Abstract
The N-terminal fragments of mutant huntingtin (mHTT) misfold and assemble into oligomers, which ultimately bundle into insoluble fibrils. Conformations unique to various assemblies of mHTT remain unknown. Knowledge on the half-life of various multimeric structures of mHTT is also scarce. Using a panel of four new antibodies named PHP1–4, we have identified new conformations in monomers and assembled structures of mHTT. PHP1 and PHP2 bind to epitopes within the proline-rich domain (PRD), whereas PHP3 and PHP4 interact with motifs formed at the junction of polyglutamine (polyQ) and polyproline (polyP) repeats of HTT. The PHP1- and PHP2-reactive epitopes are exposed in fibrils of mHTT exon1 (mHTTx1) generated from recombinant proteins and mHTT assemblies, which progressively accumulate in the nuclei, cell bodies and neuropils in the brains of HD mouse models. Notably, electron microscopic examination of brain sections of HD mice revealed that PHP1- and PHP2-reactive mHTT assemblies are present in myelin sheath and in vesicle-like structures. Moreover, PHP1 and PHP2 antibodies block seeding and subsequent fibril assembly of mHTTx1 in vitro and in a cell culture model of HD. PHP3 and PHP4 bind to epitopes in full-length and N-terminal fragments of monomeric mHTT and binding diminishes as the mHTTx1 assembles into fibrils. Interestingly, PHP3 and PHP4 also prevent the aggregation of mHTTx1 in vitro highlighting a regulatory function for the polyQ-polyP motifs. These newly detected conformations may affect fibril assembly, stability and intercellular transport of mHTT.
Introduction
Huntington’s disease (HD) is an inherited neurodegenerative disorder caused by expansion of a CAG repeat in the exon-1 of huntingtin gene, which translates into an abnormal polyglutamine (polyQ) in huntingtin protein (HTT) (1). Proteolytic processing of mutant huntingtin (mHTT) ultimately generates N-terminal fragments such as cleaved products A and B (cpA and cpB) and exon-1 (here we refer to as mHTTx1), which are amyloidogenic and assemble into various not well-characterized structures linked to impaired cell signaling, neuroinflammation and neurodegeneration (2,3). cpA is produced by the enzymatic cleavage of larger N-terminal fragments of mHTT by an aspartyl protease generating a peptide of approximately 115 amino acids (AA). cpB is estimated to be 214 AA and the pathway regulating its production remains unknown (4). Aberrant splicing of huntingtin mRNA also produces truncated transcripts in a CAG-dependent manner, which contribute to the production of mHTTx1 protein (5). Emerging studies suggest that misfolded N-terminal fragments of mHTT assemblies may be structurally and functionally diverse. For example, small soluble oligomers of mHTTx1 induce cellular stress and toxicity, whereas large bundled insoluble fibrils inhibit apoptosis but could activate necrotic pathways (6–9). Thus, neurotoxicity in HD may be triggered by collective damage induced by various assemblies of mHTT acting in different neuronal compartments. The notion that the N-terminal fragments of mHTT forms a repertoire of structures with specific biological and pathological properties in vivo merits further investigation.
Characterization of a panel of anti-HTT monoclonal antibodies (mAbs) demonstrated that mHTTx1 accumulates in different subcellular compartments (10). Whether a unique assembly of mHTTx1 interacts with a specific cellular organelle remains unknown. It is likely that each assembly of mHTTx1 may acquire ‘signature conformation(s)’, which could be identified by specific antibodies. Initially, it was predicted that novel and potentially toxic conformations may arise from the expanded polyQ in HTT. Studies using the polyQ-specific 3B5H10 mAb reported the formation of a novel epitope linked to mHTT toxicity (11). However, comparison of three anti-polyQ antibodies MW1, 3B5H10 and 1C2 did not reveal any conformation-dependent binding. All three antibodies reacted similarly to mHTT but displayed lower affinities for the normal HTT (12,13). Thus, the evolution of novel conformations by the expanded polyQ in HTT requires further investigation. More recent studies suggest that the expanded polyQ domain of HTT is static, clusteredand does not contribute to the heterogeneity of mHTTx1 assemblies (6,14,15). However, domains flanking the polyQ repeat of mHTT appear to influence misfolding and neurotoxicity. The N-terminal 17 AA domain (N17) preceding the polyQ, the polyproline (polyP) repeats and the proline-rich domain (PRD) downstream of the polyQ have been implicated in the aggregation and toxicity of mHTTx1 in cell and animal models (6,16–19). For example, the N17 domain of mHTTx1 promotes the formation of bundled aggregates, whereas the C-terminal polyP repeats including PRD favor the assembly of unbundled fibrils (6). The N17 domain also harbors the sites for several post-translational modifications, which may alter the folding of mHTTx1 (20, 21). The PRD of mHTT is the most dynamic epitope in the assembled fibrils and may contribute to misfolding and evolution of new conformations (14,15). The PRD also interacts with various cellular proteins, which may control the formation of various assemblies in vivo (22). Thus, the notion that mHTTx1 aggregates into an array of structures with potentially distinct functions is gaining momentum and remains an interesting area of investigation in HD research (6,14,15).
Using a panel of four new mAbs, we have detected novel conformations formed at the polyQ-polyP junction and within the PRD of mHTT. The epitopes at the polyQ-polyP are confined to mHTT monomers, whereas those within the PRD are predominantly exposed in fibrils, which progressively accumulate in the brains of HD mouse models, interact with vesicles and myelin sheath insulating axons and may play a role in neurotoxicity and intercellular transport of mHTT.
Results
Binding of PHP1–4 antibodies to mHTTx1 structures in vitro
We selected four clones from a mAb library based on reactivity to recombinant mHTTx1 by various assays described below and labeled as PHP1–4. The isotypes of these antibodies are IgG1κ, IgG2κ, IgG1κ and IgG1κ, respectively. An overlapping peptide array spanning the HTTx1 sequence was used to map the binding of each antibody to its target domain (10). PHP1 and PHP2 react with the peptide sequence QAQPLLPQP within the PRD of human HTT, whereas PHP3 and PHP4 recognize novel epitopes formed by QQQQQQPP AA sequence. The exact number of proline and glutamine repeats required for the binding of PHP3 and PHP4 is hard to predict, however, proline residues are critical since no reactivity is observed to polyQ peptides and binding to peptides with more than two prolines is enhanced (Supplementary Material, Fig. S1). To our knowledge, this is the first report on defined antigenic epitopes formed by the PRD and at the boundary of polyQ and polyP repeats of HTT. Although PHP1–PHP2 and PHP3–PHP4 show reactivity to similar linear peptides, all are different clones based on the AA sequence in the antigen-binding domains of variable heavy (VH) and variable light (VL) of each antibody (data not shown). Thus, we predict each PHP antibody may recognize a unique antigenic motif under optimal physiological conditions.
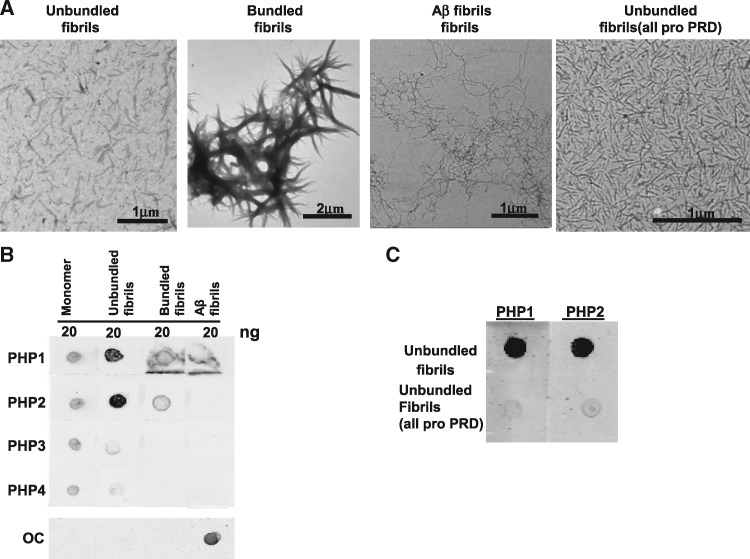
To identify structures that are recognized by PHP antibodies, we performed dot blot assays with recombinant monomers of mHTTx1 (46Q) fused to thioredoxin (TRX-mHTTx1) and in vitro-assembled fibrils that were either bundled or not. The structure of each assembly was verified by electron microscopy (EM) (Fig. 1A, leftmost two panels, respectively). We found that PHP1 and PHP2 display high reactivity for unbundled fibrils with substantially less binding to monomeric TRX-mHTTx1 or bundled fibrils (Fig.1 B). PHP3 and PHP4 showed only weak reactivity to monomers and mHTTx1 fibrils suggesting that the antibody-binding sites are not easily accessible (Fig. 1B). None of the antibodies bound to amyloid beta (Aβ) fibrils indicating specificity for mHTTx1 (Fig. 1A, third panel from left and Fig. 1B). The peptide array data suggested that QAQPLLPQP sequence is important for PHP1 and PHP2 interaction (Supplementary Material, Fig. S1). To further test this notion, we compared the reactivity of PHP1 and PHP2 to unbundled fibrils of mHTTx1 that either contain the native sequence or a version in which the PRD was mutagenized into a continuous stretch of polyP (see Materials and Methods section). The mutagenized mHTTx1 formed fibrils, which were verified by EM (Fig. 1A, right-most panel). Negligible binding was observed for the latter consistent with both antibodies binding to a region in the PRD that does not exclusively contain proline residues (Fig. 1C).
