Abstract
Gastric cancer (stomach cancer) is the fifth most common malignancy and the third leading cause of cancer-associated mortality worldwide. Identifying gastric cancer patients at an early and curable stage of the disease is essential if mortality rates for this disease are to decrease. A non-invasive blood-based test that is an indicator of gastric cancer risk would likely be of benefit in identifying gastric cancer patients at an early stage, and such a test may enhance clinical decision making. This study identified a four-gene expression signature in peripheral blood samples associated with gastric cancer. A total of 216 blood samples were collected, including those from 36 gastric cancer patients, 55 healthy controls and 125 patients with other carcinomas, and gene expression profiles were examined using an Affymetrix Gene Profiling microarray. Blood gene expression profiles were compared between patients with stomach cancer, healthy controls and patients affected with other malignancies. A four-gene panel was identified comprising purine-rich element binding protein B, structural maintenance of chromosomes 1A, DENN/MADD domain containing 1B and programmed cell death 4. The four-gene panel discriminated gastric cancer with an area under the receiver-operating-characteristic curve of 0.99, an accuracy of 95%, sensitivity of 92% and specificity of 96%. The non-invasive nature of the blood test, together with the relatively high accuracy of the four-gene panel may assist physicians in gastric cancer screening decision making.
Keywords: gastric cancer, gene expression, blood, diagnostic testing, personalized medicine, genomics
Introduction
Although the incidence of gastric cancer is on the decline, it is the fifth most common malignancy and the third leading cause of cancer-associated death worldwide, with 952,000 new cases diagnosed and 723,000 deaths occurring in 2012 (1). In China, gastric cancer ranks as the third most common cancer overall in incidence and mortality, with an even higher prevalence in rural areas (2). Gastric cancer patient survival critically depends on the stage at which the malignancy is diagnosed. The 5-year survival rate of early stage gastric cancer patients who have undergone resection is excellent, whereas survival is poor for patients with advanced disease (3,4). However, most gastric cancer patients are diagnosed only after their cancer has reached later stages, resulting in poor prognosis due to a high rate of relapse after gastrectomy (5). Thus, identifying gastric cancer patients at early and curable stages of the disease is essential for decreasing gastric cancer mortality.
Several screening techniques have been developed to facilitate detection of gastric cancer, including barium-meal photofluorography, gastric endoscopy and the serum pepsinogen test. Although it has been shown that these techniques are helpful for gastric cancer screening, none of these have become a standard or routine screening test due to the potential risks and inconvenience associated with these procedures (6–8). For example, the barium-meal photofluorography test requires dietary restriction and radiation exposure, and gastric endoscopy may result in perforation, cardiopulmonary events, aspiration pneumonia and bleeding (9). In addition, the serum pepsinogen and the barium-meal photofluorography tests are commonly plagued with false-positive results in clinical practice (9). Furthermore, the availability of endoscopic instruments and medical expertise required for mass screening remains limited, especially in rural areas (10). Thus, the challenge remains to develop a convenient, non-invasive and accurate test for detecting early stage gastric cancer.
Gene signatures or biomarkers have shown great potential in clinical applications such as disease detection, prognosis and targeted therapy. Peripheral blood includes immune cells, which dynamically respond to various physiological conditions such as lesions and cancers occurring in the body (10). In particular, blood-derived RNA biomarkers are emerging as potentially important tools in the screening and early detection of various diseases. RNA-based biomarker technology is based on the Sentinel Principle® (11), which suggests that information regarding the current state of health or disease of an organism is conveyed in the blood via interactions between circulating blood cells and the cells, tissues and organs of the body. Blood cells therefore act as sentinels that indicate the status of health or disease in the body. As blood samples may be readily obtained and with little discomfort to patients, biomarkers derived from blood RNA provide an alternative to tissue biopsies for the diagnosis and prognosis of disease.
Over the past decade, the gene expression pattern of blood cells as a valuable resource for biomarker identification and pharmacogenomics has been investigated. It has been demonstrated that RNA profiling in whole blood may be used to develop molecular signatures of disease across a broad spectrum of pathology (12–15). Blood mRNA expression profiles may identify a variety of non-hematologic disorders, such as heart failure (16), cancer (17–21), inflammatory bowel disease (22,23) and psychiatric disorders (24–26).
The present study aimed to identify gastric cancer-associated expression signatures by generating and analyzing gene expression profiles of whole blood from gastric cancer patients and corresponding controls.
Materials and methods
Ethics
This study was approved by the Ethics Committee of the Second Hospital of Anhui Medical University (IRB no. KY201405). Sample acquisition for identifying gastric cancer-specific genetic signatures was conducted between March 2014 and February 2015 at the Second Hospital of Anhui Medical University. All 216 participants including 36 gastric cancer patients, 55 healthy controls and 125 non-gastric carcinoma patients were enrolled and provided written informed consent.
