What do realtors always emphasize? Location. Location. Location. This mantra is worth repeating for those studying gut stem cell biology. During the past 18 months, the scientific community has been captivated by the importance of neighborhood (niche) in defining a cellular state of “stemness.” We have heard that adult bone marrow-derived stem cells are able to establish residence in, and supply appropriate differentiated lineages to, heart, vascular endothelium, liver, brain, and muscle (reviewed in Blau et al.; ref. 1). Elegant studies in invertebrates also have emphasized the importance of niche in regulating expression of stem cell features (e.g., ref. 2).
The article by Bjerknes and Cheng (3) that appears in this issue of PNAS introduces us to a “new” neighbor that has a significant impact on multipotential intestinal stem cells—the enteric neuron. It also presents a novel paradigm for regulating epithelial proliferation.
Glucagon-like peptide 2 (GLP-2) is produced by enteroendocrine cells—one of four cell types descended from the intestinal stem cell. Previous work had shown that administration of GLP-2 prevents damage or facilitates repair of epithelial abnormalities produced by a variety of entities, from chemotherapy to inflammatory bowel disease. There was also prior evidence that GLP-2 exerted its therapeutic effect by enhancing cell division in the proliferative units of the intestine known as crypts of Lieberkühn. However, neither the mechanism nor the direct cellular targets of GLP-2 were known.
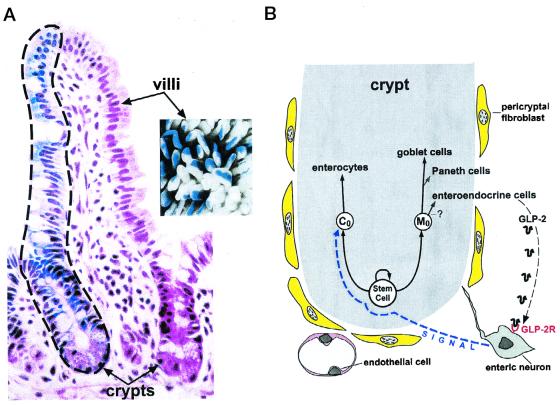
To appreciate Bjerknes and Cheng's results, it is helpful to briefly review the cellular landscape of the crypt of Lieberkühn. In adult mice and humans, there is rapid and continuous renewal of the intestinal epithelium. This renewal is driven by a multipotent stem cell that lives at or near the base of these crypts (4). In the small intestine, this stem cell gives rise to absorptive enterocytes plus three types of secretory cells: goblet, enteroendocrine, and Paneth. Paneth cells, which secrete antimicrobial peptides, digestive enzymes, and growth factors, complete their differentiation at the crypt base. The other three epithelial lineages differentiate during a rapid, highly organized upward migration from the crypt to the tips of adjacent villi where they are extruded (Fig. 1A).
Figure 1.
The intestinal stem cell niche. (A) Illustration of the stem cell hierarchy of the crypt. Chimeric mice can be generated by injecting pluripotent embryonic stem cells from one genetic background into blastocysts representing another genetic background. The small intestine of the resulting mouse contains cells representing both genetic backgrounds. The image on the left is a section prepared from the proximal small intestine of an adult chimeric mouse. The finger-shaped villus is supplied with epithelial cells from two adjacent crypts. One crypt is populated entirely by blue-colored epithelial cells representing one genetic background. The other crypt is populated entirely by nonblue cells representing the other genetic background. Crypts do not contain a mixture of cells from both genetic backgrounds. Thus, they are monoclonal, composed of epithelial cells ultimately derived from a single progenitor. This progenitor occupies the highest position in the stem cell hierarchy. The precise number of progenitor-derived multipotent stem cells that are active in each crypt is uncertain. Note that cells from each crypt migrate up the villus in an orderly fashion, forming distinct columns with discrete borders. Migration is completed in 3–5 days. The highly organized migration is illustrated further in the Inset that shows a whole-mount preparation of intestine from a chimeric mouse. Striped villi contain differentiating epithelial cells derived from monoclonal blue crypts and nonblue (white) crypts. (B) Diagrammatic representation of the stem cell niche. An active multipotent stem cell (SC) gives rise to a Co daughter that produces the enterocytic lineage, and to descendants that generate secretory lineages (goblet, Paneth, and enteroendeocrine cells; note it is uncertain whether all are derived from Mo). GLP-2 produced by a subset of enteroendocrine cells is able to stimulate proliferation of the Co daughter via interaction with enteric nervous system neurons that express the GLP-2 receptor (GLP-2R). The nature of the neuronal signal that affects Co is unknown.
