Abstract
Background
Estimating prognosis on the basis of clinicopathologic factors can inform clinical practice and improve risk stratification for clinical trials. We constructed prognostic nomograms for one-year overall survival and six-month progression-free survival in metastatic colorectal carcinoma by using the ARCAD database.
Methods
Data from 22 674 patients in 26 randomized phase III clinical trials since 1997 were used to construct and validate Cox models, stratified by treatment arm within each study. Candidate variables included baseline age, sex, body mass index, performance status, colon vs rectal cancer, prior chemotherapy, number and location of metastatic sites, tumor mutation status (BRAF, KRAS), bilirubin, albumin, white blood cell count, hemoglobin, platelets, absolute neutrophil count, and derived neutrophil-to-lymphocyte ratio. Missing data (<11%) were imputed, continuous variables modeled with splines, and clinically relevant pairwise interactions tested if P values were less than .001. Final models were internally validated via bootstrapping to obtain optimism-corrected calibration and discrimination C-indices, and externally validated on a 10% holdout sample from each trial (n = 2257).
Results
In final models, all included variables were associated with overall survival except for lung metastases, and all but total white cell count associated with progression-free survival. No clinically relevant pairwise interactions were identified. Final nomogram calibration was good (C = 0.68 for overall and C = 0.62 for progression-free survival), as was external validity (concordance between predicted >50% vs < 50% probability) and actual (yes/no) survival (72.8% and 68.2% concordance, respectively, for one-year overall and six-month progression-free survival, between predicted [>50% vs < 50% probability] and actual [yes/no] overall and progression-free survival). Median survival predictions fell within the actual 95% Kaplan-Meier confidence intervals.
Conclusions
The nomograms are well calibrated and internally and externally valid. They have the potential to aid prognostication and patient-physician communication and balance risk in colorectal cancer trials.
Advanced colorectal cancer remains a lethal disease, even though survival from the first diagnosis of metastatic disease has improved over the last 20 years, although substantial heterogeneity in survival outcomes remains. With improved treatments and understanding of tumor biology, potential prognostic factors have emerged.
Estimating survival is always difficult, even for experienced oncologists; accuracy of estimates is limited even for patients with terminal disease (1), and extrapolating results from clinical trials, where selection bias limits generalizability, is unreliable. The emergence of molecular phenotypes has further complicated prognostication, with limited data to guide clinicians on how these new biomarkers might best be integrated with established prognostic factors and incorporated in new treatment options (2).
Estimating prognosis has several advantages for clinical care. Discussion about prognosis is commonly raised by patients from the time of diagnosis; our inability to accurately predict this has been identified as an important barrier to effective physician-patient communication (3). While methods exist for estimating and communicating prognosis on the basis of medians (4) derived from clinical trial data, a more precise estimate tailored to individual patient factors is a potentially valuable tool for clinicians.
More accurate prognostication would also be helpful for designing clinical trials to evaluate new treatments. Understanding factors influencing prognosis would allow prognostic groups in randomized trials to be balanced more accurately. This may be particularly useful in smaller trials, where imbalance across arms is more likely, or in historical comparisons for rarer subtypes. Nomograms can also help identify patients suitable for clinical trials where a minimum survival estimate is required, such as the Colon Life application (5), or where a poorer prognosis may warrant treatment escalation.
Large numbers of patients are required to evaluate the relative effects of established and postulated prognostic factors. We were able to access individual patient data from the ARCAD collaborative colorectal cancer database (6), the largest collection of recent randomized phase II and III trials in advanced colorectal cancer. This allowed us to evaluate multiple postulated prognostic factors and their relative contribution on a scale not possible in individual trials or smaller pooled data sets.
To improve prognostication for clinical practice and trial design, we developed a nomogram to predict progression-free survival (PFS) and overall survival (OS) in patients commencing firstline systemic therapy for advanced or metastatic colorectal cancer from individual patient data in the ARCAD database.
Methods
Database and Candidate Variables
Data from 22 674 patients enrolled to 26 randomized clinical trials for firstline treatment of metastatic colorectal cancer since 1997 were used to construct and independently validate clinical prediction models for PFS and OS. All firstline trials with data included in the ARCAD trial database at June 30, 2016, were eligible. Trial descriptions and contributing sample sizes are shown in Supplementary Table 1 (available online). Known prognostic variables were identified and additional candidates proposed by the ARCAD project team.
Imputation of Missing Data and Construction/Validation Data Sets
Potential prognostic variables were examined for individual and joint missingness and considered for imputation. The missing-at-random assumption (that conditional on observed data, unobserved data are missing at random) was used, as most missingness was study specific (for example, a data item not consistently collected on study case-report forms for all patients in that trial). Given the large data set, independent variables with at least 35% availability across patients could be imputed. We used stochastic regression imputation and included all available variables (including outcomes and study) in the final imputation model (7,8). Independent variables missing data for more than 65% of patients (such as side of the primary tumor) were not considered candidates for imputation and modeling, with the exception of BRAF, which was included for its importance as a molecular prognostic factor (7). Patient outcome data (PFS and OS) were not imputed, and patients for whom clinical outcomes were not recorded (such as those deemed ineligible within their respective trials) were excluded from analyses.
Following imputation of missing data, the overall ARCAD database was split into a construction data set of 20 417 patients comprising a random sample of 90% of patients from each clinical trial, and a validation data set of 2257 patients comprising the remaining 10% from each trial.
Univariate Models
After imputation and using the construction data set, we examined the following variables for univariate associations with OS and PFS: age (continuous) (9), sex, body mass index (BMI; continuous) (10), performance status (PS; 0, 1, 2+), prior chemotherapy use for any reason (yes, no), KRAS or BRAF mutation, number of organs with metastatic involvement (0–1, 2+), presence vs absence of liver, lung (11), peritoneal (12), or nodal metastases, and laboratory markers including white blood cell count (WBC), platelets, hemoglobin, absolute neutrophil count, bilirubin, albumin, neutrophils, and derived neutrophil-to-lymphocyte ratio (13). For each variable and outcome of interest, univariate Cox proportional hazards regression models stratified by treatment arm within each study were fit, allowing effects to be averaged across study-specific baseline hazard functions. Continuous variables (age, BMI, and node ratio) were modeled by using restricted cubic splines to test for possible nonlinearity of their effects on the log relative hazard of outcome; where statistically significant nonlinearity was identified, splines were also used in multivariable modeling, and otherwise variables were subsequently modeled as linear on the log relative hazard scale (8,14). The proportional hazards assumption for each variable was tested using the methods of Grambsch and Therneau (15). Variables showing both statistical significance at a P value of less than .05 and clinical significance as assessed by hazard ratios were graduated to subsequent interaction testing and multivariable modeling.
Tests for Two-Way Interactions
To determine whether the effects of any covariates were dependent on other covariates, all pairs of variables showing univariate statistical significance were tested for two-way interaction. Statistically significant (P < .001) interaction and clinically differentiable effect mediation were required for subsequent consideration in final models. Higher-ordered interactions were not examined for reasons of interpretability and reproducibility.
