Abstract
Objective
Pompe disease (PD) is a progressive neuromuscular disorder that is caused by glucosidase acid alpha (GAA) deleterious mutations. Mitochondrial involvement is an important contributor to neuromuscular diseases. In this study the sequence of MT-ATP 6/8 and Cytochrome C oxidase I/II genes along with the expression levels of the former genes were compared in classic and non-classic patients.
Materials and Methods
In this case-control study, the sequence of MT-ATP 6/8 and Cytochrome C oxidase was analyzed by polymerase chain reaction (PCR)-Sanger sequencing and expression of MT-ATP genes were quantified by real time-PCR (RT-PCR) in 28 Pompe patients. The results were then compared with 100 controls. All sequences were compared with the revised Cambridge reference sequence as reference.
Results
Screening of MT-ATP6/8 resulted in the identification of three novel variants, namely T9117A, A8456C and A8524C. There was a significant decrease in MT-ATP6 expression between classic (i.e. adult) and control groups (P=0.030). Additionally, the MT-ATP8 expression was significantly decreased in classic (P=0.004) and non-classic (i.e. infant) patients (P=0.013). In total, 22 variants were observed in Cytochrome C oxidase, five of which were non- synonymous, one leading to a stop codon and another (C9227G) being a novel heteroplasmic variant. The A8302G in the lysine tRNA gene was found in two brothers in a pedigree, while a T7572C variant in the aspartate tRNA gene was observed in two brothers in another pedigree.
Conclusion
The extent of mitochondrial involvement in the classic group was more significant than in the non-classic form. Beside GAA deleterious mutations, it seems that mtDNA variants have a secondary effect on PD. Understanding, the role of mitochondria in the pathogenesis of Pompe may potentially be helpful in developing new therapeutic strategies.
Keywords: Alpha-Glucosidase, Cytochrome-C Oxidase, Mitochondria, Pompe
Introduction
Pompe disease (PD, OMIM #232300) is a progressive myopathy with an autosomal recessive mode of inheritance (1). The combined incidence of PD is generally 1 in 40,000 (2). It has two common forms (early-onset/classic and late-onset/non-classic) with differences in degree of disease severity, age of onset and organ involvement (3, 4). The patients present a broad spectrum of clinical variability such as cardiomyopathy, hypotonia and respiratory insufficiency (5, 6).
They suffer from deficiency or lack of acid alphaglucosidase enzyme (GAA) that arise as a result of various deleterious variants in GAA (1). Genotype-phenotype correlation studies among patients with the same mutation in GAA have revealed different clinical manifestations (2). It seems that this diversity may be a result of interaction of other genetic and non-genetic factors. The sign and symptoms that are observed in Pompe patients are similar to those in mitochondrial disorders.
According to previous reports, mitochondrial dysfunction can affect the neuromuscular system (7). Mitochondria (mt) are essential to aerobic respiration by producing adenosine triphosphate (ATP). The function of mt is controlled by both the mtDNA and nuclear genomes, and mtDNA variants may be affected by nuclear genome variants or vice versa (8). It is therefore possible that mtDNA genes interact with GAA. To test this hypothesis, MT-ATP6/8 and Cytochrome C oxidase I/II were screened for functional variants, and the expression level of the former genes were analyzed in early and late- onset PD patients.
Materials and Methods
In this case-control study, we recruited 28 PD patients (17 infants and 11 adults) from the Department of Neurology of both Shariati and Mofid hospitals from December 2013 to February 2015. In this study, 100 healthy controls were also recruited comprising 17 infants and 83 adults. An informed consent was obtained from each participant or a parent in the case of infants. PD was diagnosed based on clinical findings by two expert neurologists, measurement of GAA biochemical activity or detection of deleterious variants in GAA. The included patients had no family history of mitochondrial or major neuromuscular disorders. This study was approved by the Ethical Committee of Tehran University of Medical Sciences (92-02-30-23162).
DNA/RNA extraction
DNA was extracted from whole blood by using QIAamp DNA Blood Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions. Quantity and quality of DNA were checked by NanoDrop ND-1000 (NanoDrop Technologies, USA) at 260/280 nm wavelengths and running on an agarose gel (1%) respectively. Total RNAwas extracted from fresh whole blood samples by using the Hybrid-RTM Blood RNA kit and following its protocol (see http://www. tribioscience.com/files/315-150.pdf). RNA concentration and integrity were measured by NanoDrop and agarose gel respectively prior to cDNA synthesis. Presence of sharp bands for both 18S and 28S rRNA was checked. Purified RNA was then stored at -80°C.
cDNA synthesis
Total RNA was used to synthesize cDNA by using the cDNA synthesis kit (Fermentas, Germany) according to manufacturer’s instructions. Briefly, 2 µg of total RNA, 1 µL of oligo dT and random hexamer primers and 8 µL nuclease-free water were mixed in a sterile, nuclease- free tube and placed on ice. After incubation at 65°C for 5 minutes, it was chilled on ice and 4 µL of 5X reaction buffer, 1 µL of RiboLock RNase Inhibitor (20 U/µL), 2 µL of 10 mM dNTP mix, 1 µL of RevertAid M-MuLV RT (200 U/µL) were added.
