Abstract
TCP proteins, the plant-specific transcription factors, are involved in the regulation of multiple aspects of plant development among different species, such as leaf development, branching, and flower symmetry. However, thus far, the roles of TCPs in legume, especially in nodulation are still not clear. In this study, a genome-wide analysis of TCP genes was carried out to discover their evolution and function in Medicago truncatula. In total, 21 MtTCPs were identified and classified into class I and class II, and the class II MtTCPs were further divided into two subclasses, CIN and CYC/TB1. The expression profiles of MtTCPs are dramatically different. The universal expression of class I MtTCPs was detected in all organs. However, the MtTCPs in CIN subclass were highly expressed in leaf and most of the members in CYC/TB1 subclass were highly expressed in flower. Such organ-specific expression patterns of MtTCPs suggest their different roles in plant development. In addition, most MtTCPs were down-regulated during the nodule development, except for the putative MtmiR319 targets, MtTCP3, MtTCP4, and MtTCP10A. Overexpression of MtmiR319A significantly reduced the expression level of MtTCP3/4/10A/10B and resulted in the decreased nodule number, indicating the important roles of MtmiR319-targeted MtTCPs in nodulation. Taken together, this study systematically analyzes the MtTCP gene family at a genome-wide level and their possible functions in nodulation, which lay the basis for further explorations of MtmiR319/MtTCPs module in association with nodule development in M. truncatula.
Keywords: Medicago truncatula, TCP genes, miR319, expression analysis, nodule development
Introduction
TCP proteins, a small family of plant-specific transcription factors, was first described in 1999 and named after its initial members TEOSINTE BRANCHED1 (TB1) in maize (Zea mays), CYCLOIDEA (CYC) in snapdragon (Antirrhinum majus), and PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) in rice (Oryza sativa) (Luo et al., 1996; Doebley et al., 1997; Kosugi and Ohashi, 1997; Cubas et al., 1999). All TCP proteins contain a highly conserved TCP domain which is a 59-amino-acid non-canonical basic-helix-loop-helix (bHLH) motif at the N-terminus and functions in DNA binding, protein–protein interaction, and protein nuclear localization (Kosugi and Ohashi, 2002; Aggarwal et al., 2010; Martin-Trillo and Cubas, 2010; Dhaka et al., 2017). So far, TCP genes have been identified in various plant species including Arabidopsis, rice, tomato, tobacco, and strawberry as shown in Supplementary Figure S1 (Palatnik et al., 2003; Li et al., 2005; Aguilar-Martinez et al., 2007; Ori et al., 2007; Yao et al., 2007; Schommer et al., 2008; Nag et al., 2009; Giraud et al., 2010; Koyama et al., 2010b; Sarvepalli and Nath, 2011; Yang et al., 2012; Ma et al., 2014; Parapunova et al., 2014; Wang et al., 2015b; Chen et al., 2016; Lin et al., 2016; Ma et al., 2016; Wei et al., 2016; Zhou et al., 2016; Du et al., 2017; Xie et al., 2017).
The TCP family can be classified into two classes: class I (also known as PCF class or TCP-P class) and class II (also known as TCP-C class) according to the amino acid sequences of the TCP domain (Kosugi and Ohashi, 2002; Navaud et al., 2007; Martin-Trillo and Cubas, 2010). The class II can be further divided into two unlike subclasses: CIN and CYC/TB1 (Crawford et al., 2004; Howarth and Donoghue, 2006; Martin-Trillo and Cubas, 2010). The difference between the class I and class II TCP proteins is a four-amino-acid deletion in the TCP domain in class I. Besides the TCP domain, all members of the CYC/TB1 TCP proteins contain an arginine-rich R domain with unknown function, which is speculated to facilitate protein–protein interaction (Cubas et al., 1999).
Class I TCPs mainly participate in promoting cell proliferation and plant growth, such as the PCF1/PCF2 in rice and TCP20 in Arabidopsis (Kosugi and Ohashi, 1997; Li et al., 2005). Most loss-of-function mutants of class I TCP genes do not exhibit obvious phenotypic defects due to the genetic redundancy. In Arabidopsis, class I genes TCP14 and TCP15 have redundant functions in the regulation of internode length, leaf shape, seed germination, and endoreduplication (Kieffer et al., 2011; Uberti-Manassero et al., 2012; Resentini et al., 2015). TCP20 appears to participate in different developmental processes, such as jasmonic acid (JA) biosynthesis and leaf senescence (Li et al., 2005; Danisman et al., 2012, 2013). Meanwhile, TCP9 and TCP19 act redundantly with TCP20 and positively regulate leaf senescence through the JA signaling pathway (Danisman et al., 2012, 2013). TCP16 mainly expresses in developing microspores and plays a crucial role at the early stage of pollen development (Takeda et al., 2006). Moreover, TCP7, TCP8, TCP22, and TCP23 show similar expression patterns in young leaves and have genetic redundancy in regulation of leaf development (Aguilar-Martinez and Sinha, 2013).
MicroRNAs (MiRNAs) are small non-coding RNAs (20–24 nucleotides in length) that can complementarily bind to their target mRNAs and reduce their expression levels (Reinhart et al., 2002; Voinnet, 2009; Rogers and Chen, 2013). Recent evidences indicate that miRNAs are essential genetic regulators and play vital roles in plant development (Chen, 2008). In the CIN subclass, five Arabidopsis TCP genes TCP2, TCP3, TCP4, TCP10, and TCP24 are the targets of miR319 and have been involved in regulation of cell division to control leaf morphogenesis (Palatnik et al., 2003; Bresso et al., 2017). In tomato, high levels of miR319 or low LA activity cause an excess of cell expansion, resulting in the super compound leaves (Ori et al., 2007). Meanwhile, the CYC/TB1 subclass TCPs involve in floral dorsoventral asymmetry development and shoot branching, such as CYC in Lotus japonicus, snapdragon and Pisum sativum (Luo et al., 1996, 1999; Almeida and Galego, 2005; Feng et al., 2006; Wang et al., 2008, 2010; Li et al., 2010; Weng et al., 2011; Xu et al., 2013) and TB1 in maize (Doebley et al., 1995). In Arabidopsis, two orthologs of maize TB1, BRANCHED1 (BRC1, TCP18) and BRANCHED2 (BRC2, TCP12), are expressed in axillary buds. Both of them are involved in suppressing axillary bud outgrowth and loss-of-function in these genes results in the increased shoot branching (Aguilar-Martinez et al., 2007; Finlayson, 2007). TCP1, a member of the CYC/TB1 subclade, is implicated in the control of brassinosteroid (BR) biosynthesis and the regulation of longitudinal elongation in plant (Guo et al., 2010; Koyama et al., 2010b).
Medicago truncatula is a model species for legume genetics and functional genomics study. However, the TCP family in M. truncatula has not been characterized, and their roles are still unknown, especially in nodulation. In this study, a global analysis of the TCP gene family in M. truncatula was carried out. Twenty-one MtTCP genes were identified, and their phylogenetic relationship, gene structure, protein motifs, chromosomes locations, and transcriptional levels in different organs were analyzed. Furthermore, we found that the expression patterns of MtTCP genes are different during the nodule development. Based on the time course of MtTCPs expression in nodules, MtTCP3, MtTCP4, and MtTCP10A, three of four target genes of MtmiR319, were significantly up-regulated after inoculation with rhizobia. Overexpression of MtmiR319A led to the decreased nodule number, which further confirms the significant roles of MtmiR319-targeted MtTCPs in nodulation. Therefore, this study provides detailed information of the MtTCPs classification and throws some light into the function of MtTCPs involved in nodule development in M. truncatula.
Materials and Methods
Plant Material and Root Nodule Induction
For nodule induction, wild-type (ecotype R108) and 35S:MtmiR319A seeds were transferred to plastic seedling holes containing a 3:1 ratio of pearlite/sand under a 16-h/8-h light/dark at 22°C and 60% relative humidity. The S. meliloti 1021 strain harboring the lacZ reporter gene (Boivin et al., 1990) was cultured in TY medium supplemented with 6 mmol l-1 calcium chloride, 200 μg ml-1 streptomycin, and 10 μg ml-1 tetracycline. The S. meliloti culture was shaken at 220 rpm and 28°C overnight. The S. meliloti growth was monitored by measuring optical density at a wavelength of 600 nm (OD600) until the OD600 value reached to 1.0. Five-day-old seedling was inoculated with 5 ml of S. meliloti 1021 strain suspension with OD600 of 0.1 (adjusted OD600 value from 1.0 to 0.1 by dilution). At 3 weeks post-inoculation, seedling roots, nodule number, and the fresh weight of shoot/root/nodule were analyzed. For nodulation time course assays, nodules in wild-type were harvested at 7, 14, and 21 days post inoculation (dpi). For LacZ staining, LacZ activity of nodules was performed as previously described (Boivin et al., 1990).
