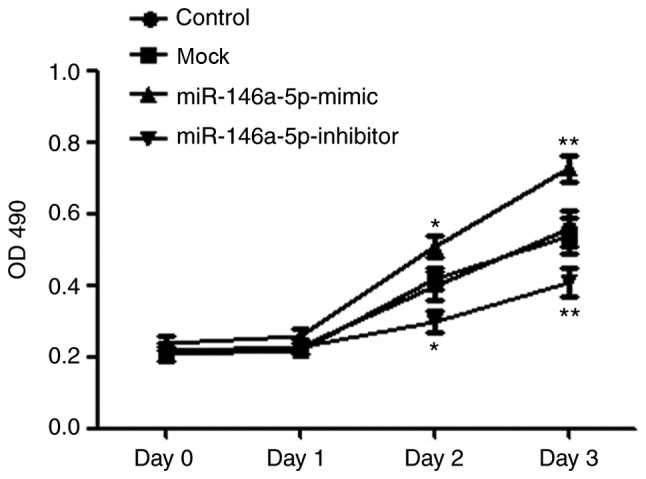
Effects on the proliferation of MCF-7 following transfection of mimic miR-146a-5p (*P<0.05 vs. the blank control group) and inhibitor miR-146a-5p (**P<0.01 vs. the blank control group). miR-146a-5p, microRNA-146a-5p; OD, optical density.
Detection of miR-146a-5p target genes
Through further analysis of bioinformatic methods, it was determined that the 3′UTR of tumor suppressor gene BRCA1 existed at the acting site of miR-146a-5p (Fig. 4). Furthermore, the interacting sequence was highly conserved between species. All examples indicated that BRCA1 may be the target gene of miR-146a-5p. The pGL3 plasmid was recombined with luciferase expression of BRCA1 3′UTR. The luciferase results demonstrated that the luciferase activity (0.40±0.04) of transfected miR-146a-5p group was significantly lower, compared with the control group transfected with mock (Fig. 5, P<0.01), indicating that BRCA1 was the target gene of miR-146a-5p.
Figure 4.

Bioinformatics analysis revealed that the 3′untranslated region of tumor suppressor gene BRCA1 existed at the acting site of miR-146a-5p. miR-146a-5p, microRNA-146a-5p.
Figure 5.
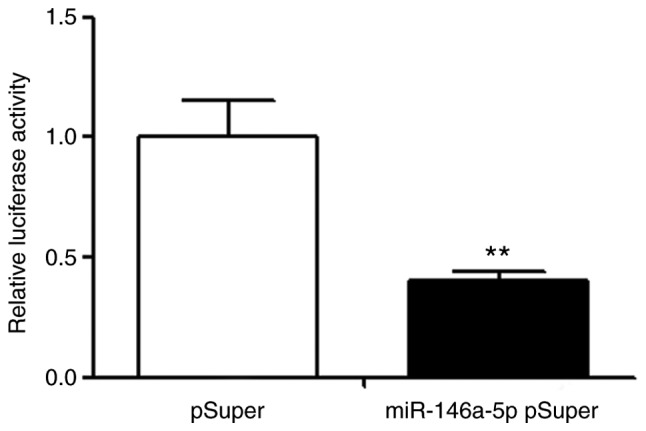
The luciferase activity of transfected miR-146a-5p group was significantly reduced compared with the control group. **P<0.01.
Discussion
miRNA is a type of non-coding small single stranded RNA containing 20–24 nucleotides involved in gene transcription and expression (19). miRNA is not encoding protein, but has the ability to direct degrade the target mRNA or inhibit its translation by complete or incomplete complementary combination with the target mRNA (20). Numerous studies have demonstrated that miRNA is involved in the process of tumor growth, metastasis and angiogenesis by regulating the expression of oncogenesis, migration and other associated genes (21–23).
In 2005, Iorio et al (24) first reported the change of miRNAs expression profile in human breast cancer and determined that there were 29 kinds of miRNAs with an expression disorder, in which the expression of miRNA-21 and miRNA-155 were significantly upregulated, whilst the expression of miRNA-10b, miRNA-125b and miRNA-145 were significantly reduced. Additionally, a number were associated with the clinical and pathological features of breast cancer, including estrogen and progesterone receptor expression and vascular invasion. In 2007, Blenkiron et al (25) determined that there were 133 kinds of miRNA expression in normal breast tissue and/or cancer tissues, a number of which were associated with molecular subtype of breast cancer. Following this, the role of miRNAs in breast cancer gained more attention.
In the previous work, it was determined that the expression of miR-146a-5p in breast cancer tissue was significantly higher, compared with paraneoplastic tissue (16). It was confirmed through RT-qPCR in the present study and also discovered that the expression of miR-146a-5p in breast cancer cell line MCF-7 was also significantly higher, compared with control cells. High expression of miR-146a-5p in MCF-7 could significantly promote the proliferation, and low expression of miR-146a-5p could significantly inhibit it. BRCA1 was preliminarily confirmed as the target gene of miR-146a-5p by bioinformatics prediction and fluorescence reporter gene detection.
