Abstract
Background
Investigating the genetic and environmental causes of variation in genome-wide average DNA methylation (GWAM), a global methylation measure from the HumanMethylation450 array, might give a better understanding of genetic and environmental influences on methylation.
Methods
We measured GWAM for 2299 individuals aged 0 to 90 years from seven twin and/or family studies. We estimated familial correlations, modelled correlations with cohabitation history and fitted variance components models for GWAM.
Results
The correlation in GWAM for twin pairs was ∼0.8 at birth, decreased with age during adolescence and was constant at ∼0.4 throughout adulthood, with no evidence that twin pair correlations differed by zygosity. Non-twin first-degree relatives were correlated, from 0.17 [95% confidence interval (CI): 0.05–0.30] to 0.28 (95% CI: 0.08–0.48), except for middle-aged siblings (0.01, 95% CI: −0.10–0.12), and the correlation increased with time living together and decreased with time living apart. Spouse pairs were correlated in all studies, from 0.23 (95% CI: 0.3–0.43) to 0.31 (95% CI: 0.05–0.52), and the correlation increased with time living together. The variance explained by environmental factors shared by twins alone was 90% (95% CI: 74–95%) at birth, decreased in early life and plateaued at 28% (95% CI: 17–39%) in middle age and beyond. There was a cohabitation-related environmental component of variance.
Conclusions
GWAM is determined in utero by prenatal environmental factors, the effects of which persist throughout life. The variation of GWAM is also influenced by environmental factors shared by family members, as well as by individual-specific environmental factors.
Keywords: Epigenomics, DNA methylation, twin study
Key Messages
This study is the largest collaboration of twin and/or family studies on DNA methylation.
This study is the first to investigate the genetic and environmental influences on genome-wide average DNA methylation (GWAM), a global methylation measure and a strong risk factor for cancers.
This study provides important evidence that GWAM is determined in utero by prenatal environmental factors, the effects of which persist throughout the whole life.
The variation of GWAM across the lifespan is also influenced by environmental factors shared by family members, as well as by individual-specific environmental factors.
Introduction
DNA methylation, mainly occurring at cytosine-guanine dinucleotide (CpG) where cytosine is converted to 5-methylcytosine (5meC), has been proposed to play a critical role in the aetiology of complex traits and diseases.1,2 DNA methylation has been found to be associated with environmental and lifestyle-related disease risk factors and diseases, such as body mass index,3 smoking,4 maternal plasma folate5 and cancers.6–9
Global DNA methylation refers to the level of 5meC content relative to total cytosine. The global methylation can be accurately measured by labour- and resource-intensive high-performance liquid chromatography (HPLC),10 or can be estimated by quantifying methylation of DNA repetitive elements such as long interspersed numerical elements (LINE) and Alu elements.11 With the technology of genome-wide methylation profiling, the Illumina Infinium HumanMethylation450 (HM450) BeadChip array can be used to quantify a measure of global methylation, which is usually calculated as the mean or median methylation value from CpGs across the genome.6–9 Being enriched for gene-associated CpGs, particularly those surrounding CpG-rich islands,12 the global methylation measure based on this assay is potentially highly relevant to gene function.
The global methylation measure based on the HM450 assay in peripheral blood has been found to be negatively associated with increased cancer risks: the mean beta-value across CpGs has been found to be associated breast cancer, 6,7 and the median M-value has been found to be associated with mature B-cell neoplasms8 and urothelial cell carcinoma.9 Assessed by the odds per adjusted standard deviation (OPERA),13 which enables comparison of the strengths of risk gradients in differentiating cases from controls, this global methylation measure is among the stronger known risk-discriminating factors for cancers: the OPERAs were estimated to be 1.5 ∼ 1.6 for breast cancer and 1.4 for superficial urothelial cell carcinoma. In comparison, the OPERAs for breast cancer risk factors are ∼1.5 for polygenic risk scores,14 ∼1.4 for mammographic density adjusted for age and body mass index and ∼1.2 for mutations in BRCA1 and BRCA2,13 and the OPERA is ∼1.5 for methylation levels at CpGs15 and epigenetic clock16 for lung cancer.