Identification and Phylogenetic Analysis of the TCP Genes in M. truncatula
To identify all TCP genes in M. truncatula, we use the 24 known TCP genes from Arabidopsis and 21 TCP genes from rice to perform protein to protein BLAST in Medicago truncatula resource website1 (Martin-Trillo and Cubas, 2010; Shi et al., 2016). TCP genes in Arabidopsis were downloaded from TAIR2 online. TCP genes in rice were downloaded from the Rice Genome Annotation Project3 online. Based on the searched TCP chromosome locus, we downloaded the genome sequence from Medicago truncatula Genome Database4. The BLASTP parameters are E-value = 10, top hits number = 30. In total, 27 MtTCPs sequences with annotations were obtained. As each of Medtr2g090960, Medtr1g038650, and Medtr2g006150 has two splice variants, and Medtr1g101810 has four splice variants. We selected splice variant 1 for study, and other splice variants were excluded. Therefore, 21 independent MtTCP genes were identified in M. truncatula. Then, the MtTCP sequences were confirmed in the Plant Transcription Factor Database (PlantTFDB5) online. To study the phylogenetic relationships among TCP genes in M. truncatula, Arabidopsis, and rice, 21 identified MtTCP proteins in M. truncatula and 24 Arabidopsis AtTCP proteins and 21 rice OsTCP proteins were used to generate the phylogenetic tree. Multiple sequence alignments were executed using CLUSTALW6 online. Then, the phylogenetic trees were constructed using MEGA7.1 by the Neighbor Joining (NJ) method with 1000 bootstrap replicates in p-distance model. Since the TCP family in Arabidopsis has been identified and studied well, we named MtTCPs based on the phylogenetic relationship with AtTCPs and Blast analysis against the Arabidopsis genome in TAIR.
Gene Structure Analysis of MtTCP Genes
To analyze the gene structures and exon–intron organization of the MtTCP genes, we downloaded the MtTCP genes genomic sequences and structural information from the Medicago truncatula Genome Database. Then, the MtTCP genomic sequences and CDS sequences were aligned using the gene structure display server 2.0 (GSDS) online7 to generate the diagrams of exon–intron structures.
Conserved Domains and Motif Analysis in MtTCP Proteins
Conserved motifs in MtTCP proteins were analyzed with the online Multiple Em for Motif Elicitation (MEME) program8. The optimized MEME parameters were as follows: repetition number, any; maximum motif width, 200; minimum motif width, 6; and maximum motif number, 20. Multiple protein sequences alignment was carried out with Jalview software9.
RNA Isolation, Gene Expression Analysis, and Statistical Analysis
To detect the expression patterns of TCP genes in M. truncatula, total RNA was extracted from the young leaves (folded leaves developed on the first and second internodes), mature leaves (fully expanded leaves developed on the older internodes), roots, stems, flowers, shoot buds, pods, and nodules at the different stages using the Trizol-RT Reagent (Molecular Research Center, INC), according to the manufacturer’s instructions. RNA quantitative and qualitative measurements were achieved using Nanodrop 2000 Spectrophotometer (NanoDrop Technologies, United States). Three micrograms of total RNA from each organs/tissue was reverse-transcribed into cDNA using the Roche RNA Reverse Transcription Kit (Roche, United States). The qRT-PCR was performed on Bio-Rad CFX ConnectTM using Roche SYBR-green fluorescence dye (FastStart Essential DNA Green Master). The relative expression levels were calculated using 2-ΔΔCT method. For all qRT-PCR analysis, triplicate biological samples were collected. The MtUBI gene was selected as internal control for normalization. T-test was used to estimate if the difference is significant in analysis of gene expression level, fresh weight, and nodule number.
Subcellular Localization of MtTCPs and Cloning of the MtmiR319A Gene
For subcellular localization, PCR was performed to amplify the coding sequences of MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B genes using gene-specific primers which are listed in Supplementary Table S1. Then, the PCR products were purified and cloned into pEarleyGate 103 (Earley et al., 2006) using the Gateway LR reaction (Invitrogen). The Agrobacterium tumefaciens EHA105 strain harboring relevant plasmids was transformed into tobacco epidermal cells. The MtTCP-GFP fusion proteins were examined using a confocal laser scanning microscope LSM 700 (Zeiss). To construct the MtmiR319A overexpression vector, MtmiR319A genomic sequence was PCR amplified and transferred into pEarleyGate 100. The leaves of the wild-type were transformed with the EHA105 strain harboring the MtmiR319A overexpression vector.
Identification of the MtmiR319 and Prediction of the MtmiR319 Target Genes
The mature MtmiR319 sequences were obtained on miRBase Database10 online. To predict MtmiR319 target sites, full length of MtTCPs coding sequences was analyzed using the psRNATarget (A Plant Small RNA Target Analysis11) online. The mature miR319 sequences of rice were obtained on miRBase Database. The mature miR319 sequences of Arabidopsis were downloaded on TAIR12 online and confirmed on miRBase Database.
Results
Identification of TCP Genes in M. truncatula
To identify TCP proteins in M. truncatula, the TCP protein sequences of Arabidopsis and rice were used to BLAST search against the Medicago truncatula Genome Database13 and Plant Transcription Factor Database (PlantTFDB14). A total of 21 putative MtTCP sequences were obtained, which all contained the conserved TCP domain. The length of the protein sequences of 21 MtTCPs ranged from 205 to 489 amino acids. The gene locus, exon number, amino acid length, molecular weight (Mw), isoelectric point (pI), type, and chromosome location of MtTCPs are listed in Table 1. Based on the gene locus data, these MtTCPs were unevenly located on different chromosomes. Five MtTCPs were located on Chr1. Four MtTCPs were located on Chr4. Furthermore, the Chr2 and Chr7 chromosomes each had three MtTCPs. The other MtTCPs were distributed on Chr3, Chr5, Chr6, and Chr8, respectively (Table 1).
Table 1.
TCP gene family in M. truncatula.