Study population
A total of 36 blood samples from gastric cancer patients were obtained from 27 male and 9 female adult patients (age range, 21–89 years; mean age, 63±10 years) and were collected before the patients had undergone any form of treatment including gastrectomy, radio/chemo-therapy or surgery. Patients enrolled for gastroendoscopy donated blood before the gastroendoscopy and were categorized after pathological examination. Healthy control samples comprised 55 blood samples from subjects with no pathology at gastroendoscopy (30 males and 25 females; mean age, 31±9 years). In addition, 125 blood samples from patients with non-gastric carcinomas (91 males and 34 females; mean age, 56±12 years) were collected before any form of treatment and were categorized after pathologist reports were reviewed. The non-gastric carcinomas included 33 lung cancers, 30 liver cancers, 21 prostate cancers, 20 nasopharyngeal carcinomas, 12 breast cancers, 8 oral cancers and 1 colorectal cancer.
Blood collection, RNA isolation and RNA quality control
Peripheral whole blood (2.5 ml) was collected in PaxGene Blood RNA tubes (PreAnalytix GmbH, Hombrechtikon, Switzerland). Total RNA was then isolated as described previously (11). RNA quality was accessed using 2100 Bioanalyzer RNA 6000 Nano Chips (Agilent Technologies, Inc., Santa Clara, CA, USA). All the samples for microarray analysis met the following quality criteria: RNA integrity number ≥7.0 and 28S:18S rRNA ≥1.0. RNA quantity was determined by a NanoDrop 1000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Microarray hybridization
Whole blood RNA from the 216 samples, including 36 gastric cancer, 55 healthy controls and 125 non-gastric carcinomas, was analysed by microarray hybridization in accordance with the manufacturer's protocol for Gene Profiling Array cGMP U133 P2 (Affymetrix; Thermo Fisher Scientific, Inc.). A total of 200 ng of each RNA sample was used for cDNA synthesis (GeneChip 3′IVT PLUS Reagent kit; Thermo Fisher Scientific, Inc.) and hybridization using the accessory reagents of the Affymetrix microarray, according to the manufacturer's protocols. Gene expression profiles of RNA samples were processed using Affymetrix Expression Console software (version 1.4.1; Affymetrix; Thermo Fisher Scientific, Inc.) and normalized by the MAS5 normalization method (27), in which the global signal intensity value was adjusted to 500 for each microarray to make it possible to compare the profiling variations among microarrays (27).
Microarray data analysis
To identify candidate genes for gastric cancer, probe sets were selected from 54,675 probe sets on the Affymetrix Gene Profiling cGMP U133 P2 microarray, according to criteria of reliability, repeatability and linearity. A total of 4,303 probe sets were selected for further analysis, which presented in all the samples with intensities ranging from 100 to 10,000. These probe sets were also present within the MAQC list for Affymetrix U133 P2 microarray, as reported by the MAQC Consortium (28) and were repeatable within ±15% in whole blood technical replicates (unpublished data). All microarray procedures were performed by our laboratory at the National Engineering Center for Biochip at Shanghai (Shanghai, China).
A total of 216 samples was divided into a training set of 31 gastric cancer, 33 controls and 99 non-gastric carcinoma (163 samples) and a test set of 5 gastric cancer, 22 controls and 26 non-gastric carcinoma (53 samples), which were used to generate and to validate a predictive model of gastric cancer. The methodology for data mining was based on the self-training logistic regression algorithm developed and reported previously (12,29,30). The area under the receiver-operating characteristic (ROC) curve (AUC) was calculated to characterize the ability of each probe set to distinguish gastric cancer from healthy controls and non-gastric carcinomas. First, of the 4,303 probe sets, the top 60 probe sets exhibiting the highest correlation coefficient values when compared with gastric cancer (calculated using the CORREL function of Microsoft Excel 2010 software) were selected as the primary probe sets. Subsequently, another 60 probe sets (of the 4,303 probe sets) exhibiting high correlation coefficient values with the primary 60 probe sets were selected as secondary probe sets. A total of 60 primary and 60 secondary probe sets were paired one-by-one to achieve a data matrix containing a possible 14,280 pairs of probe sets. The pair combinations were then generated from the 14,280 pairs and were used to calculate the ROC AUC values for each pair combination in order to find the combination with the highest ROC AUC.
To reduce the risk of overfitting the data in the ROC AUC calculation, each combination was limited to 2 or 3 pairs. As the number of 3-pair combinations from 14,280 pairs was extremely large (4.9×1011) and would make the screening process tedious and inefficient, the process of identifying top combinations was accelerated using a Monte Carlo algorithm (31). First a training fold was generated, made up of repeated random selections of gastric cancer samples, controls, and non-gastric cancer samples; that is, each set in the training fold was independently generated from the total samples and contained half of the samples of each category of samples (18 gastric cancer; 27 healthy controls and 63 non-gastric cancer), and this process was repeated 1,000 times to produce 1,000 randomly selected sets. Three pairs from the total 14,280 pairs were then randomly selected and combined continuously, until a 3-pair combination repetition occurred. A total of >70,000 3-pair combinations were generated during this process. The 3-pair combination was used as a basic unit to predict gastric cancer in the 1,000 sample cohorts of the training fold. By comparing the ROC AUC value of each combination for predicting gastric cancer, the final 3-pair combination with the best ability to distinguish gastric cancer from healthy controls and from non-gastric carcinomas was identified. The ROC AUC values of each 3-pair combination in diagnosing gastric cancer were calculated using Commercial MedCalc statistical software (MedCalc Software bvba, Ostend, Belgium), according to DeLong methodology (32).