Bjerknes and Cheng had already shown that the multipotent crypt stem cell gives rise to two types of long-lived daughters. One daughter is committed to produce columnar-shaped enterocytes (this daughter was called Co). The other daughter was designated Mo and yields mucus-producing goblet cells (5). In their report in this issue of PNAS, they use a clever assay, with a great name for those who do gut research (SPASM, specific progenitor assay by somatic mutation), to show that GLP-2 treatment specifically stimulates proliferation of the Co progenitor. In SPASM, mice belonging to the inbred Swiss–Webster (SWR) strain are treated with N-nitroso-N-ethylurea (NEU) to induce mutations in a small percentage of intestinal stem cells or their immediate daughters. SWR mice are Dlb1−/−. It appears that Dlb1 is the same as Galgt2, which encodes an N-acetylgalactosaminyltransferase (6). NEU-induced mutation of Dlb1−/− intestinal epithelial cells to Dlb1+/− allows them to acquire the ability to produce carbohydrates containing terminal GalNAcα(1–3)GalNAc epitopes recognized by the lectin Dolichos bifluorus agglutinin (DBA). When SWR intestinal epithelial stem cells, or their immediate daughters, are mutated from Dlb1−/− to Dlb1+/−, they, and all of their differentiating progeny, form a cluster or clone. The clones appear as rare but distinctive DBA-positive groups of cells within the DBA-negative, Dlb1−/− SWR epithelium.
Three weeks after NEU treatment, Bjerknes and Cheng administered GLP-2 to SWR mice. Ten days later, they scored the size (number of cells) and cellular composition of DBA+ clones scattered along the length of treated intestines. Animals that received NEU but not GLP-2 served as controls. GLP-2 increased the average size of DBA+ clones composed only of enterocytes (called columnar clones because each originates from a Co progenitor with a mutated Dlb1 locus). The size of DBA+ clones composed of both enterocytes and goblet cells (called stem cell clones because they originate from stem cells with a mutated Dlb1 locus) was also significantly increased. However, the increased size of stem cell clones was caused by an expansion of Co-derived enterocytes: the number of goblet cells in these clones was similar to the number in stem cell clones of control animals. Consistent with these findings, the average size of DBA+ clones composed of goblet cells alone (mucous clones from mutated Mo progenitors) was the same as in controls.
Having shown that GLP-2 specifically stimulates proliferation of Co progenitors, Bjerknes and Cheng go on to demonstrate that the GLP-2 receptor is expressed in a subset of enteric neurons and not in the crypt epithelium. The enteric nervous system (ENS) is a complex network of interacting neurons and supporting glial cells that regulates intestinal motility, blood flow, and secretion. ENS neurons are located in two distinct anatomic regions: the myenteric plexus and the submucosal plexus. Myenteric plexus neurons send most of their axonal projections to the muscle layers of the intestine. Submucosal plexus neurons send the majority of their projections to the subepithelial region, including the area surrounding crypts. Bjerknes and Cheng report that GLP-2 treatment produces a rapid induction of c-Fos expression in enteric neurons, followed by c-Fos induction in the region of the crypt containing dividing descendants of the multipotent stem cell. Blocking neuronal voltage-gated sodium channels with tetrodotoxin suppresses the crypt epithelial c-Fos response to GLP-2. Based on these findings, they propose a model where GLP-2 signals enteric neurons through their GLP-2 receptors to produce an as yet unknown signal that stimulates Co enterocyte progenitors (Fig. 1B).
These results provide us with an intriguing new paradigm for regulating epithelial growth that can be described in the following general terms. Two different cell lineages are derived from the same stem cell. A factor is produced by members of one lineage. (In this case, an important function of the enteroendocrine lineage is to sense the nutrient environment in the gut lumen). This factor is able to stimulate production of the second lineage. The function of the second lineage complements that of the first (enterocytes absorb nutrients). The two epithelial lineages are “connected” to one another through (enteric) neurons. It will be interesting to see whether a comparable apparatus functions in other renewing epithelia.
Other Niche Players
The microvasculature that surrounds the crypt has recently been identified as a new and influential player in the intestinal stem cell niche. The extensive denuding of intestinal epithelium that occurs after large doses of radiation for abdominal cancers is known as the GI syndrome. The syndrome has been attributed to apoptosis of the crypt's multipotent stem cell. However, a study just published by Paris and coworkers (7) challenges this belief. They used a mouse model to show that radiation first triggers rapid endothelial apoptosis, which later leads to epithelial death. Their conclusions are based on two key observations: loss of epithelial stem cells did not occur when endothelial apoptosis was blocked by giving basic fibroblast growth factor (an endothelial survival factor) or if the mice were homozygous for a null allele of asmase. This gene encodes the acid sphingomyelinase required to produce a critical ceramide intermediate in the endothelial apoptotic pathway.