Figure 1.
PHP1 and PHP2 bind to preformed recombinant mHTTx1. (A) Electron micrographs of unbundled, bundled, Aβ-42, and unbundled (all pro) fibrils left to right, respectively. (B) Soluble recombinant TRX-HTTx1 (46Q), unbundled and bundled fibrils of mHTTx1, as well as Aβ fibrils were spotted on nitrocellulose paper and processed for reactivity to PHP1–4 and OC (specific for Aβ-42 fibrils) antibody as a positive control. Fluorescently labeled secondary antibody (IRDye-800 anti-mouse IgG) was used as secondary antibody. (C) Dot blot of PHP1 and PHP2 binding to unbundled fibrils (5 ng) containing the native PRD or an all pro-PRD (see sequence in Materials and Methods section) region. Results were reproduced with multiple preparations.
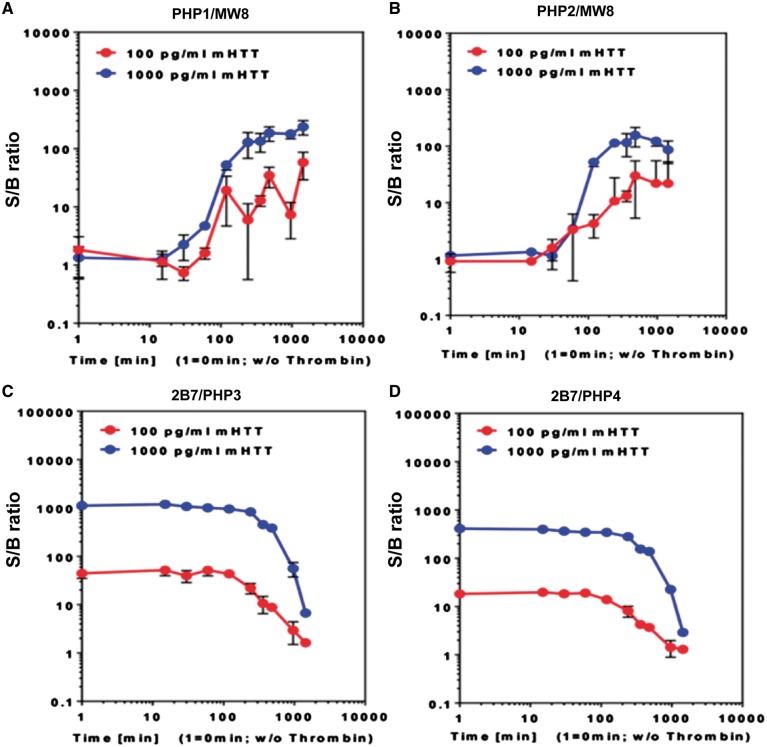
To further examine the interactions of PHP antibodies with mHTTx1 assemblies, we used a sensitive immunoassay based on the SMC Errena immunoassay platform (23). mHTTx1 (46Q) fused to maltose-binding protein (MBP) remains soluble but aggregates once MBP is removed. We performed a time-course fibril assembly experiment initiated by removing MBP from mHTTx1. Products were captured at different time points by PHP1 and PHP2 antibodies and detected by MW8, which recognizes an epitope at the C-terminus of mHTTx1 and has an affinity for fibrils (10,24). We found that PHP1 and PHP2 bind to mHTTx1 structures assembled within ∼30 min after the cleavage of MBP with a peak binding at ∼6 h post aggregation (Fig. 2A and B, respectively). The reactivity of PHP1 and PHP2 towards mHTTx1 assemblies was confirmed by using PHP1 and PHP2 antibodies for both capturing and detecting fibrils in the SMC Errena immunoassays. These assays generated profiles very similar to those observed with MW8 as detection antibody. Consistent with the dot blot assays (Fig. 1B), weak binding was detected when soluble MBP-mHTTx1 was used as antigen (Fig. 2, Supplementary Material, Fig. S2). Previous studies demonstrated that MW8 antibody is reactive to some assembled structures of mHTTx1 in FRET assays (24). Thus, the binding sites for MW8, PHP1 and PHP2 mAbs may be exposed on similar assemblies of mHTTx1 (2,10). PHP1 and PHP2 also captured misfolded species of mHTT in the brain lysates of symptomatic het Q175 HD mice, which express full-length HTT with175Qs (25) (Supplementary Material, Fig. S3). In similar assays, PHP3 and PHP4 reacted with monomeric MBP-mHTTx1 fusion protein captured by 2B7 antibody specific to the N17 domain of HTT (24). However, binding of PHP3 and PHP4 to soluble mHTTx1 diminished gradually as fibril assembly proceeded (Fig. 2C and D). The low binding of PHP3 and PHP4 for assembled structures of mHTTx1 was further verified by SMC Errena immunoassays using PHP3 and PHP4 antibodies for capturing and MW8 for detecting fibrils. The binding of PHP3 and PHP4 to assembled fibrils was minimal supporting the preference of these mAbs for soluble mHTTx1 (Supplementary Material, Fig. S4).
Figure 2.
PHP1 and PHP2 detect early assemblies of mHTTx1. (A and B) SMC Erenna immunoassays demonstrating capture of misfolded mHTTx1 by PHP1 and PHP2 antibodies over time and detected by the MW8. (C and D) present binding of PHP3 and PHP4 to monomeric soluble MBP-HTTx1 captured by B27, which is lost after initiation of aggregation. S/B ratio was determined by dividing the EP sample values by the EP values from assay buffer.
PHP1 and PHP2 bind to mHTTx1 fibrils assembled in HEK-293 cells
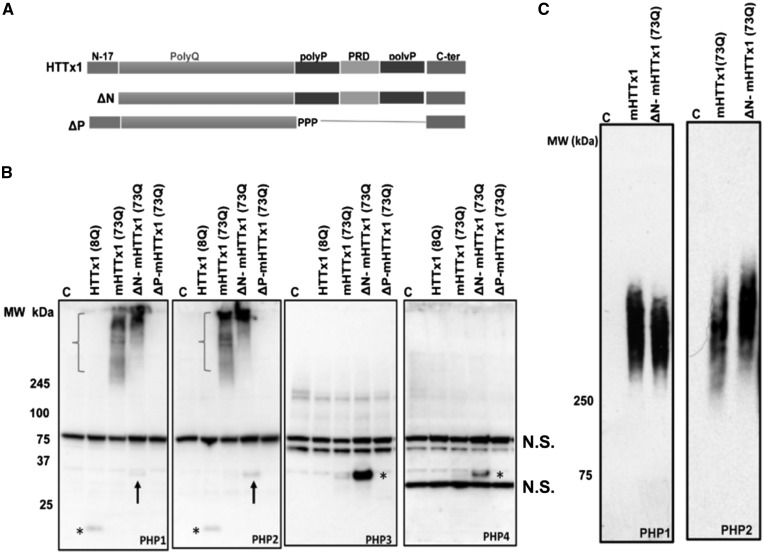
We expressed WT (8Q) and various constructs of mHTTx1 (73Q) (Fig. 3A) in HEK-293 cells and examined the binding of PHP antibodies to monomeric or assemblies by SDS-PAGE western blotting (WB). PHP1 and PHP2 displayed weak binding to WT HTTx1 but showed no binding to the monomeric mHTTx1 (Fig. 3B, left two panels, asterisks). Minimal reactivity was also detected with mHTTx1 lacking the N17 domain (ΔN) (Fig. 3B, left two panels, arrows). However, PHP1 and PHP2 bound to fibrils formed by mHTTx1 and ΔN constructs (Fig. 3B, left two panels, brackets). Agarose-SDS gel analysis, which is useful for resolving various assemblies of mHTTx1 (6), revealed that PHP1 and PHP2 bind to a heterogeneous mixture of mHTTx1 and ΔN-mHTTx1 assemblies (Fig. 3C). PHP3 and PHP4 only reacted with monomeric soluble ΔN mHTTx1 (Fig. 3B, right two panels, asterisks). The enhanced binding of PHP3 and PHP4 to the ΔN construct of mHTTx1 suggests that removal of N17 domain may stabilize or better expose the conformations detected by these antibodies. The low concentration of monomeric mHTTx1 due to rapid aggregation may also be responsible for the low reactivity of PHP1–4 in the HEK-293 cell lysates. Similar to PHP1 and PHP2, MW8 also reacted with mHTTx1 assemblies in HEK-293 cell lysates further suggesting the PRD and the epitope at C-terminus of mHTTx1 (the binding site for MW8) are exposed on the assembled mHTTx1 fibrils (Supplementary Material, Fig. S5A).