| Name | Locus | CDS | Extrons | Length (aa) | MW (kDa) | pI | Type | Chromosomes location |
|---|---|---|---|---|---|---|---|---|
| MtTCP1A | Medtr6g017055 | 1230 | 2 | 409 | 46,096.18 | 9.436 | CYC/TB1 | chr6:6710788..6713192 (+) |
| MtTCP1B | Medtr7g018500 | 1125 | 2 | 374 | 42,704.88 | 6.966 | CYC/TB1 | chr7:6031315..6033168 (+) |
| MtTCP3 | Medtr2g078200 | 885 | 1 | 294 | 32,176.38 | 7.084 | CIN | chr2:32505911..32507609 (+) |
| MtTCP4 | Medtr8g463380 | 1302 | 1 | 433 | 47,448.71 | 6.953 | CIN | chr8:22326723..22329800 (+) |
| MtTCP5A | Medtr3g026050 | 1158 | 1 | 385 | 43,448.1 | 7.716 | CIN | chr3:7969037..7970349 (+) |
| MtTCP5B | Medtr4g109660 | 921 | 1 | 306 | 33,901.83 | 9.198 | CIN | chr4:45588798..45589718 (+) |
| MtTCP7 | Medtr1g038650 | 708 | 1 | 235 | 25,831.56 | 8.251 | PCF | chr1:14248871..14250840 (-) |
| MtTCP9 | Medtr8g033070 | 1002 | 1 | 333 | 36,244.28 | 9.491 | PCF | chr8:12681955..12683583 (+) |
| MtTCP10A | Medtr2g090960 | 990 | 1 | 329 | 36,325.62 | 6.322 | CIN | chr2:39075847..39078457 (-) |
| MtTCP10B | Medtr4g079580 | 1008 | 1 | 335 | 37,656.05 | 6.327 | CIN | chr4:30767465..30768533 (-) |
| MtTCP11 | Medtr1g063870 | 618 | 1 | 205 | 21,958.6 | 7.447 | PCF | chr1:28041642..28042798 (+) |
| MtTCP12 | Medtr1g103380 | 1470 | 2 | 489 | 56,235 | 7.966 | CYC/TB1 | chr1:46794627..46797920 (-) |
| MtTCP13 | Medtr7g015010 | 1056 | 1 | 351 | 39,686.79 | 8.564 | CIN | chr7:4515349..4517203 (+) |
| MtTCP14 | Medtr5g039600 | 1251 | 1 | 416 | 44,227.9 | 6.726 | PCF | chr5:17427951..17429938 (-) |
| MtTCP15 | Medtr4g108370 | 1314 | 1 | 437 | 47,447.01 | 7.818 | PCF | chr4:44959266..44961490 (-) |
| MtTCP17 | Medtr6g015350 | 1002 | 1 | 333 | 37,706.17 | 6.671 | CIN | chr6:5010976..5013085 (+) |
| MtTCP18 | Medtr4g111935 | 1167 | 2 | 388 | 44,497.4 | 8.813 | CYC/TB1 | chr4:46279251..46280786 (-) |
| MtTCP19 | Medtr1g101810 | 1104 | 1 | 367 | 38,729.49 | 5.251 | PCF | chr1:45965265..45966764 (+) |
| MtTCP20 | Medtr7g028160 | 855 | 1 | 284 | 30,714.79 | 8.664 | PCF | chr7:9483450..9485316 (-) |
| MtTCP21 | Medtr1g114380 | 774 | 1 | 257 | 27,430.59 | 9.492 | PCF | chr1:51614461..51616519 (+) |
| MtTCP22 | Medtr2g006150 | 1464 | 1 | 487 | 51,820.53 | 6.581 | PCF | chr2:468724..471446 (-) |
Phylogenetic Analysis and Classification of TCP Genes in M. truncatula
To evaluate the evolutionary and phylogenetic relationships among the TCP transcription factor families among species, a total of 70 TCP protein sequences, including 21 MtTCPs, 24 AtTCPs, and 21 OsTCPs, 1 ZmTCP (maize, TB1), and 3 AmTCPs (A. majus, AmCIN/AmCYC/AmDICH) were collected to construct an unrooted phylogenetic tree using MEGA7.1 by the NJ method with 1000 bootstrap replicates. According to its classification in Arabidopsis, the MtTCPs could also be classified into two TCP classes. Class I contained nine members of MtTCPs. The rest of MtTCPs were grouped as Class II which can be further divided into two subclasses: the CIN (eight members) and the CYC/TB1 (four members) (Figures 1, 2A). Then, the TCP domains of each MtTCP were identified and used for further phylogenetic analysis (Supplementary Figure S2). The phylogenetic trees derived from full-length protein sequences and TCP domains are essentially the same, indicating the evolutionary conservation among these TCPs.
FIGURE 1.
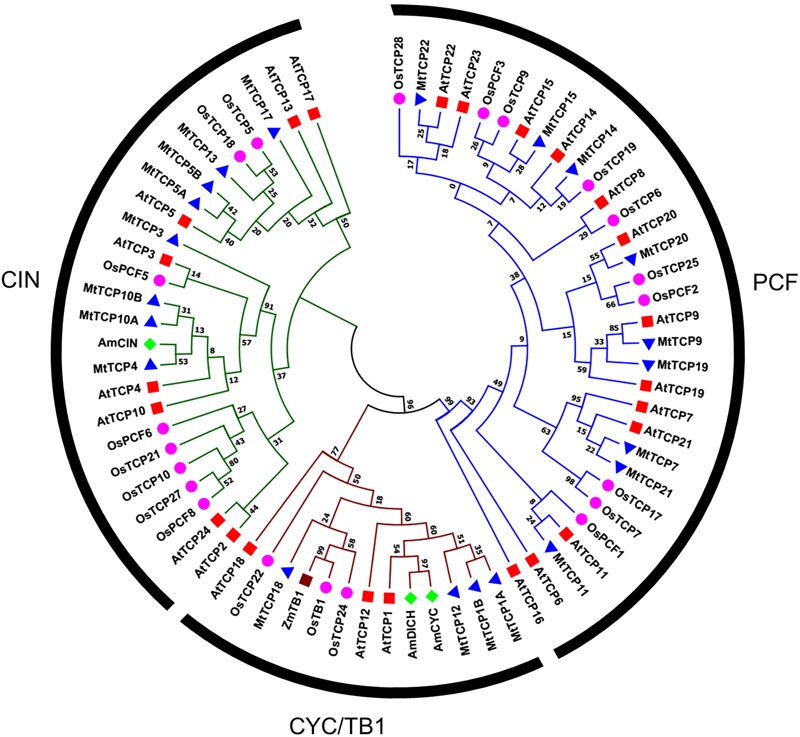
Phylogenetic relationships of TCP transcription factors from M. truncatula (MtTCP), Arabidopsis (AtTCP), rice (OsTCP), maize (ZmTB1), and A. majus (AmCIN/AmCYC/AmDICH). The phylogenetic tree was constructed using 70 full-length protein sequences from M. truncatula (21), Arabidopsis (24), rice (21), maize (1), and A. majus (3) by the Neighbor-Joining method in MEGA 7.1 with 1000 bootstrap replicates. The branched lines of the subtrees are colored to indicate different TCP subclasses.
FIGURE 2.
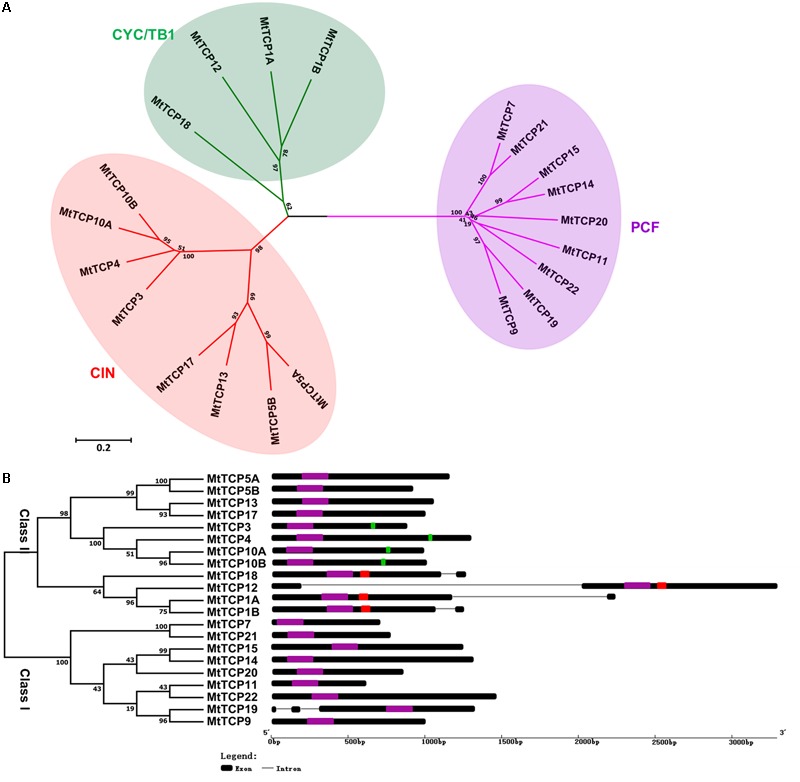
Phylogenetic tree and exon/intron structural analysis of all TCP transcription factors in M. truncatula. (A) Phylogenetic tree was generated based on the full-length protein sequences of all MtTCP genes. Multiple protein sequences were aligned using Clustal W and the phylogenetic trees were constructed using MEGA7.1 by the Neighbor-Joining method and the bootstrap test was performed with 1000 replicates. (B) The exon/intron organization of MtTCP genes in M. truncatula. Exons and introns of MtTCP genes were indicated by black rounded rectangles and black lines, respectively. The TCP domain, R domain, and MtmiR319 recognition sequence are indicated in purple, red, and green rounded rectangles, respectively. The scale was referred to the lengths of the genes.