The genetic and environmental causes of variation in this global methylation measure are unknown. To investigate these causes and get a better understanding of genetic and environmental influences on methylation, we used data from seven twin and/or family studies of genetically related and genetically unrelated individuals across different stages of the lifespan, from birth to old age, and studied the mean beta-value across the genome, the genome-wide average DNA methylation (GWAM). Note that, although the average of the heritability estimates of the individual CpGs covered by the HM450 assay has been estimated to be approximately 20% under the assumptions of the classic twin model,17–19 this is not the same as the heritability of the average methylation level, the GWAM.
Methods
Subjects
We used data for 2299 individuals aged from 0 to 90 years from 816 families in seven twin and/or family studies (Table 1). These studies, listed in the order of the mean age of monozygotic (MZ) twins, are the Peri/postnatal Epigenetic Twins Study (PETS),20 Brisbane Systems Genetic Study (BSGS),19 Korean Healthy Twin Study (KHTS),21 Australian Mammographic Density Twins and Sisters Study (AMDTSS),22 Multiple Tissue Human Expression Resource (MuTHER) Study,17 Older Australian Twins Study (OATS)23 and Melbourne Collaborative Cohort Study (MCCS).24 Details of these studies are described in the Supplementary material, available at IJE online.
Table 1.
Characteristics of subjects within each studya
| Characteristics | PETS (birth) | PETS (18 months) | BSGS | KHTS | AMDTSS | MuTHER | OATS | MCCS | |
|---|---|---|---|---|---|---|---|---|---|
| Families | 14 | 10 | 177 | 97 | 130 | 246 | 108 | 43 | |
| Individuals | MZ twins | 18 | 12 | 134 | 182 | 132 | 186 | 216 | – |
| DZ twins | 10 | 8 | 222 | – | 132 | 306 | – | – | |
| Twins' siblings | – | – | 119 | 64 | 215 | – | – | – | |
| Parents or spouses | – | – | 139 | 136 | – | – | – | 86 | |
| Age (years) | MZ twins | 0 | 1.5 | 13.9±1.9 | 39.2±6.9 | 55.6±8.4 | 57.4±9.3 | 71.1±6.0 | – |
| DZ twins | 0 | 1.5 | 13.2±2.0 | – | 57.0±7.2 | 61.0±9.3 | – | – | |
| Twins' siblings | – | – | 15.4±2.8 | 38.3±10.8 | 56.6±8.0 | – | – | – | |
| Parents or spouses | – | – | 46.6±5.6 | 62.8±9.3 | – | – | – | 59.8±6.6 | |
| Sex | Male | 16 | 12 | 313 | 188 | – | – | 80 | 43 |
| Female | 12 | 8 | 301 | 194 | 479 | 492 | 136 | 43 | |
| Tissue | Buccal cells | Buccal cells | Whole blood | Whole blood | Whole blood | Adipose | Whole blood | Whole blood | |
| GWAMb | MZ twins | 43.39±0.31 | 43.68±0.36 | 50.57±0.29 | 53.59±1.53 | 52.95±0.32 | 48.10±0.08 | 51.56±0.59 | – |
| DZ twins | 43.19±0.29 | 43.04±0.16 | 50.60±0.32 | – | 52.99±0.32 | 48.09±0.08 | – | – | |
| Twins' siblings | – | – | 50.53±0.29 | 52.41±1.63 | 52.98±0.32 | – | – | – | |
| Parents or spouses | – | – | 50.31±0.31 | 52.33±1.59 | – | – | – | 53.18±0.50 | |
| GWAM by agec | – | – | −0.08 (0.01) | 0.003 (0.02) | 0.02 (0.02) | 0.01 (0.004) | −0.05 (0.05) | −0.08 (0.09) | |
| GWAM by sexb | Male | 43.28±0.12 | 43.23±0.30 | 50.48±0.33 | 52.68±1.69 | – | – | 51.50±0.57 | 53.09±0.47 |
| Female | 43.37±0.46 | 43.71±0.45 | 50.56±0.32 | 53.15±1.64 | – | – | 51.60±0.60 | 53.27±0.52 |
PETS, Peri/postnatal Epigenetic Twins Study; BSGS, Brisbane Systems Genetic Study; KHTS, Korean Healthy Twin Study; AMDTSS, Australian Mammographic Twins and Sisters Study; MuTHER, Multiple Tissue Human Expression Resource Study; OATS, Older Australian Twins Study; MCCS, Melbourne Collaborative Cohort Study; MZ, monozygotic; DZ, dizygotic; GWAM, genome-wide average DNA methylation.
