Abstract
We present the description of Sanguibacter massiliensis sp. nov., Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Neobittarella massiliensis gen. nov. and Miniphocibacter massiliensis gen. nov., new bacterial species isolated by culturomics from human stool samples.
Keywords: Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Miniphocibacter massiliensis gen. nov., Neobittarella massiliensis gen. nov., Sanguibacter massiliensis sp. nov
As part of the effort aiming to describe the human gut microbiota by the means of culturomics [1], we report in the present study the isolation of four new bacterial species and two new bacterial genera. Before initiation of the study, approval from the Institut Federatif de Recherche IFR48 (Marseille, France) was obtained under number 09-022, along with signed informed consent provided by the donors.
All stool samples were independently diluted with phosphate-buffered saline and incubated in a blood culture bottle supplemented with 5% sheep's blood and 5% filtered rumen. A 30-day follow-up was done in order to assess the bacterial growth, and matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and 16S rRNA gene sequencing were used for strain identification, as previously described [2], [3]. Because the organisms' spectra were missing from the MALDI-TOF MS database, we were unable to identify them. Thus, 16S rRNA gene sequencing was applied. A similarity threshold of less than 98.65% between the isolated strains and the phylogenetically closest species with standing in nomenclature was adapted to delimitate a new species, and a divergence of more than 5% was adapted to delimitate a new genus [4].
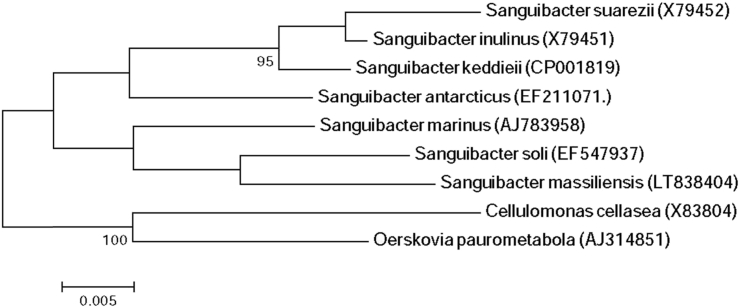
Strain Marseille-P3815 was isolated on Columbia medium supplemented with 5% sheep's blood (COS; bioMérieux, Marcy l’Étoile, France) from the stool samples of a 12-year-old healthy Pygmy girl after 2 days' incubation under anaerobic conditions at 37°C. This strain is a Gram-positive rod, catalase positive but oxidase negative. Its colonies have a diameter ranging between 0.2 to 0.9 mm, with its cells' average size being 1.0 × 0.4 μm. It has a smooth and grey appearance after 48 hours of growth on COS medium. Strain Marseille-P3815 exhibited a 96.96% sequence similarity with Sanguibacter inulinus strain ST50 (NR_029277.1). It was thus classified as a new species, Sanguibacter massiliensis (ma.ssi.lien′sis, L. adj. neut., massiliensis, from ‘Massilia,’ the antic name of Marseille, France, where the strain was isolated) (Fig. 1). The strain Marseille-P3815T is the type strain of the species Sanguibacter massiliensis.
Fig. 1.
Phylogenetic tree showing position of Sanguibacter massiliensis strain Marseille-P3815 towards its closest species. CLUSTALW was used for alignment and MEGA software for phylogenetic inferences generation with maximum likelihood method. After 500 repeats, bootstrap values are shown on nodes, with only values above 90% kept.