Gene Structure, Conserved Motifs, and Recognition Sequence of miR319
To get the understanding of the diversification of MtTCPs, the structure and number of exon/intron of MtTCPs were analyzed (Figure 2B). We investigated the exon/intron organization of individual MtTCPs by aligning the CDS sequences and corresponding genomic DNA sequences. The class I MtTCPs displayed a conserved exon–intron organization: eight of nine MtTCPs had no intron, only MtTCP19 possessed two introns. However, the class II MtTCPs showed different number of introns. All the CYC/TB1 subclass MtTCPs contained one more intron than those of CIN subclass genes (Figure 2B).
To obtain a better understanding of the evolutionary relationships of the TCP proteins in M. truncatula, the motifs of MtTCPs were analyzed (Figure 3). In total, 20 motifs were identified in MtTCPs using the online MEME tool. As expected, all the 21 MtTCPs displayed a highly conserved TCP domain. Analysis of the phylogenetic tree and the alignment of the TCP domains showed that MtTCP proteins can be divided into two classes (Figures 2B, 4A and Supplementary Figure S3), as for all species so far. The conserved R domain was only found in MtTCP1A, MtTCP1B, MtTCP12, and MtTCP18, which are the members of CYC/TB1 subclass in Class II (Figures 2B, 4B and Supplementary Figure S3).
FIGURE 3.

Motif composition of MtTCP proteins. The motifs in Medicago TCP proteins were indicated using MEME online tool. The motifs in the MtTCP proteins are denoted by rectangles with different colored boxes. The conserved TCP domains were shown by red boxes. The positions and lengths of motifs in the MtTCP proteins were identified by the colored boxes.
FIGURE 4.
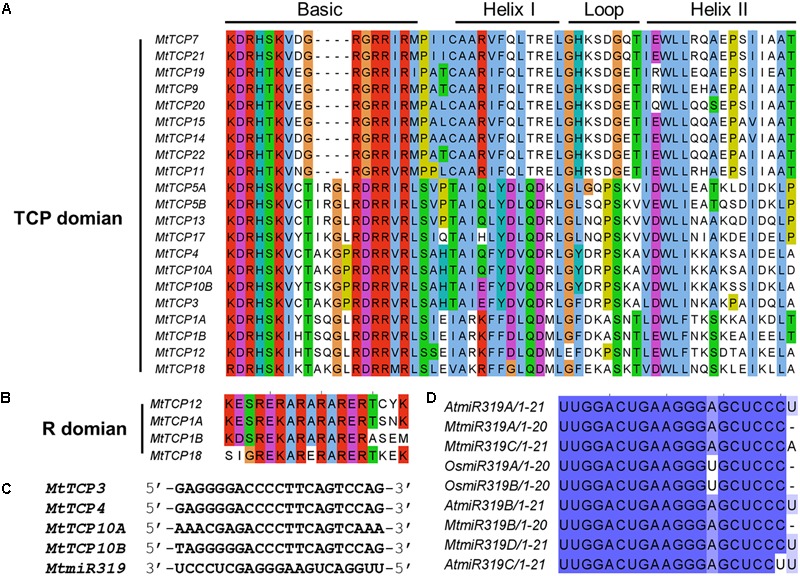
Multiple sequence alignment of MtTCP transcription factors and the MtmiR319-targeted MtTCPs. (A) Alignment of TCP domain of the 21 TCP proteins in M. truncatula. Amino acids that are conserved throughout are shaded in different colors. Conserved domains, including Basic, Helix I, Loop, and Helix II, are shown at the top. (B) Alignment of R-domain of the CYC/TB1 subclass members. Conserved amino acids are shaded in different colors. Multiple sequence alignment was carried out using ClustalW and visualized with Jalview software. (C) Alignment of putative target regions for MtmiR319 binding in CIN subclass MtTCP genes. (D) Mature miR319 sequences in rice, Arabidopsis, and M. truncatula. Shaded zones show the conserved sequences.
In Arabidopsis, five members of the Class II MtTCPs subfamily (AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24) are post-transcriptionally regulated by AtmiR319 (Palatnik et al., 2003). In M. truncatula, the evolutionarily closest homologs of these Arabidopsis genes are MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B, which contained a putative recognition site of MtmiR319 (Figures 2B, 4C). We searched the miRBase Database and found four MtmiR319 (MtmiR319A-D) in M. truncatula genome (Supplementary Figure S4). The alignment of multiple miR319 mature sequences showed that the miR319-TCP regulation module was highly conserved among species (Figure 4D). Although mismatches existed at 3′ of the MtmiR319 and 5′ of the targeted MtTCP mRNA, core target sequences were conserved. These data suggest that MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B probably play the roles in an MtmiR319-regulated manner in M. truncatula.
Distinct Expression Profiles of MtTCPs in M. truncatula
To investigate the tissue-specific expression profiles of MtTCP genes, their relative expression levels in different organs, including young leaves, mature leaves, roots, stems, flowers, shoot buds, and pods, were analyzed by quantitative real-time PCR (qRT-PCR). As indicated in Figure 5, some Class I MtTCP genes were differentially expressed in the different organs, while other Class I MtTCP genes showed similar expression patterns among different organs. For example, MtTCP9, MtTCP11, and MtTCP19 were highly expressed in root and flower, whereas MtTCP7, MtTCP20, and MtTCP21 were expressed at very low level in all organs examined. This finding implies that those Class I MtTCP genes probably play different roles during plant growth and development. Most CYC/TB1 subclass MtTCPs (MtTCP1A, MtTCP1B, and MtTCP12) showed relatively weak expression in the root, stem, leaf, and pod, but highly expressed in flower, implying their specific roles in flower development. While another CYC/TB1 subclass MtTCP gene (MtTCP18) was highly expressed in leaf, flower, and shoot bud, but lowly expressed in root, stem, and pod. In contrast, the expression levels of all the MtmiR319 target CIN subclass MtTCPs (MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B) were very high in young leaf, indicating that they may play an important role in leaf development. The non-MtmiR319 target CIN MtTCPs, MtTCP13, and MtTCP17 showed similar expression patterns. Both of them were strongly expressed in leaf, flower, shoot bud, and pod, but weakly expressed in root and stem, implying their similar roles in plant developmental processes. MtTCP5A and MtTCP5B were highly expressed in root, leaf, and flower, but weakly expressed in stem. The qRT-PCR data suggest that MtTCPs may be involved in different aspects of plant growth and development. However, further studies are still needed to uncover the functional divergence of MtTCP genes in M. truncatula.
FIGURE 5.
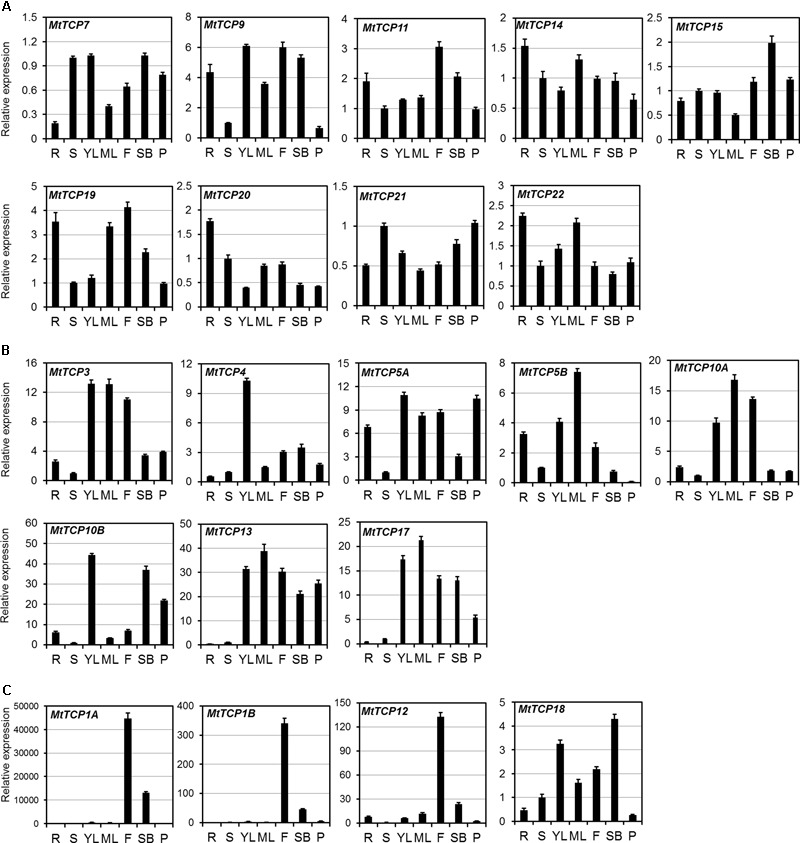
The expression patterns of 21 MtTCP genes in different organs. (A) The expression level of MtTCPs of Class I (A), Class II CIN clade (B), and CYC/TB1 clade (C) in roots (R), stems (S), young leaves (YL), mature leaves (ML), flowers (F), shoot buds (SB), and pods (P), were detected by qRT-PCR. The relative expression level was normalized using MtUBI. Values are the mean and SD of three replicates.