Categorical variables are presented as counts, and continuous variables are presented as mean±SD.
GWAM is presented as the percentage of methylation, that is beta-value × 100. Reported GWAM was adjusted for batch effects through a linear mixed effects model in the PETS, BSGS, KHTS, MuTHER and OATS.
The linear regression coefficient (standard error) between GWAM and age. Reported as the change in percentage of methylation per 10-year increment in age. The regressions in the KHTS, OATS and MCCS were adjusted for study design or sampling factors.
DNA methylation measurement
DNA was extracted from buccal cells in the PETS, from adipose tissue in the MuTHER and from whole blood in the other studies. Each study measured DNA methylation using the HM450 assay and performed data processing independently. Details are described in the Supplementary material. GWAM was calculated as the average beta-value across autosomal CpGs (Supplementary Table 1, available as Supplementary data at IJE online). In the PETS, methylation in cord blood mononuclear cells was also measured for 17 newborn monozygotic twin pairs and nine newborn dizygotic (DZ) twin pairs, by the Illumina Infinium HumanMethylation27 BeadChip array.25 GWAM was also calculated for these twins.
Statistical methods
Within each study, we performed a two-stage adjustment on GWAM to minimize batch effects and to adjust for the effects of covariates, as described in the Supplementary material. It has been suggested that retaining group differences in batch effects adjustment might be problematic.26 To avoid such potential bias, no group difference was retained in the batch effects adjustment stage. Residuals from the adjustments were used in subsequent analyses.
We estimated the familial correlations in the residuals for different pairs of family members in each study (Supplementary Table 2, available as Supplementary data at IJE online) using a multivariate normal model.27,28 Sensitivity analyses were performed to examine the robustness of results to adjustment for cell mixture and to CpG selections, as described in the Supplementary material. The correlation in GWAM from cord blood mononuclear cells was also estimated for the 26 newborn pairs in the PETS, and was compared with the correlation from buccal cells for newborns to examine tissue heterogeneity.
We modelled the familial correlation as a function of cohabitation history using the combined data from all studies. To account for the different distributions of GWAM across studies, a standardised normal Z-score [mean = 0, standard deviation (SD) = 1] was calculated from the residuals within each study and used in the modelling. In the modelling, each study had its own mean and covariance functions for the Z-score. According to the familial correlation estimates in each age range, and following previous theoretical and empirical studies of familial covariance as a function of cohabitation status,29–31 we fitted a model in which the pair correlation increases or decreases with cohabitation history. Details are described in the Supplementary material.
We fitted variance components models using the combined data from all studies (Z-score as above). We assumed that the residual variance can be partitioned into four variance components: , the effects of additive genetic factors; , the effects of environmental factors shared by twins alone and assumed to be shared to the same extent within MZ and DZ pairs; , the effects of environmental factors shared by all family members (including twins) and assumed to be shared to the same extent within all pairs; and , the effects of individual-specific environmental factors and measurement error. According to the results of familial correlation modelling, we fitted the variance components dependent on cohabitation history. Details are described in the Supplementary material.
The correlations and variance components were estimated using the program FISHER.32 Other statistical analyses were performed using R [https://www.R-project.org/]. Inference was based on asymptotic likelihood theory, and the likelihood ratio test was used for comparisons between nested models.
Results
Table 1 shows the characteristics of subjects in each study. GWAM decreased with age in the BSGS and increased with age in the MuTHER, and there was no evidence that GWAM changed with age in the other studies. GWAM was higher for females than for males in the PETS at 18 months, and in BSGS, KHTS and OATS.
Table 2 shows the familial correlation estimates in GWAM within each study. MZ and DZ pairs were correlated in all studies. There was no evidence from any study for a difference in GWAM correlation according to zygosity (all P > 0.09). Combining MZ and DZ pairs, the correlation for twin pairs was about 0.8 both at birth and at age 18 months, and about 0.4 in adulthood. Non-twin first-degree relatives were correlated, from 0.17 [95% confidence interval(CI): 0.05–0.30] to 0.28 (95% CI: 0.08–0.48), except for middle-aged sisters in the AMDTSS (0.01; 95% CI: −0.10–0.12) whose separation time was the longest. Spouse pairs were correlated, from 0.23 (95% CI: 0.3–0.43) to 0.31 (95% CI: 0.05–0.52), in all studies.