The mixture was centrifuged briefly and incubated for 5 minutes at 25°C followed by 60 minutes at 42°C. The reaction terminated by heating at 70°C for 5 minutes and stored at -80°C until further use.
Variant detection
Polymerase chain reaction (PCR) was performed with primers specific to MT- ATP6/8, Cytochrome C oxidase and their flanking sequences (Table 1) (8).
Table 1.
Comparison of MT-ATP6/8 gene variants in PD and control groups
| Infant/Adult | Nucleotide | Locus | Amino acid change | R/N.R | Hm/Ht | Pompe | Control | P value |
|---|---|---|---|---|---|---|---|---|
| + | + | |||||||
| Adult | C8406T | MT-ATPase8 | p.T14I | R | Hm | 1 | 0 | 0.219 |
| A8456C | MT-ATPase6/8 | p.T31P | N.R | Hm | 1 | 0 | 0.219 | |
| G9039A | MT-ATPase6 | p.M171I | R | Hm | 1 | 0 | 0.219 | |
| G9055A | MT-ATPase6 | p.A177T | R | Hm | 1 | 3 | 0.99 | |
| Infant | A8502T | MT-ATPase8 | p.N46I | R | Hm | 1 | 0 | 0.219 |
| C8562T | MT-ATPase6 | p.P66L | R | Hm | 2 | 0 | 0.047 | |
| C8562T | MT-ATPase8 | p.P66P | R | Hm | 2 | 0 | 0.047 | |
| C8684T | MT-ATPase6 | p.T53I | R | Hm | 1 | 6 | 0.650 | |
| G8697A | MT-ATPase6 | p.M57I | R | Hm | 1 | 0 | 0.219 | |
| T9117A | MT-ATPase6 | p.I197I | N.R | Ht | 1 | 0 | 0.219 | |
| C9129T | MT-ATPase6 | p.I201I | R | Hm | 1 | 0 | 0.219 | |
| Both | A8524C | MT-ATPase8 | p.P53P | N.R | Ht | 2 | 0 | 0.047 |
| A8701G | MT-ATPase6 | p.T59A | R | Hm | 2 | 6 | 0.556 | |
| A8860G | MT-ATPase6 | p.T112A | R | Hm | 24 | 81 | 0.235 | |
R and N.R; Denote reported and not-reported respectively, Hm and Ht; Denote homoplasmy and heteroplasmy respectively, and PD; Pompe disease.
The PCR reaction included 50 ng of genomic DNA, 1 µL of each primer (10 pmol), 0.2 mM of each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2 and 1 U of Taq polymerase (CinnaGen, Inc, Iran). Cycling conditions for all PCR reactions were an initial denaturation at 94°C for 4 minutes, followed by 35 cycles of denaturation at 94°C for 35 seconds, annealing at 56° for 35 seconds, extension at 72°C for 35 seconds, and a final extension at 72°C for 5 minutes. PCR-amplified fragments were sequenced by Macrogen (South Korea) using the same PCR primers in both directions along with a series of overlapping primers to cover all regions of interest for more accurate results. Finch TV version 1.4 (Geospiza, Inc., USA) was used to analyze the chromatograms and were then checked using BLAST (https://blast.ncbi.nlm.nih.gov). The results were compared with the revised Cambridge reference sequence MITOMAP (www.mitomap.org) and the 1000 Genome databases. Presence of variants was also checked in controls which were selected from different Iranian ethnicities. Furthermore, the effect of missense variants on protein structure was assessed by Polyphen-2 (http://genetics.bwh.harvard.edu/pph2) and CADD (Combined Annotation Depedent Depletion; http:// cadd.gs.washington.edu/score) scores.
MT-ATP 6/8 expression analysis
Expression levels of MT-ATP 6/8 were quantified using a quantitative PCR (qPCR) assay. In brief, expression values were normalized relative to a housekeeping gene (ß-actin) to calculate the relative gene expression based on the 2-ΔΔCt method (9). Details of primers are given in Table 2.
qPCR reactions were performed on a Corbett 6000 PCR-Real-time Detection System with a total volume of 20 µl reaction mixture, containing 1 µl DNA template (50 ng), 10 µl SYBR Green PCR Master Mix (Takara, Japan), 8 µl nuclease- free water and 0.5 µl of each primer (10 pmol). The cycling conditions were an initial denaturation step at 95ºC for 30 seconds followed by 40 cycles of a denaturation step at 95ºC for 12 seconds and an annealing step at 58ºC for 35 seconds. Melting curve analysis was used to validate the specificity and identity of the PCR product for each primer pair. Each sample was run in duplicate to ensure the reliability of the results. Also, a non-template control was included in each qPCR run.
Statistical analysis
Quantitative variables, in the form of frequency, such as participant characteristics and mitochondrial involvement were described as mean ± SD. Fisher’s exact test was used to compare frequency of mitochondrial involvement in PD and control groups. P<0.05 was considered statistically significant. All analyses were implemented in SPSS version 16 (IBM, USA). Bonferroni’s multiple-testing correction was used to adjust the significance level (a). For frequency comparison of the identified 14 variants, a was set to 0.0036. For differential expression, given that two genes were compared, a was set to 0.025.