MtmiR319-Targeted MtTCPs Play the Vital Roles in Nodule Development
Roots play an important role in plant growth and development, such as nutrient and water uptake and the establishment of symbiotic interactions (Lopez-Bucio et al., 2003). It is important for legume species, since it can adapt to low nitrogen (N) by symbiosis with nitrogen-fixing rhizobia in nodules (Graham et al., 1981; Carroll et al., 1985; Caetanoanolles and Gresshoff, 1990). To elucidate the roles of MtTCPs in nodule development, the qRT-PCR was conducted to measure their relative expression levels at the different stages of nodule development (Figure 6). Nodules were harvested at 7, 14, and 21 dpi inoculated with S. meliloti 1021 strain, roots without inoculation as control (Supplementary Figure S5). The expression levels of most MtTCPs were down-regulated at different developmental stages of nodules, compared with those in root of 0 dpi. However, the expression levels of MtmiR319-targeted genes, MtTCP3, MtTCP4, and MtTCP10A, were significantly increased. This finding suggests that MtmiR319-MtTCPs module is probably involved in nodule development.
FIGURE 6.
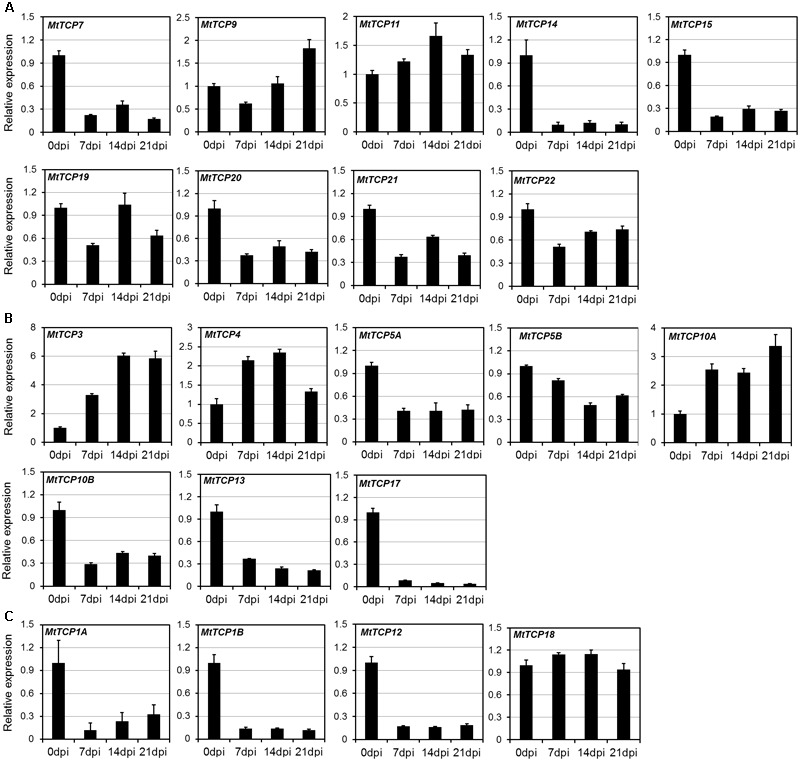
Expression pattern of MtTCPs in different developmental stages of nodule using qRT-PCR. The relative expression level of MtTCPs of Class I (A), Class II CIN clade (B), and CYC/TB1 clade (C) in 0, 7, 14, and 21 days post inoculation (dpi) were detected by qRT-PCR. The expression profiles were normalized using MtUBI. Values are the mean and SD of three replicates.
To further evaluate the possible function of MtmiR319-MtTCPs module, the subcellular localization of the MtmiR319-targeted MtTCPs was performed. MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B were, respectively, fused with GFP, and transformed into tobacco epidermal cells. Based on the observation of green fluorescence signal, these MtmiR319-target MtTCPs were located in the cell nucleus, suggesting that they are functional transcription factors (Figures 7A,B). Then, the MtmiR319A sequence was introduced into wild-type plants under the regulation of the cauliflower mosaic virus 35S promoter and the stable transgenic plants were obtained. The expression levels of MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B were significantly down-regulated in the 35S:MtmiR319A plants, indicating that these four MtTCPs are the targets of MtmiR319A (Figures 7C,D). 35S:MtmiR319A transgenic plant displayed downward-curled leaves with pronounced serrations on the leaf margin (Figures 7E,F), indicating that MtmiR319-target MtTCPs play the conserved roles in leaf development, similar to their functions in Arabidopsis.
FIGURE 7.
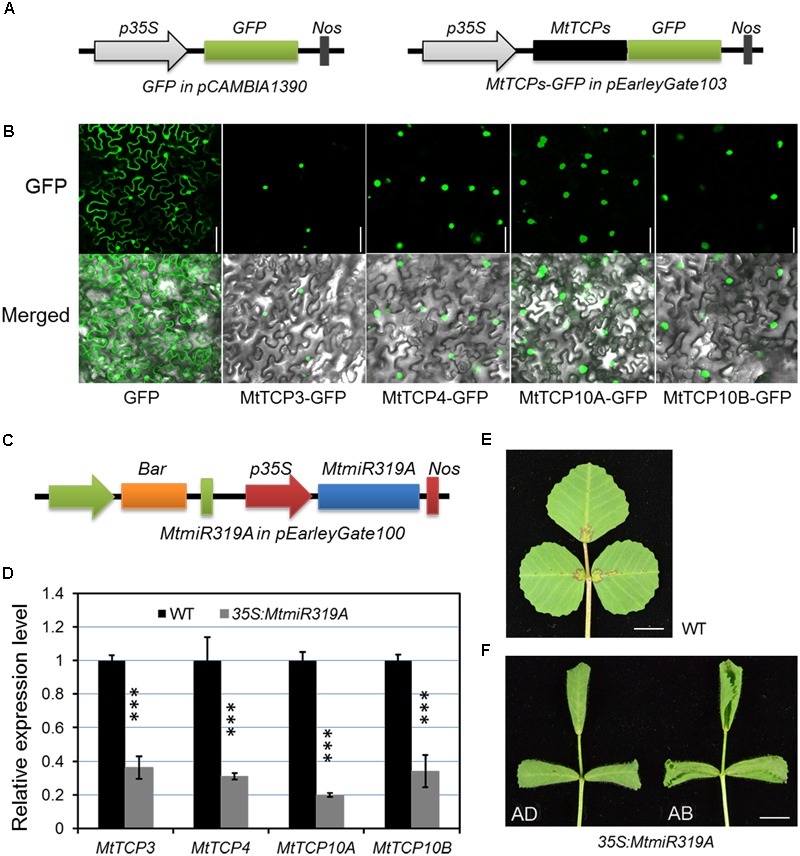
Subcellular localization of the four MtmiR319-targeted MtTCP genes and phenotypic effects of the 35S:MtmiR319A. (A) Schematic illustration of vectors used in subcellular localization analysis. Black box represents the MtTCP genes. Green boxes represent green fluorescence protein (GFP). The MtmiR319-targeted MtTCP genes were cloned to pEarleyGate 103 vector, in which GFP was fused at the C-terminus. (B) The subcellular localization of four MtmiR319-target MtTCPs-GFP fusion proteins. Free GFP as control. Bar = 100 μm. (C) Schematic illustration of vectors used for MtmiR319A overexpression. (D) Relative expression level of the target genes in the T1 generation of 35S:MtmiR319A plants. Transcript levels were measured by qRT-PCR. Values are the means and SD of three biological replicates. T = 2 and ∗∗∗P < 0.001. (E) The leaf of wild-type. (F) The leaf of 35S:MtmiR319A plant. AD, adaxial side of leaf; AB, abaxial side of leaf. Bars = 5 mm in (E,F).