Figure 3.
PHP antibodies bind to HTTx1 assemblies formed in HEK-293 cells. (A) Schematic depiction of constructs used for transfection in part (B). (B) WB analysis of lysates from HEK-293 cells transfected with various constructs of HTTx1. Membranes were probed with PHP1–4 antibodies. Asterisks in the left two panels indicate faint detection of WT HTTx1 (8Q) by PHP1 and PHP2, respectively. Arrows in the left two panels point at weak reactivity of PHP1 and PHP2 to ΔN-mHTTx1. Brackets indicate the assemblies of mHTTx1 and ΔN-mHTTx1 detected by PHP1 and PHP2. PHP3 and PHP4 only detected monomeric mHTTx1 highlighted by asterisks in the right-most two panels, respectively. Part (C) shows examination of HEK-293 cell lysate expressing mHTTx1 and ΔN-mHTTx1 by agarose-SDS gel electrophoresis and probing with PHP1 and PHP2. All experiments were repeated three times. N.S., non-specific bands.
PHP1 and PHP2 react with mHTT fibrils in HD mice
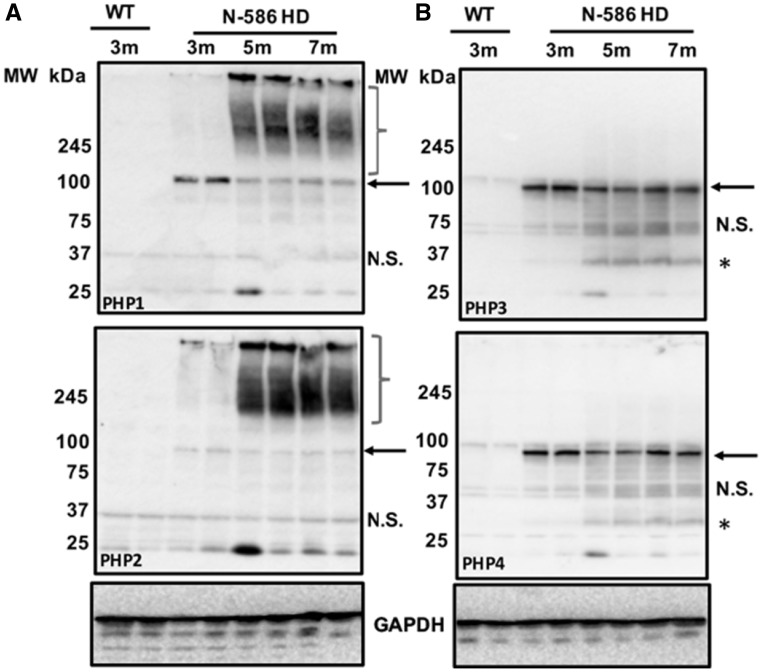
To confirm that PHP antibodies recognize mHTT assemblies in vivo, we examined brain lysates of two HD mouse models by WB. N-586 HD mice express the 586 N-terminal AAs of mHTT with 82Qs, which is proteolytically processed to generate smaller N-terminal fragments (26). PHP1 recognized monomeric mutant N-586 in brain lysates obtained from 3-month (3m) old animals, whereas PHP2 displayed weak binding (Fig. 4A, top and bottom panels, respectively, arrows). Similar to cell culture experiments (Fig. 3), PHP1 and PHP2 bound to high-molecular weight fibrils assembled in the brains of N-586 HD mice (Fig. 4A, brackets). Notably, mHTT assemblies were minimal in the 3-month-old brain lysates but accumulated as the animals aged and developed neurological symptoms (Fig. 4A, 3m compared with 5m and 7m) (26). Surprisingly, MW8 did not react to mHTT fibrils in the brain lysates of N-586 HD mice, whereas MW7, which is specific to polyP repeat of HTT (10), recognized high-molecular weight structures (Supplementary Material, Fig. S5A and B). Since MW8 binds to the exposed C-terminus of mHTTx1 (2,10), it is likely the levels of mHTTx1 fibrils that are minimal in the N-586 HD mice. PHP1 and PHP2 reacted with mHTT fibrils in lysates extracted from the brains of het Q175 HD mice (Supplementary Material, Fig. S6A and B). In this model, PHP1- and PHP2-reactive assemblies also accumulated progressively with age, which was further confirmed by immunoprecipitation from cortical lysates (Supplementary Material, Fig. S7A). SDS-agarose gel analyses of brain lysates of het Q175 HD mice also confirmed the binding of PHP2 antibody to a heterogeneous mixture of mHTT fibrils, which are more abundant in the 9-month-old cortical lysates (Supplementary Material, Fig. S7B). Thus, PHP1- and PHP2-reactive mHTT fibrils accumulate in the brains of symptomatic HD mice (25,26). Although PHP1 and PHP2 showed some reactivity to the monomeric N-586 fragments, they did not recognize full-length mHTT in the brain lysates of het Q175 HD mice on WB (Fig. 4A, Supplementary Material, Fig. S7A). A likely explanation is that proteolytic cleavage of full-length mHTT may expose or ‘mature’ the conformations recognized by PHP1 and PHP2 in the PRD. Alternatively, binding of PHP1 and PHP2 to PRD of HTT may be influenced by the polyQ length (82Qs versus 175Qs in the animal models used) or biochemical processing of brain lysates. PHP3 and PHP4 reacted with N-586 fragment and full-length mHTT monomer. Cleaved products of soluble mHTT were also detected by PHP3 and PHP4 (Fig. 4B, top and bottom panels, respectively, Supplementary Material, Fig. S6C and D). In these experiments, PHP3 and PHP4 did not recognize the endogenous mouse HTT suggesting that the conformations may be unique to human mHTT.
Figure 4.
PHP antibodies bind to mHTT assemblies in the brains of HD mice. (A) WB analysis of cortical lysates of N-586 HD mice harvested at 3, 5 and 7 months (m) of age and probed with PHP1 and PHP2 (top and bottom panels, respectively). Cortical lysates of 3-month-old WT mice were used as controls. Arrows point to monomeric N-586 (82Q) detected by each antibody. Brackets are the mHTT assemblies present in the brain lysate of 5-month and 7-month-old N-586 HD mice, which are less abundant in the 3-month olds. Part (B) is WB analysis of the same lysates probed by PHP3 and PHP4. Both antibodies react with monomeric N-586 fragment and some smaller products highlighted by asterisks. N.S., non-specific bands. The house keeping protein glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. Lysates obtained from brains of offspring from various cohorts revealed similar results.
PHP1- and PHP2-reactive mHTT assemblies are present in various neuronal organelles
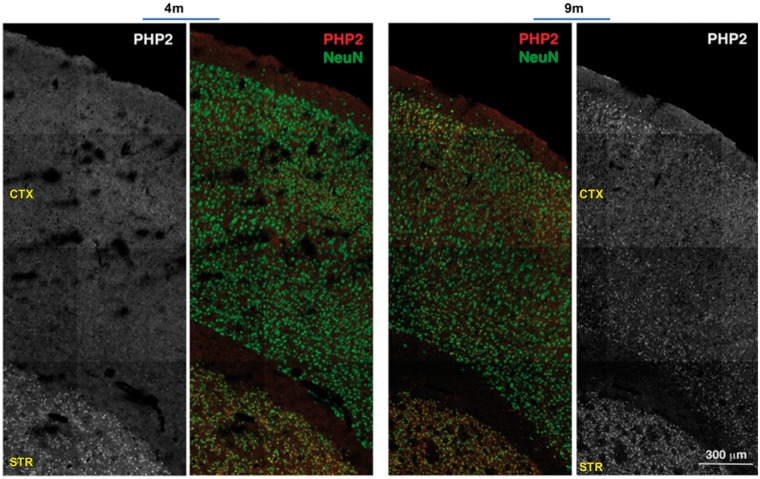
We further examined the reactivity of PHP antibodies to mHTT assemblies in the brain sections of HD mice by immunohistochemistry (IHC). Since PHP1 and PHP2 displayed similar immunostaining pattern, we focused on the characterization of PHP2 in the het Q175 HD mouse line. PHP2 recognized mHTT assemblies in the striatum and to a lesser extent in the cortical layers of 4-month-old (4m) het Q175 HD brains. Similar to WB results, PHP2-reactive mHTT assemblies were more abundant in the 9-month old (9m) brain sections when compared with 4-month old (4m) and were located in the cytoplasm, nuclei and neuropils (Fig. 5A, Supplementary Material, Fig. S8). In the cortical layers of N-586 HD brains, PHP1- and PHP2-reactive mHTT assemblies were found within cell bodies but some were also lined-up in neuropils, which did not overlap with neuronal marker MAPII (Supplementary Material, Fig. S9). The lined-up arrangement of mHTT fibrils remains to be investigated.