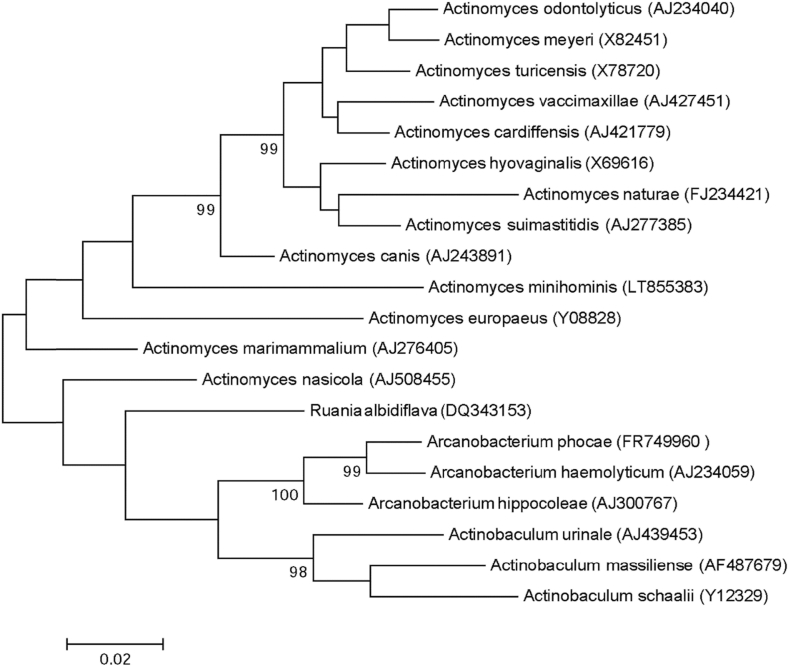
Strain Marseille-P3850 was isolated on COS (bioMérieux) from the stool samples of a 12-year-old healthy Pygmy girl after 5 days' incubation under anaerobic conditions at 37°C. This strain is a Gram-positive rod, catalase and oxidase negative. Its colonies have a diameter ranging between 0.3 to 1 mm, with its cells' average size being 1.0 × 0.5 μm. It has a smooth and grey appearance after 48 hours of growth on COS medium. Strain Marseille-P3850 exhibited a 93.36% 16S rRNA gene sequence similarity with Actinomyces marimammalium strain CCUG 41710 (NR_025395.1). Thus it is classified as a new species, Actinomyces minihominis (mini.ho.min′is, L. adj. neut., minihominis, referring to the Pygmy people from whom the strain was isolated and who are characterized by their small size) (Fig. 2). The strain Marseille-P3850T is the type strain of the species Actinomyces minihominis.
Fig. 2.
Phylogenetic tree showing position of Actinomyces minihominis strain Marseille-P3850 towards its closest species. CLUSTALW was used for alignment and MEGA software for phylogenetic inferences generation with maximum likelihood method. After 500 repeats, bootstrap values are shown on nodes, with only values above 90% kept.
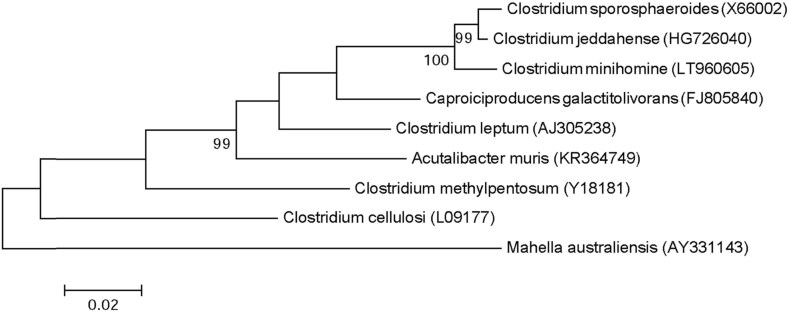
Strain Marseille-P4642 was isolated on COS (bioMérieux) from the stool samples of a 39-year-old healthy Pygmy man after 10 days' incubation under anaerobic conditions at 37°C. This strain is a Gram-positive bacilli, catalase and oxidase negative. Its colonies have a diameter ranging between 0.05 to 1 mm, with its cells' average size being 2.2 × 0.43 μm. It has a smooth and grey appearance after 48 hours of growth on COS medium. Strain Marseille-P4642 exhibited a 98.13% 16S rRNA gene sequence similarity with Clostridium jeddahense strain JCD (NR_144697.1); thus, it is classified as a new species, Clostridium minihomine (mini.ho.min′e, L. adj. neut., minihomine, to refer to the Pygmy people from whom the strain was isolated and who are characterized by their small size) (Fig. 3). The strain Marseille-P4642T is the type strain of the species Clostridium minihomine.
Fig. 3.
Phylogenetic tree showing position of Clostridium minihomine strain Marseille-P4042 towards its closest species. CLUSTALW was used for alignment and MEGA software for phylogenetic inferences generation with maximum likelihood method. After 500 repeats, bootstrap values are shown on nodes, with only values above 95% kept.