Due to the fact that the sample size used in the present study was small, it is possible that the genes with the highest ROC AUC values selected for gastric cancer detection may have derived from random chance. In order to avoid this, we performed a 2-fold cross-validation (iterated 1,000 times). In brief, all the samples were randomly and equally distributed into the training fold and the test fold. A predictive model was constructed based on selected genes and using logistic regression, according to the performance of the model in the training fold. The model was then used to predict the rest of the samples in the test fold. This was repeated 1,000 times. In order to test whether the selected genes were derived due to random chance, the model was tested twice. The first time the model was used to predict the sample cohort with the true cancer/control status and the second time the model was used to predict the sample cohort with the status randomly reassigned (null set).
Results
For the present study, blood samples were collected from 216 individuals, including 36 gastric cancer patients (27 males and 9 females; mean age, 63±10 years), 55 healthy controls (30 males and 25 females; mean age, 31±9 years) and 125 non-gastric carcinomas (91 males and 34 females; mean age, 56±12 years). The clinical characteristics of the gastric cancer patients are described in Fig. 1A-D, including age, tumor location, tumor node metastasis (TNM) status and pathological classification.
Figure 1.
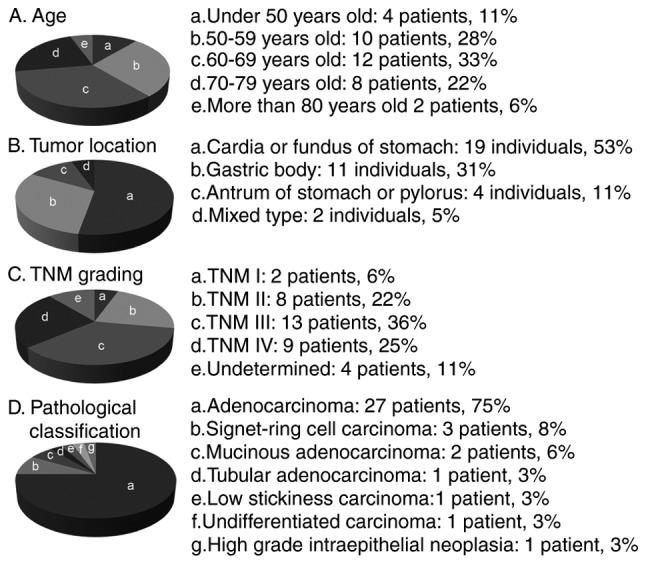
Clinical characteristics of patients with gastric cancer. There were 27 males (75%) and 9 females (25%).
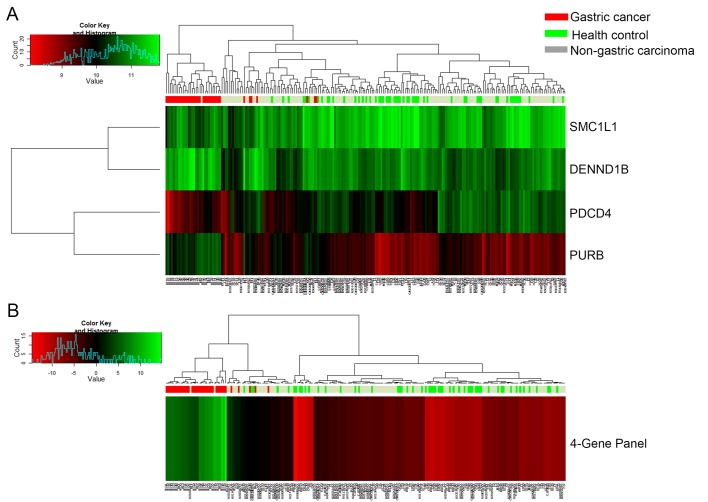
Four candidate genes for detecting gastric cancer were identified via a self-training logistic regression model, including purine-rich element binding protein B (PURB), structural maintenance of chromosomes 1A (SMC1L1), DENN/MADD domain containing 1B (DENND1B) and programmed cell death 4 (PDCD4). Two of the genes (PURB and DENND1B) were overexpressed (2.2- and 1.5-fold, respectively) in gastric cancer samples when compared with healthy control samples, while the other two genes (SMC1L1 and PDCD4) were underexpressed (−1.6- and −1.7-fold, respectively; data not shown). The linear fold changes in the expression of the four genes were statistically significantly different (P<0.001) between the gastric cancer patient samples and the healthy control samples (data not shown). Fig. 2A and B demonstrate, through hierarchical cluster diagrams, the performance of each candidate gene and of the four-gene panel for gastric cancer differentiation for a total of 216 samples (36 gastric cancer, 55 controls and 125 non-gastric carcinomas). It was demonstrated that the majority of the 36 gastric cancer samples were clustered together and separated from healthy controls and non-gastric carcinoma samples. Table I demonstrates the performance of the four candidate genes (PURB, SMC1L1, DENND1B and PDCD4) selected from the microarray analysis based on 163 samples in the training set cohort (31 gastric cancer, 33 controls and 99 non-gastric carcinoma). The remaining 53 of the 216 samples were set aside as independent samples for validation, as a test set cohort. The four-gene panel had an AUC of 0.99, with 95% accuracy, 90% sensitivity, 96% specificity for healthy controls and non-gastric carcinoma in the training set.
Figure 2.