Figure 5.
PHP2-reactive assemblies accumulate in the striatum and cortical layers. (A) Low magnification of a composite demonstrating the reactivity of PHP2 to mHTT assemblies in the brain sections (striatum and cortical layers) of 4-month (4m) and 9-month (9m) old het Q175 HD mice. The neuronal marker NeuN (green) was used to stain neurons. PHP2 staining of assemblies is in red. Black and white panels for the PHP2 staining is shown to demonstrate the intensity of staining for PHP2 masked by the NeuN staining. Images were captured by a confocal microscope (Zeiss LSM 710). IHC results were reproduced with offspring from various cohorts. CTX, cortex; STR, striatum.
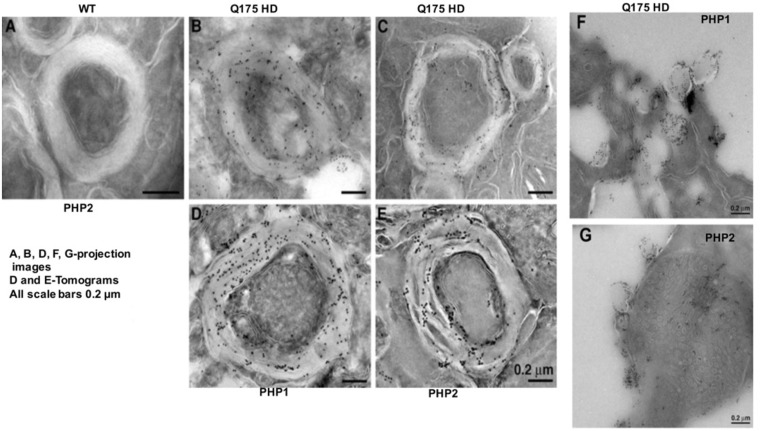
To better understand the properties of PHP1- and PHP2-reactive mHTT assemblies, we performed electron microscopic analysis of brain sections obtained from the striatum of 9-month-old het Q175 HD mice using immunogold labeling. These studies revealed that PHP1 and PHP2 bind to mHTT assemblies associated with myelin sheath (Fig. 6B–E). mHTT assemblies were found within and on the inner and outer leaflets of myelin and between axons (Supplementary Material, Fig. S10). Notably, PHP1 and PHP2 also reacted with mHTT assemblies associated with vesicle-like structures; further supporting their association with neuronal membranes (Fig. 6F and G). These findings do not exclude the presence of PHP-reactive mHTT assemblies in other neuronal organelles since IHC showed that they are found in somas and neuropils (Fig. 5, Supplementary Material, Figs. S8 and S9). However, detection of PHP-reactive mHTT assemblies in myelin is intriguing and highlights the possibility of fibrils disrupting myelination. By IHC we did not detect any specific reactivity of PHP3 and PHP4 antibodies to mHTT. One possibility is that the epitopes are buried within soluble mHTT in neurons or masked by other neuronal proteins. Some diffused nuclear binding to the brain sections of het Q175 HD mice was observed under florescent microscope; however, the authenticity of the binding remains to be investigated considering that the majority of monomeric mHTT is localized to the cytoplasm.
Figure 6.
PHP-1 and PHP2-reactive mHTT assemblies associate with myelin. (A) Electron micrograph of brain sections of 9-month-old het Q175 HD mice showing the presence of PHP1 and PHP2 reactive fibrils in the myelin sheath. Panels (A)–(C) are projection images and panels (D) and (E) are tomograms. Panels (F) and (G) show the association of PHP1 and PHP2-reactive fibrils with vesicle-like structures. EM studies were reproduced with sections obtained from the brain of several het Q175 HD mice.
PHP antibodies block the fibril assembly and seeding of mHTTx1
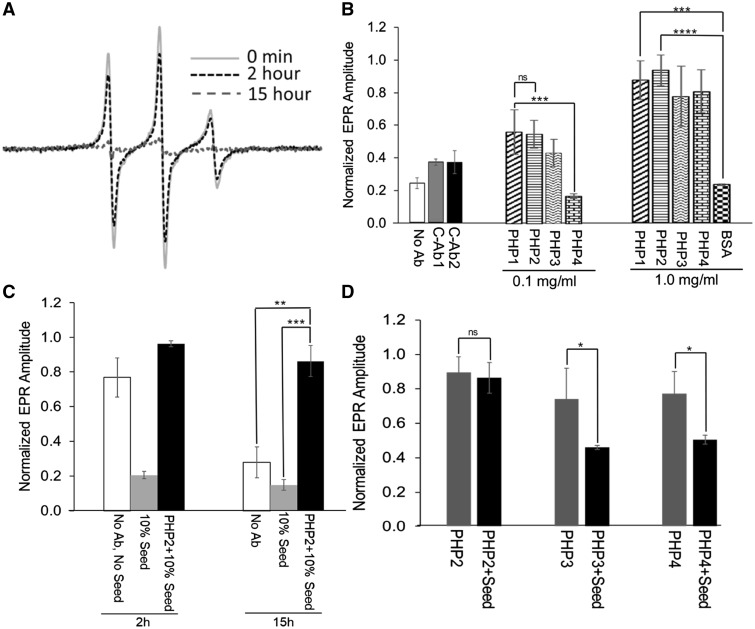
To better understand the influence of QAQPLLPQP and QQQQQQPP epitopes on mHTTx1 folding and assembly, we applied electron paramagnetic resonance (EPR) to examine the aggregation of mHTT in the presence of each antibody. EPR combined with site-directed spin labeling can monitor the mobility of spin-labeled AA. EPR can be used to follow the fibril formation of amyloidogenic proteins over time since the mobility decreases severely as the intrinsically disordered monomers assemble into fibrils (27). To initiate fibril assembly, soluble recombinant thioredoxin-mHTTx1 [TRX-mHTTx1 (46Q)] protein labeled with spin label R1 at position 15, was treated with enterokinase to remove the TRX fusion partner. Aggregation of mHTTx1 decreased spin label mobility and thus reduced the EPR signal amplitudes over a period of 15 h (Fig. 7A). When PHP antibodies (PHP1–4) were included in the fibril assembly assays, the reduction of the EPR amplitudes was significantly attenuated in a concentration-dependent manner indicating that the antibodies inhibited fibril formation (Fig. 7B). PHP1 and PHP2 were able to bind to fibrillar species (Figs 1, 3 and 4) and thus, it is possible that some of these species facilitate seeding. To test whether these antibodies can act on seeds, we investigated seeding-dependent aggregation using sonicated preformed fibrils in the presence of PHP2. In the absence of PHP2, seeds strongly enhanced misfolding as monitored by EPR (Fig. 7C), whereas the addition of PHP2 completely inhibited seeding. PHP3 and PHP4, which do not preferentially recognize fibrillar species, exhibited only marginal inhibitory activity towards the seeded reaction compared to the unseeded one (Fig. 7D).
Figure 7.
