Abstract
Objectives
In this study, we characterize a concurrent disseminated infection with a virulent hypermucoviscous (HMV) Klebsiella pneumoniae and an OXA-181-producing XDR K. pneumoniae from a patient with recent hospitalization in India. During exposure to meropenem therapy, the highly susceptible HMV K. pneumoniae became resistant to carbapenems, consistent with the acquisition of blaOXA-181.
Methods
Twelve K. pneumoniae isolates were recovered from the patient and the hospital room environment over a 3 month hospitalization. Phenotypic and molecular studies were completed to characterize the isolates. Oxford Nanopore and Illumina MiSeq WGS were performed to study phylogeny (MLST and SNPs), plasmids and virulence genes and demonstrate changes in the organism’s resistome that occurred over time.
Results
WGS revealed that the HMV K. pneumoniae belonged to ST23 and harboured an IncH1B virulence plasmid, while the XDR K. pneumoniae belonged to ST147 and possessed two MDR plasmids (IncR and IncFII), the blaOXA-181-bearing ColKP3 plasmid and chromosomal mutations conferring the XDR phenotype. Sequential isolates demonstrated plasmid diversification (fusion of the IncR and IncFII plasmids), mobilization of resistance elements (ompK35 inactivation by ISEcp1-blaCTX-M-15 mobilization, varying numbers of resistance genes on plasmid scaffolds) and chromosomal mutations (mutations in mgrB) leading to further antibiotic resistance that coincided with antibiotic pressure. Importantly, the HMV strain in this study was unable to preserve the carbapenem-resistant phenotype without the selective pressure of meropenem.
Conclusions
To the best of our knowledge, we are the first to report a carbapenem-resistant HMV K. pneumoniae strain in the USA. Ultimately, this case demonstrates the role of antibiotic pressure in the acquisition and loss of important genetic elements.
Introduction
Klebsiella pneumoniae is responsible for significant morbidity and mortality in hospitalized patients.1 Two trends in K. pneumoniae pathogenicity have been apparent for several years. The first is the emergence of hypervirulent variants of K. pneumoniae causing life-threatening, community-acquired and metastatic infections in otherwise healthy patients. This was initially described in and localized to the Asian Pacific Rim. More recently, sporadic cases have been reported worldwide.2 These variants harbour a virulence plasmid bearing the capsular polysaccharide regulator genes (rmpA/A2) and several siderophore gene clusters leading to a hypermucoviscous (HMV) phenotype that can be detected by a positive string test.3,4 The second trend is the acquisition of MDR plasmids in non-HMV or ‘classic’ K. pneumoniae, exemplified by several hospital outbreaks of carbapenemase-producing K. pneumoniae causing high morbidity and mortality.5,6 Rarely are HMV K. pneumoniae isolates also carbapenemase-producing and this occurrence has yet to be observed in the USA.3 We report the molecular characterization of strains isolated over a 3 month period from a complex case of a concurrent disseminated infection with an HMV K. pneumoniae and an OXA-181 XDR K. pneumoniae in a patient recently hospitalized in India. We demonstrate the evolution of strains under selective antibiotic pressure.
Patient and methods
Patient—clinical context
The patient is a 44-year-old man with liver cirrhosis who recently returned to the USA after a 2 month visit to India, which included a 7 day hospitalization for injuries related to a fall. Shortly after returning to the USA, he was hospitalized with fevers and right upper quadrant pain and was found to have a highly susceptible K. pneumoniae bacteraemia and right-sided pneumonia with an associated uncomplicated parapneumonic effusion causing respiratory distress. His parapneumonic effusion was drained via a chest tube and he was treated with intravenous cefepime for 4 days, intravenous ampicillin/sulbactam for 8 days, oral ciprofloxacin for 4 days and oral amoxicillin/clavulanate for 10 days.