Table 2.
Familial correlation estimates (95% confidence intervals) of genome-wide average DNA methylation within each study
| Pairs | PETS (birth) | PETS (18 months) | BSGS | KHTS | AMDTSS | MuTHER | OATS | MCCS |
|---|---|---|---|---|---|---|---|---|
| MZ pairs | 0.82 | 0.82 | 0.58 | 0.42 | 0.42 | 0.23 | 0.31 | – |
| (0.75-0.87) | (0.74-0.87) | (0.47-0.66) | (0.25-0.59) | (0.26-0.56) | (0.05-0.39) | (0.16-0.45) | ||
| DZ pairs | 0.85 | 0.89 | 0.40 | – | 0.40 | 0.45 | – | – |
| (0.79-0.89) | (0.85-0.92) | (0.28-0.51) | (0.24-0.54) | (0.35-0.53) | ||||
| Twin pairs combined | 0.83 | 0.84 | 0.46 | – | 0.43 | 0.36 | – | – |
| (0.78-0.86) | (0.80-0.88) | (0.37-0.53) | (0.32-0.53) | (0.27-0.45) | ||||
| Sibling pairs | – | – | 0.28 | 0.28 | 0.01 | – | – | – |
| (0.15-0.40) | (0.08-0.48) | (−0.10-0.12) | ||||||
| Parent-offspring pairs | – | – | 0.26 | 0.17 | – | – | – | – |
| (0.15-0.35) | (0.05-0.30) | |||||||
| Spouse pairs | – | – | 0.26 | 0.23 | – | – | – | 0.31 |
| (0.04-0.46) | (0.03-0.43) | (0.05-0.52) |
PETS, Peri/postnatal Epigenetic Twins Study; BSGS, Brisbane Systems Genetic Study; KHTS, Korean Healthy Twin Study; AMDTSS, Australian Mammographic Twins and Sisters Study; MuTHER, Multiple Tissue Human Expression Resource Study; OATS, Older Australian Twins Study; MCCS, Melbourne Collaborative Cohort Study; MZ, monozygotic; DZ, dizygotic.
From the sensitivity analyses, the familial correlations were robust to adjustment for cell mixture (Supplementary Table 3, available as Supplementary data at IJE online). Similar results were observed when GWAM was based on CpGs common to the seven studies or on non-noisy CpGs (Supplementary Tables 4 and 5, available as Supplementary data at IJE online), or on CpGs located in gene bodies or promoters (Supplementary Tables 6 and 7, available as Supplementary data at IJE online). The correlations in GWAM from cord blood mononuclear cells for PETS newborns were 0.80 (95% CI: 0.75–0.84) for MZ pairs, 0.80 (95% CI: 0.72–0.86) for DZ pairs and 0.80 (95% CI: 0.76–0.84) for twin pairs combined, similar to the correlations in GWAM from buccal cells.
From the modelling of correlation as a function of cohabitation history, there was no difference between MZ and DZ pairs (P = 0.08), so we combined MZ and DZ pairs to model the correlation for twin pairs. For similar reasons, we combined sibling pairs and parent-offspring pairs to model the correlation for non-twin first-degree relatives (P = 0.91 for comparison). The estimate of the correlation when non-twin pairs started living together, ε, was 0.08 (95% CI: −0.44–0.60) for non-twin first-degree relatives, and 0.10 (95% CI: −0.18–0.37) for spouse pairs. Neither estimate was different from zero, so we set them to zero.