Monitoring mHTTx1 misfolding and inhibition by PHP antibodies using EPR. (A) Continuous wave EPR spectra of mHTTx1 spin labeled at position 15 obtained at the indicated time points. Misfolding causes line broadening and a concomitant reduction in amplitude. (B) Summary of EPR amplitudes of the same spin-labeled derivative used in (A) after 15 h of incubation in the absence (no Ab) or presence of controls or PHP1–4 antibodies with the indicated concentrations. The addition of PHP1–4 results in a dose-dependent protection from signal loss due to misfolding. The control Abs (C-Ab1 and C-Ab2, not against HTT 1 mg/ml) and control protein (BSA, 1 mg/ml) did not affect mHTTx1 misfolding significantly. Panel (C) illustrates the effect of seeded misfolding in the presence (black bars) or absence (gray bars) of PHP2 antibody at 1 mg/ml at the indicated time points. The white bars (no Ab, no seed) are the control which did not contain added seeds and antibodies. (D) Comparison of EPR amplitude of the unseeded (gray bar) and seeded (black bar) reaction for mHTTx1 misfolding after 15 h in the presence of PHP2–4 antibodies. PHP2 significantly inhibits the seeded reaction, whereas the inhibitory effects of PHP3 and PHP4 were marginal. The error bar represents the standard deviation of the mean from the three individual measurements. (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
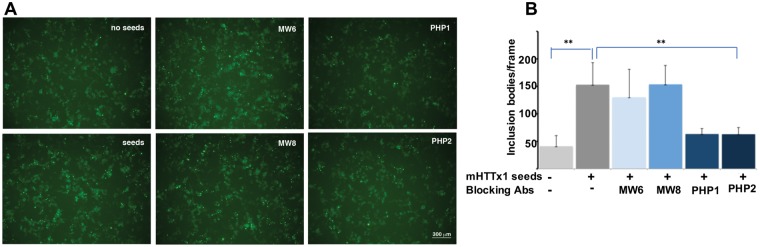
Some assemblies of mHTT are present in the cerebrospinal fluids of HD patients, which promote seeding of monomeric mHTTx1 in vitro and in cell models (28,29). We asked whether PHP1 and PHP2 antibodies decrease seeding of mHTTx1 in a cell culture model (29). Cell lysates containing mHTTx1–73Q fibrils (Fig. 3C) were added to HEK-293 cells expressing subthreshold levels of mHTTx1-EGFP (103Q) with minimal aggregation. We found that treatment of cells expressing mHTTx1-EGFP with the lysates containing mHTTx1–73Q assemblies accelerates the aggregation of intracellular mHTTx1-EGFP (Fig. 8A and B). To confirm that PHP1 and PHP2-reactive mHTTx1 structures are indeed responsible for the seeding effects, we preincubated the seeding lysates with several antibodies such as polyQ-specific MW6, which recognizes soluble mHTT, MW8 specific for the C-terminus of mHTTx1, and PHP1 or PHP2. Preincubation with MW6 or MW8 had no effects, whereas addition of PHP1 or PHP2 antibodies to the seeding lysates significantly reduced the assembly of mHTTx1-EGFP fibrils (Fig. 8A and B). Thus, PHP1 and PHP2 bind to mHTTx1 species, which may have the propensity for cell-to-cell transmission and the ability to induce seeding.
Figure 8.
Cell-based assays examining the effects of PHP1 and PHP2 on seeding of mHTTx1. (A) Fifty μg of lysates obtained from HEK-293 transfected with mHTTx1 (Q73) (Fig. 3B) were treated with DNase and RNase (1 μg each) and added to HEK-293 cells in six well dishes transfected with subthreshold level of mHTTx1-EGFP (0.2 μg plasmid DNA) to reduce spontaneous aggregation. For antibody interference, lysate was preincubated with each indicated antibody for 2 h rocking at RT and then added to cells. Plates were examined 12 h after treatment. Random pictures were taken at low magnification and quantified for aggregated bodies in each frame. All experiments were repeated three times. (B) The average of six microscope fields for each condition is presented in (C). Data shown are means ± SD. **P <0.001.
Discussion
The N-terminal fragments of mHTT are predicted to acquire a spectrum of conformations as it misfolds and assembles into oligomers, fibrils and inclusion bodies (6–8,30). Distinguishing between different assemblies of mHTT in vivo is critical for studying their biological properties and ultimately identifying the elusive neurotoxic species. Using PHP1 and PHP2 mAbs, we detected epitopes within the PRD of mHTT fibrils in vitro and in cellular and mouse models of HD. Moreover, the PHP-reactive fibrils accumulate in various neuronal organelles including myelin and vesicle-like structures. PHP3 and PHP4 mAbs recognized novel epitopes formed at the border of polyQ-polyP in monomeric full-length and N-terminal fragments of mHTT. The newly discovered motifs recognized by PHP1–4 play a role in the fibril assembly and seeding of amyloidogenic N-terminal fragments of mHTT. These findings and the generated mAbs will help to better understand the epitopes involved in the assembly of mHTT fibrils and may lead to identifying unique structures linked to specific cellular compartments with novel pathological phenotypes.
We previously reported that the PRD of mHTT serves several biological functions such as regulating protein stability, aggregation and neurotoxicity (31,32). Recent studies suggest that PRD contributes to the heterogeneity of mHTTx1 assemblies, raising the possibility that the impact of PRD on the biological and pathological activities of mHTTx1 may be mediated by conformations evolved during misfolding and aggregation process (14,15). Here we identified PHP1- and PHP2-reactive epitopes within the PRD, which may biologically be important. Based on differences in the AA sequence of the antigenic binding domains of each antibody (VH and VL), we envision that PHP1 and PHP2 may recognize distinct but overlapping antigenic motifs, which are exposed in the assembled mHTT fibrils. However, our assays lack the sensitivity to determine the unique properties or each antibody and remains to be investigated. It is likely that mHTT fibrils detected by PHP1 and PHP2 are heterogeneous (Figs 1–4). Heterogeneity in amyloidogenic proteins such as Aβ are linked to distinct clinical phenotypes in Alzheimer’s disease (AD) (33). Structural diversity in fibrils also contributes to distinct biological and pathological functions in Tau and α-synuclein proteins implicated in Taupathies and Parkinson’s disease (PD), respectively (34,35). We predict that the intrinsically disordered N-terminal fragments of mHTT may also assemble into a spectrum of structures and those identified by PHP1 and PHP2 may represent only a fraction of all misfolded mHTT. Supporting this hypothesis is the reactivity of PHP1 and PHP2 but not MW8 to mHTT fibrils in N-586 HD mice, whereas all three antibodies recognize mHTTx1 assemblies in cell culture (Figs 3 and 4, and Supplementary Material, Fig. S5). To expand on this hypothesis, it is essential to develop a repertoire of oligomeric- and fibril-specific antibodies and identify mHTT assemblies that are different from those detected by PHP1 and PHP2 in vivo. Such studies are in progress and will facilitate investigating the biological and pathological functions of various assembled structures of mHTT.
Age-dependent accumulation of PHP1- and PHP2-reactive mHTT fibrils in HD mouse models and overlap with neurological symptoms supports the notion that such structures may pathologically be relevant. The interaction of mHTT with cellular organelles including nuclei, mitochondria and other membranous structures may disrupt neuronal physiology, impair organelles’ architectures and disrupt neuroprotective pathways (3). A novelty of the current studies was discovering the association of PHP1- and PHP2-reactive mHTT assemblies within myelin sheath and membranous-like vesicles in the brains of HD animals (Fig. 6, and Supplementary Material, Fig. S10). Recent studies indicate that fibrils of mHTTx1-GFP interact with the membranes of endoplasmic reticulum causing architectural deformities and toxicity (36). We speculate that the presence of PHP1- and PHP2-reactive mHTT in myelin sheath may promote axonal defects and/or impair the physiology of oligodendrocytes. Mice expressing mHTT exclusively in oligodendrocyte show deficit in myelin formation and animals develop severe neurological symptoms with accelerated death (37). Moreover, myelin defects precede the appearance of aberrant cognitive and motor symptoms in HD mouse models and in HD patients (3,38,39). Although it remains to be confirmed, mHTT aggregates are less abundant in oligodendrocytes when compared with neurons (38). We predict that PHP1- and PHP2-reactive mHTT assemblies in myelin sheath are of neuronal origin and highlight the interesting possibility of impairing oligodendrocytes’ functions non-cell-autonomously.
The interaction of PHP1- and PHP2-reactive assemblies with membranes may also facilitate their transport to neighboring cells and contribute to the propagation of mHTT in the CNS. mHTT is released into intercellular space through endosomal/lysosomal secretory vesicles (40). Moreover, assemblies of mHTTx1 are transported along the axons of primary neurons and secreted into culture medium (41). It is likely that not all assemblies of mHTT are capable of associating with membranes or being transported to other cells. Notably, PHP1 and PHP2 antibodies block seeding of mHTTx1 in vitro and in a cell model (Figs 7 and 8); further suggesting the propensity of such structures for intercellular spread and propagation. The mechanism of how PHP-reactive assemblies associate with myelin and vesicle-like structures remains to be investigated. However, it is known that the N17 domain of monomeric HTT interacts with membranes in vitro and influences aggregation and toxicity in cell models (42–44). Thus, it may be that the assembly of PHP1- and PHP2-reactive fibrils is regulated by membranes. Alternatively, preformed PHP1- and PHP2-reactive fibrils of mHTT may directly bind to membranes (45). Insertion in membranes and myelin may also stabilize mHTT fibrils by physical constraints thus, impeding the formation of bundled fibrils and inclusion bodies. Overall, mHTT assemblies identified by PHP1 and PHP2 mAbs exemplify viable biomarkers for exploring various aspects of HD pathogenesis including neurotoxicity, pathways for spreading within the CNS, as well as myelin and oligodendrocytes impairments. Experiments are in progress to examine these and to further determine whether the interaction of PHP-reactive fibrils with myelin precedes the development of motor behaviors in animal models.