Results
Screening of MT-ATP6/8 in patients resulted in the identification of 14 variants, of which three were novel variants (Fig .1). Of the total, 7 variants were in the classic group, 4 in the non-classic group and 3 were shared between the two groups (Table 1). There were four synonymous and ten non synonymous variants in MT-ATPase6/8 (Table 1). The variants not previously reported in MITOMAP (WWW.mitomap.org) and other variant databases (e.g. the 1000 Genome database), were checked in controls. Frequencies of A8524C and C8562T were significantly different between patients and controls (P=0.047). A8860G, A8524C and C8562T were observed in 85.71, 7.14 and 7.14% of patients, respectively. The Polyphen-2 and CADD scores of missense variants showed in the Table 2.
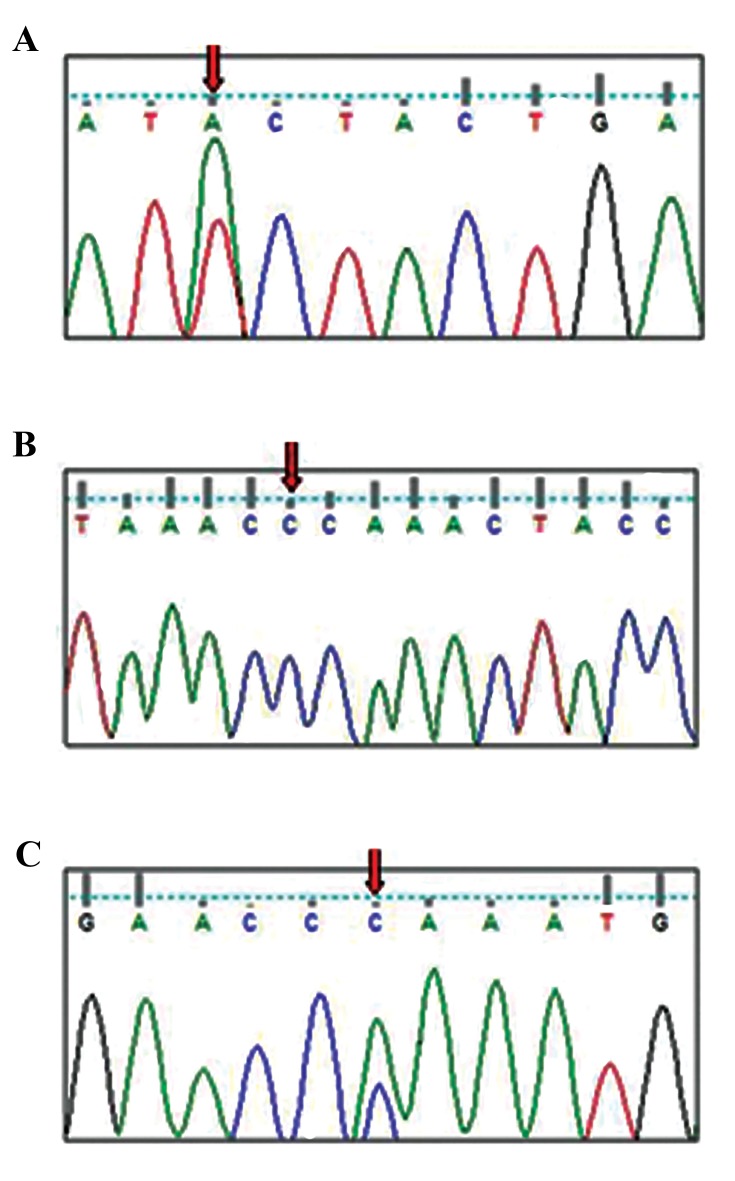
Fig.1.
Chromatogram of the three novel variants in Mt.ATPase6/8 genes. A. T9117A, p.I197I, Heteroplasmy, B. A8456C, p.T31P, Homoplasmy, and C. A8524C, p.P53P, Heteroplasmy.
Table 2.
Predicted effect of missense variants on protein structure
| Gene | Nucleotide position | Polyphen-2 score | CADD score | Prediction effect |
|---|---|---|---|---|
| MT-CO2 | 7805 | 0.00 | 0.01 | Benign |
| MT-CO2 | 7859 | 0.00 | 0.50 | Benign |
| MT-ATP8 | 8406 | 0.08 | 0.16 | Benign |
| MT-ATP8 | 8456 | 0.02 | 3.82 | Benign |
| MT-ATP8 | 8502 | 0.99 | 18.47 | Damaging |
| MT-ATP6 | 8701 | 0.00 | 0.09 | Benign |
| MT-ATP6 | 9039 | 0.89 | 18.04 | Damaging |
| MT-ATP6 | 9055 | 0.84 | 22.60 | Damaging |
| MT-CO3 | 9336 | 0.00 | 0.01 | Benign |
| MT-CO3 | 9949 | 0.99 | 23.60 | Damaging |
| MT-CO3 | 9963 | 1.00 | 23.70 | Damaging |
MT-ATP6/8 genes expression
A significant decrease was observed for MT-ATP6 expression inclassic group compared with the control group. In addition, MT-ATP8 expression was significantly decreased in the classic (P=0.004) and non-classic (P=0.013) patient groups compared with their controls.