To better characterize their roles in nodule development, wild-type and MtmiR319A-overexpressing plants were inoculated with S. meliloti 1021 strain. After 3 weeks of inoculation, the MtmiR319A-overexpressing plants exhibited reduced shoot fresh weight, nodule numbers, and decreased ratio of nodule/root fresh weight compared with those in wild-type (Figures 8A–D and Supplementary Figure S6). However, there was no obvious difference in nodule size and shape between wild-type and MtmiR319A-overexpressing plants (Supplementary Figure S7). After 3 weeks of inoculation, the nodules were harvested for histological studies. The longitudinal sections of nodules were stained for β-galactosidase activity to visualize the bacteria, since the S. meliloti 1021 strain carried a constitutively expressed hemA/lacZ fusion. The staining in the distal zone of wild-type nodules was much darker than that of MtmiR319A-overexpressing nodules, implying less functional zone of nitrogen fixation exists in MtmiR319A-overexpressing nodules (Figures 8E,F). These results indicate that MtmiR319/MtTCPs module is essential not only for nodule number, but also for nodule symbiosis.
FIGURE 8.
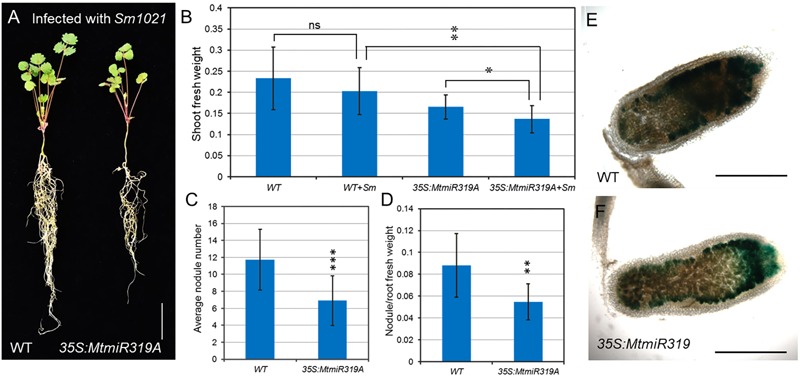
MtmiR319-targeted MtTCP genes are involved in symbiotic root nodule development. (A) Phenotype of wild-type and 35S:MtmiR319A plants after 3 weeks inoculated with S. meliloti strains 1021. Bar = 1 cm. (B) The shoot fresh weight of 3-week-old wild-type and 35S:MtmiR319A plants with inoculation (+Sm) or without inoculation. (C) The average of nodule number in wild-type and 35S:MtmiR319A plants. (D) The ratio of nodule/root fresh weight in wild-type and 35S:MtmiR319A plants. (E,F) The 21 dpi nodules in wild-type and 35S:MtmiR319A plants were sliced into sections for β-galactosidase staining. Bar = 2 mm in (E,F). Values are the means ± SD (n = 20). T = 2, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Discussion
TCP family transcription factors play important roles in multiple processes during plant growth and development, such as leaf development, flower development, phytohormone biosynthesis, endoreduplication, circadian clock, and shoot lateral branching (Palatnik et al., 2003; Takeda et al., 2003; Aguilar-Martinez et al., 2007; Damerval et al., 2007; Ori et al., 2007; Nag et al., 2009; Guo et al., 2010; Koyama et al., 2010a; Wang et al., 2010, 2015a,b; Sarvepalli and Nath, 2011; Shleizer-Burko et al., 2011; Danisman et al., 2012; Li et al., 2012; Aguilar-Martinez and Sinha, 2013; Li and Zachgo, 2013; Niwa et al., 2013; Tao et al., 2013; Das Gupta et al., 2014; Schommer et al., 2014; Lopez et al., 2015; Lucero et al., 2015; Yang et al., 2015; Alvarez et al., 2016; Danisman, 2016; Nicolas and Cubas, 2016; Uberti-Manassero et al., 2016; Wei et al., 2016; Bresso et al., 2017; Koyama et al., 2017). In this study, 21 MtTCPs were identified in M. truncatula, which were distributed on eight chromosomes with different densities.
Generally, genes within same class/clade shared the similar gene structure. Through exon–intron organization analysis of MtTCP family members, except for MtTCP19, the class I and CIN clade TCP genes show similar exon–intron structure that possessed only a single exon. While all members in CYC/TB1 clade display similar exon–intron organization with one exon and one intron. Such exon–intron structure similarity/variation might be caused by single intron loss or gain during the process of gene structure evolution. Protein sequence motif analysis revealed that MtTCPs within the same class/clade exhibit similar motif distribution. Moreover, TCP domains are highly conserved in all members of the MtTCP family, suggesting that the MtTCPs may share similar DNA binding capacity (Kosugi and Ohashi, 1997; Cubas et al., 1999).
It is noted that all class I MtTCPs showed more widespread and less tissue-/organ-specific expression patterns and had relatively high expression levels in all organs. These results indicated that class I MtTCPs might play various regulatory roles at multiple developmental stages.
The class II CYC/TB1 clade TCPs are mainly participated in the developmental regulation of axillary meristems, which gives rise to either lateral branches or flowers. This clade included AtTCP1/12/18 and MtTCP1A/1B/12. MtTCP1A/1B/12 were grouped into a subclade and closely related to AtTCP1, although AtTCP12 was also grouped into this subclade. AtTCP1, the ortholog gene of CYC in A. majus, is involved in the regulation of longitudinal elongation of multiple organs, such as petioles, rosette leaves, and inflorescent stems. AtTCP1 shows high expression level in inflorescence stem, leaf, and petiole (Koyama et al., 2010b). However, MtTCP1A, MtTCP1B, and MtTCP12 exhibited high transcriptional levels in flower and shoot bud, which is different with that in AtTCP1. This finding implies that they may function redundantly in flower and young leaf development in M. truncatula. BRANCHED1 (BRC1, also named AtTCP18) and BRANCHED2 (BRC2, also named AtTCP12), two orthologous genes of TB1 in maize, were transcribed at high levels in axillary meristems or buds (Aguilar-Martinez et al., 2007; Finlayson, 2007; Niwa et al., 2013). AtTCP12 shows a lower transcription level and mild mutant phenotype than its homolog AtTCP18 (Aguilar-Martinez et al., 2007; Finlayson, 2007). The phylogenetically closest gene of AtTCP18 in M. truncatula is MtTCP18. MtTCP18 displayed relatively high expression level in shoot buds. This result suggested that MtTCP18 is likely to execute similar roles with AtTCP18 in axillary bud development and branching control.
The class II TCP gene CIN is expressed downstream of the cell cycle arrest front progression and acts as a repressor of cell proliferation in leaves (Nath et al., 2003). In Arabidopsis, the miR319-targeted TCP genes, AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24, regulate leaf morphogenesis, leaf senescence, and petal growth (Palatnik et al., 2003; Schommer et al., 2008, 2014; Nag et al., 2009; Sarvepalli and Nath, 2011; Li et al., 2012; Bresso et al., 2017; Koyama et al., 2017; Samad et al., 2017). In M. truncatula, four closest MtTCPs (MtTCP3, MtTCP4, MtTCP10A, and MtTCP10B) have a putative recognition site of MtmiR319. All the MtmiR319-targeted MtTCPs were highly expressed in leaves. Meanwhile, MtTCP3 and MtTCP10A also exhibited high expression levels in flowers. However, the non-MtmiR319 target MtTCP genes, such as MtTCP13 and MtTCP17, also exhibited similar expression pattern with those of MtmiR319-targeted MtTCPs, implying the different regulation mechanism controlling their expression.