Model Construction
Multivariable Cox proportional hazards models for OS and PFS were formulated from all variables and two-way interactions demonstrating statistically and clinically significant associations with their respective end points, where clinical significance was achieved if the effect of one variable (eg, hazard ratio) differed in a clinically meaningful way across levels of the other variable in the interaction. After backwards stepwise elimination, final models included all main effects and pairwise interactions remaining statistically (P < .05) and clinically significant after adjustment. Nomograms (calculators) based on the final models were constructed for the likelihood of PFS at six months and OS at one year. All statistical tests were two-sided, and all imputation, analyses, and figures were produced using “rms,” part of R statistical software (Vienna, Austria), version 3.2.1 (16).
Internal Validation
Final models for OS and PFS were internally validated using bootstrapping resampling of the construction data set (with 1000 bootstrap samples per model) to obtain optimism-corrected discrimination via the concordance index for survival data and calibration plots (8,14).
External Validation
External validation was performed by comparing the predicted six-month PFS and one-year OS probabilities of patients from the 10% validation set and the observed outcomes of the same patients. For each end point, the median ARCAD-based prediction across patients was compared with the observed Kaplan-Meier estimate (and its confidence interval) for the same patients and time point, overall and within patient subgroups. As another measure of external validation, rates of correct prediction, that is, the concordance of observed (event, no event) and predicted (using 50% predicted probability as a dichotomizing threshold) six-month PFS and one-year OS status across validation set patients and subgroups were also computed.
Results
Descriptive Statistics
The rate of missingness across all independent variables and patients combined was less than 11%, and the distribution of each variable was maintained with multiple imputation (Table 1). Patients were primarily male (61.6%), with a median age of 62 years (interquartile range = 55 to 69 years). More than half (53.5%) of patients had performance status of 0, 69.0% had colon-only primary tumors, 57.4% had two or more sites of metastatic disease, and 78.9% had never received chemotherapy for any reason.
Table 1.
Demographics and disease characteristics of patients used for nomogram construction: Pre-imputation and postimputation*
| Characteristic | Pre-imputation No. (%) | Postimputation No. (%) |
|---|---|---|
| Age, y | ||
| Mean (SD) | 61 (11) | 61 (11) |
| Median (IQR) | 62 (55–69) | 62 (55–69) |
| Missing | 7 (0.0) | 0 (0.0) |
| Sex | ||
| Male | 13 954 (61.6) | 13 965 (61.6) |
| Female | 8702 (38.4) | 8709 (38.4) |
| Missing | 18 (0.1) | 0 (0.0) |
| Body mass index | ||
| Mean (SD) | 26 (5) | 26 (5) |
| Median (IQR) | 25 (23–29) | 25 (23–29) |
| Missing | 1525 (6.7) | 0 (0.0) |
| Performance status | ||
| 0 | 11 997 (53.5) | 12 123 (53.5) |
| 1 | 9496 (42.3) | 9595 (42.3) |
| 2+ | 948 (4.2) | 956 (4.2) |
| Missing | 233 (1.0) | 0 (0.0) |
| Tumor location | ||
| Colon | 11 826 (69.0) | 15 691 (69.2) |
| Rectum | 5030 (29.3) | 6615 (29.2) |
| Both | 283 (1.7) | 368 (1.6) |
| Missing | 5535 (24.4) | 0 (0.0) |
| No. of metastatic sites (organs) | ||
| 0–1 | 7611 (42.6) | 9409 (41.5) |
| 2+ | 10 235 (57.4) | 13 265 (58.5) |
| Missing | 4828 (21.3) | 0 (0.0) |
| Liver metastases | ||
| Yes | 14 422 (77.9) | 17 632 (77.8) |
| No | 4088 (22.1) | 5042 (22.2) |
| Missing | 4164 (18.4) | 0 (0.0) |
| Lung metastases | ||
| Yes | 6647 (37.2) | 8559 (37.7) |
| No | 11 242 (62.8) | 14 115 (62.3) |
| Missing | 4785 (21.1) | 0 (0.0) |
| LN metastases | ||
| Yes | 6140 (38.9) | 8845 (39.0) |
| No | 9643 (61.1) | 13 829 (61.0) |
| Missing | 6891 (30.4) | 0 (0.0) |
| Peritoneal metastases | ||
| Yes | 1624 (15.8) | 4261 (18.8) |
| No | 8626 (84.2) | 18 413 (81.2) |
| Missing | 12 424 (54.8) | 0 (0.0) |
| Prior chemotherapy | ||
| Yes | 4331 (21.1) | 4779 (21.1) |
| No | 16 206 (78.9) | 17 895 (78.9) |
| Missing | 2137 (9.4) | 0 (0.0) |
| KRAS status | ||
| Mutant | 3033 (38.3) | 8924 (39.4) |
| Wild-type | 4896 (61.7) | 13 750 (60.6) |
| Missing | 14 745 (65.0) | 0 (0.0) |
| BRAF status | ||
| Mutant | 388 (8.1) | 1921 (8.5) |
| Wild-type | 4421 (91.9) | 20 753 (91.5) |
| Missing | 17 865 (78.8) | 0 (0.0) |
| White blood cells, ×109/L | ||
| Mean (SD) | 8.4 (3.4) | 8.5 (3.4) |
| Median (IQR) | 7.8 (6.3–9.7) | 7.8 (6.4–9.7) |
| Missing | 4442 (19.6) | 0 (0.0) |
| Platelets, ×109/L | ||
| Mean (SD) | 335 (128) | 334 (127) |
| Median (IQR) | 310 (245–398) | 309 (245–398) |
| Missing | 1899 (8.4) | 0 (0.0) |
| Albumin, g/L | ||
| Mean (SD) | 39 (6) | 39 (6) |
| Median (IQR) | 40 (36–43) | 39 (36–42) |
| Missing | 14 695 (64.8) | 0 (0.0) |
| Hemoglobin, g/dL | ||
| Mean (SD) | 12.4 (1.7) | 12.4 (1.7) |
| Median (IQR) | 12.4 (11.2–13.6) | 12.4 (11.1–13.6) |
| Missing | 7618 (33.6) | 0 (0.0) |
| Absolute neutrophil count, ×109/L | ||
| Mean (SD) | 5.7 (2.7) | 5.6 (2.6) |
| Median (IQR) | 5.1 (3.9–6.8) | 5.2 (4.0–6.6) |
| Missing | 6480 (28.6) | 0 (0.0) |
| Bilirubin, mg/dL | ||
| Mean (SD) | 0.63 (0.94) | 0.63 (0.92) |
| Median (IQR) | 0.50 (0.34–0.69) | 0.50 (0.34–0.69) |
| Missing | 3021 (13.3) | 0 (0.0) |
| Neutrophil to lymphocyte ratio | ||
| Mean (SD) | 2.4 (1.4) | 2.5 (2.3) |
| Median (IQR) | 2.1 (1.5–2.8) | 2.0 (1.4–2.9) |
| Missing | 8516 (37.6) | 0 (0.0) |
| Total | 22 674 (100) | 22 674 (100) |
IQR = interquartile range; LN = lymph node.
Single Variable Models and Two-Way Interaction Testing
All variables demonstrated some degree of statistical and clinical significance in univariate models for PFS and OS; therefore, all variables were carried forward for potential inclusion in the final multivariable models. However, no statistically significant and clinically relevant interactions were identified for either end point, where clinical relevance was judged via examination of spline plots for continuous variables and hazard ratios for categorical variables across subgroups (data not shown).