BRCA1 is a tumor suppressor gene, which frequently mutated in hereditary breast cancer (26). BRCA1 serves an important role in DNA repair, cell cycle control, transcriptional activation, ubiquitin and other processes (27,28). In the present study, it was determined that the proliferation of MCF-7 cells was enhanced following high expression of miR-146a-5p, whilst the proliferation of MCF-7 cells decreased following low expression of miR-146a-5p. It was hypothesized that the decreased expression of BRCA1, in consequence of the high expression of miR-146a-5p, weakened the anti-tumor ability and caused the increase of tumor cell proliferation. Following miR-146a-5p low expression, the expression of BRCA1 was increased, which caused the inhibition of tumor and the decrease of the proliferation ability.
In conclusion, high expression of miR-146a-5p in breast cancer and breast cancer cell lines may regulate the proliferation of MCF-7 by regulating the expression of BRCA1.
Acknowledgements
Not applicable.
Funding
The present study was supported by the natural science foundation of Science and Technology Commission of Shanghai Municipality (grant no. 14ZR1408800).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GW, HJ and YY designed this study. JZ, DJ, HZ, DY and LQ performed the experiments and interpreted the data. GW and HJ analyzed and interpreted the data. GW and HJ performed the histological examination of the breast and were major contributors towards the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Changhai Hospital Scholastic Ethics Committee approved the research. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
All subjects signed informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rodriguez-Barrueco R, Nekritz EA, Bertucci F, Yu J, Sanchez-Garcia F, Zeleke TZ, Gorbatenko A, Birnbaum D, Ezhkova E, Cordon-Cardo C, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31:553–566. doi: 10.1101/gad.292318.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y, Dong L, Zhang J, Zhao Y, Li Z. Recent advances in microRNA detection. Analyst. 2018 doi: 10.1039/C7AN02001E. [DOI] [PubMed] [Google Scholar]
- 4.Rapado-González Ó, Majem B, Muinelo-Romay L, Álvarez-Castro A, Santamaría A, Gil-Moreno A, López-López R, Suárez-Cunqueiro MM. Human salivary microRNAs in Cancer. J Cancer. 2018;9:638–649. doi: 10.7150/jca.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev Biol. 2016;410:1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Harding RL, Velleman SG. MicroRNA regulation of myogenic satellite cell proliferation and differentiation. Mol Cell Biochem. 2016;412:181–195. doi: 10.1007/s11010-015-2625-6. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa R, Leyland R, Meyer-Hermann M, Lu D, Turner M, Arbore G, Phan TG, Brink R, Vigorito E. MicroRNA-155 controls affinity-based selection by protecting c-MYC+ B cells from apoptosis. J Clin Invest. 2016;126:377–388. doi: 10.1172/JCI82914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito Y, Nakaoka T, Saito H. microRNA-34a as a therapeutic agent against human cancer. J Clin Med. 2015;4:1951–1959. doi: 10.3390/jcm4111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, Rehimi H. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12:1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou LK, Yu Y, Xie YG, Wang J, Mao JF, Zhang B, Wang X, Cao XC. miR-340 and ZEB1 negative feedback loop regulates TGF-β-mediated breast cancer progression. Oncotarget. 2016;7:26016–26026. doi: 10.18632/oncotarget.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimono Y, Mukohyama J, Nakamura S, Minami H. MicroRNA regulation of human breast cancer stem cells. J Clin Med. 2015;5 doi: 10.3390/jcm5010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi RU, Miyazaki H, Ochiya T. The roles of micrornas in breast cancer. Cancers(Basel) 2015;7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Teng Y, Liu Q. MicroRNA-153 regulates NRF2 expression and is associated with breast carcinogenesis. Clin Lab. 2016;62:39–47. doi: 10.7754/Clin.Lab.2015.150518. [DOI] [PubMed] [Google Scholar]
- 16.Meshkat M, Tanha HM, Naeini MM, Ghaedi K, Sanati MH, Meshkat M, Bagheri F. Functional SNP in stem of mir-146a affects Her2 status and breast cancer survival. Cancer Biomark. 2016;17:213–222. doi: 10.3233/CBM-160633. [DOI] [PubMed] [Google Scholar]
- 17.Majer A, Blanchard AA, Medina S, Booth SA, Myal Y. Claudin 1 expression levels affect miRNA dynamics in human basal-like breast cancer cells. DNA Cell Biol. 2016;35:328–339. doi: 10.1089/dna.2015.3188. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Schanza LM, Seles M, Stotz M, Fosselteder J, Hutterer GC, Pichler M, Stiegelbauer V. MicroRNAs associated with von hippel-lindau pathway in renal cell carcinoma: A comprehensive review. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18112495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagiya T. MicroRNAs: Potential biomarkers and therapeutic targets for alveolar bone loss in periodontal disease. Int J Mol Sci. 2016;17:E1317. doi: 10.3390/ijms17081317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA processing and human cancer. J Clin Med. 2015;4:1651–1667. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 25.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schayek H, Korach H, Laitman Y, Bernstein-Molho R, Friedman E. Mutational analysis of candidate genes in Israeli male breast cancer cases. Breast Cancer Res Treat. 2018 doi: 10.1007/s10549-018-4765-3. [DOI] [PubMed] [Google Scholar]
- 27.Drost R, Jonkers J. Opportunities and hurdles in the treatment of BRCA1-related breast cancer. Oncogene. 2014;33:3753–3763. doi: 10.1038/onc.2013.329. [DOI] [PubMed] [Google Scholar]
- 28.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.