Heat map of gene expression and hierarchical cluster diagram showing (A) the performance of four separate candidate genes and (B) four-gene panel for clustering 163 samples in the training set, including 31 gastric cancers, 33 healthy controls and 99 non-gastric carcinoma samples. The dendrogram was generated using the heatmap function in R, using default settings for the clustering algorithm.
Table I.
Performance of the four-gene panel for gastric cancer diagnosis.
| Training set | Test set | |||||
|---|---|---|---|---|---|---|
| Characteristic | Gastric cancer | Healthy controls | Non-GC controls | Gastric controls | Healthy controls | Non-GC controls |
| Positive prediction | 28 | 0 | 5 | 5 | 0 | 3 |
| Negative prediction | 3 | 33 | 94 | 0 | 22 | 23 |
| Sensitivity | 90% | 100% | ||||
| Specificity | 96% | 94% | ||||
| Accuracy | 95% | 94% | ||||
| ROC AUC | 0.99 | 0.99 | ||||
GC, gastric cancer; ROC AUC, area under the receiver operating characteristic curve.
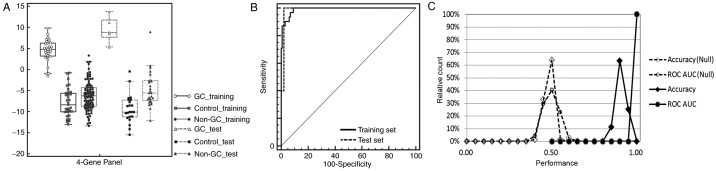
Mathematical predictive models built on the training set were then used to evaluate the completely independent samples in the test set cohort (total 53: gastric cancer, 5; controls, 22; and non-gastric carcinoma, 26). The performance of the predictive model for the test set had characteristics similar to that of the training set: 0.99 of ROC AUC, 94% accuracy, 100% sensitivity, 94% specificity for healthy controls and non-gastric carcinoma (Table I). Three of the 26 non-gastric cancer samples, including 1 breast cancer, 1 colorectal cancer and 1 lung cancer in the training set were predicted as positives; the reason for these false-positive results requires further study in larger cohorts. When results for the training set and test set were combined, the four-gene panel had an accuracy of 95%, sensitivity of 92% and specificity of 96% for gastric cancer diagnosis. Fig. 3A demonstrates the performance of the four-gene panel as a box-whisker plot based on logistic regression analysis. Fig. 3B demonstrates the high ROC AUC value of >0.9 for gastric cancer diagnosis.
Figure 3.
Performance of four-gene panel for gastric cancer detection. (A) Box-whisker plot to display the logistic regression scores in gastric cancer samples, healthy controls and other non-gastric carcinoma samples in the training set and test set. (B) Logistic regression scores were calculated from a self-trained logistic regression model. ROC, receiver operating characteristic. (C) Two-fold cross validation for predicting true disease state and random assigned disease state (null) for 1,000 iterations.
To rule out the possibility that this four-gene panel was due merely to chance 2-fold cross-validations were performed, in which samples were randomly and equally divided into training and test folds. The samples in the training folds were used to define coefficients and thresholds for building the predictive model based on logistic regression analysis, and the resulting predictive model was used to make predictions of the samples in the test folds. This process was repeated 1,000 times to evaluate gastric cancer detection performance for the sample cohorts using the measured gene expression data, first with the true cancer/control status and then with the status randomly reassigned (null set).
It was demonstrated that the distributions of the accuracy and AUC ROC values over the 1,000 iterations resulted in 2 well-separated curves for gastric cancer, with the ‘true disease state’ results forming a group with an accuracy and ROC AUC >90%, whereas the ‘null-set’ with the randomly reassigned disease status cluster had an accuracy and ROC AUC of ~50% (Fig. 3C). There was no significant overlap between the two clusters, from which it was concluded that the observed performance of the 4-gene panel is unlikely to be the result of random chance.
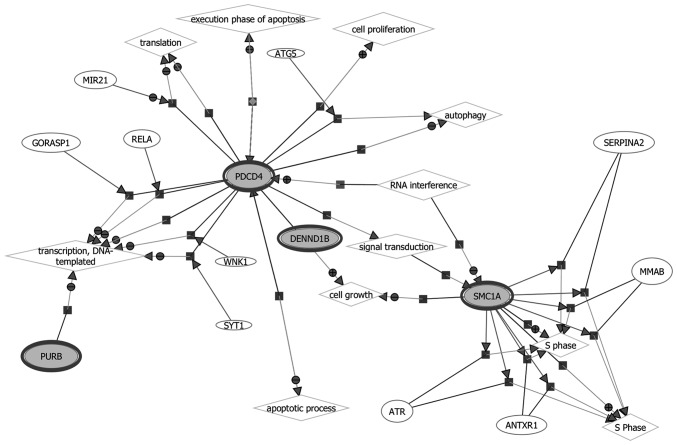
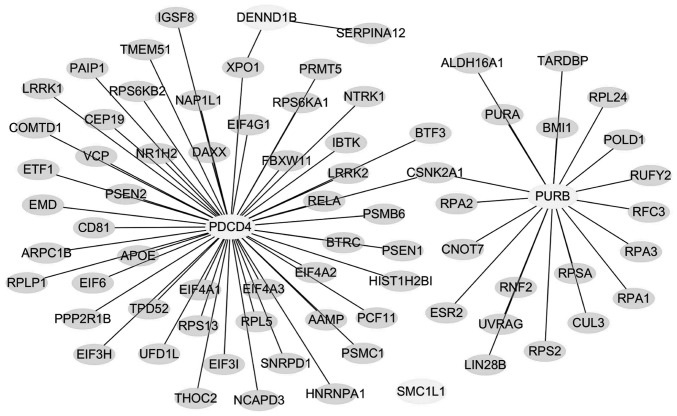
The biological functions and gene networks of the selected 4 genes (PURB, SMC1L1, DENND1B and PDCD) were investigated, as demonstrated in Fig. 4. Since there was an insufficient number of genes in the gastric cancer signature to conduct pathway analysis directly, the four genes in the blood-based gene signature were assessed for their known protein interactions, and a protein-protein interaction (PPI) network was constructed as demonstrated in Fig. 5. It was identified that 3 genes (PURB, DENND1B and PDCD) were connected through a protein-protein network. The pathways of PURB, DENND1B and PDCD were enriched with the proteins in the network according to their biological functions. SMC1L1 by contrast was not found to be connected in the protein network, and the reason for this is unknown. Table II presents the closely associated network canonical pathways using hypergeometric distribution analysis.