Final Multivariable Models
Patient and disease variables statistically significantly associated with lower survival in multivariable modeling included young or old age (P < .001), male sex (HR = 1.05, 95% CI = 1.01 to 1.09, P = .02), low BMI (P < .001), and worsened performance status (PS1/PS0 HR = 1.31, 95% CI = 1.25 to 1.43; PS2+/PS0 HR = 1.73, 95% CI = 1.53 to 1.84, P < .001) (Table 2). Prior chemotherapy for any reason was also associated with a 15% increased risk of death (HR = 1.15, 95% CI = 1.10 to 1.20, P < .001). KRAS mutant status was associated with a higher likelihood of death during follow-up (HR = 1.35, 95% CI = 1.30 to 1.39, P < .001); similarly, BRAF mutant status was associated with a higher risk of death (HR = 2.21, 95% CI = 2.09 to 2.34, P < .001). The presence of two or more metastatic sites was associated with higher risk of death than zero or one metastatic sites (HR = 1.20, 95% CI = 1.16 to 1.26, P < .001), as was the presence of liver metastases (HR = 1.20, 95% CI = 1.15 to 1.26, P < .001), lymph node metastases (HR = 1.15, 95% CI = 1.10 to 1.19, P < .001), and peritoneal metastases (HR = 1.19, 95% CI = 1.13 to 1.23, P < .001). Among the baseline laboratory markers considered, higher levels of platelets (P < .001), WBC (P = .02), and neutrophils (P < .001) were associated with a higher risk of death, while elevated hemoglobin (P < .001) and albumin (P < .001) were associated with lower risk (Table 2). Primary tumor site (colon vs rectum), presence vs absence of lung metastases, and baseline derived neutrophil-to-lymphocyte ratio (dNLR) were not associated with OS after adjustment for other factors.
Table 2.
Final multivariable Cox models associated with nomogram for overall survival and progression-free survival
| Variable | OS |
PFS |
||||||
|---|---|---|---|---|---|---|---|---|
| Coefficient | SE | HR (95% CI) | P* | Coefficient | SE | HR (95% CI) | P* | |
| Age, y | <.001 | .03 | ||||||
| −0.0012 | 0.0017 | † | −0.0036 | 0.0015 | † | |||
| 0.0078 | 0.0020 | 0.0045 | 0.0018 | |||||
| Sex | .01 | .04 | ||||||
| Female | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Male | 0.0442 | 0.0186 | 1.05 (1.01 to 1.09) | 0.0342 | 0.0167 | 1.04 (1.00 to 1.06) | ||
| Body mass index, kg/m2 | † | <.001 | .001 | |||||
| −0.0236 | 0.0046 | −0.0133 | 0.0042 | † | ||||
| 0.0167 | 0.0055 | 0.0101 | 0.0049 | |||||
| Performance status | <.001 | <.001 | ||||||
| 0 | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| 1 | 0.2663 | 0.0184 | 1.31(1.25 to 1.34) | 0.1599 | 0.0166 | 1.17 (1.13 to 1.20) | ||
| 2+ | 0.5471 | 0.0407 | 1.73 (1.58 to 1.84) | 0.3358 | 0.0395 | 1.40 (1.29 to 1.49) | ||
| Prior chemotherapy | <.001 | <.001 | ||||||
| No | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Yes | 0.1358 | 0.0237 | 1.15 (1.10 to 1.20) | 0.1124 | 0.0211 | 1.12 (1.08 to 1.17) | ||
| KRAS mutation status | <.001 | <.001 | ||||||
| Wild-type | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Mutant | 0.3000 | 0.0181 | 1.35 (1.30 to 1.39) | 0.2623 | 0.0163 | 1.30 (1.25 to 1.33) | ||
| BRAF mutation status | <.001 | <.001 | ||||||
| Wild-type | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Mutant | 0.7922 | 0.0304 | 2.21 (2.09 to 2.34) | 0.6125 | 0.0285 | 1.85 (1.73 to 1.92) | ||
| Platelets, ×109/L | <.001 | <.001 | ||||||
| 0.0012 | 0.0002 | † | 0.0009 | 0.0002 | † | |||
| −0.0013 | 0.0003 | −0.0009 | 0.0002 | |||||
| White blood cells, ×109/L | 0.0063 | 0.0027 | 1.01 (1.00 to 1.01) | .01 | − | − | − | ‡ |
| Hemoglobin, g/dL | −0.0449 | 0.0063 | 0.96 (0.94 to 0.97) | <.001 | −0.0229 | 0.0056 | 0.98 (0.97 to 0.99) | <.001 |
| Albumin, g/L | <.001 | <.001 | ||||||
| −0.0481 | 0.0032 | † | −0.0273 | 0.0031 | † | |||
| 0.0097 | 0.0036 | 0.0097 | 0.0032 | |||||
| Absolute neutrophil count, ×109/L | <.001 | <.001 | ||||||
| 0.1129 | 0.0069 | † | 0.0274 | 0.0033 | 1.03 | |||
| −0.0767 | 0.0118 | |||||||
| Bilirubin, mg/dL | <.001 | <.001 | ||||||
| 0.4842 | 0.0758 | † | 0.2376 | 0.0679 | † | |||
| −0.5016 | 0.0831 | −0.2332 | 0.0745 | |||||
| No. of met sites | <.001 | <.001 | ||||||
| 0–1 | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| 2+ | 0.1859 | 0.0224 | 1.20 (1.16 to 1.26) | 0.1103 | 0.0247 | 1.12 (1.07 to 1.17) | ||
| Liver metastases | <.001 | <.001 | ||||||
| No | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Yes | 0.1811 | 0.0240 | 1.20 (1.15 to 1.26) | 0.1304 | 0.0230 | 1.14 (1.09 to 1.19) | ||
| Lymph node metastases | <.001 | <.001 | ||||||
| No | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Yes | 0.1375 | 0.0214 | 1.15 (1.10 to 1.19) | 0.0740 | 0.0206 | 1.08 (1.04 to 1.12) | ||
| Peritoneal metastases | <.001 | .01 | ||||||
| No | – | – | 1.00 (reference) | – | – | 1.00 (reference) | ||
| Yes | 0.1706 | 0.0248 | 1.19 (1.13 to 1.23) | 0.0586 | 0.0242 | 1.06 (1.02 to 1.11) | ||
| Lung metastases | − | − | − | ‡ | <.001 | |||
| No | – | – | 1.00 (reference) | |||||
| Yes | 0.1029 | 0.0204 | 1.11 (1.07 to 1.15) | |||||
P values were calculated using Wald chi-square tests; the P values are two-sided. Only variables that contributed statistically significantly to final models are included in the table. Derived neutrophil to lymphocyte ratio and tumor location were significant on univariate analyses but did not contribute to the final model. CI = confidence interval; HR = hazard ratio; OS = overall survival; PFS = progression-free survival.
Single hazard ratio not available due to nonlinear effect for these continuous variables.
Variables did not statistically significantly contribute to their respective models.