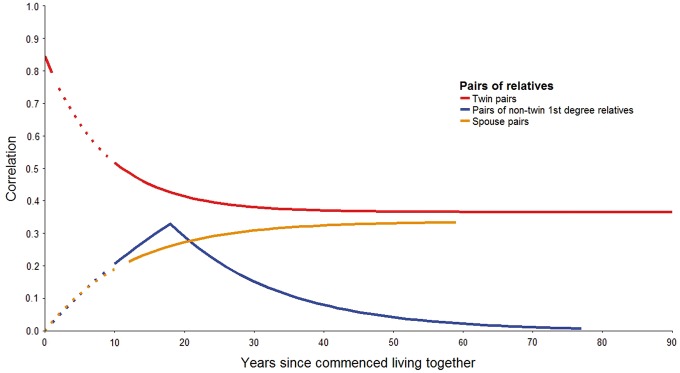
Figure 1 (and Supplementary Table 8, available as Supplementary data at IJE online) show the results from the modelling. The twin pair correlation decreased (λtwin = 0.12; 95% CI: 0.04–0.19) from 0.85 (95% CI: 0.74–0.94) at birth to 0.37 (95% CI: 0.30–0.44) in old age. The correlation for non-twin first-degree relatives increased with time living together (λ1st = 0.03; 95% CI: 0.003–0.06) and decreased with time living apart (ν= 0.06; 95% CI: 0.004–0.12). The spouse-pair correlation increased with time living together (λspouse = 0.08; 95% CI: 0.01–0.15).
Figure 1.
Familial correlations in genome-wide average DNA methylation with cohabitation history. The plot shows results from modelling the familial correaltion using the combined data (Z-score) from seven studies. Solid lines were based on the combined data. Dotted lines were theoretical lines extrapolated by the data, for which there were no data for the corresponding cohabitation duration.
From the fitted variance components model (Supplementary Table 9 and Supplementary Figure 1, available as Supplementary data at IJE online), the variance explained by additive genetic factors () was estimated to be −7% (95% CI: −25%–10%). The variance explained by environmental factors shared by twins alone () was 90% (95% CI: 74%–95%) at birth, decreased during adolescence and plateaued at about 28% (95% CI: 17%–39%) in adulthood. The variance explained by environmental factors shared by all family members () increased with time living together to about 26% (95% CI: 9%–44%) and decreased with time living apart. The variance explained by individual-specific environmental factors () increased with age, especially during adolescence.
Discussion
We investigated the influences of unmeasured genetic and environmental factors on variation in a global DNA methylation measure, GWAM derived from the HM450 assay. The repeatability of GWAM measurement has previously been estimated to be 0.74.33 Assuming this repeatability applied to our studies, the twin pair correlations at birth and 18 months of about 0.8 (with tight confidence intervals) were at the limit of repeatability. Therefore, taking into account measurement error, both MZ and DZ pairs were almost perfectly correlated at birth, and were still substantially correlated in old age (∼0.4/0.74 = 0.54).
Our study suggests that GWAM is determined before birth. Plausible reasons for the non-differential and high correlations in GWAM for newborn MZ and DZ pairs are: shared environmental factors in utero, maternal factors before and during pregnancy and paternal factors. We found no difference in MZ and DZ pair correlations at birth in the PETS and therefore no evidence for a role of genetic factors at birth. Although such a role cannot be discounted, it is unlikely to be substantial given the standard error of 0.04 for the difference between our high MZ and DZ pair correlations. Therefore, the main source of variation in GWAM at birth would appear to be shared prenatal environmental factors affecting infants in utero. Several prenatal environmental factors, such as maternal smoking in pregnancy34 and maternal plasma folate,5 have been found to be associated with CpG-specific DNA methylation for newborns. Specific prenatal environmental factors associated with GWAM remain unknown.
The twin pair correlations were substantial even in old age, and this suggests that the intrauterine effects persist during the whole life course. Assume that an individual’s GWAM at birth has a direct correlation with the GWAM in old age. Under an autoregressive model for longitudinal data,27 and taking into account the twin pair correlation at birth of 0.85 from the correlation modelling, the correlation for twin pairs in old age of 0.37 implies that the correlation between an individual’s GWAMs at birth and in old age must be about (0.37/0.85)1/2 = 0.66. Therefore, an individual’s GWAM at birth is a substantial predictor of his/her GWAM in old age. The empirical longitudinal correlation in GWAM needs to be investigated.
Our study found that the twin pair correlations decreased in childhood, which suggests that individual environmental effects increase during childhood. This may be due to ‘epigenetic drift’,35 which is in effect the role of non-genetic factors inducing variation in methylation levels. However, given that the twin pair correlations were relatively stable in adult life, our study provides evidence that ‘epigenetic drift’ may be manifest in early life but not in middle age and beyond.