Figure 4.
Gene network of biological processes involving the four-gene panel, PURB, SMC1L1 (SMC1A), DENND1B and PDCD4 (all in bold ovals). PURB, purine-rich element binding protein B; SMC1L1 (SMC1A), structural maintenance of chromosomes 1A; DENND1B, DENN/MADD domain containing 1B; PDCD4, programmed cell death 4.
Figure 5.
Protein-protein interaction network of the four gastric-cancer-specific genes in peripheral blood. The network indicates that PURB, DENND1B and PDCD4 are connected, while SMC1L1 was not.
Table II.
Closely associated canonical pathways using a hypergeometric distribution analysis.
| Canonical pathway | Genes in category | Percent in the observed list | Percent in the genome | Fold of over-representation | Odds ratio | P-value |
|---|---|---|---|---|---|---|
| RNA transport | 10 | 0.20 | 0.03 | 7.93 | 10.33 | 3.28×10−7 |
| Mismatch repair | 5 | 0.10 | 0.00 | 26.04 | 36.63 | 9.89×10−7 |
| DNA replication | 5 | 0.10 | 0.01 | 16.64 | 21.22 | 1.02×10−5 |
| Nucleotide excision repair | 5 | 0.10 | 0.01 | 13.61 | 16.84 | 2.80×10−5 |
| Homologous recombination | 4 | 0.08 | 0.00 | 17.11 | 21.47 | 7.58×10−5 |
| Ribosome | 6 | 0.12 | 0.02 | 7.90 | 9.41 | 9.63×10−5 |
| Shigellosis | 4 | 0.08 | 0.01 | 7.85 | 8.99 | 1.58×10−3 |
| Neurotrophin signaling pathway | 5 | 0.10 | 0.02 | 4.72 | 5.31 | 3.90×10−3 |
| mRNA surveillance pathway | 4 | 0.08 | 0.01 | 5.77 | 6.46 | 4.86×10−3 |
| Wnt signaling pathway | 5 | 0.10 | 0.03 | 3.99 | 4.45 | 7.87×10−3 |
| Oocyte meiosis | 4 | 0.08 | 0.02 | 4.28 | 4.70 | 1.38×10−2 |
| Circadian rhythm | 2 | 0.04 | 0.00 | 10.89 | 12.34 | 1.42×10−2 |
| Spliceosome | 4 | 0.08 | 0.02 | 3.77 | 4.12 | 2.09×10−2 |
| Ribosome biogenesis in eukaryotes | 3 | 0.06 | 0.01 | 4.49 | 4.86 | 2.87×10−2 |
Discussion
It is well recognized that a minimally invasive, accurate diagnostic test would be of major importance in reducing mortality from gastric cancer. Efforts have been made to identify biomarkers that may be of clinical use in detecting early stage gastric cancer, including high throughput genetic, epigenetic, transcriptomic and proteomic technologies (33,34). Other investigators have studied circulating tumor cells in late-stage gastric cancer (35). By contrast, this study aimed to identify a tumor-independent gene expression biological signature for gastric cancer in peripheral blood, a potential ‘liquid biopsy’. Such a blood-based signature, if clinically verified, may potentially provide an objective, non-invasive, tissue-independent test for the detection of early gastric cancer.
The expression signature of peripheral blood cells that the present study has identified in gastric cancer patients may be a result of the interaction between peripheral blood cells and gastric cancer lesions. This hypothesis is supported by a previous study by Sakai et al (36), who reported that the peripheral blood cells of hepatocellular carcinoma patients shared common gene expression alterations with tumor-infiltrating mononuclear inflammatory cells, providing clues to the source of gene expression alteration in peripheral blood cells of gastric cancer patients.
Although the gastric cancer samples used in the present study were limited in number, the prediction performance was evaluated by distributing the samples into traditional training and test sets. To reduce any possible bias that may occur due to the limited number of gastric cancer samples (for example, a single data point skewing the results, particularly for samples in the test set), the samples were randomly distributed using a random sampling process with fixed proportions in the training set (28/33) and in the test set (5/33). The four-gene panel was used to predict outcomes across the two sample sets (the training and test sets) via random sampling and cross-validation for 1,000 iterations. The prediction performances of the four-gene panel were consistent throughout the 1,000-times cross-validation process, suggesting high reliability and reproducibility of prediction results, despite the small number of gastric cancer samples in the test set.