In addition to being in close contact with enteric neurons and blood vessels, adult crypts are surrounded by a layer of specialized mesenchymal cells known as pericryptal fibroblasts (or subepithelial myofibroblasts). Tritiated thymidine labeling studies have indicated that like their overlying epithelial cells pericryptal fibroblasts continuously migrate upward from the crypt base (8). This puts them in strategic position to establish and maintain instructive communications with stem cells and their descendants.
There is already a large body of genetic evidence that mesenchymal epithelial cross-talk is critical for proper gut development. Participants include platelet-derived growth factor (PDGF) A and its receptor, PDGF-Rα (required for formation of villi; ref. 9), the winged helix transcription factor Fkh6 (10), the homeodomain transcription factor Nkx2–3 (11), as well as Hox and ParaHox cluster genes (12), Sonic hedgehog, and bone morphogenetic proteins (13). The role pericryptal fibroblasts play in maintaining the stem niche in fully formed adult crypts remains unclear: are they participants in a critical mesenchymal-epithelial dialogue, and if so what are the mediators? Very little is known about the repertoire of genes expressed in pericryptal fibroblasts. The time is ripe for a pericryptal fibroblast genome anatomy project where these cells are retrieved from their native environment without significant perturbation (e.g., by laser capture microdissection; ref. 14), and their mRNA transcripts are profiled. The results would be a first step toward determining their impact on gut stem cell biology.
Intraepithelial lymphocytes (IELs) and microbes may be additional modulators. The IEL population of the gut is equivalent in size to the entire population of peripheral lymphocytes residing in the spleen. Genetically engineered T cell receptor (TCR) δ subunit-deficient mice that lack crypt γδ TCR+ IELs have reduced epithelial proliferation (15). The intestine is home to a diverse collection of indigenous microbes known as the microbiota. Comparisons of rodents with and without a microbiota (i.e., germ-free versus conventionally raised) indicate that components of this microbial society affect proliferative status in the crypt, expression of epithelial-based members of the angiogenin family factors, as well as enteric nervous system activity (16, 17).
What We Know About the Properties of Crypt Stem Cells
Alas, there is scant information about the molecular features of the multipotent crypt cell itself, or the factors that regulate specification of its descendant lineages. A recent study of Hes1 knockout mice indicated that, as in other systems, Notch signaling is an important contributor to lineage specification (18). Transcriptional activation of Hes1, a mammalian homolog of Drosophila Hairy and Enhancer of split genes, occurs immediately downstream of Notch. Hes1 encodes a basic helix–loop–helix (bHLH) transcriptional repressor that opposes the activity of various bHLH transcriptional activators. In the sensory epithelium of the inner ear, Hes1 negatively regulates hair cell differentiation from progenitors by antagonizing Math1, the mouse homolog of the Drosophila Atonal gene (19). Hes1 is also required for repressing expansion of neuronal precursors during neurogenesis (20). Targeted disruption of the Hes1 gene increases enteroendocrine and goblet cells and reduces enterocytes in the fetal mouse intestine (18), suggesting that Hes1 normally functions to negatively regulate specification of enteroendocrine/goblet cell lineages and to positively regulate specification of enterocytes.
There is a critical need for convenient biological assays that allow intestinal stem cells to be identified based on their ability to give rise to descendant lineages ex vivo. Without such clonigenic assays, characterization of the cellular and molecular factors required to sustain growth or survival of stem cells, or to generate descendant lineages, will continue to be a slow and haphazard process. Thus, it is encouraging to read reports such as the one by Whitehead et al. (21) describing a technique for establishing in vitro growth of undifferentiated epithelial cells from disaggregated normal human and mouse colonic crypts.
These days it may not be necessary to take gut stem cells out of their niche before gaining insights about their expressed genes. An alternative is to “purify” an expression profile in silico. For example, in the mouse stomach, lineage tracing studies using tritiated thymidine have identified a presumptive multipotent stem cell and its immediate committed daughters (22). We have used DNA microarrays to profile gene expression in (i) normal adult mouse stomachs where these presumptive stem cells and committed lineage progenitors represent a small fraction of the total epithelial population, (ii) embryonic day 18.5 stomachs where stem cells and committed progenitors predominate, and (iii) stomachs from adult transgenic mice where an engineered ablation of acid-producing parietal cells results in a marked amplification of the stem cells and lineage progenitors (23). We first generated a list of mRNAs enriched in both parietal cell-ablated and embryonic day 18.5 stomachs (i.e., increased in ii and iii relative to i). This list was then cross-indexed to the publicly available hematopoietic stem cell database (http://stemcell.princeton.edu/; ref. 24), producing several dozen likely candidates for preferential expression in gastric stem cells (unpublished work).