We found evidence that GWAM is influenced by environmental factors shared within households, the effects of which increase with the cohabitation duration of family members (including spouse pairs) and attenuate when they live apart. Similar cohabitation-related effects of shared environmental factors have been found for other traits, such as blood lead level29 and bone mineral density.36
We did not find evidence that genetic factors explain variation in GWAM, given that we did not find difference between MZ and DZ pair correlations in any study or from the correlation modelling of the combined data, and the estimate of was zero if not negative. Genetic variants influencing methylation at specific loci are called methylation quantitative trait loci (meQTL). Several studies have examined meQTL; however, only a small proportion (10–15%) of CpGs has been found to be associated with meQTL.37–39 Given the small proportion, it is plausible that the variation in the average methylation level mixing of half million CpGs is not explained by genetic factors to an extent detectable by this study. Note that, given the confidence interval of , we cannot exclude a small genetic component of variance.
Given that GWAM has been found to be associated with risks of breast cancer, mature B-cell neoplasms and urothelial cell carcinoma, our results are consistent with hypothesis that risks of these cancers are initiated in utero.40,41 The developmental origins of health and disease (DOHaD) hypothesis considers that epigenome reprogramming during the fetal development period is one possible biological mechanism.42,43 We hypothesize that prenatal factors might influence risks of these cancers by altering the GWAM of the fetus. Identification of the prenatal factors associated with a newborn’s GWAM might open the possibility for risk-reducing interventions before birth. Our observation that the influences of individual-specific environmental factors increased during adolescence implies that early life is also important for intervention application. Consistently, early life is recognized as a critical window of vulnerability to breast carcinogens: commencing during fetal life and accelerating at puberty, the developing breast is exquisitely sensitive to carcinogens during periods of rapid fibro-glandular tissue proliferation.44 There is also evidence that the period between puberty and first completed pregnancy is a critical window for carcinogenic exposures.45
Our study has several strengths. First, to our knowledge our study is the first to investigate the influences of unmeasured genetic and environmental factors on global methylation using the HM450 assay. Previous studies focused on individual CpGs covered by this assay.17–19 Second, to our knowledge our study is the most comprehensive collaboration of twin studies on DNA methylation. Third, we included individuals from birth to 90 years of age, to obtain evidence across the lifespan. Fourth, we used a variety of family designs that provided contrasts in terms of shared genes and sharing of environment, and we used an optimal statistical analysis based on likelihood theory and flexible and realistic modelling.
The main limitation of our study is the potential heterogeneity across studies due to different populations, tissues, and aspects of DNA methylation measurement (e.g. methylation data normalization). For tissue heterogeneity, the familial correlations from buccal cells and from cord blood mononuclear cells were similar for newborn twins, which suggests there is little difference in the degree of resemblance for newborn twin pairs regardless of GWAM being measured using blood or buccal cells. Other limitations were that cohabitation history was not collected by some studies (although our assumption that separation occurs on average around age 18 years is based on empirical evidence), and the reliance on cross-sectional data; future studies that follow relatives prospectively are warranted.
We conclude that GWAM is determined before birth, possibly by prenatal environmental factors acting in utero, the effects of which persist during the whole life. Variation in GWAM is also influenced by individual-specific environmental factors, especially in early life, as well as by environmental factors shared by cohabiting family members, including spouse pairs.
Supplementary Data
Supplementary data are available at IJE online.
Funding
Data from some studies were obtained from public data repositories. However, we would like to acknowledge the funding for all studies. The PETS was supported by grants from the Australian National Health and Medical Research Council (NHMRC) (grant numbers 437015 and 607358 to J.C. and R.S.), the Bonnie Babes Foundation (grant number BBF20704 to J.M.C.), the Financial Markets Foundation for Children (grant number 032-2007 to J.M.C.) and by the Victorian Government’s Operational Infrastructure Support Program. The BSGS was supported by NHMRC grants 1010374, 496667 and 1046880. The KHTS was supported by a fund (2014-E71004-00) by Research of Korea Centers for Disease Control, and Prevention and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (grant number NRF-2017R1A2B2002136). The AMDTSS was facilitated through the Australian Twin Registry, a national research resource in part supported by a Centre for Research Excellence Grant from the NHMRC (grant number 1079102). The AMDTSS was supported by NHMRC (grant numbers 1050561 and 1079102), Cancer Australia and National Breast Cancer Foundation (grant number 509307). The MuTHER was funded by a programme grant from the Wellcome Trust (081917/Z/07/Z), and receives support from the National Institute for Health Research BioResource Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The OATS was facilitated through the Australian Twin Registry, a national research resource in part supported by a Centre of Research Excellence Grant from the NHMRC (grant number 1079102). OATS was funded by the NHMRC/Australian Research Council Strategic Award Grant 401162 and the NHMRC Project Grants 1045325 and 613608. The MCCS was made possible thanks to the funding obtained from the NHMRC (including project grant numbers 1011618, 1026892, 1026522, 1050198, 623206 and 1043616, and program grant numbers 209057 and 1074383).