Patient and disease variables statistically significantly associated with worse PFS in multivariable models included young or old age (P = .04), male sex (HR = 1.04, 95% CI = 1.00 to 1.06, P = .04), low BMI (P < .001), and poorer performance status (PS1/PS0 HR = 1.17, 95% CI = 1.13 to 1.20; PS2+/PS0 HR = 1.40, 95% CI = 1.29 to 1.49, P < .001) (Table 2). Prior chemotherapy was associated with 12% higher risk of disease progression or death (P < .001) during follow-up (HR = 1.12, 95% CI = 1.08 to 1.17). KRAS mutant status was also associated with 30% higher likelihood of progression or death (HR = 1.30, 95% CI = 1.25 to 1.33, P < .001); similarly, BRAF mutant status was associated with an 85% higher chance of progression or death (HR = 1.85, 95% CI = 1.73 to 1.92, P < .001). Presence of two or more metastatic sites, compared with zero or one, was associated with a 12% higher risk of progression (HR = 1.12, 95% CI = 1.07 to 1.17, P < .001), and having lung metastases (HR = 1.11, 95% CI = 1.07 to 1.15, P < .001), liver metastases (HR = 1.14, 95% CI = 1.09 to 1.19, P < .001), lymph node metastases (HR = 1.08, 95% CI = 1.04 to 1.12, P < .001), and peritoneal metastases (HR = 1.06, 95% CI = 1.02 to 1.11, P = .02) were each statistically significantly associated with higher risk of progression. Among the baseline laboratory markers considered, higher levels of platelets (P < .001), neutrophils (P < .001), or bilirubin (P < .001) were associated with higher risk of progression, while elevated hemoglobin (P < .001) and albumin (P < .001) were associated with lower risk (Table 2). Primary tumor location (colon vs rectum), baseline WBC, and baseline dNLR were not associated with PFS after adjustment for other variables.
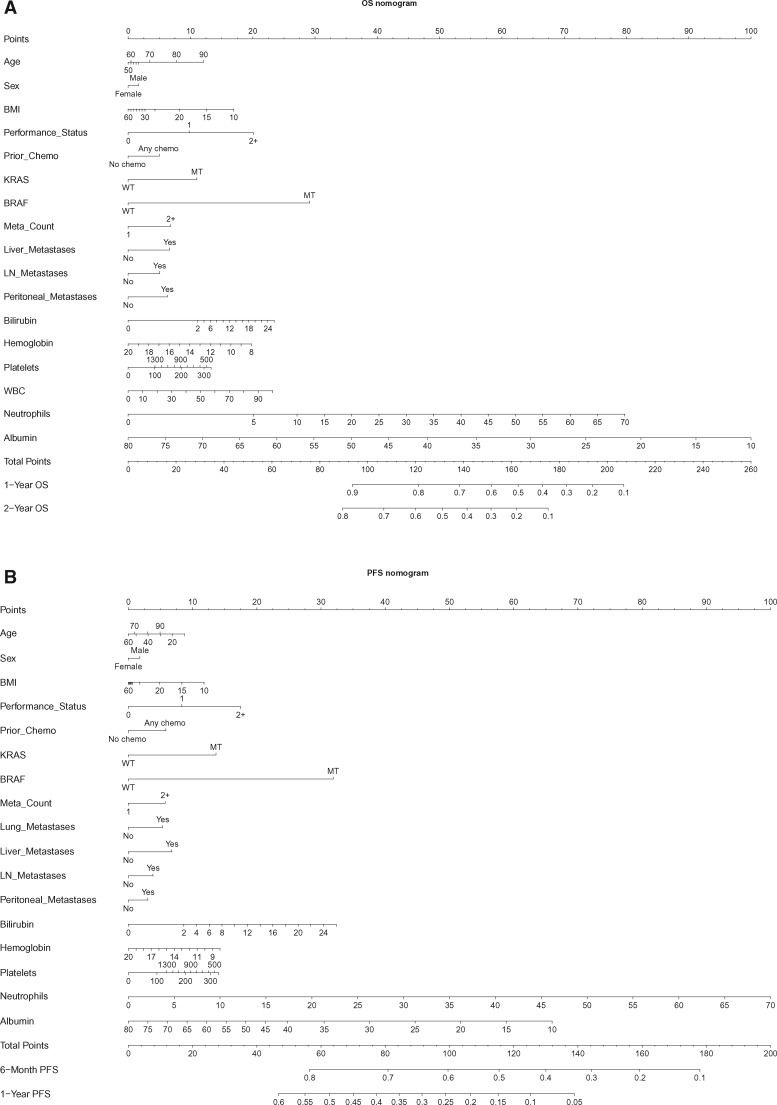
While familiarity with nomograms is not required to use the web-based tools, brief instructions are provided in the Supplementary Materials (available online). From Figure 1, the relative prognostic importance of each variable for each outcome may be readily gauged; for example, levels of baseline neutrophils and albumin have the largest impact on OS risk, while sex has the smallest (but still clinically relevant) impact.
Figure 1.
Nomograms for (A) overall survival and (B) progression-free survival. See the Supplementary Materials (available online) for instructions for use. BMI = body mass index (mg/kg2); BRAF = BRAF gene status; KRAS = Kirsten rat sarcoma gene; Meta count = number of metastatic sites; MT = mutant; OS = overall survival; Performance status = Eastern Cooperative Oncology Group/World Health Organization performance status (0,1,2); PFS = progression-free survival; Prior chemo = previous (adjuvant) chemotherapy; WT = wild-type.
Internal Validation
The final model for OS had an adjusted concordance index (C) of 0.68, and the model for PFS yielded a C of 0.62. Calibration of observed vs predicted one-year OS and six-month PFS was strong across the spectrum of ordered risk groups (Figure 2).
Figure 2.

Calibration plots for (A) overall survival (OS) and (B) progression-free survival (PFS) nomograms.
External Validation
The one-year survival of the validation set of patients had high concordance: 72.8%. When median (across patients) one-year OS predictions obtained from the ARCAD calculator were compared with the observed Kaplan-Meier one-year OS rates, predictions fell within 5% of the actual rates, both overall and within most of the subgroups defined by those variables appearing in the ARCAD calculators (Table 3), although the calculator trended toward overestimation of survival to a small degree. In most patient subgroups, predictions fell within the 95% confidence intervals of the Kaplan-Meier rates, demonstrating strong agreement.
Table 3.