MT-Cytochrome C oxidase
Screening of MT-Cytochrome C oxidase led to the observation of 22 variants which C9227G sequence being a new heteroplasmic variant (Fig .2). Fifteen and five variants were identified in the classic and non-classic groups respectively, while two variants were shared between the two groups. Furthermore, 5 variants were non-synonymous, 15 synonymous with one stop codon (Table 3). The predicted effect of missense variants on protein structure are given in Table 2.
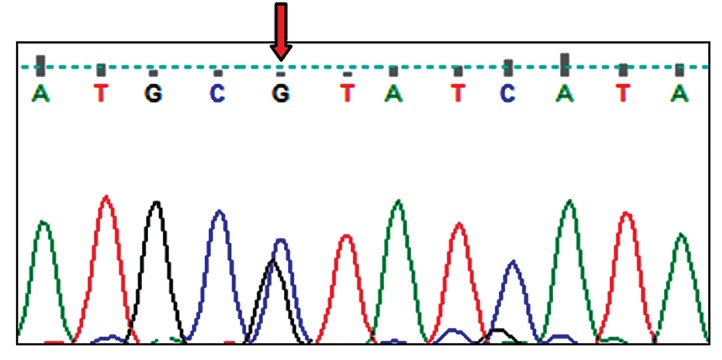
Fig.2.
Chromatogram of C9227G as a novel heteroplasmic variant in the Mt. Cytochrome C oxidase gene.
Table 3.
Frequency distribution of Mt-Cytochrome C oxidase variants in pompe and controls
| Infant/Adult | Nucleotide | Locus | Amino acid change | R/N.R | Hm/Ht | Pompe | Control | P value |
|---|---|---|---|---|---|---|---|---|
| + | + | |||||||
| Adult | A7933G | Cyto co2 | p.L116L | R | Hm | 1 | 1 | 0.388 |
| C7939T | Cyto co2 | p.F118F | R | Hm | 1 | 0 | 0.219 | |
| A8170G | Cyto co2 | p.Gln195Gln | R | Hm | 1 | 0 | 0.219 | |
| T9581C | Cyto co3 | p.Asn125Asn | R | Hm | 1 | 0 | 0.219 | |
| G9986A | Cyto co3 | p.Gly260Gly | R | Hm | 1 | 1 | 0.388 | |
| Infant | T7645C | MT co2 | p.L20L | R | Hm | 1 | 1 | 0.388 |
| G7805A | MT-CO2 | p.Val74IL | R | Hm | 1 | 0 | 0.219 | |
| G7859A | MT-CO2 | p.D92N | R | Hm | 1 | 0 | 0.219 | |
| T7861C | MT-CO2 | p.D92D | R | Hm | 1 | 1 | 0.388 | |
| C7945T | MT-CO2 | p.S120S | R | Hm | 1 | 0 | 0.219 | |
| C7990A | MT-CO2 | p.L135L | R | Hm | 1 | 0 | 0.219 | |
| A8014T | MT-CO2 | p.V143V | R | Hm | 1 | 0 | 0.219 | |
| C8137T | MT-CO2 | p.F184F | R | Hm | 1 | 2 | 0.520 | |
| G8206A | MT CO2 | p.Met207Met | R | Hm | 1 | 3 | 0.990 | |
| C9227G | Cyto co3 | p.Ala7Ala | N.R | h.t | 1 | 0 | 0.219 | |
| A9336G | Cyt co3 | p.Met44Val | R | Hm | 1 | 0 | 0.219 | |
| T9530C | Cyto co3 | p.P108P | R | Hm | 1 | 1 | 0.388 | |
| C9776T | cyto co3 | p.Asp190Asp | R | Hm | 1 | 0 | 0.219 | |
| T9949G | Cyto co3 | p.Val248Gly | R | Hm | 1 | 0 | 0.219 | |
| T9963G | MT-Co3 | p.Tyr253Asp | R | Hm | 1 | 0 | 0.219 | |
| Both | G8269A | MT-CO2 | p.X228X | R | Hm | 3 | 0 | 0.219 |
| T9540C | cyto co3 | p.L112L | R | Hm | 3 | 0 | 0.219 | |
R and N.R; Denote reported and not reported, respectively and Hm and Ht; Denotes homoplasmy and heteroplasmy, respectively.