S.L. is supported by the Australian Postgraduate Award (international), International Postgraduate Research Scholarship and the Richard Lowell Travelling Scholarship from the University of Melbourne. T.L.N. is supported by an NHMRC Post-Graduate Scholarship and the Richard Lowell Travelling Scholarship from the University of Melbourne. J.L.H. is a Senior Principal Research Fellow of NHMRC and a Distinguished Visiting Professor at Seoul National University. A.F.M. and G.W.M. are supported by the NHMRC Fellowship Scheme (1083656 and 1078399).
Supplementary Material
Acknowledgements
We would like to thank the participants and research team members in each included study. The OATS research team would like to thank its Chief Investigators including Julian Trollor, Henry Brodaty, Nicholas Martin, Katherine Samaras and Teresa Lee.
Author Contributions
S.L. and J.L.H. initiated and designed the study. S.L. performed statistical analyses. S.L. and J.L.H. wrote the first draft of the manuscript. All authors participated in the manuscript revision and have read and approved the final manuscript. Study contribution: PETS–J.M.C. and R.S.; BSGS–A.F.M. and G.W.M.; KHTS–E.K., Y.M.S. and J.S.; AMDTSS-S.L., E.M.W., J.E.J., T.L.N., J.S., G.S.D., M.C.S., G.G.G. and J.L.H.; MuTHER–T.D.S.; OATS–N.J.A., K.A.M., A.T., M.J.W., D.A. and P.S.S.; MCCS–E.M.W., J.E.J., P.A.D., R.L.M., M.C.S., G.G.G. and J.L.H.
Conflict of interest: The authors have no conflict of interest to disclose with respect to this manuscript.
References
- 1. Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 2010;465:721–27. [DOI] [PubMed] [Google Scholar]
- 2. Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148–59. [DOI] [PubMed] [Google Scholar]
- 3. Dick KJ, Nelson CP, Tsaprouni L. et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet 2014;383:1990–98. [DOI] [PubMed] [Google Scholar]
- 4. Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H.. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet 2011;88:450–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joubert BR, den Dekker HT, Felix JF. et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 2016;7:10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Severi G, Southey MC, English DR. et al. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res Treat 2014;148:665–73. [DOI] [PubMed] [Google Scholar]
- 7. van Veldhoven K, Polidoro S, Baglietto L. et al . Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin Epigenetics 2015;7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong Doo N, Makalic E, Joo JE. et al. Global measures of peripheral blood-derived DNA methylation as a risk factor in the development of mature B-cell neoplasms. Epigenomics 2016;8:55–66. [DOI] [PubMed] [Google Scholar]
- 9. Dugue PA, Brinkman MT, Milne RL. et al. Genome-wide measures of DNA methylation in peripheral blood and the risk of urothelial cell carcinoma: a prospective nested case-control study. Br J Cancer 2016;115:664–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuo KC, McCune RA, Gehrke CW, Midgett R, Ehrlich M.. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res 1980;8:4763–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004;32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sandoval J, Heyn H, Moran S. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011;6:692–702. [DOI] [PubMed] [Google Scholar]
- 13. Hopper JL. Odds per adjusted standard deviation: comparing strengths of associations for risk factors measured on different scales and across diseases and populations. Am J Epidemiol 2015;182:863–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dite GS, MacInnis RJ, Bickerstaffe A. et al. Breast cancer risk prediction using clinical models and 77 independent risk-associated SNPs for women aged under 50 years: Australian Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev 2016;25:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baglietto L, Ponzi E, Haycock P. et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer 2017;140:50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S.. DNA methylation age of blood predicts future onset of lung cancer in the Women's Health Initiative. Aging (Albany NY) 2015;7:690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grundberg E, Meduri E, Sandling JK. et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet 2013;93:876–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Dongen J, Nivard MG, Willemsen G. et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun 2016;7:11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McRae AF, Powell JE, Henders AK. et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol 2014;15:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martino D, Loke YJ, Gordon L. et al. Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol 2013;14:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sung J, Cho SI, Lee K. et al . Healthy Twin:a twin-family study of Korea - protocols and current status. Twin Res Hum Genet 2006;9:844–48. [DOI] [PubMed] [Google Scholar]