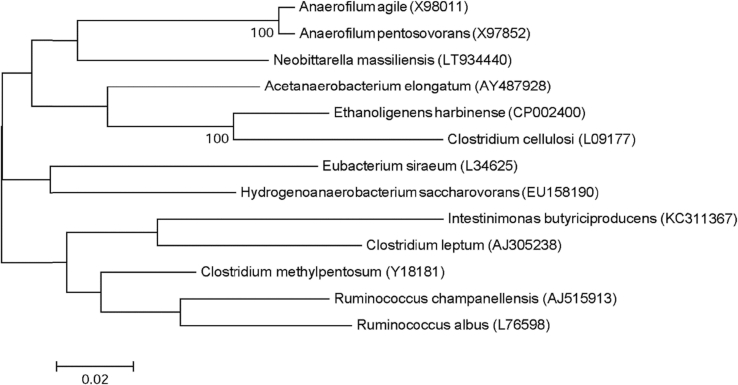
Strain Marseille-P4047 was isolated on COS medium (bioMérieux) from the stool samples of a healthy Senegalese male subject after 5 days' incubation under anaerobic conditions at 37°C. This strain is a Gram-positive coccobacilli, catalase and oxidase negative. Its colonies have a diameter ranging between 0.04 to 0.9 mm, with its cells' average size being 1.6 × 0.6 μm. It has a shiny grey appearance after 48 hours of growth on COS medium. Strain Marseille-P4047 exhibited an 89.22% 16S rRNA gene sequence similarity with Acetanaerobacterium elongatum strain Z7 (NR_042930.1); thus, it is classified as a new genus, Neobitarella (neo.bita.rel′la, L. adj. fem., in honor of microbiologist Fadi Bittar). Neobitarella massiliensis is the type species of the new genus Neobittarella (ma.ssi.lien′sis, L. adj. fem., massiliensis, from ‘Massilia,’ the antic name of Marseille, France, where the strain was isolated) (Fig. 4). The strain Marseille-P4047T is the type strain of the species Neobittarella massiliensis.
Fig. 4.
Phylogenetic tree showing position of Neobittarella massiliensis strain Marseille-P4047 towards its closest species. CLUSTALW was used for alignment and MEGA software for phylogenetic inferences generation with maximum likelihood method. After 500 repeats, bootstrap values are shown on nodes, with only values above 95% kept.
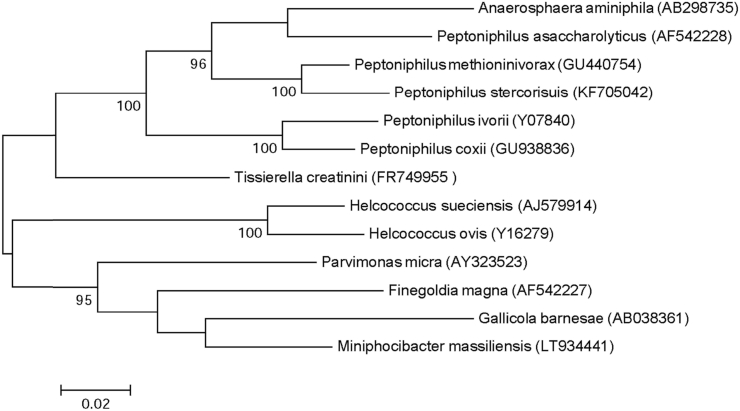
Strain Marseille-P4678 was isolated on COS (bioMérieux) from the stool samples of a 39-year-old healthy Pygmy man after 10 days' incubation under anaerobic conditions at 37 °C. This strain is a Gram-positive cocci, catalase positive and oxidase negative. Its colonies have a diameter ranging between 0.02 to 0.08 mm, with its cells' average diameter being 0.6 μm. It has a smooth and grey appearance after 48 hours of growth on COS medium. Strain Marseille-P4047 exhibited an 89.69% 16S rRNA gene sequence similarity with Parvimonas micra strain 3119B (NR_036934.1); it is thus classified as a new genus, Miniphocibacter (mini.phoci.bacter, L. adj. masc., miniphocibacter composed of mini, referring to the small size of the Pygmy people from whom the strain was isolated, and phoci, referring to Phocae, the Latin name of the city that the founders of Marseille came from). Miniphocibacter massiliensis is the type species of the genus Miniphocibacter (ma.ssi.lien′sis, L. adj. masc., massiliensis, from ‘Massilia,’ the antic name of Marseille, France, where the strain was isolated) (Fig. 5). The strain Marseille-P4678T is the type strain of the species Miniphocibacter massiliensis.
Fig. 5.
Phylogenetic tree showing position of Miniphocibacter massiliensis strain Marseille-P4678 towards its closest species. CLUSTALW was used for alignment and MEGA software for phylogenetic inferences generation with maximum likelihood method. After 500 repeats, bootstrap values are shown on nodes, with only values above 95% kept.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of the reported organisms are available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequences of Sanguibacter massiliensis sp. nov., Actinomyces minihominis sp. nov., Clostridium minihomine sp. nov., Neobittarella massiliensis gen. nov. and Miniphocibacter massiliensis gen. nov. were deposited in GenBank under the following accession numbers respectively: LT838404, LT855383, LT960605, LT934440 and LT934441.
Deposit in a culture collection
Strain Marseille-P3815, strain Marseille-P3850, strain Marseille-P4642, strain Marseille-P4047 and strain Marseille-P4678 were deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number the following numbers respectively: P3815, P3850, P4642, P4047 and P4678.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]