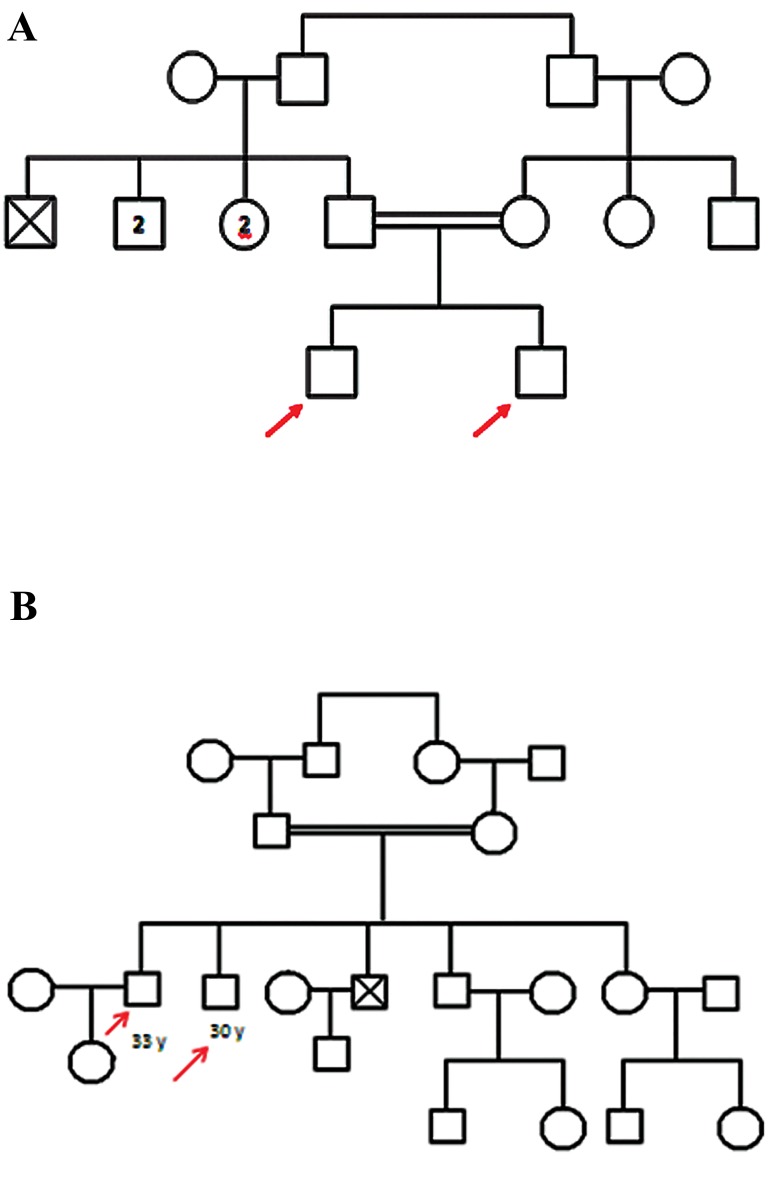
Moreover, based on the analysis of the sequence of lysine tRNA and aspartate tRNA genes, the A8302G variant was found in the former gene in two brothers in a pedigree (Fig .3A) and the T7572C variant was found in the latter gene in two brothers in another pedigree (Fig .3B). Variants C15904T and G15928A were also found in two infants in the threonine and tyrosine tRNA genes, respectively.
Fig.3.
Pedigree diagram. A. Pedigree of a family with a A8302G variant in siblings and B. Pedigree of a family with a T7572C variant in siblings.
Discussion
PD is a heterogeneous neuromuscular disorder. Patients suffer from myopathy, hypotonia and other neuromuscular manifestations (5). Some tissues such as the nerve and muscle are more susceptible to mitochondrial dysfunction since these tissues are highly dependent on oxidative phosphorylation (10). According to previous studies, mitochondrial abnormality has been observed in PD patients (11-21). It is therefore possible that mitochondrial variants have a secondary role in PD. In this study, MT-ATP6/ 8 and Cytochrome C oxidase genes of 28 PD patients were screened. The last complex in the mitochondria, MT-ATP synthase, plays an important role in the production of ATP. It has 14 subunits, of which 2, namely MT-ATP6/8 are encoded by mtDNA (21). Variants in these genes may result in ATP production impairment in some vulnerable tissues such as the muscle (22).
The role of complex V variants has already been seen in the increase of free radicals. They can affect gene expression as a secondary factor. In these genes, some amino acids are conserved and any change could be potentially pathogenic (23, 24). Fourteen variants were found in MT-ATP6/8 of which three were novel. One was a non-synonymous variant, however, it had a predicted benign effect on protein structure.
MT-ATP6/8 expression decreased in the classic group and the number of variants in this group was more than the non-classic group. These are consistent with the severity of symptoms in the classic group. The missense variants A8502T, G9039A and G9055A replace Asparagine to Isoleucine, Methionine to Isoleucine and Alanine to Threonine respectively which all have a predicted damaging effect on protein structure. A8502T was reported by Gurses in 44 patients with Epilepsy (25). Asparagine is a polar amino acid which can form hydrogen bonds and acts as a neurotransmitter with Glutamate, while leucine is a non-polar and hydrophobic amino acid. This replacement may thus affect protein function as predicted by Polyphen 2.
With respect to G8697A, methionine is a conserved amino acid. It has a sulfur in its structure that tends to form beta-sheets and despite owning hydrophobic properties, it can interact with some electrophilic regions. In contrast, isoleucine participates in alpha-helix structure and plays a role in ligand binding to protein. Such a replacement may change the structure and function of MT-ATP6. This variant was also observed in other studies (23, 26, 27). MT-ATP6/8 genes were also studied in other neurodegenerative diseases such as Huntington (26), Friedreich’s ataxia and multiple sclerosis (MS) (28).