The binding of PHP3 and PHP4 to epitopes formed by the polyQ-polyP junction further adds to repertoire of novel conformations formed by the mHTTx1. Little is known about the interactions of polyQ with polyP domains and how they may influence the misfolding of mHTTx1. Thus, PHP3 and PHP4 mAbs are useful reagents to perform structure–function analyses and further dissect the involvement of structures formed by polyQ-polyP junction in the biology of mHTT. It is relevant that binding of PHP3 and PHP4 to monomeric mHTTx1 diminishes as the fibrils assemble (Fig. 2). One possibility is that conformations formed by the combination of polyQ and polyP repeats are dynamic and disappear as the mHTTx1 misfolds and ensues aggregation. Alternatively, such conformations could be buried within the fibrils and are less accessible to binding. Interestingly, PHP3 and PHP4 block the fibril assembly of mHTTx1 in vitro highlighting a potential role for polyQ/polyP junction in the biogenesis of mHTT assemblies and further supporting that multiple epitopes downstream of the polyQ domain are critical for the biogenesis of misfolded mHTT structures (6,14,15).
In summary, we have identified new antigenic motifs linked to monomers and fibril assemblies of mHTT. The application of PHP1–4 antibodies to monitor the fibril assembly of mHTT and to enrich for assemblies from various sources will help to better understand their properties and to identify structures, which may influence neurotoxicity in HD. Considering the recent findings on the release of mHTT assemblies in biological fluids of HD patients and the ability to spread by a ‘prion-like’ mechanism (28,29,46), testing of PHP antibodies as immunotherapies to interfere with seeding and/or spreading of mHTT structures within the CNS and developing assays to screen for small molecules to impede mHTT fibril assembly may reveal interesting results.
Materials and Methods
Animal work
The N-586 (82Q) breeding pairs were provided by Dr Christopher Ross, and heterozygous (het) Q7/Q175 knock-in HD mouse brains were obtained from the Jackson Laboratory. All mice experiments and care complied with federal regulations and were reviewed and approved by the California Institute of Technology Institutional Animal Care and Use Committee (IACUC).
Immunization and antibody production
Immunization and hybridoma production was carried out as described previously (10). Recombinant purified WT HTTx1 (20Q) fused to glutathione S transferase (GST) was used as an antigen. Initially clones were screened based on reactivity to lysates obtained from HEK-293 cells expressing various constructs of HTT by WB analysis. Four reactive clones described here were labeled as PHP1–4 in honor of the late professor Paul H. Patterson.
Antibodies
2B7, MW6 and MW8 antibodies have been reported previously (10,23). MAPII was purchased from ThermoFisher and rabbit anti-NeuN (ABN78) was obtained from EMD Millipore.
Epitope mapping
To determine the epitopes recognized by the new mAbs, we utilized arrays of dot blots that contain overlapping 14mer peptides synthesized from the first 91 AA of normal human Huntingtin (HTT; containing a 23 polyQ domain) (10). That is, the first dot contains the peptide corresponding to AA 1–14, the second dot contains the peptide corresponding to 4–17, etc. Two rows of peptide dot blots are shown for each with the upper row corresponding to the peptides shown at the top of the figure, and the lower row corresponding to the peptides shown at the bottom of the array in Supplementary Material, Figure S1. Also, included in the array are corresponding peptides from the mouse HTT sequence in order to confirm that binding of antibodies is unique to human HTT (spot #27–32). The mouse peptides are shown at the bottom right of Supplementary Material, Figure S1.
Purification of fusion protein HTTx1
The fusion protein human HTTx1 was expressed and purified as described previously (14,27). Briefly, the overnight cultures of pET32a-HTTx1 (46Q) transformed BL21 cells were grown to A600 nm = 0.7–0.8 at a dilution of 50 folds in Lauria Broth (LB) medium and further grown at 37°C. The expression was achieved using IPTG and subsequent purification was done using affinity chromatography followed by ion exchange chromatography.
Fibril formation
Unbundled fibrils were prepared by cleaving the TRX-mHTTx1 (46Q) fusion protein using 1 unit of EKmax per 280 µg of HTT. The protein concentration was kept at 25 μM in 20 mM Tris, 150 mM Nacl pH 7.4 buffer. Prior to cleavage reaction the samples were treated with 5% molar ratio HTTx1 (Q46) seeds. The final reaction mixture was incubated at 4°C for 2 days. The bundled fibrils were prepared in a similar manner, except the pre-seeded samples were incubated at 37°C for several days until bundled fibrils were observed.
Proline C-terminus fibrils
The plasmids were reconstructed from parent construct to make all proline C-terminus HTTx1 by GenScript. Primary sequence of the modified poly PRD HTTx1 is listed: MATLEKLMKA FESLKSF (Q)46-P(38) GGAVAEEGL HRGHHHHHH. The expression and purification of the fusion protein was achieved using previously reported methods (27). The fibril formation was induced similarly, using EK max digestion. The final protein concentration in the fibrils was estimated using spin-labeled derivative.
Aβ42 fibrils
Aβ42 fibrils were prepared using a standard protocol (47). Briefly, the lyophilized peptide was dissolved in DMSO at a concentration of 5 mM. The aliquots were then transferred to 10 mM HCl to a final concentration of 100 µM peptide. The samples were incubated for 24 h at 37°C.
Dot blots
A total of 1.5 µl of desired samples were blotted onto nitrocellulose membrane (VWR) and incubated with desired primary antibody at 1–500 dilutions. Fluorescently labeled secondary antibody (IRDye-800 anti-Mouse IgG, 1: 10 000 dilution; LI-COR) was used to visualize binding via a LI-COR Odyssey Infrared Imaging system.
Huntingtin protein aggregation
To perform aggregation kinetics, 10 µM MBP-tagged HTT exon1-Q46 were mixed with 150 µg/ml bovine thrombin protease (Sigma) and 2 mM CaCl2 in a total volume of 1400 µl 50 mM Tris (pH 8.0), 150 mM NaCl. Samples were incubated for 24 h at 37°C. Aliquots were analyzed before and at multiple time points after initiation of aggregation by protease addition. Samples were immediately snap-frozen at each time point and stored at −20°C.
Huntingtin protein detection assay
Huntingtin protein was detected by the SMC Erenna immunoassay system by Singulex (now EMD Millipore) using the previously described protocol (23) with the following modifications. In short, samples were diluted into dilution buffer containing 6% BSA, 0.8% Triton X-100, 750 mM NaCl. Samples were added to a 96-conical assay plate (catalog P-96-450V-C; Axygen). Capture antibodies (2B7, MW8 or PHP1-4) coupled to magnetic particles was added, and the plate was sealed and incubated under shaking at room temperature for the indicated time points. After washing, the fluorophore-labeled detection antibodies (MW8 or PHP1-4) was added to the plate and incubated at room temperature for 1 h. Plates were washed, and the antibody-antigen complex was transferred to a new 96-conical assay plate to eliminate non-specific binding events to the plastic. After washing, elution buffer and the eluted detection antibody was transferred to a Nunc 384-well analysis plate (catalog 264573; Sigma-Aldrich) and neutralized with buffer (1 M Tris, pH 9). The analysis plate was spun down to eliminate foaming and bubble formation, sealed and subsequently analyzed with the SMC Erenna immunoassays. Event photons (EP) values were taken as absolute signal intensity values. Signal-to-background (S/B) ratio was determined by dividing the EP sample values by the EP values from assay buffer.
Transfections
HEK-293 cells were transfected with the indicated constructs in Figure 3A (6) by calcium phosphate precipitation. Cells were harvested 36 h post-transfection, lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% sodium deoxycholate, 1%SDS, 1%NP40, 2m MEDTA, pH 7.4 with protease inhibitor cocktail) (Roche) and examined by SDS-PAGE and WB analysis using PHP antibodies as the primary. All HRP-conjugated secondary antibodies were used at 1: 10 000 dilutions.