The root nodule symbiosis is a complex biological process that is controlled by several transcription factors, such as NSP1/2, ERN1, and IPN2 (Schauser et al., 1999; Andriankaja et al., 2007; Middleton et al., 2007; Yano et al., 2008; Zhu et al., 2008; Hirsch et al., 2009; Wang et al., 2013; Battaglia et al., 2014; Kang et al., 2014; Singh et al., 2014; Soyano and Hayashi, 2014). Plant hormone cytokinin (CK) plays an essential role in nodulation. CK signaling is required for cell divisions that initiate nodule development (Murray et al., 2007; Tirichine et al., 2007). Previous reports suggest that downregulation of the expression level of MtLOG1, a CK riboside 5′-monophosphate phosphoribohydrolase, reduces the nodule number, supporting the positive effect of CK on nodulation (Lohar et al., 2004; Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). However, the nodule number also decreases in 35S: MtLOG1 plants, indicating the CK homeostasis is important for nodulation (Mortier et al., 2014). TCPs promote leaf growth and development by modification of responses or sensitivity to CK (Steiner et al., 2012, 2016; Efroni et al., 2013). Overexpression of MtmiR319A led to the downregulation of the targeted MtTCPs. The transgenic plants exhibited small plants size with reduced nodule number and fresh weight during rhizobial infection, which is similar to the plants with CK deficiency. Therefore, it is possible that MtmiR319/MtTCPs module play a significant role in nodule development by regulating the CK pathway.
Conclusion
In this study, we executed genome-wide analyses and identified TCP genes in M. truncatula. Those MtTCP genes were placed on eight chromosomes with different densities. We characterized MtTCP genes expression profiles in different tissues/organs and developmental stages of nodule, suggesting MtTCP genes could play vital roles in M. truncatula growth and development. In addition, the nuclear-localized signal of four MtTCP-GFP fusion proteins indicated that the MtTCPs were functional transcription factors. Remarkably, overexpression of MtmiR319A in M. truncatula leads to the decreased nodule number and nodule weight, demonstrating that MtmiR319/MtTCPs module might be involved in the regulation of nodule development. Further study is needed to illuminate the molecular mechanism that MtTCPs genes involved in leaf and nodule development.
Author Contributions
HfW and CZ designed the research. HfW and HwW carried out most of the research and analyzed the data. RL and YX performed some nodule analysis experiments. ZL contributed analytical tools. HfW and CZ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MB and handling Editor declared their shared affiliation.
Funding. This work was supported by grants from the Ministry of Science and Technology of China (2015CB943500), the National Natural Science Foundation of China (31671507 and 31371235), and the 1000-Talents Plan from China for young researchers (1011.2.206.99.99).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00774/full#supplementary-material
References
- Aggarwal P., Das Gupta M., Joseph A. P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22 1174–1189. 10.1105/tpc.109.066647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez J. A., Poza-Carrion C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 458–472. 10.1105/tpc.106.048934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez J. A., Sinha N. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 4:406. 10.3389/fpls.2013.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J., Galego L. (2005). Flower symmetry and shape in Antirrhinum. Int. J. Dev. Biol. 49 527–537. 10.1387/ijdb.041967ja [DOI] [PubMed] [Google Scholar]
- Alvarez J. P., Furumizu C., Efroni I., Eshed Y., Bowman J. L. (2016). Active suppression of a leaf meristem orchestrates determinate leaf growth. Elife 5:e15023 10.7554/eLife.15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja A., Boisson-Demier A., Frances L., Sauviac L., Jauneau A., Barker D. G., et al. (2007). AP2-ERF transcription factors mediate nod factor-dependent mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19 2866–2885. 10.1105/tpc.107.052944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M., Ripodas C., Clua J., Baudin M., Aguilar O. M., Niebel A., et al. (2014). A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol. 164 1430–1442. 10.1104/pp.113.230896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin C., Camut S., Malpica C. A., Truchet G., Rosenberg C. (1990). Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2 1157–1170. 10.1105/tpc.2.12.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresso E. G., Chorostecki U., Rodriguez R. E., Palatnik J. F., Schommer C. (2017). Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 176 1694-1708. 10.1104/pp.17.00823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetanoanolles G., Gresshoff P. M. (1990). Early induction of feedback regulatory responses governing nodulation in soybean. Plant Sci. 71 69–81. 10.1016/0168-9452(90)90069-Z [DOI] [Google Scholar]
- Carroll B. J., Mcneil D. L., Gresshoff P. M. (1985). Isolation and properties of soybean [Glycine-Max (L) Merr] mutants that nodulate in the presence of high nitrate concentrations. Proc. Natl. Acad. Sci. U.S.A. 82 4162–4166. 10.1073/pnas.82.12.4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chen Y. Q., Ding A. M., Chen H., Xia F., Wang W. F., et al. (2016). Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 15:7728 10.4238/gmr.15027728 [DOI] [PubMed] [Google Scholar]
- Chen X. M. (2008). MicroRNA metabolism in plants. RNA Interference 320 117–136. 10.1007/978-3-540-75157-1_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford B. C., Nath U., Carpenter R., Coen E. S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135 244–253. 10.1104/pp.103.036368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18 215–222. 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Damerval C., Le Guilloux M., Jager M., Charon C. (2007). Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae. Plant Physiol. 143 759–772. 10.1104/pp.106.090324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7:1930. 10.3389/fpls.2016.01930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S., van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159 1511–1523. 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S., van Dijk A. D., Bimbo A., van der Wal F., Hennig L., de Folter S., et al. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64 5673–5685. 10.1093/jxb/ert337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta M., Aggarwal P., Nath U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204 901–912. 10.1111/nph.12963 [DOI] [PubMed] [Google Scholar]
- Dhaka N., Bhardwaj V., Sharma M. K., Sharma R. (2017). Evolving Tale of TCPs: new paradigms and old lacunae. Front. Plant Sci. 8:479. 10.3389/fpls.2017.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). Teosinte branched1 and the origin of maize - evidence for epistasis and the evolution of dominance. Genetics 141 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Du J., Hu S., Yu Q., Wang C., Yang Y., Sun H., et al. (2017). Genome-wide identification and characterization of BrrTCP transcription factors in Brassica rapa ssp. rapa. Front. Plant Sci. 8:1588. 10.3389/fpls.2017.01588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K. M., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Efroni I., Han S. K., Kim H. J., Wu M. F., Steiner E., Birnbaum K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24 438–445. 10.1016/j.devcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X. Z., Zhao Z., Tian Z. X., Xu S. L., Luo Y. H., Cai Z. G., et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. U.S.A. 103 4970–4975. 10.1073/pnas.0600681103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson S. A. (2007). Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 48 667–677. 10.1093/pcp/pcm044 [DOI] [PubMed] [Google Scholar]
- Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22 3921–3934. 10.1105/tpc.110.074518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693. 10.1105/tpc.106.043778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. H., Leonard R. T., Menge J. A. (1981). Membrane-mediated decrease in root exudation responsible for phosphorus inhibition of vesicular-arbuscular mycorrhiza formation. Plant Physiol. 68 548–552. 10.1104/pp.68.3.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E. B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22 1161–1173. 10.1105/tpc.109.069203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Munoz A., Heckmann A. B., Downie J. A., Oldroyd G. E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21 545–557. 10.1105/tpc.108.064501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D. G., Donoghue M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103 9101–9106. 10.1073/pnas.0602827103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Chu X. J., Wang C., Xiao A. F., Zhu H., Yuan S. L., et al. (2014). A MYB coiled-coil transcription factor interacts with NSP2 and is involved in nodulation in Lotus japonicus. New Phytol. 201 837–849. 10.1111/nph.12593 [DOI] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68 147–158. 10.1111/j.1365-313X.2011.04674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619. 10.1105/tpc.9.9.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 337–348. 10.1046/j.1365-313X.2002.01294.x [DOI] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010a). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22 3574–3588. 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Sato F., Ohme-Takagi M. (2010b). A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 74 2145–2147. 10.1271/bbb.100442 [DOI] [PubMed] [Google Scholar]
- Koyama T., Sato F., Ohme-Takagi M. (2017). Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 175 874–885. 10.1104/pp.17.00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. X., Potuschak T., Colon-Carmona A., Gutierrez R. A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102 12978–12983. 10.1073/pnas.0504039102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zachgo S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76 901–913. 10.1111/tpj.12348 [DOI] [PubMed] [Google Scholar]