Results of external validation of the ARCAD nomograms for overall survival (OS) and Progression-Free Survival (PFS), with comparison of six-month PFS and one-year OS predictions*
| Group | No. | 1-y OS |
6-mo PFS |
||||
|---|---|---|---|---|---|---|---|
| Observed, % | Predicted, % | % delta: predicted–observed | Observed, % | Predicted, % | % delta: predicted–observed | ||
| K-M (95% CI) | K-M (95% CI) | ||||||
| Overall | 2257 | 69.8 (67.9 to 71.7) | 71.9 | 2.1 | 66.7 (64.8 to 68.7) | 64.5 | −2.2 |
| Age, y | |||||||
| <70 | 1641 | 72.1 (69.9 to 74.3) | 73.2 | 1.1 | 67.4 (65.2 to 69.8) | 65.0 | −2.4 |
| 70+ | 616 | 63.6 (59.8 to 67.6) | 69.0 | 5.4 | 64.5 (60.7 to 68.4) | 63.0 | −1.5 |
| Sex | |||||||
| Male | 1385 | 70.8 (68.4 to 73.3) | 72.5 | 1.7 | 68.1 (65.7 to 70.7) | 65.4 | −2.7 |
| Female | 872 | 68.1 (65.0 to 71.3) | 71.1 | 3.0 | 64.2 (61.1 to 67.6) | 63.4 | −0.8 |
| Performance status | |||||||
| 0 | 1183 | 77.1 (74.7 to 79.6) | 77.0 | −0.1 | 71.5 (69.0 to 74.2) | 68.2 | −3.3 |
| 1 | 964 | 64.2 (61.2 to 67.4) | 66.4 | 0.2 | 63.0 (60.0 to 66.2) | 61.0 | −2.0 |
| 2+ | 110 | 39.3 (31.1 to 49.6) | 45.0 | 5.7 | 45.9 (37.4 to 56.2) | 49.9 | 4.0 |
| Body mass index, kg/m2 | |||||||
| <25 | 1029 | 65.3 (62.4 to 68.3) | 69.1 | 3.8 | 64.6 (61.7 to 67.6) | 63.0 | −1.6 |
| 25+ | 1228 | 73.2 (70.8 to 75.8) | 74.0 | 0.8 | 68.4 (65.8 to 71.1) | 66.1 | −2.3 |
| Prior chemotherapy | |||||||
| No | 1824 | 69.5 (67.4 to 71.7) | 71.3 | 1.8 | 66.5 (64.4 to 68.8) | 64.2 | −2.3 |
| Yes | 433 | 70.6 (66.4 to 75.1) | 75.4 | 4.8 | 67.6 (63.2 to 72.2) | 65.7 | −1.9 |
| BRAF status | |||||||
| Wild-type | 2063 | 71.8 (69.9 to 73.8) | 73.2 | 1.4 | 68.7 (66.7 to 70.8) | 65.5 | −3.2 |
| Mutant | 194 | 47.7 (41.0 to 55.4) | 52.1 | 4.4 | 45.8 (39.2 to 53.4) | 49.6 | 3.8 |
| KRAS status | |||||||
| Wildtype | 1374 | 71.0 (68.6 to 73.5) | 74.9 | 3.9 | 68.6 (66.2 to 71.2) | 68.0 | −0.6 |
| Mutant | 883 | 67.9 (64.8 to 71.1) | 68.3 | 0.4 | 63.7 (60.5 to 67.0) | 61.1 | −2.6 |
| Platelets, ×109/L | |||||||
| <310 | 1132 | 75.8 (73.3 to 78.4) | 76.2 | 0.4 | 71.5 (68.9 to 74.2) | 67.8 | −3.7 |
| 310+ | 1125 | 63.4 (60.6 to 66.3) | 66.6 | 3.2 | 61.9 (59.1 to 64.8) | 61.4 | −0.5 |
| White blood cells, ×109/L | |||||||
| <8.0 | 1199 | 76.6 (74.2 to 79.0) | 76.4 | −0.2 | 71.3 (68.7 to 73.9) | 67.4 | −3.9 |
| 8.0+ | 1058 | 61.8 (58.9 to 64.9) | 65.5 | 3.7 | 61.3 (58.4 to 64.4) | 61.1 | −0.2 |
| Hemoglobin, g/dL | |||||||
| <12.4 | 1132 | 62.7 (59.9 to 65.6) | 66.8 | 6.1 | 62.1 (59.3 to 65.0) | 61.6 | −0.5 |
| 12.4+ | 1125 | 76.9 (74.4 to 79.4) | 75.9 | −1.0 | 71.4 (68.8 to 74.1) | 67.3 | −4.1 |
| Albumin, g/L | |||||||
| <40.0 | 1208 | 60.8 (58.1 to 63.7) | 65.7 | 4.9 | 60.1 (57.4 to 63.0) | 60.9 | 0.8 |
| 40.0+ | 1049 | 80.0 (77.6 to 82.5) | 77.9 | −2.1 | 74.2 (71.5 to 76.9) | 68.5 | −5.7 |
| ANC, ×109/L | |||||||
| <5.2 | 1172 | 76.3 (73.9 to 78.8) | 76.7 | 0.4 | 71.2 (68.7 to 73.9) | 67.5 | −3.7 |
| 5.2+ | 1085 | 62.4 (59.5 to 65.4) | 65.6 | 3.2 | 61.6 (58.8 to 64.6) | 61.3 | −0.3 |
| Bilirubin, mg/dL | |||||||
| < 0.50 | 1115 | 71.8 (69.1 to 74.5) | 73.3 | 1.5 | 67.7 (64.9 to 70.5) | 65.2 | −2.5 |
| 0.50+ | 1.142 | 67.8 (65.1 to 70.6) | 70.8 | 3.0 | 65.7 (63.0 to 68.6) | 63.9 | −1.8 |
| No. of metastasis sites | |||||||
| 0–1 | 965 | 75.2 (72.5 to 78.1) | 76.8 | 1.6 | 70.0 (67.1 to 73.0) | 69.1 | −0.9 |
| 2+ | 1292 | 65.6 (63.1 to 68.3) | 67.6 | 2.0 | 64.2 (61.6 to 66.9) | 61.5 | −2.7 |
| Liver metastasis | |||||||
| No | 495 | 72.9 (69.0 to 77.0) | 76.2 | 3.3 | 68.1 (64.0 to 72.3) | 67.4 | −0.7 |
| Yes | 1762 | 68.8 (66.7 to 71.1) | 70.6 | 1.8 | 66.3 (64.1 to 68.6) | 63.9 | −2.4 |
| Lung metastasis | |||||||
| No | 1410 | 70.2 (67.9 to 72.7) | 72.5 | 2.3 | 66.3 (63.9 to 68.9) | 66.2 | −0.1 |
| Yes | 847 | 69.0 (65.9 to 72.2) | 70.9 | 1.9 | 67.4 (64.3 to 70.7) | 62.3 | −5.1 |
| Lymph node metastases | |||||||
| No | 1385 | 71.7 (69.3 to 74.1) | 73.9 | 2.2 | 68.9 (66.5 to 71.4) | 66.2 | −2.7 |
| Yes | 872 | 66.6 (63.5 to 69.8) | 67.9 | 1.3 | 62.9 (59.8 to 66.3) | 62.1 | −0.8 |
| Peritoneal | |||||||
| No | 1818 | 71.1 (69.0 to 73.3) | 73.4 | 2.3 | 67.3 (65.2 to 69.5) | 65.4 | −1.9 |
| Yes | 439 | 64.0 (59.5 to 68.8) | 65.8 | 1.8 | 64.3 (59.9 to 69.0) | 60.9 | −3.4 |
Validation based on 2257 patients comprising a 10% holdout sample from each trial. CI = confidence interval; HR = hazard ratio; K-M = Kaplan-Meier; OS = overall survival; PFS = progression-free survival.
Strong external validation results were observed for PFS, with 68.2% concordance of predicted and observed six-month PFS status. The median predicted six-month PFS rates obtained from the ARCAD calculator were within 5% of the corresponding actual rates, overall and within most patient subgroups (Table 3). Predictions fell within the 95% confidence intervals for the actual Kaplan-Meier rates in most subgroups, again showing strong predictive accuracy for most types of patients.
Discussion
Using the ARCAD database, we were able to develop internally and externally valid nomograms that were accurate for both PFS and OS. They highlight the relative contribution of baseline clinicopathologic variables to survival estimates using information that is generally available in the clinic at the time of diagnosis of metastatic disease. The large amount of data used to develop these nomograms allowed assessment of a variety of potential prognostic factors and their relative contributions to survival outcomes.