Western blots of brain lysates
Flash frozen cortical and striatal brain tissues were collected as follows: brains free from skull were immersed in cold PBS briefly and placed in dish on top of ice pack for dissection. Under dissection microscope with ventral side up two coronal cuts were made, one cut to remove olfactory bulbs, the other cut at optical chiasm. Separate cortex and striatum, collect them in Eppendorf tubes, flash frozen in liquid nitrogen and store in −80°C until ready to use. Cortical or striatal tissues were extracted with total extraction buffer: RIPA buffer and sonicated briefly before centrifugation at 14 300 rpm at 4°C for 30 min using tabletop centrifuge. Protein fractions were run with either 4–15% or –20% polyacrylamide Criterion gels (Bio-Rad Laboratories) and transferred overnight, blots were blocked with 5% milk in PBS and blotted with PHP (1: 1000) Abs overnight. Blots incubated with HRP-conjugated goat anti-mouse secondary Ab and developed with ECL substrate (ThermoFisher, SuperSignal West Dura substrate 34705) and X-ray film. Agarose gel electrophoresis of samples to confirm the heterogeneous nature of reactive products was carried out as described previously (6,48).
Cryosections and immunohistochemistry
WT and HD mice were perfused with PBS followed by 4% paraformaldehyde and processed for cryosectioning by standard procedures as described previously (10). Frozen 18 um thick brain sections were obtained using cryostat (Leica CM3050) and mounted onto slides (Fisher Superfrost Plus), were dried overnight and stored slides at −20°C. Slides were thawed and dried and washed in PBS briefly before antigen retrieval. Sections were immersed in antigen retrieval solution (Dako Target retrieval solution, S1699) inside a plastic slide-staining dish (Tissue Tek) and incubated at 95°C water bath for 25 min. Slides were cooled in a dish and washed in PBS before applying primary antibodies. Primary Abs were diluted in 2% BSA, 10% normal goat serum in PBS with 0.4% Triton X-100. Sections were incubated with primary Abs at room temperature overnight. Slides were then washed with PBS, three times, 10 min each, before adding secondary Abs. After 1 h of incubation, sections were then washed and mounted with mounting medium (ThermoFisher, prolong gold antifade mounting solution). Sections were observed under epi-fluorescence (Nikon Diaphot 300) or a confocal microscope (Zeiss LSM 710).
Electron microscopy of mHTTx1 and Aβ fibrils
A total of 10 µl of preformed fibril sample was absorbed onto EM grid (150 mesh copper) for 5 min. Grids were then negatively stained by adding 10 μl of 1% uranyl acetate solution for 12 min and dried at room temperature. Images were acquired using a Gatan digital camera equipped with a JOEL JEM-1400 electron microscope (JOEL, Peabody, MA) at 100 kV.
Immuno-electron microscopy
Brain tissue was and fixed with 4% paraformaldehyde (PFA) in PBS. Tissue was excised and infiltrated with 2.1 M sucrose in 0.1 M sodium cacodylate trihydrate for 12 h. Small (1 mm3) cubes of tissue were prepared with a scalpel, affixed to aluminum sectioning stubs and frozen quickly in liquid nitrogen. Frozen specimens were placed in the cryostage of a UC6/FC6 cryoultramicrotome (Leica Microsystems, Vienna) set to −110°C. Thin (100 nm) cryosections were cut using a diamond knife (Diatome Ltd., Switzerland) and transferred to Formvar-coated, carbon-stabilized 100-mesh copper-rhodium EM grids (Electron Microscopy Sciences, Port Washington PA). The sections were blocked with 10% calf serum in PBS for 30 min, then labeled with PHP1, PHP2 diluted in PBS with 5% calf serum for 2 h, followed by colloidal gold (15 nm) conjugated anti-mouse secondary antibodies (BBI Solutions, Cardiff UK) for 2 h. Sections were simultaneously negative-stained with 1% uranyl acetate and embedded with 1% methylcellulose, then air-dried in wire loops.
Grids were placed in a dual-axis tomography specimen holder (Model 2040, E.A. Fischione Instruments, Inc. Export PA) and imaged with a Tecnai TF30ST transmission electron microscope (ThermoFisher Scientific, Hillsboro OR) at 300 keV. Images were recorded digitally with a US1000 CCD camera (Gatan, Inc. Pleasanton CA) using the Serial EM software package (49). Digital images, montages and tomographic datasets were calculated and analyzed with the IMOD software package (50,51).
Effect of antibodies on misfolding of HTTx1: EPR study
The Trx-mHTTx1 (46Q) fusion protein harboring a cys residue at position 15 was reacted with MTSL spin label to yield the new spin-labeled side chain R1 as described in previous publications (27). The concentration of the spin-labeled protein was ∼15 uM in 20 mM Tris, 150 mM Nacl pH 7.4 buffer. Misfolding of mHTTx1 was initiated by the addition of Ekmax (Invitrogen), which cleaves off the stabilizing TRX moiety, as described in the fibril preparation section. The continuous wave EPR spectra were recorded using a Bruker EMX EPR spectrometer at 100 G and optimized power and modulation settings as in previous publications (14,27). Control experiments showed that Ekmax cleavage was not affected by antibodies.
Cell lysate seeding assays
HEK-293 cells were transfected with mHTTx1-EGFP (103Q) in 6-well plates at subthreshold concentration (0.2 μg of plasmid DNA) for spontaneous aggregation using Lipofectamine (Thermo Fisher Scientific, Carlsbad, CA). Twelve h post transfection, cells were treated with 50 μg of DNase/RNAse-treated lysates obtained from HEK-293 cells expressing mHTTx1 (73Qs) (Fig. 3B) and incubated for additional 12 h. For testing the effects of antibodies on seeding, 50 μg of lysate containing the seeding material was prec-incubated with 5 μg of each antibody and rocked at RT for 2 h before addition to target cells expressing mHTTx1-EGFP (103Q). Samples were examined by a Nikon florescent microscope and images were captured. The number of inclusion foci were counted in six microscope fields for each condition and averaged.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to Dr Judith Frydman and Dr Koning Shen (Stanford University) for providing the HTTx1 constructs. We are also thankful to Dr Christopher Ross (Johns Hopkins University) for providing breeding pairs of the N-586 HD mice.
Conflict of Interest statement. None declared.
Funding
This work was supported by the NINDS grant (NS074374-01A1) awarded to A.K. and P.H.P., awards (A-8456, A-10468) from CHDI to A.K., NIH grant (NS084345) and CHDI award (A-12640) to R.L.
References
- 1. Huntington’s Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2. Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L.M., Trottier Y.. et al. (2010) Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem., 285, 8808–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R.. et al. (2015) Huntington’s disease. Nat. Rev. Dis. Primers, 1, 15005.. [DOI] [PubMed] [Google Scholar]
- 4. Kegel K.B., Sapp E., Alexander J., Reeves P., Bleckmann D., Sobin L., Masso N., Valencia A., Jeong H., Krainc D.. et al. (2010) Huntingtin cleavage product A forms in neurons and is reduced by gamma-secretase inhibitors. Mol. Neurodegener., 5, 58.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L.M., Roos R.A.C., Howland D.. et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U.S.A., 110, 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen K., Calamini B., Fauerbach J.A., Ma B., Shahmoradian S.H., Lachapel I.L.S., Chiu W., Lo D., Frydman J. (2016) Control of the structural landscape and neuronal proteotoxicity of mutant huntingtin by domains flanking the polyQ tract. Elife, 5, 18065.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leitman J., Hartl F.U., Lederkremer G.Z. (2013) Soluble forms of polyQ-expanded huntingtin rather than large aggregates cause endoplasmic reticulum stress. Nat. Commun., 4, 2753.. [DOI] [PubMed] [Google Scholar]
- 8. Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature, 431, 805–810. [DOI] [PubMed] [Google Scholar]
- 9. Ramdzan Y.M., Trubetskov M.M., Ormsby A.R., Newcombe E.A., Sui X., Tobin M.J., Bongiovanni M.N., Gras S.L., Dewson G., Miller J.M.L.. et al. (2017) Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep., 19, 919–927. [DOI] [PubMed] [Google Scholar]
- 10. Ko J., Ou S., Patterson P.H. (2001) New anti-huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins. Brain Res. Bull., 56, 319–329. [DOI] [PubMed] [Google Scholar]
- 11. Miller J., Arrasate M., Brooks E., Libeu C.P., Legleiter J., Hatters D., Curtis J., Cheung K., Krishnan P., Mitra S.. et al. (2011) Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol., 7, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein F.A., Zeder-Lutz G., Cousido-Siah A., Mitschler A., Katz A., Eberling P., Mandel J.L., Podjarny A., Trottier Y. (2013) Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10. Hum. Mol. Genet., 22, 4215–4223. [DOI] [PubMed] [Google Scholar]