The blood-based four-gene signature identified in the present study may be the first reported, and comprised the genes PURB, SMC1L1, DENND1B and PDCD4. The four-gene panel was obtained by analyzing blood cell expression profiles from 36 gastric cancer patients, 55 healthy controls and 125 patients with non-gastric carcinomas. The logistic regression scores of gastric cancer, healthy controls and non-gastric cancer carcinoma samples in the training set (31 gastric cancers, 33 controls and 99 non-gastric carcinomas) and the test set (5 gastric cancers, 22 controls and 26 non-gastric carcinomas) were analyzed. The logistic regression scores for each sample were calculated through a self-trained logistic regression model. The samples were predicted as gastric cancer if their logistic regression scores were ≥0. The four-gene panel showed high performance in discriminating gastric cancer from healthy controls and from non-gastric carcinomas with an accuracy of 95% and ROC AUC value of 0.99 (95% confidence interval) by combining the results of the training and test sets.
There have been no previous reports associating PURB, SMC1L1 and DENND1B with gastric cancer. However, the involvement of PDCD4 in gastric cancer has been extensively examined. PDCD4 is associated with tumor cell promotion, progression and metastasis and is regarded as a tumor suppressor and a potential molecular target for the diagnosis and treatment of certain tumors. It has been demonstrated that PDCD4 expression in human digestive tract cancers such as gastric, colorectal and pancreatic cancers was significantly downregulated compared with PDCD4 expression in normal digestive tract tissues (37–39). In addition, the degree of PDCD4 downregulation was associated with the degree of differentiation of hepatocellular and gastric cancer cells (36,37).
The PPI network contained 75 genes closely associated with PURB, DENND1B and PDCD, which may be enriched to 14 canonical pathways. Hypergeometric distribution analysis against canonical pathways demonstrated that these genes are closely associated with pathways involved in RNA transport, replication and repair, oocyte meiosis and Wnt signaling. Through the closely associated genes presented in the PPI network, the four gastric cancer-specific genes found in this study were directly or indirectly involved in important biological tumorigenic processes such as regulation of cell growth and death, DNA replication and mismatch repair (Table II).
Although SMC1L1, DENND1B and PURB have not been directly associated with gastric cancer, these genes exhibit biological relevance in other types of carcinoma, including colorectal, cervical, pancreatic and renal cancer. SMC1L1 is represented in cell cycle and oocyte meiosis pathways, which are responsible for the structural maintenance of chromosome 1A and provide nucleotide binding sites. SMC1L1 has not yet been associated with gastric cancer; however, this gene is reportedly associated with biological processes involved in cervical and colorectal cancer. It has been reported that SMC1L1 expression is upregulated in cervical cancer relative to normal cervical tissue (40). The SMC1L1 protein was also identified in gastric tissue and acts as a CDX2-binding protein, and CDX2 is regarded as a tumor suppressor for colorectal cancer (41). The expression of SMC1L1 for the S phase checkpoint protein was upregulated by treatment with ferulic acid (FA), whereas FA served a protective role in the development of colon cancer (42). With respect to the DENND1B (DENN/MADD domain containing 1B), its single nucleotide polymorphisms have been significantly associated with risk for pancreatic cancer (43) and have been associated with genetically inherited renal cancer (44). PURB is responsible for encoding functionally cooperative proteins in the Pur family. It has been reported that the deletion of PURB in patients with acute myelogenous leukemia was significantly higher (>5-fold higher) than statistically expected (45).
In conclusion, the four-gene panel identified from peripheral blood was able to differentiate gastric cancer from healthy controls and non-gastric carcinomas, and shows biological plausibility in cancer pathogenesis. The biological signature identified in this study differed from conventional tumor-derived cancer biomarkers. The candidate four-gene signature identified in the present study likely reflects subtle alterations in blood gene expression, serving as a systemic response to disease and possibly acting to maintain homeostasis or mediating disease pathology.
Although the findings of the present study require further validation using larger cohorts, this study suggests the possibility of detecting gastric cancer using gene expression profiles derived from blood. As a non-invasive, blood-based test, the gene signature may be of benefit to healthcare providers to help assess the requirement for increased monitoring of patients, or to suggest the requirement for further, more invasive and expensive procedures to confirm gastric cancer in an individual patient. The results of this study and other research demonstrate the potential for mining the dynamic genome to identify multiple disease signatures using quantitative RNA expression analysis of a single blood sample.
Acknowledgements
The authors would like to thank Mr. Wang Min who performed the Affymetrix studies, and Mr. Guangdong Duan, who helped analyze the gene pathways. Ms Isolde Prince helped with the editing of the manuscript.