The largest contributions to PFS and OS come from those factors previously established as prognostic in other data sets. Albumin, and other markers of inflammation combined, contributed statistically significantly, along with performance status. While tumor factors, including mutation status (BRAF, KRAS), were included in the final model, a substantial proportion of prognostic information is contributed by patient factors: for example, sex, performance status, low BMI (17), and laboratory values. This highlights the importance of considering prognostic biomarkers beyond the immediate tumor environment.
Limitations of this work are acknowledged, including the generalizability and availability of baseline prognostic factors within the database. The included clinical trial populations did not represent the full spectrum of patients in the clinic. Although trials of reduced-intensity treatment (18) and more poorly performing populations were included, the generalizability of the nomograms beyond the types of patients included in the database is unknown. Although the models are well calibrated and accurate, they could be updated in the future by including additional biomarkers found to be prognostic. Other potentially prognostic variables, such as blood-based tumor markers at baseline (for example, carcinoembryonic antigen), could not be included as sufficient data were not collected. Tumor location within the colon (sidedness), in particular, was not included, although tumor site (colon or rectum) was considered, but was not statistically significant. While this limitation is acknowledged, and additional analyses including tumor location would be of interest, the overall impact of adding this to the current model is likely to be limited. Although sidedness may be a surrogate for tumor (19) and patient biology (20), primary tumor location has been established as prognostic mainly in retrospective subgroup analyses of patients with all RAS wild-type tumors receiving firstline systemic therapies (21,22). The effect on RAS-mutant tumors has not been examined, and relatively few patients in chemotherapy-alone arms were included. None of the analyses to date evaluating the prognostic effect of sidedness in advanced colorectal cancer (23) have adjusted for the comprehensive set of prognostic variables established here. Restricting analyses to only those patients for whom sidedness was known would have substantially reduced the numbers and limited the ability to evaluate a comprehensive list of prognostic factors. We intend to develop future versions of the model, potentially incorporating additional factors, including sidedness, as appropriate.
These nomograms were intended to be purely prognostic, and, as such, we assume that treatment has been delivered according to best practice in a patient cohort eligible for clinical trials. No evaluation of the predictive effect on treatment response was intended. This model cannot therefore estimate outcomes in the absence of systemic therapy, at commencement of later lines of therapy, or with different treatment types. Nor is it intended to be used to evaluate outcomes from different therapies or to select between them. Although an online calculator is planned to make these nomograms more readily available, clinicians need to consider these caveats when counseling patients on likely outcomes of treatment for individual patients.
The ability to more accurately predict individual outcomes is a key factor in personalizing therapy for metastatic colorectal carcinoma. The developed nomograms are able to accurately describe outcomes for patients with metastatic colorectal carcinoma who are about to commence firstline therapy and are the most comprehensive developed to date. The models highlight key clinical and pathological factors associated with prognosis and their relative contributions.
The proposed nomograms are well calibrated and internally and externally valid. These tools use easily accessible clinicopathologic information in patients with metastatic colorectal carcinoma before commencement of firstline systemic therapy. They have the potential to aid prognostication and patient/physician communication and balance risk in randomized trials in metastatic colorectal carcinoma. Development of a web-based tool is underway.
Funding
ARCAD Foundation grant. This work was supported by the National Institutes of Health (CTSA Grant Number KL2 TR000136 from the National Center for Advancing Translational Science [NCATS]). This work was supported in part by National Health and Medical Research Council (NHMRC) Program grant 1037786.
Notes
Authors: Katrin M. Sjoquist, Lindsay A. Renfro, R. John Simes, Niall C. Tebbutt, Stephen Clarke, Matthew T. Seymour, Richard Adams, Timothy S. Maughan, Leonard Saltz, Richard M. Goldberg, Hans-Joachim Schmoll, Eric Van Cutsem, Jean-Yves Douillard, Paulo M. Hoff, Joel Randolph Hecht, Christophe Tournigand, Cornelis J. A. Punt, Miriam Koopman, Herbert Hurwitz, Volker Heinemann, Alfredo Falcone, Rainer Porschen, Charles Fuchs, Eduardo Diaz-Rubio, Enrique Aranda, Carsten Bokemeyer, Ioannis Souglakos, Fairooz F. Kabbinavar, Benoist Chibaudel, Jeffrey P. Meyers, Daniel J. Sargent, Aimery de Gramont, John R. Zalcberg; on behalf of the Fondation Aide et Recherche en Cancerologie Digestive Group (ARCAD).
Affiliations of authors: NHMRC Clinical Trials Centre, University of Sydney, Sydney, Australia (KMS, RJS); Cancer Care Centre, St George Hospital, Kogarah, NSW, Australia (KMS); Mayo Clinic, Rochester, MN (LAR, JPM, DJS); Austin Health, Heidelberg, Victoria, Australia (NCT); Royal North Shore Hospital, St Leonards, Australia (SC); Cancer Research UK Clinical Centre, Leeds, UK (MTS); Cardiff University and Velindre Cancer Centre, Cardiff, UK (RA); St James’s Hospital and University of Leeds, Leeds, UK (TSM); Memorial Sloan Kettering Cancer Center, New York, NY (LS); West Virginia University Cancer Institute, Morgantown, WV (RMG); Martin-Luther-University, Halle, Germany (HJS); University Hospital Leuven, Leuven, Belgium (EVC); European Society for Medical Oncology (ESMO) Chief Medical Officer (CMO), Institut de Cancérologie de l'Ouest (ICO) René Gauducheau, Saint-Herblain, France (JYD); Instituto do Cancer do Estado de Sao Paulo, Universidade de Sao Paolo, Sao Paolo, Brazil (PMH); David Geffen School of Medicine at University of California at Los Angeles, Los Angeles, CA (JRH, FFK); University of Paris Est Creteil, Paris, France (CT); Assistance Hopitaux Publique de Paris Henri-Mondor Hospital, Creteil, France (CT); Department of Medical Oncology, Academic Medical Center, University of Amsterdam, Amsterdam, the Netherlands (CJAP); University Medical Center Utrecht, Utrecht University, the Netherlands (MK); Duke University Medical Center, Durham, NC (HH); University of Munich, Department of Medical Oncology and Comprehensive Cancer Center, Munich, Germany (VH); Department of Oncology, University of Pisa, Pisa, Italy (AF); Klinikum Bremen-Ost Klinik fur Innere Medizin, Bremen, Germany (RP); Dana-Farber Cancer Institute, Boston, MA (CF); Department of Oncology, Hospital Clínico San Carlos, CIBERONC Instituto de Salud Carlos III, Madrid, Spain (EDR); Department of Medical Oncology IMIBIC, Reina Sofía Hospital, University of Córdoba, CIBERONC Instituto de Salud Carlos III, Córdoba, Spain (EA); University Hospital, Hamburg-Eppendorf, Germany (CB); University of Crete, Heraklion, Greece (IS); Franco-British Institute, Levallois-Perret, France (BC, AdG); School of Public Health and Preventative Medicine, Monash University, Melbourne, Australia (JRZ).
The authors thank Rhana Pike, from the NHMRC Clinical Trials Centre, who assisted with the manuscript.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
This work is dedicated to Daniel J. Sargent (deceased September 2016).