- 22. Li S, Wong EM, Joo JE. et al. Genetic and environmental causes of variation in the difference between biological age based on DNA methylation and chronological age for middle-aged women. Twin Res Hum Genet 2015;18:720–26. [DOI] [PubMed] [Google Scholar]
- 23. Sachdev PS, Lammel A, Trollor JN. et al. A comprehensive neuropsychiatric study of elderly twins: the Older Australian Twins Study. Twin Res Hum Genet 2009;12:573–82. [DOI] [PubMed] [Google Scholar]
- 24. Giles GG, English DR.. The Melbourne Collaborative Cohort Study. IARC Sci Publ 2002;156:69–70. [PubMed] [Google Scholar]
- 25. Gordon L, Joo JE, Powell JE. et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res 2012;22:1395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nygaard V, Rodland EA, Hovig E.. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 2016;17:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hopper JL, Mathews JD.. A multivariate normal model for pedigree and longitudinal data and the software ‘FISHER’. Aust J Stat 1994;36:153–76. [Google Scholar]
- 28. Hopper JL, Mathews JD.. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet 1982;46:373–83. [DOI] [PubMed] [Google Scholar]
- 29. Hopper JL, Mathews JD.. Extensions to multivariate normal models for pedigree analysis. II. Modeling the effect of shared environment in the analysis of variation in blood lead levels. Am J Epidemiol 1983;117:344–55. [DOI] [PubMed] [Google Scholar]
- 30. Lange K. Cohabitation, convergence, and environmental covariances. Am J Med Genet 1986;24:483–91. [DOI] [PubMed] [Google Scholar]
- 31. Eaves LJ, Long J, Heath AC.. A theory of developmental change in quantitative phenotypes applied to cognitive development. Behav Genet 1986;16:143–62. [DOI] [PubMed] [Google Scholar]
- 32. Lange K, Weeks D, Boehnke M.. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 1988;5:471–72. [DOI] [PubMed] [Google Scholar]
- 33. Dugue PA, English DR, MacInnis RJ. et al. Reliability of DNA methylation measures from dried blood spots and mononuclear cells using the HumanMethylation450k BeadArray. Sci Rep 2016;6:30317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joubert BR, Felix JF, Yousefi P. et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 2016;98:680–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fraga MF, Ballestar E, Paz MF. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 2005;102:10604–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hopper JL, Green RM, Nowson CA. et al. Genetic, common environment, and individual specific components of variance for bone mineral density in 10- to 26-year-old females: a twin study. Am J Epidemiol 1998;147:17–29. [DOI] [PubMed] [Google Scholar]
- 37. McClay JL, Shabalin AA, Dozmorov MG. et al. High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biol 2015;16:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lemire M, Zaidi SH, Ban M. et al. Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci. Nat Commun 2015;6:6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith AK, Kilaru V, Kocak M. et al. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics 2014;15:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trichopoulos D. Hypothesis: does breast cancer originate in utero? Lancet 1990;335:939–40. [DOI] [PubMed] [Google Scholar]
- 41. Marshall GM, Carter DR, Cheung BB. et al. The prenatal origins of cancer. Nat Rev Cancer 2014;14:277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Waterland RA, Michels KB.. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007;27:363–88. [DOI] [PubMed] [Google Scholar]
- 43. Wadhwa PD, Buss C, Entringer S, Swanson JM.. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med 2009;27:358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fenton SE, Reed C, Newbold RR.. Perinatal environmental exposures affect mammary development, function, and cancer risk in adulthood. Annu Rev Pharmacol Toxicol 2012;52:455–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colditz GA, Bohlke K, Berkey CS.. Breast cancer risk accumulation starts early: prevention must also. Breast Cancer Res Treat 2014;145:567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.