Competing interests
Dr Choong-Chin Liew is Chair and Founder and Ma Jun is employed by Golden Health Diagnostics Inc., China, who funded this research.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.1186/s40880-015-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JM, Ryu WS, Kim JH, Park SS, Kim SJ, Kim CS, Mok YJ. Prognostic factors for advanced gastric cancer: Stage-stratified analysis of patients who underwent curative resection. Cancer Res Treat. 2006;38:13–18. doi: 10.4143/crt.2006.38.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canyilmaz E, Soydemir G, Serdar L, Uslu GH, Sahbaz A, Colak F, Kandaz M, Bahat Z, Yoney A. Evaluation of prognostic factors and survival results in gastric carcinoma: Single center experience from Northeast Turkey. Int J Clin Exp Med. 2014;7:2656–2666. [PMC free article] [PubMed] [Google Scholar]
- 5.Hundahl SA, Phillips JL, Menck HR. The National Cancer Data Base Report on poor survival of U.S. gastric carcinoma patients treated with gastrectomy: Fifth Edition American Joint Committee on Cancer staging, proximal disease, and the ‘different disease’ hypothesis. Cancer. 2000;88:921–932. doi: 10.1002/(SICI)1097-0142(20000215)88:4<921::AID-CNCR24>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 6.Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, et al. Screening for gastric cancer in Asia: Current evidence and practice. Lancet Oncol. 2008;9:279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- 7.Hamashima C, Shibuya D, Yamazaki H, Inoue K, Fukao A, Saito H, Sobue T. The Japanese guidelines for gastric cancer screening. Jpn J Clin Oncol. 2008;38:259–267. doi: 10.1093/jjco/hyn016. [DOI] [PubMed] [Google Scholar]
- 8.Tashiro A, Sano M, Kinameri K, Fujita K, Takeuchi Y. Comparing mass screening techniques for gastric cancer in Japan. World J Gastroenterol. 2006;12:4873–4874. doi: 10.3748/wjg.v12.i30.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leja M, You W, Camargo MC, Saito H. Implementation of gastric cancer screening-the global experience. Best Pract Res Clin Gastroenterol. 2014;28:1093–1106. doi: 10.1016/j.bpg.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JY, Karsa L, Herrero R. Prevention strategies for gastric cancer: A global perspective. Clin Endosc. 2014;47:478–489. doi: 10.5946/ce.2014.47.6.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liew CC. Method for the detection of gene transcripts in blood and uses there of U.S. Patent Application No. 2002000268730. 1999 [Google Scholar]
- 12.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. Peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Burczynski ME. Transcriptional profiling of peripheral blood in oncology. In: Burczynski ME, Rockett JC, editors. Surrogate tissue analysis: Genomic, proteomic and metabolomic approaches. Taylor and Francis; Boca Raton: 2006. pp. 47–63. (edition) [Google Scholar]
- 14.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat Rev Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 15.Ponnampalam SN, Kamaluddin NR, Zakaria Z, Matheneswaran V, Ganesan D, Haspani MS, Ryten M, Hardy JA. A blood-based gene expression and signaling pathway analysis to differentiate between high and low grade gliomas. Oncol Rep. 2017;37:10–22. doi: 10.3892/or.2016.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanburen P, Ma J, Chao S, Mueller E, Schneider DJ, Liew CC. Blood gene expression signatures associate with heart failure outcomes. Physiol Genomics. 2011;43:392–397. doi: 10.1152/physiolgenomics.00175.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall KW, Mohr S, Khettabi FE, Nossova N, Chao S, Bao W, Ma J, Li XJ, Liew CC. Blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer. 2010;126:1177–1186. doi: 10.1002/ijc.24910. [DOI] [PubMed] [Google Scholar]
- 18.Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374–3380. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]
- 19.Liong ML, Lim CR, Yang H, Chao S, Bong CW, Leong WS, Das PK, Loh CS, Lau BE, Yu CG, et al. Blood-based biomarkers of aggressive prostate cancer. PLoS One. 2012;7:e45802. doi: 10.1371/journal.pone.0045802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twine NC, Stover JA, Marshall B, Dukart G, Hidalgo M, Stadler W, Logan T, Dutcher J, Hudes G, Dorner AJ, et al. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res. 2003;63:6069–6075. [PubMed] [Google Scholar]
- 21.Baine MJ, Chakraborty S, Smith LM, Mallya K, Sasson AR, Brand RE, Batra SK. Transcriptional profiling of peripheral blood mononuclear cells in pancreatic cancer patients identifies novel genes with potential diagnostic utility. PLoS One. 2011;6:e17014. doi: 10.1371/journal.pone.0017014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burakoff R, Chao S, Perencevich M, Ying J, Friedman S, Makrauer F, Odze R, Khurana H, Liew CC. Blood-based biomarkers can differentiate ulcerative colitis from Crohn's disease and noninflammatory diarrhea. Inflamm Bowel Dis. 2011;17:1719–1725. doi: 10.1002/ibd.21574. [DOI] [PubMed] [Google Scholar]
- 23.Burakoff R, Hande S, Ma J, Banks PA, Friedman S, Makrauer F, Liew CC. Differential regulation of peripheral leukocyte genes in patients with active Crohn's disease and Crohn's disease in remission. J Clin Gastroenterol. 2010;44:120–126. doi: 10.1097/MCG.0b013e3181a9ef53. [DOI] [PubMed] [Google Scholar]