The C8684T variant in MT-ATP6 changes threonine to isoleucine. The former is a polar amino acid while the latter is non-polar. This change may affect the tertiary structure of the protein and its interaction with the ATP molecule. This variant was also observed in ataxia and autism (29). The C8684T was observed in MS and Huntington’s (23), and G8697A in MS (23) and ataxia telangiectasia (27). The C8562T is a synonymous variant which has been reported in patients with ataxia (29). A8456C and C8406T have benign effects. A8860G was found in 85.71% of patients. In addition, the variants G8697A and A8701G were previously reported in cardiomyopathy. This variant has also been reported in neurodegenerative diseases (30).
It is possible that the cardiomyopathy observed in PD patients is the result of the presence of these variants, by dysregulating Mt.ATPase6/8 expression that was observed in this study. According to the previous study promoters are located in the D-loop region, this area is a hot spot therefor D-loop variants may change the sequence of promoters and binding affinity of transcription factors to enhancer or silencer elements (13, 17, 18). It seems that these variants along with other genetic and environmental factors are involved in development of PD.
Mitochondrial DNA encodes 3 subunits for cytochrome C oxidase. It is the last enzyme in the mitochondrial respiratory chain and is responsible for electron transfer from the cytochrome to oxygen (31). Impairment of cytochrome C oxidase is clinically highly heterogeneous. It starts at any age and includes a diverse range of myopathy to severe multi-organ involvement (32).Genetic defects that affect the structure and function of this gene are usually severe and often lead to fatal metabolic disorders. Such disorders usually occur before the age of 2 and involve tissues such as the heart, muscle and liver, however, its manifestation in adulthood is with less severity. Severity of this disorder can vary even in family members.
In the case of early onset cardiac muscle involvement is usually associated with hypertrophic cardiomyopathy, however, in late onset cases, myopathy and hypotonia are observed (33). It seems that the pattern of manifestations associated with MT-cytochrome C oxidase is similar to PD. Variants T9540C, T7645C and G8269A observed in this study were identical to those found in the study by Mkaouar-Rebai et al. (34) on patients with myopathy. These variants are likely to be related to myopathy and hypotonia symptoms of PD. The two variants T9949G and T9963G change valine to glycine and tyrosine to aspartic acid respectively. Polyphen 2 predicted these variants to have a damaging effect on protein structure. Both variants were in the classic group, thus suggesting that they may lead to more severe phenotypes in this group.
The observed A8302G variant in lysine tRNA has been previously reported in encephalopathy (35). Also, Govindaraj et al. (36) reported this mutation in three patients with Madras motor neuron disease (MMND).
The variant T7572C located on the T arm of the gene encoding aspartate tRNA was previously reported by Reddy et al. (37) in myelodysplastic syndrome, however, Li et al. (38) found it as a neutral polymorphism, showing that this variant did not affect the secondary structure of the corresponding tRNA by using RNA fold.
G15928A, which is located at the anticodon stem of the tRNAThr, is shown to be a neutral polymorphism (38). The C15904T variant is also shown to be a polymorphism which occurs in the general population with low frequency (39). The C15904T and G15928A variants were observed in patients with encephalopathy by Houshmand et al. (40). Overall, validation of the function of each variant is recommended.
Conclusion
The extent of mitochondrial involvement in the classic group was more severe than the non-classic group. According to these findings, it seems that mtDNA variants have a secondary role in PD. Understandingthe role of mitochondria in the pathogenesis of PD may potentially lead to the development of new therapeutic strategies.
Acknowledgments
We thank all the participants in this study. This study was funded by Tehran University of Medical Sciences, Tehran, Iran (Grant Number: 23162). The authors declare no conflict of interest.
Author’s Contributions
F.B., S.M.A., M.H., M.H.M.; Conceptualization, investigation, methodology, supervision, validation, visualization, writing-review and editing. F.B.; Data curation. F.B., M.H.; Formal analysis. F.B., S.M.A., M.H.; Project administration. F.B., S.M.A.; Software and writing-original draft. All authors read and approved the final manuscript.