One application of this in silico “profile surfing” approach is to identify candidate cell surface markers that might be used to retrieve rare stem cells from complex cellular populations. These comparisons can also be extended as more and more searchable datasets of genes expressed in defined cellular populations are deposited in the public domain. For example, C-GAP (Cancer Genome Anatomy Project; http://cgap.nci.nih.gov/) collects libraries of expressed sequence tags from normal and neoplastic tissues and provides software tools that investigators can use to generate customized datasets of genes preferentially expressed in gastrointestinal cancers versus normal tissues. These lists can then be compared with lists of genes preferentially expressed in normal gut epithelial progenitors.
The study of Bjerknes and Cheng (3) emphasizes the importance of obtaining a broad cellular and molecular view of the intestinal stem cell neighborhood. One way of achieving this goal will be to mobilize stakeholders in this scientific community to profile gene expression in cellular members of the niche. Generating well-annotated and searchable databases from these studies would set the stage for subsequent bioinformatic exercises. The results should provide us with a new understanding of neighborhood resources and testable hypotheses about the rules that govern interactions among neighbors.
Footnotes
See companion article on page 12497.
References
- 1.Blau H M, Brazelton T R, Weimann J M. Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- 2.Xie T, Spradling A C. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- 3.Bjerknes M, Cheng H. Proc Natl Acad Sci USA. 2001;98:12497–12502. doi: 10.1073/pnas.211278098. . (First Published September 25, 2001; 10.1073/pnas.211278098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjerknes M, Cheng H. Am J Anat. 1982;166:76–92. [Google Scholar]
- 5.Bjerknes M, Cheng H. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 6.Mohlke K L, Purkayastha A A, Westrick R J, Smith P L, Petryniak B, Lowe J B, Ginsburg D. Cell. 1999;96:111–120. doi: 10.1016/s0092-8674(00)80964-2. [DOI] [PubMed] [Google Scholar]
- 7.Paris F, Fuks Z, Kang A, Capodieci P, Juan G, Ehleiter D, Haimovitz-Friedman A, Cordon-Cardo C, Kolesnick R. Science. 2001;293:293–297. doi: 10.1126/science.1060191. [DOI] [PubMed] [Google Scholar]
- 8.Marsh M N, Trier J S. Gastroenterology. 1974;67:636–645. [PubMed] [Google Scholar]
- 9.Karlsson L, Lindahl P, Heath J K, Betsholtz C. Development (Cambridge, UK) 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- 10.Kaestner K H, Silberg D G, Traber P G, Schutz G. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 11.Pabst O, Zeigerdt R, Arnold H-H. Development (Cambridge, UK) 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 12.Beck F, Tata F, Chawengsaksophak K. BioEssays. 2000;22:431–441. doi: 10.1002/(SICI)1521-1878(200005)22:5<431::AID-BIES5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Ramalho-Santos M, Melton D A, McMahon A. Development (Cambridge, UK) 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- 14.Emmert-Buck M R, Bonner R F, Smith P D, Chuaqui R F, Zhuang Z, Goldstein S R, Weiss R A, Liotta L A. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 15.Komano H, Fujiura Y, Kawaguchi M, Matsumoto S, Hashimoto Y, Obana S, Mombaerts P, Tonegawa S, Yammamoto H, Itohara S, et al. Proc Natl Acad Sci USA. 1995;92:6147–6151. doi: 10.1073/pnas.92.13.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Husebye E, Hellstrom P M, Sundler F, Chen J, Midtvedt T. Am J Physiol. 2001;280:G368–G380. doi: 10.1152/ajpgi.2001.280.3.G368. [DOI] [PubMed] [Google Scholar]
- 17.Hooper L V, Wong M H, Thelin A, Hansson L, Falk P G, Gordon J I. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 18.Jensen J, Pedersen E E, Galante P, Hald J, Heller R S, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen O. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J L, Shou J, Guillemot F, Kageyama R, Gao W-Q. Development (Cambridge, UK) 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- 20.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitehead R H, Demmler K, Rockman S P, Watson N K. Gastroenterology. 1999;117:858–865. doi: 10.1016/s0016-5085(99)70344-6. [DOI] [PubMed] [Google Scholar]
- 22.Karam S M, Leblond C P. Anat Rec. 1993;236:259–279. doi: 10.1002/ar.1092360202. [DOI] [PubMed] [Google Scholar]
- 23.Syder A J, Guruge J L, Li Q, Oleksiewicz C, Lorenz R G, Karam S M, Falk P G, Gordon J I. Mol Cell. 1999;3:263–274. doi: 10.1016/s1097-2765(00)80454-2. [DOI] [PubMed] [Google Scholar]
- 24.Phillips R L, Ernst R E, Brunk B, Ivanova N, Mahan M A, Deanehan J K, Moore K A, Overton G C, Lemischka I R. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]