- 24.Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, Glatt SJ, Liew CC. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: A preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- 25.Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sásik R, Khanlou N, Han M, Liew CC, Tsuang MT. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glatt SJ, Stone WS, Nossova N, Liew CC, Seidman LJ, Tsuang MT. Similarities and differences in peripheral blood gene-expression signatures of individuals with schizophrenia and their first-degree biological relatives. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:869–887. doi: 10.1002/ajmg.b.31239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speedet TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MAQC Consortium, corp-author. Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, et al. The MicroArray quality control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han M, Liew CT, Zhang HW, Chao S, Zheng R, Yip KT, Song ZY, Li HM, Geng XP, Zhu LX, et al. Novel, blood-based five-gene panel biomarker set for the detection of colorectal cancer. Clin Cancer Res. 2008;14:455–460. doi: 10.1158/1078-0432.CCR-07-1801. [DOI] [PubMed] [Google Scholar]
- 30.Burakoff R, Pabby V, Onyewadume L, Odze R, Adackapara C, Wang W, Friedman S, Hamilton M, Korzenik J, Levine J, et al. Blood-based biomarkers used to predict disease activity in Crohn's disease and ulcerative colitis. Inflamm Bowel. Dis. 2015;21:1132–1140. doi: 10.1097/MIB.0000000000000340. [DOI] [PubMed] [Google Scholar]
- 31.Chao S, Cheng C, Liew CC. Mining the dynamic genome: A method for identifying multiple disease signatures using quantitative RNA expression analysis of a single blood sample. Microarrays (Bassel) 2015;4:671–689. doi: 10.3390/microarrays4040671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Dou M, Yao X, Tang H, Li Z, Zhao X. Potential biomarkers in diagnosis of human gastric cancer. Cancer Invest. 2016;34:115–122. doi: 10.3109/07357907.2015.1114122. [DOI] [PubMed] [Google Scholar]
- 34.Kalnina Z, Meistere I, Kikuste I, Tolmanis I, Zayakin P, Line A. Emerging blood-based biomarkers for detection of gastric cancer. World J Gastroenterol. 2015;21:11636–11653. doi: 10.3748/wjg.v21.i41.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumura N, Zembutsu H, Yamaguchi K, Sasaki K, Tsuruma T, Nishidate T, Denno R, Hirata K. Identification of novel molecular markers for detection of gastric cancer cells in the peripheral blood circulation using genome-wide microarray analysis. Exp Ther Med. 2011;2:705–713. doi: 10.3892/etm.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai Y, Honda M, Fujinaga H, Tatsumi I, Mizukoshi E, Nakamoto Y, Kaneko K. Common transcriptional signature of tumor-infiltrating mononuclear inflammatory cells and peripheral blood mononuclear cells in hepatocellular carcinoma patients. Cancer Res. 2008;68:10267–10279. doi: 10.1158/0008-5472.CAN-08-0911. [DOI] [PubMed] [Google Scholar]
- 37.Tu H, Sun H, Lin Y, Ding J, Nan K, Li Z, Shen Q, Wei Y. Oxidative stress upregulates PDCD4 expression in patients with gastric cancer via miR-21. Curr Pharm Des. 2014;20:1917–1923. doi: 10.2174/13816128113199990547. [DOI] [PubMed] [Google Scholar]
- 38.Ma G, Zhang H, Dog M, Zheng X, Ozaki I, Matsuhashi S, Guo K. Downregulation of programmed cell death 4 (PDCD4) in tumorigenesis and progression of human digestive tract cancers. Tumour Biol. 2013;34:3879–3885. doi: 10.1007/s13277-013-0975-9. [DOI] [PubMed] [Google Scholar]
- 39.Yu H, Zeng J, Liang X, Wang W, Zhou Y, Sun Y, Liu S, Li W, Chen C, Jia J. Helicobacter pylori promotes epithelial-mesenchymal transition in gastric cancer by downregulating programmed cell death protein 4 (PDCD4) PLoS One. 2014;9:e105306. doi: 10.1371/journal.pone.0105306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Dürst M, Schneider A, Pothuri B, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: Identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer. 2007;46:373–384. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- 41.Jeong MS, Hwang EY, Kim HT, Yoo M, Jang SB. Purification of caudal-related homeodomain transcription factor and its binding characterization. J Microbiol Biotechnol. 2009;19:1557–1564. doi: 10.4014/jmb.0905.05021. [DOI] [PubMed] [Google Scholar]
- 42.Janicke B, Hegardt C, Krogh M, Onning G, Akesson B, Cirenajwis HM, Oredsson SM. The antiproliferative effect of dietary fiber phenolic compounds ferulic acid and p-coumaric acid on the cell cycle of Caco-2 cells. Nutr Cancer. 2011;63:611–622. doi: 10.1080/01635581.2011.538486. [DOI] [PubMed] [Google Scholar]
- 43.Cotterchio M, Lowcock E, Bider-Canfield Z, Lemire M, Greenwood C, Gallinger S, Gallinger S, Hudson T. Association between variants in atopy-related immunologic candidate genes and pancreatic cancer risk. PLoS One. 2015;10:e0125273. doi: 10.1371/journal.pone.0125273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montaño B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2012;2:120071. doi: 10.1098/rsob.120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lezon-Geyda K, Najfeld V, Johnson EM. Deletions of PURA, at 5q31, and PURB, at 7p13, in myelodysplastic syndrome and progression to acute myelogenous leukemia. Leukemia. 2001;15:954–962. doi: 10.1038/sj.leu.2402108. [DOI] [PubMed] [Google Scholar]