References
- 1.Hobson Webb L, Proia AD, Thurberg BL, Banugaria S, Prater SN, Kishnani PS. Autopsy findings in late-onset Pompe disease: a case report and systematic review of the literature. Mol Genet Metab. 2012;106(4):462–469. doi: 10.1016/j.ymgme.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Pittis MG, Donnarumma M, Montalvo AL, Dominissini S, Kroos M, Rosano C, et al. Molecular and functional characterization of eight novel GAA mutations in Italian infants with Pompe disease. Hum Mutat. 2008;29(6):E27–E36. doi: 10.1002/humu.20753. [DOI] [PubMed] [Google Scholar]
- 3.Kishnani PS, Beckemeyer AA, Mendelsohn NJ. The new era of Pompe disease: advances in the detection, understanding of the phenotypic spectrum, pathophysiology, and management. Am J Med Genet C Semin Med Genet. 2012;160C(1):1–7. doi: 10.1002/ajmg.c.31324. [DOI] [PubMed] [Google Scholar]
- 4.Kishnani PS, Steiner RD, Bali D, Berger K, Byrne BJ, Case LE, et al. Pompe disease diagnosis and management guideline. Genet Med. 2006;8(5):267–288. doi: 10.1097/01.gim.0000218152.87434.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kishnani P, Hwu W, Mandel H, Nicolino M, Yong F, Corzo D. A retrospective, multinational, multicenter study on the natural history of infantile-onset Pompe disease. Pediatrics. 2006;148(5):671–676. doi: 10.1016/j.jpeds.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Schuller A, Wenninger S, Strigl-Pill N, Schoser B. Toward deconstructing the phenotype of late-onset Pompe disease. Am J Med Genet C Semin Med Genet. 2012;160C(1):80–88. doi: 10.1002/ajmg.c.31322. [DOI] [PubMed] [Google Scholar]
- 7.Nardin RA, Johns DR. Mitochondrial dysfunction and neuromuscular disease. Muscle Nerve. 2001;24(2):170–191. doi: 10.1002/1097-4598(200102)24:2<170::aid-mus30>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147–147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.DiMauro S, Bonilla E, Davidson M, Hirano M, Schon EA. Mitochondria in neuromuscular disorders. Biochim Biophys Acta. 1998;1366(1-2):199–210. doi: 10.1016/s0005-2728(98)00113-3. [DOI] [PubMed] [Google Scholar]
- 11.Engel AG, Dale AJ. Autophagic glycogenosis of late onset with mitochondrial abnormalities: light and electron microscopic observations. Mayo Clin Proc. 1968;43(4):233–279. [PubMed] [Google Scholar]
- 12.Hudgson P, Fulthorpe JJ. The pathology of type II skeletal muscle glycogenosis A light and electron-microscopic study. J Pathol. 1975;116(3):139–147. doi: 10.1002/path.1711160303. [DOI] [PubMed] [Google Scholar]
- 13.Verity MA. Infantile Pompe’s disease, lipid storage, and partial carnitine deficiency. Muscle Nerve. 1991;14(5):435–440. doi: 10.1002/mus.880140509. [DOI] [PubMed] [Google Scholar]
- 14.Fernandeza R, Fernandezb JM, Cerverac C, Teijeiraa S, Teijeiroa A, Domınguezd C, et al. Adult glycogenosis II with paracrystalline mitochondrial inclusions and Hirano bodies in skeletal muscle. Neuromuscul Disord. 1999;9(3):136–143. doi: 10.1016/s0960-8966(98)00117-5. [DOI] [PubMed] [Google Scholar]
- 15.Laforet P, Nicolino M, Eymard PB, Puech JP, Caillaud C, Poenaru L, et al. Juvenile and adult-onset acid maltase deficiency in France: genotype-phenotype correlation. Neurology. 2000;55(8):1122–1128. doi: 10.1212/wnl.55.8.1122. [DOI] [PubMed] [Google Scholar]
- 16.Selak MA, de Chadarevian JP, Melvin JJ, Grover WD, Salganicoff L, Kaye EM. Mitochondrial activity in Pompe’s disease. Pediatr Neurol. 2000;23(1):54–57. doi: 10.1016/s0887-8994(00)00145-4. [DOI] [PubMed] [Google Scholar]
- 17.Thurberg BL, Lynch Maloney C, Vaccaro C, Afonso K, Tsai AC, Bossen E, et al. Characterization of pre- and post-treatment pathology after enzyme replacement therapy for Pompe disease. Lab Invest. 2006;86(12):1208–1220. doi: 10.1038/labinvest.3700484. [DOI] [PubMed] [Google Scholar]
- 18.Lewandowska E, Wierzba-Bobrowicz T, Rola R, Modzelewska J, Stępień T, Ługowska A, et al. Pathology of skeletal muscle cells in adult-onset glycogenosis type II (Pompe disease): ultrastructural study. Folia Neuropathol. 2008;46(2):123–133. [PubMed] [Google Scholar]
- 19.Herzog A, Hartung R, Reuser AJ, Hermanns P, Runz H, Karabul N, et al. A cross-sectional single-centre study on the spectrum of Pompe disease, German patients:molecular analysis of the GAA gene, manifestation and genotype-phenotype correlations. Orphanet J Rare Dis. 2012;7:35–35. doi: 10.1186/1750-1172-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroos M, Hoogeveen-Westerveld M, Van der ploeg A, Reuser AJ. The genotype-phenotype correlation in Pompe disease. Am J Med Genet C Semin Med Genet. 2012;160C(1):59–68. doi: 10.1002/ajmg.c.31318. [DOI] [PubMed] [Google Scholar]