Earlier versions of this work were presented in poster format at the GI Cancers Symposium and ASCO annual meeting in 2015: Sjoquist KM, Renfro LA, Simes J, et al. Nomograms for overall survival (OS) and progression-free survival (PFS) in metastatic colorectal cancer (metastatic colorectal carcinoma): Construction from 19,678 ARCAD patients. In: ASCO Meeting Abstracts. 2015:33:659. Sjoquist KM, Bokemeyer C, Renfro LA, et al. Calculators for overall survival (OS) and progression-free survival (PFS) in metastatic colorectal cancer (metastatic colorectal carcinoma): Construction from 19,678 ARCAD patients. In: ASCO Meeting Abstracts. 2015;33:3555.
Disclaimers: Katrin M. Sjoquist: Honoraria: Pfizer; Research Funding (institution): Bayer; Travel, Accommodations, Expenses: Ipsen, Amgen. Lindsay A. Renfro: No relationships to disclose. R. John Simes: Research funding (institution): Bayer, Schering Pharma, Roche, Merck Serono. Niall C. Tebbutt: Consulting or Advisory Role: Merck Serono, Roche, Amgen. Stephen Clarke: Consulting or Advisory Role: Merck, Ipsen, Bayer, AstraZeneca/MedImmune; Speakers' Bureau: Merck. Matthew T. Seymour: Research Funding: Amgen (Inst), IntegraGen (Inst). Richard Adams: Honoraria: Merck Serono; Consulting or Advisory Role: Merck Serono, Sanofi, Bristol-Myers Squibb, Amgen; Research Funding: Merck Serono (Inst); Travel, Accommodations, Expenses: Bristol-Myers Squibb. Timothy S. Maughan: Research/consulting: Vertex. Leonard Saltz: Consulting or Advisory Role: Genentech, Eli Lilly, McNeil (I), AbbVie; Research Funding: Taiho Pharmaceutical. Richard M. Goldberg: Honoraria: Immunocare Therapies, Immunovative Therapies, Forty Seven, Merck, Pfizer, Sirtex Medical, Taiho Pharmaceutical, Targovax, Merrimack Pharmaceuticals, Merck Serono, Roche; Research Funding: Sanofi (Inst), Bayer AG (Inst), Immunomedics (Inst), Merck (Inst), Bristol-Myers Squibb (Inst); Travel, Accommodations, Expenses: Sanofi, Merck KGaA, Baxter, Amgen. Hans-Joachim Schmoll: Honoraria: Roche, GlaxoSmithKline, Servier; Consulting or Advisory Role: Roche, Bayer AG, GlaxoSmithKline; Research Funding: Roche, GlaxoSmithKline; Travel, Accommodations, Expenses: Roche, Bayer AG, Servier, GlaxoSmithKline. Eric Van Cutsem: Research Funding: Amgen (Inst), Bayer AG (Inst), Boehringer Ingelheim (Inst), Celgene (Inst), Eli Lilly (Inst), Merck Serono (Inst), Merck Sharp and Dohme (Inst), Novartis (Inst), Ipsen (Inst), Roche (Inst), Sanofi (Inst). Jean-Yves Douillard: No relationships to disclose. Paulo M. Hoff: No relationships to disclose. Joel Randolph Hecht: Consulting or Advisory Role: Amgen, Roche, Cornerstone Pharmaceuticals, Celgene, Forty Seven, Boston Biomedical, ARMO BioSciences, Lexicon, Symphogen; Research Funding: Amgen (Inst), Immunomedics (Inst), Merrimack Pharmaceuticals (Inst), Symphogen (Inst), Taiho Pharmaceutical (Inst), OncoMed (Inst). Christophe Tournigand: Honoraria: Roche, Sanofi, Bayer AG, Eli Lilly; Consulting or Advisory Role: Eli Lilly, Sandoz; Research Funding: Roche; Travel, Accommodations, Expenses: Roche, Sanofi. Cornelis J. A. Punt: Consulting or Advisory Role: Nordic Pharma, Servier J. Randolph Hecht; Consulting or Advisory Role: Amgen, Roche, Cornerstone Pharmaceuticals, Celgene, Forty Seven, Boston Biomedical, ARMO BioSciences, Lexicon, Symphogen; Research Funding: Amgen (Inst), Immunomedics (Inst), Merrimack Pharmaceuticals (Inst), Symphogen (Inst), Taiho Pharmaceutical (Inst), OncoMed (Inst). Miriam Koopman: Consulting or Advisory Role: Amgen, Bayer, Merck, Roche; Speakers' Bureau: Bayer; Research Funding: Bayer, Merck, Roche, Servier; Travel, Accommodations, Expenses: Amgen, Bayer, Roche (all more than 3 years ago). Herbert Hurwitz: Honoraria: Genentech, Eli Lilly/ImClone Systems; Consulting or Advisory Role: Genentech, Bristol-Myers Squibb, Eli Lilly, Novartis, Incyte, TRACON Pharmaceuticals, Acceleron Pharma, GlaxoSmithKline, OncoMed; Research Funding: Genentech (Inst), GlaxoSmithKline (Inst), Novartis (Inst), TRACON Pharmaceuticals (Inst), Bristol-Myers Squibb (Inst), Regeneron (Inst), Eli Lilly (Inst), MacroGenics (Inst), National Cancer Institute (Inst). Volker Heinemann: Honoraria: Roche, Celgene, Amgen, Sanofi, Merck, Sirtex Medical, Baxalta; Consulting or Advisory Role: Merck, Amgen, Roche, Sanofi, Boehringer Ingelheim, Celgene, Sirtex Medical, Baxalta; Research Funding: Merck (Inst), Amgen (Inst), Roche (Inst), Sanofi (Inst), Celgene (Inst), Boehringer Ingelheim (Inst), Sirtex Medical (Inst), IntegraGen (Inst), Taiho Pharmaceutical (Inst), Bayer AG (Inst); Travel, Accommodations, Expenses: Merck, Roche, Sirtex Medical, Amgen, Baxalt. Alfredo Falcone: Honoraria: Amgen, Bayer, Roche, Merck, Servier, Lilly; Consulting or Advisory Role: Amgen, Bayer, Roche, Merck, Servier, Lilly; Research Funding Amgen, Bayer, Merck, Roche, Servier. Rainer Porschen: Stock or Other Ownership: Fresenius Medical Care; Honoraria: Falk Foundation; Consulting or Advisory Role: Lilly, Sanofi; Travel, Accommodations, Expenses: Falk Foundation, Roche, Sanofi, Lilly. Charles S. Fuchs: Consulting or Advisory Role: Genentech, Eli Lilly, Sanofi, Bayer AG, Celgene, Merck, Entrinsic Health Solutions, Five Prime Therapeutics, Agios Pharmaceuticals. Eduardo Diaz-Rubio: Consulting or Advisory Role: Roche, Merck Serono, Amgen, Bayer AG, Merck Sharp and Dohme, Genomica; Speakers’ Bureau: Servier, Merck Sharp and Dohme; Research Funding: Roche (Inst), Merck Serono (Inst), Amgen, AstraZeneca (Inst) Rainer Porschen; Stock or Other Ownership: Fresenius Medical Care; Honoraria: Falk Foundation, Roche, Sanofi; Consulting or Advisory Role: Eli Lilly, Sanofi; Travel, Accommodations, Expenses: Falk Foundation, Roche Pharma AG, Sanofi, Eli Lilly. Enrique Aranda: Honoraria/consulting: Amgen, Bayer, Celgene, Merck, Roche, Sanofi. Carsten Bokemeyer: Honoraria: Merck KGaA, Sanofi, Roche, Bayer AG, AstraZeneca, Servier, Bristol-Myers Squibb; Consulting or Advisory Role: Eli Lilly/ImClone Systems, Merck Serono, Sanofi, Mundipharma, Bayer AG Schering Pharma, Hexal; Travel, Accommodations, Expenses: Merck Serono, Sanofi. Ioannis (John) Souglakos: No relationships to disclose. Fairooz F. Kabbinavar: Employment: Genentech