- 13. Owens G.E., New D.M., West A.P., Bjorkman P.J. (2015) Anti-PolyQ antibodies recognize a short PolyQ stretch in both normal and mutant huntingtin exon 1. J. Mol. Biol., 427, 2507–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Isas J.M., Langen R., Siemer A.B. (2015) Solid-state nuclear magnetic resonance on the static and dynamic domains of huntingtin exon-1 fibrils. Biochemistry, 54, 3942–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin H.K., Boatz J.C., Krabbendam I.E., Kodali R., Hou Z., Wetzel R., Dolga A.M., Poirier M.A., van der Wel C.A. (2017) Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nat. Commun., 8, 15462.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duennwald M.L., Jagadish S., Muchowski P.J., Lindquist S. (2006) Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. U.S.A., 103, 11045–11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crick S.L., Ruff K.M., Garai K., Frieden C., Pappu R.V. (2013) Unmasking the roles of N-and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. U.S.A., 110, 20075–20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. André E.A., Braatz E.M., Liu J.P., Zeitlin S.O. (2017) Generation and characterization of knock-in mouse models expressing versions of huntingtin with either an N17 or a combined PolyQ and proline-rich region deletion. J. Huntingtons Dis., 6, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu X., Cantle J.P., Greiner E.R., Lee C.Y., Barth A.M., Gao F., Park C.S., Zhang Z., Sandoval-Miller S., Zhang R.L.. et al. (2015) N17 Modifies mutant huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron, 85, 726–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daldin M., Fodale V., Cariulo C., Azzollini L., Verani M., Martufi P., Spiezia M.C., Deguire S.M., Cherubini M., Macdonald D.. et al. (2017) Polyglutamine expansion affects huntingtin conformation in multiple Huntington’s disease models. Sci. Rep., 7, 5070.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiki A., DeGuire S.M., Ruggeri F.S., Sanfelice D., Ansaloni A., Wang Z.M., Cendrowska U., Burai R., Vieweg S., Pastore A.. et al. (2017) Mutant exon1 huntingtin aggregation is regulated by T3 phosphorylation-induced structural changes and crosstalk between T3 phosphorylation and acetylation at K6. Angew. Chem. Int. Ed. Engl., 56, 5202–5207. [DOI] [PubMed] [Google Scholar]
- 22. Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E. (1998) Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet., 7, 1463–1474. [DOI] [PubMed] [Google Scholar]
- 23. Wild E.J., Boggio R., Langbehn D., Robertson N., Haider S., Miller J.R., Zetterberg H., Leavitt B.R., Kuhn R., Tabrizi S.J.. et al. (2015) Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J. Clin. Investig., 125, 1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baldo B., Paganetti P., Grueninger S., Marcellin D., Kaltenbach L.S., Lo D.C., Semmelroth M., Zivanovic A., Abramowski D., Smith D.. et al. (2012) TR-FRET-based duplex immunoassay reveals an inverse correlation of soluble and aggregated mutant huntingtin in huntington's disease. Chem. Biol., 19, 264–275. [DOI] [PubMed] [Google Scholar]
- 25. Loh D.H., Kudo T., Truong D., Wu Y., Colwell C.S. (2013) The Q175 mouse model of Huntington's disease shows gene dosage- and age-related decline in circadian rhythms of activity and sleep. PLoS One, 8, e69993.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waldron-Roby E., Ratovitski T., Wang X., Jiang M., Watkin E., Arbez N., Graham R.K., Hayden M.R., Hou Z., Mori S.. et al. (2012) Transgenic mouse model expressing the caspase 6 fragment of mutant huntingtin. J. Neurosci., 32, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bugg C.W., Isas J.M., Fischer T., Patterson P.H., Langen R. (2012) Structural features and domain organization of huntingtin fibrils. J. Biol. Chem., 287, 31739–31746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morozova O.A., Gupta S., Colby D.W. (2015) Prefibrillar huntingtin oligomers isolated from HD brain potently seed amyloid formation. FEBS Lett., 589, 1897–1903. [DOI] [PubMed] [Google Scholar]
- 29. Tan Z., Dai W., Van Erp T.G., Overman J., Demuro A., Digman M.A., Hatami A., Albay R., Sontag E.M., Potkin K.T.. et al. (2015) Huntington’s disease cerebrospinal fluid seeds aggregation of mutant huntingtin. Mol. Psychiatry, 20, 1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sahoo B., Arduini I., Drombosky K.W., Kodali R., Sanders L.H., Greenamyre J.T., Wetzel R. (2016) Folding landscape of mutant huntingtin exon1: diffusible multimers, oligomers and fibrils, and no detectable monomer. PLoS One, 11, e0155747.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Southwell A.L., Khoshnan A., Dunn D.E., Bugg C.W., Lo D.C., Patterson P.H. (2008) Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J. Neurosci., 28, 9013–9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Southwell A.L., Ko J., Patterson P.H. (2009) Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington's disease. J. Neurosci., 29, 13589–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qiang W., Yau W.M., Lu J.X., Collinge J., Tycko R. (2017) Structural variation in amyloid-β fibrils from Alzheimer's disease clinical subtypes. Nature, 541, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaufman S.K., Sanders D.W., Thomas T.L., Ruchinskas A.J., Vaquer-Alicea J., Sharma A.M., Miller T.M., Diamond M.I. (2016) Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron, 92, 796–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peelaerts W., Bousset L., Van der Perren A., Moskalyuk A., Pulizzi R., Giugliano M., Van den Haute C., Melki R., Baekelandt V. (2015) α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature, 522, 340–344. [DOI] [PubMed] [Google Scholar]
- 36. Bäuerlein F.J., Saha I., Mishra A., Kalemanov M., Martínez-Sánchez A., Klein R., Dudanova I., Hipp M.S., Hartl F.U., Baumeister W.. et al. (2017) In situ architecture and cellular interactions of polyQ inclusions. Cell, 171, 179–187. [DOI] [PubMed] [Google Scholar]
- 37. Huang B., Wei W., Wang G., Gaertig M.A., Feng Y., Wang W., Li X.J., Li S. (2015) Mutant huntingtin downregulates myelin regulatory factor-mediated myelin gene expression and affects mature oligodendrocytes. Neuron, 85, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith G.A., Rocha E.M., McLean J.R., Hayes M.A., Izen S.C., Isacson O., Hallett P.J. (2014) Progressive axonal transport and synaptic protein changes correlate with behavioral and neuropathological abnormalities in the heterozygous Q175 KI mouse model of Huntington's disease. Hum. Mol. Genet., 23, 4510–4527. [DOI] [PubMed] [Google Scholar]
- 39. Teo R.T.Y., Hong X., Yu-Taeger L., Huang Y., Tan L.J., Xie Y., To X.V., Guo L., Rajendran R., Novati A.. et al. (2016) Structural and molecular myelination deficits occur prior to neuronal loss in the YAC128 and BACHD models of Huntington disease. Hum. Mol. Genet., 25, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trajkovic K., Jeong H., Krainc D. (2017) Mutant huntingtin is secreted via a late endosomal/lysosomal unconventional secretory pathway. J. Neurosci., 37, 9000–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brahic M., Bousset L., Bieri G., Melki R., Gitler A.D. (2016) Axonal transport and secretion of fibrillar forms of α-synuclein, Aβ42 peptide and HTTExon 1. Acta Neuropathol., 131, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suopanki J., Götz C., Lutsch G., Schiller J., Harjes P., Herrmann A., Wanker E.E. (2006) Interaction of huntingtin fragments with brain membranes–clues to early dysfunction in Huntington's disease. J. Neurochem., 96, 870–884. [DOI] [PubMed] [Google Scholar]
- 43. Burke K.A., Kauffman K.J., Umbaugh C.S., Frey S.L., Legleiter J. (2013) The interaction of polyglutamine peptides with lipid membranes is regulated by flanking sequences associated with huntingtin. J. Biol. Chem., 288, 14993–15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. (2007) Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet., 16, 2600–2615. [DOI] [PubMed] [Google Scholar]
- 45. Monsellier E., Bousset L., Melki R. (2016) α-Synuclein and huntingtin exon1 amyloid fibrils bind laterally to the cellular membrane. Sci. Rep., 6, 19180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jeon I., Cicchetti F., Cisbani G., Lee S., Li E., Bae J., Lee N., Li L., Im W., Kim M.. et al. (2016) Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol., 132, 577–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stine W.B. Jr, Dahlgren K.N., Krafft G.A., LaDu M.J. (2003) In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J. Biol. Chem., 278, 11612–11622. [DOI] [PubMed] [Google Scholar]
- 48. Halfmann R., Lindquist S. (2008) Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp., 17, e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mastronarde D.N. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol., 152, 36–51. [DOI] [PubMed] [Google Scholar]
- 50. Kremer J.R., Mastronarde D.N., McIntosh J.R. (1996) Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol., 116, 71–76. [DOI] [PubMed] [Google Scholar]
- 51. Mastronarde D.N. (2008) Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J. Microsc., 230, 212–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.