- 21.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 22.Tzen C, Wu TY, Liu HF. Sequence polymorphism in the coding region of mitochondrial genome encompassing position 8389-8865. Forensic Sci Int. 2001;120(3):204–209. doi: 10.1016/s0379-0738(01)00389-9. [DOI] [PubMed] [Google Scholar]
- 23.Ghaffarpour M, Mahdian R, Fereiduni F, Kamalidehghan B, Moazami N, Houshmand M. The mitochondrial ATPase6 gene is more susceptible to mutation than the ATPase8 gene in breast cancer patients. Cancer Cell Int. 2014;14(1):21–21. doi: 10.1186/1475-2867-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzen CY, Wu TY. Evolutionalanalysis in determining pathogenic versus nonpathogenic mutations of ATPase 6 in human mitochondriopathy. Ann N Y Acad Sci. 2005;1042:19–24. doi: 10.1196/annals.1338.002. [DOI] [PubMed] [Google Scholar]
- 25.Gurses C, Azakli H, Alptekin A, Cakiris A, Abaci N, Arikan M, et al. Mitochondrial DNA profiling via genomic analysis in mesial temporal lobe epilepsy patients with hippocampal sclerosis. Gene. 2014;538(2):323–3277. doi: 10.1016/j.gene.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 26.Kasraie S, Houshmand M, Banoei MM, Ahari SE, Panahi MS, Shariati P, et al. Investigation of tRNA(Leu/Lys) and ATPase 6 genes mutations in Huntington’s disease. Cell Mol Neurobiol. 2008;28(7):933–938. doi: 10.1007/s10571-008-9261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.KulawIec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009;8(14):1378–1385. doi: 10.4161/cbt.8.14.8751. [DOI] [PubMed] [Google Scholar]
- 28.Ahari S, Houshmand M, Kasraie S, Moin M, Bahar M, Shafa Shariat Panahi M, et al. Point mutations on mitochondrial DNA in Iranian patients with friedreich’s ataxia. Iran J Child Neurol. 2007;2(1):41–45. [Google Scholar]
- 29.Houshmand M, Kasraie S, Etemad Ahari S, Moin M, Bahar M, Zamani A. Investigation of tRNA and ATPase 6/8 gene mutations in Iranian ataxia telangiectasia patients. Arch Med Sci. 2011;7(3):523–527. doi: 10.5114/aoms.2011.23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houshmand M, Montazeri M, Kuchekian N, Noohi F, Nozar G, Zamani A. Is 8860 variation a rare polymorphism or associated as a secondary effect in HCM disease? Arch Med Sci. 2011;7(2):242–246. doi: 10.5114/aoms.2011.22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siskova A, Wihelm J. The effects of hyperoxia, hypoxia, and ischemia/ reperfusion on the activity of cytochrome oxidase from the rat retina. Physiol Res. 2001;50(3):267–273. [PubMed] [Google Scholar]
- 32.Shoubridge EA. Cytochrome c oxidase deficiency. Am J Med Genet. 2001;106(1):46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 33.Pecina P, Houstkova H, Hansíkova H, Zeman J, Houstek J. Genetic defects of cytochrome c oxidase assembly. Physiol Res. 2004;53(Suppl 1):S213–S223. [PubMed] [Google Scholar]
- 34.Mkaouar-Rebai E, Ben Mahmoud A, Chamkha I, Chabchoub I, Kammoun T, Hachicha M, et al. A novel MT-CO2 m.8249G>A pathogenic variation and the MT-TW m.5521G>A mutation in patients with mitochondrial myopathy. Mitochondrial DNA. 2014;25(5):394–399. doi: 10.3109/19401736.2013.803086. [DOI] [PubMed] [Google Scholar]
- 35.Sternberg D, Chatzoglou E, Laforet P, Fayet G, Jardel C, Blondy P, et al. Mitochondrial DNA transfer RNA gene sequence variations in patients with mitochondrial disorders. Brain. 2001;124(Pt 5):984–994. doi: 10.1093/brain/124.5.984. [DOI] [PubMed] [Google Scholar]
- 36.Govindaraj P, Nalini A, Krishna N, Sharath A, Khan NA, Tamang R, et al. Mitochondrial DNA variations in Madras motor neuron disease. Mitochondrion. 2013;13(6):721–728. doi: 10.1016/j.mito.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy PL, Shetty VT, Dutt D, York A, Dar S, Mundle SD, et al. Increased incidence of mitochondrial cytochrome c-oxidase gene mutations in patients with myelodysplastic syndromes. Br J Haematol. 2002;116(3):564–575. doi: 10.1046/j.0007-1048.2001.03323.x. [DOI] [PubMed] [Google Scholar]
- 38.Li LJ, Huang H, Hu YR, Chen H, Chen X, Yang J, et al. Mitochondrial transfer RNA mutations and male infertility. Biomed Res. 2017;28(11):4908–4912. [Google Scholar]
- 39.van den Ouweland JM, Bruining GJ, Lindhout D, Wit JM, Veldhuyzen BF, Maassen JA. Mutations in mitochondrial tRNA genes: non-linkage with syndromes of Wolfram and chronic progressive external ophthalmoplegia. Nucleic Acids Res. 1992;20(4):679–682. doi: 10.1093/nar/20.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houshmand M, Larsson NG, Holme E, Oldfors A, Tulinius MH, Andersen O. Automatic sequencing of mitochondrial tRNA genes in patients with mitochondrial encephalomyopathy. Biochim Biophys Acta. 1994;122(1):49–55. doi: 10.1016/0925-4439(94)90058-2. [DOI] [PubMed] [Google Scholar]