Alfredo Falcone; Consulting or Advisory Role: Amgen, Genentech, Bayer AG, Servier, Merck Serono; Research Funding: Amgen (Inst), Roche (Inst), Bayer AG (Inst), Servier (Inst), Merck Serono (Inst). Benoist Chibaudel: No relationships to disclose. Jeffrey P. Meyers: No relationships to disclose. Daniel J. Sargent: Consulting or Advisory Role: AbbVie, Acerta Pharma, ARIAD Pharmaceuticals, Astellas Pharma, AstraZeneca/MedImmune, Biothera, Celldex, Exelixis, Genentech, Incyte, Kyowa Hakko Kirin, Medivation, Merck, Merrimack Pharmaceuticals, Nektar, Novartis, Pharmacyclics, Pique, Spiration, Xbiotech; Research Funding: Celgene (Inst), Genentech (Inst); Travel, Accommodations, Expenses: Celgene. Aimery de Gramont: No relationships to disclose. John R. Zalcberg: Honoraria: Bayer, Roche, Amgen, Pfizer, Specialised Therapeutics, Merck Serono; Consulting or Advisory Role: Bayer, Roche, Amgen, Pfizer, Specialised Therapeutics, Merck Serono; Research Funding (Institution): Novartis, Bayer, Amgen, Merck Serono, Roche, BristolMyers Squibb, Pfizer, AstraZeneca, Shire; Travel, Accommodations, Expenses: Ipsen, Merck Serono.
Supplementary Material
References
- 1. Glare P, Virik K, Jones M, et al. A systematic review of physicians' survival predictions in terminally ill cancer patients. BMJ. 2003;327(7408):195.http://dx.doi.org/10.1136/bmj.327.7408.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Temel JS, Shaw AT, Greer JA.. Challenge of prognostic uncertainty in the modern era of cancer therapeutics. J Clin Oncol. 2016;34(30):3605–3608. [DOI] [PubMed] [Google Scholar]
- 3. Mack JW, Smith TJ.. Reasons why physicians do not have discussions about poor prognosis, why it matters, and what can be improved. J Clin Oncol. 2012;30(22):2715–2717.http://dx.doi.org/10.1200/JCO.2012.42.4564 [DOI] [PubMed] [Google Scholar]
- 4. Kiely BE, Soon YY, Tattersall MHN, et al. How long have i got? Estimating typical, best-case, and worst-case scenarios for patients starting first-line chemotherapy for metastatic breast cancer: A systematic review of recent randomized trials. J Clin Oncol. 2011;29(4):456–463.http://dx.doi.org/10.1200/JCO.2010.30.2174 [DOI] [PubMed] [Google Scholar]
- 5. Pietrantonio F, Miceli R, Rimassa L, et al. Estimating 12-week death probability in patients with refractory metastatic colorectal cancer: The Colon Life nomogram. Ann Oncol. 2016;28(3):555–561. [DOI] [PubMed] [Google Scholar]
- 6. de Gramont A, Haller DG, Sargent DJ, et al. Toward efficient trials in colorectal cancer: The ARCAD Clinical Trials Program. J Clin Oncol. 2010;28(4):527–530.http://dx.doi.org/10.1200/JCO.2009.25.2544 [DOI] [PubMed] [Google Scholar]
- 7. Barras D. BRAF mutation in colorectal cancer: An update. Biomarkers Cancer. 2015;7(suppl 1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrell FE. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis (Springer Series in Statistics). Switzerland: Springer International Publishing AG; 2010. [Google Scholar]
- 9. Lieu C, Renfro L, De Gramont A, et al. The impact of young age on survival in patients with metastatic colorectal cancer: Analysis from the ARCAD Clinical Trials Program. Eur J Cancer. 2013:S483–S483. Abstract 49 [Google Scholar]
- 10. Renfro LA, Loupakis F, Adams RA, et al. Body mass index is prognostic in metastatic colorectal cancer: Pooled analysis of patients from first-line clinical trials in the ARCAD Database. J Clin Oncol. 2016:34(2):144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henriques J, Vernerey D, de Gramont A, et al. Prognosis of lung metastases in patients with metastatic colorectal cancer: An ARCAD meta analysis. Ann Oncol. 2016;27(suppl 2):ii122. [Google Scholar]
- 12. Franko J, Shi Q, Meyers JP, et al. Prognostic value of isolated peritoneal versus other metastatic sites in colorectal cancer (CRC) patients treated by systemic chemotherapy: Findings from 9,265 pts in the ARCAD database. J Clin Oncol. 2016;34(4_suppl):656-. [Google Scholar]
- 13. Proctor MJ, McMillan DC, Morrison DS, et al. A derived neutrophil to lymphocyte ratio predicts survival in patients with cancer. Br J Cancer. 2012;107(4):695–699.http://dx.doi.org/10.1038/bjc.2012.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steyerberg E. Clinical Prediction Models: A Practical Approach to Development, Validation, And Updating. New York, NY: Springer Science and Business Media; 2008. [Google Scholar]
- 15. Grambsch PM, Therneau TM.. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):515–526.http://dx.doi.org/10.1093/biomet/81.3.515 [Google Scholar]
- 16. Team RC. R: A Language and Environment for Statistical Computing. http://www.R-project.org. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 17. Renfro LA, Loupakis F, Adams RA, et al. Body mass index is prognostic in metastatic colorectal cancer: Pooled analysis of patients from first-line clinical trials in the ARCAD database. J Clin Oncol. 2016;34(2):144–150.http://dx.doi.org/10.1200/JCO.2015.61.6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tebbutt NC, Wilson K, Gebski VJ, et al. Capecitabine, bevacizumab, and mitomycin in first-line treatment of metastatic colorectal cancer: Results of the Australasian Gastrointestinal Trials Group randomized phase III MAX study. J Clin Oncol. 2010;28(19):3191–3198.http://dx.doi.org/10.1200/JCO.2009.27.7723 [DOI] [PubMed] [Google Scholar]
- 19. Stintzing S, Wirapati P, Lenz H-J, et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. J Clin Oncol. 2017;35(15_suppl):3510–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flemer B, Lynch DB, Brown JMR, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–643.http://dx.doi.org/10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with ras wild-type metastatic colorectal cancer: Retrospective analyses of the crystal and fire-3 trials. JAMA Oncol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials†. Ann Oncol. 2017;28(8):1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic survival associated with left-sided vs right-sided colon cancer: A systematic review and meta-analysis. JAMA Oncol. 2017;3(2):211–219.http://dx.doi.org/10.1001/jamaoncol.2016.4227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.