- Li X., Zhuang L. L., Ambrose M., Rameau C., Hu X. H., Yang J., et al. (2010). Genetic analysis of ele mutants and comparative mapping of ele1 locus in the control of organ internal asymmetry in garden pea. J. Integr. Plant Biol. 52 528–535. 10.1111/j.1744-7909.2010.00949.x [DOI] [PubMed] [Google Scholar]
- Li Z., Li B., Shen W. H., Huang H., Dong A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71 99–107. 10.1111/j.1365-313X.2012.04973.x [DOI] [PubMed] [Google Scholar]
- Lin Y. F., Chen Y. Y., Hsiao Y. Y., Shen C. Y., Hsu J. L., Yeh C. M., et al. (2016). Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67 5051–5066. 10.1093/jxb/erw273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar D. P., Schaff J. E., Laskey J. G., Kieber J. J., Bilyeu K. D., Bird D. M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 38 203–214. 10.1111/j.1365-313X.2004.02038.x [DOI] [PubMed] [Google Scholar]
- Lopez J. A., Sun Y., Blair P. B., Mukhtar M. S. (2015). TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 20 238–245. 10.1016/j.tplants.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J., Cruz-Ramirez A., Herrera-Estrella L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6 280–287. 10.1016/S1369-5266(03)00035-9 [DOI] [PubMed] [Google Scholar]
- Lucero L. E., Uberti-Manassero N. G., Arce A. L., Colombatti F., Alemano S. G., Gonzalez D. H. (2015). TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 84 267–282. 10.1111/tpj.12992 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99 367–376. 10.1016/S0092-8674(00)81523-8 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 794–799. 10.1038/383794a0 [DOI] [PubMed] [Google Scholar]
- Ma J., Wang Q., Sun R., Xie F., Jones D. C., Zhang B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4:6645. 10.1038/srep06645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ma J., Fan D., Li C., Jiang Y., Luo K. (2016). Genome-wide identification of TCP family transcription factors from Populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 6:32795. 10.1038/srep32795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Middleton P. H., Jakab J., Penmetsa R. V., Starker C. G., Doll J., Kalo P., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction. Plant Cell 19 1221–1234. 10.1105/tpc.106.048264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V., Wasson A., Jaworek P., De Keyser A., Decroos M., Holsters M., et al. (2014). Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 202 582–593. 10.1111/nph.12681 [DOI] [PubMed] [Google Scholar]
- Murray J. D., Karas B. J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104. 10.1126/science.1132514 [DOI] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 22534–22539. 10.1073/pnas.0908718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B. C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299 1404–1407. 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Herve C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65 23–33. 10.1007/s00239-006-0174-z [DOI] [PubMed] [Google Scholar]
- Nicolas M., Cubas P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33 33–41. 10.1016/j.pbi.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Niwa M., Daimon Y., Kurotani K., Higo A., Pruneda-Paz J. L., Breton G., et al. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25 1228–1242. 10.1105/tpc.112.109090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X. L., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Parapunova V., Busscher M., Busscher-Lange J., Lammers M., Karlova R., Bovy A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. 10.1186/1471-2229-14-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. 10.1101/gad.1004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resentini F., Felipo-Benavent A., Colombo L., Blazquez M. A., Alabadi D., Masiero S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8 482–485. 10.1016/j.molp.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Rogers K., Chen X. M. (2013). Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell 25 2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A. F. A., Sajad M., Nazaruddin N., Fauzi I. A., Murad A. M. A., Zainal Z., et al. (2017). MicroRNA and transcription factor: key players in plant regulatory network. Front. Plant Sci. 8:565. 10.3389/fpls.2017.00565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K., Nath U. (2011). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67 595–607. 10.1111/j.1365-313X.2011.04616.x [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. 10.1038/46058 [DOI] [PubMed] [Google Scholar]
- Schommer C., Debernardi J. M., Bresso E. G., Rodriguez R. E., Palatnik J. F. (2014). Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 7 1533–1544. 10.1093/mp/ssu084 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chetelat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Guy K. M., Wu W., Fang B., Yang J., Zhang M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16:85. 10.1186/s12870-016-0765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N. (2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138 695–704. 10.1242/dev.056770 [DOI] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microb. 15 139–152. 10.1016/j.chom.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Soyano T., Hayashi M. (2014). Transcriptional networks leading to symbiotic nodule organogenesis. Curr. Opin. Plant Biol. 20 146–154. 10.1016/j.pbi.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T. S., Kieffer M., et al. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24 96–108. 10.1105/tpc.111.093518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Livne S., Kobinson-Katz T., Tal L., Pri-Tal O., Mosquna A., et al. (2016). The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171 1485–1494. 10.1104/pp.16.00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Amano K., Ohto M. A., Nakamura K., Sato S., Kato T., et al. (2006). RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 61 165–177. 10.1007/s11103-006-6265-9 [DOI] [PubMed] [Google Scholar]
- Takeda T., Suwa Y., Suzuki M., Kitano H., Ueguchi-Tanaka M., Ashikari M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33 513–520. 10.1046/j.1365-313X.2003.01648.x [DOI] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., et al. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25 421–437. 10.1105/tpc.113.109223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L. H., Radutoiu S., Albrektsen A. S., Sato S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107. 10.1126/science.1132397 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Coscueta E. R., Gonzalez D. H. (2016). Expression of a repressor form of the Arabidopsis thaliana transcription factor TCP16 induces the formation of ectopic meristems. Plant Physiol. Biochem. 108 57–62. 10.1016/j.plaphy.2016.06.031 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Lucero L. E., Viola I. L., Vegetti A. C., Gonzalez D. H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63 809–823. 10.1093/jxb/err305 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 136 669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wang C., Zhu H., Jin L. P., Chen T., Wang L. X., Kang H., et al. (2013). Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Plant Mol. Biol. 82 97–111. 10.1007/s11103-013-0042-3 [DOI] [PubMed] [Google Scholar]
- Wang H., Mao Y., Yang J., He Y. (2015a). TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 6:436. 10.3389/fpls.2015.00436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yang X., Xu M., Lin X., Lin T., Qi J., et al. (2015b). A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Mol. Plant 8 1795–1808. 10.1016/j.molp.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y., Luo D. (2010). LjCYC genes constitute floral dorsoventral asymmetry in Lotus japonicus. J. Integr. Plant Biol. 52 959–970. 10.1111/j.1744-7909.2010.00926.x [DOI] [PubMed] [Google Scholar]
- Wang Z., Luo Y. H., Li X., Wang L. P., Xu S. L., Yang J., et al. (2008). Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc. Natl. Acad. Sci. U.S.A. 105 10414–10419. 10.1073/pnas.0803291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Hu Y., Cui M. Y., Han Y. T., Gao K., Feng J. Y. (2016). Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 7. 10.3389/fpls.2016.01937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Tian Z., Feng X., Li X., Xu S., Hu X., et al. (2011). Petal development in Lotus japonicus. J. Integr. Plant Biol. 53 770–782. 10.1111/j.1744-7909.2011.01072.x [DOI] [PubMed] [Google Scholar]
- Xie Q., Liu X., Zhang Y., Tang J., Yin D., Fan B., et al. (2017). Identification and characterization of microRNA319a and its putative target gene, PvPCF5, in the bioenergy grass switchgrass (Panicum virgatum). Front. Plant Sci. 8:396. 10.3389/fpls.2017.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Luo Y., Cai Z., Cao X., Hu X., Yang J., et al. (2013). Functional diversity of CYCLOIDEA-like TCP genes in the control of zygomorphic flower development in Lotus japonicus. J. Integr. Plant Biol. 55 221–231. 10.1111/j.1744-7909.2012.01169.x [DOI] [PubMed] [Google Scholar]
- Yang X., Pang H. B., Liu B. L., Qiu Z. J., Gao Q., Wei L., et al. (2012). Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24 1834–1847. 10.1105/tpc.112.099457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhao X. G., Li C. Q., Liu J., Qiu Z. J., Dong Y., et al. (2015). Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of Petrocosmea spp. of the family gesneriaceae. Plant Physiol. 169 2138–2151. 10.1104/pp.15.01181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Yoshida S., Muller J., Singh S., Banba M., Vickers K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U.S.A. 105 20540–20545. 10.1073/pnas.0806858105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Ma H., Wang J., Zhang D. B. (2007). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49 885–897. 10.1111/j.1744-7909.2007.00509.x [DOI] [Google Scholar]
- Zhou Y. Z., Xu Z. D., Zhao K., Yang W. R., Cheng T. R., Wang J., et al. (2016). Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front. Plant Sci. 7:1301. 10.3389/fpls.2016.01301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Chen T., Zhu M. S., Fang Q., Kang H., Hong Z. L., et al. (2008). A novel ARID DNA-Binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol. 148 337–347. 10.1104/pp.108.119164 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