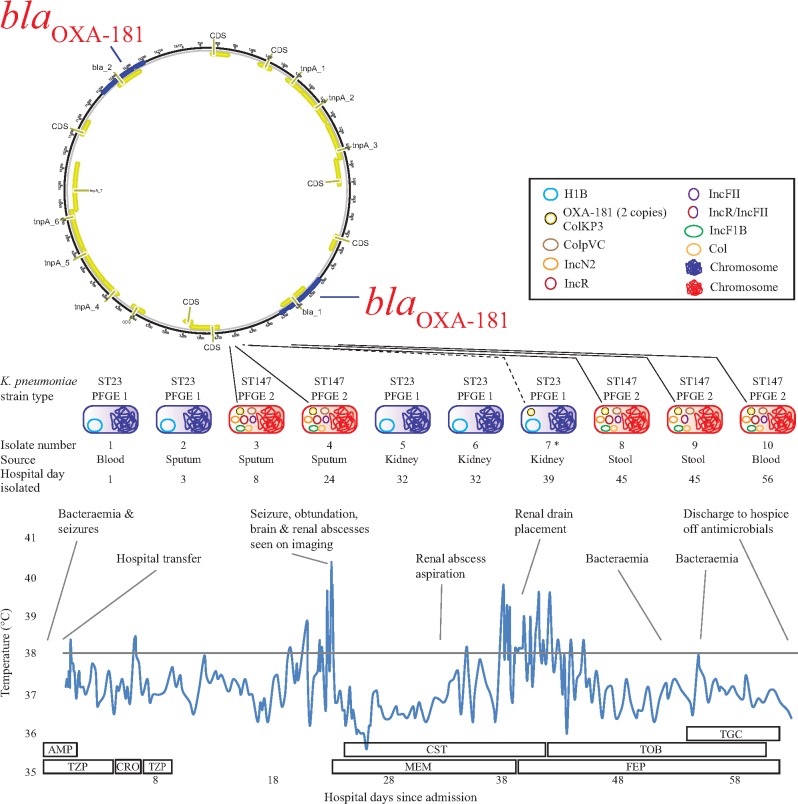
Twenty days after antibiotic cessation, he re-presented with fevers, seizures and respiratory distress. Blood cultures were again positive with a highly susceptible K. pneumoniae. CT imaging of his head demonstrated mild hyperattenuated lesions reported as intraparenchymal haemorrhages in the right thalamus and parietal lobe. On hospital day 3, a K. pneumoniae isolate that tested intermediate to cefazolin, cefoxitin and tetracycline was isolated from the patient’s sputum. On hospital day 8, an XDR K. pneumoniae resistant to all tested antibiotics on the standard antimicrobial susceptibility testing (AST) panel was recovered from a further sputum sample. Due to persistent fevers and seizures, repeat brain imaging was performed. A 3.6 cm brain abscess was found in his right thalamus, which was unamenable to drainage. CT imaging of his abdomen demonstrated a 5.4 cm renal abscess, which was drained. Multiple bacterial cultures from blood, sputum, stool and renal abscess sampled during his admission displayed an unusual pattern of antibiotic susceptibilities, fluctuating from highly susceptible to MDR and XDR (Table 1). In addition, some of the K. pneumoniae isolates were HMV, as defined by positive string test [Table 1 and Figure S1 (available as Supplementary data at JAC Online)]. Due to ongoing clinical deterioration, with XDR K. pneumoniae bacteraemia, despite multiple weeks of antibiotic therapy, the patient was discharged to hospice care. A summary of his clinical course, fever curve, antibiotic coverage and bacterial isolates from his second hospitalization is found in Figure 1. Remarkably, at the time of writing this manuscript, he is living at home and remains infection-free and off antibiotics for over 18 months from the time of his transfer to hospice care. Consent from the patient was obtained to present the case report.
Table 1.
Antibiotic susceptibility profiles and characteristics of the K. pneumoniae strains isolated throughout the patient’s hospital course
| Isolate #, source and hospital day of isolation |
|||||||
|---|---|---|---|---|---|---|---|
| #1 blood (day 1) | #2 sputum (day 3); #5 renal abscess (day 32); #6 renal abscess (day 32) | #3 sputum (day 8); #4 sputum (day 24); #10 blood (day 56) | #7 renal abscess (day 39) | #8 and #9 stool surveillance (day 45); #11 and #12 environmental isolates | E. coli transformant with ColKP3 plasmid bearing OXA-181 | electrocompetent NEB 5-alpha E. coli with no plasmid | |
| String test | positive | positive | negative | positive | negative | negative | negative |
| MLST | ST23 | ST23 | ST147 | ST23 | ST147 | NA | NA |
| mCIM | negative | negative | positive | positive | positive | positive | negative |
| AST results - MIC (mg/L) and breakpoint interpretation or epidemiological cut-off value | |||||||
| ampicillin/sulbactam | 8/4 (S) | 8/4 (S) | >16/8 (R) | >16/8 (R) | >16/8 (R) | >16/8 (R) | 4/2 (S) |
| piperacillin/tazobactam | 4/4 (S) | 4/4 (S) | >64/4 (R) | >64/4 (R) | >64/4 (R) | 32/4 (I) | ≤2/4 (S) |
| cefazolin | ≤1 (S) | 4 (I) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | 2 (S) |
| cefoxitin | ≤4 (S) | 16 (I) | >16 (R) | >16 (R) | >16 (R) | ≤4 (S) | ≤4 (S) |
| ceftriaxone | ≤1 (S) | ≤1 (S) | >32 (R) | 2 (I) | >32 (R) | ≤1 (S) | ≤1 (S) |
| ceftazidime | ≤0.5 (S) | ≤0.5 (S) | >16 (R) | >16 (R) | >16 (R) | ≤0.5 (S) | ≤0.5 (S) |
| cefepime | ≤1 (S) | ≤1 (S) | >16 (R) | ≤1 (S) | >16 (R) | ≤1 (S) | ≤1 (S) |
| aztreonam | ≤2 (S) | ≤2 (S) | >16 (R) | ≤2 (S) | >16 (R) | ≤2 (S) | ≤2 (S) |
| meropenem | ≤0.5 (S) | ≤0.5 (S) | >8 (R) | 4 (R) | >8 (R) | ≤0.5 (S) | ≤0.5 (S) |
| ertapenem | ≤0.25 (S) | ≤0.25 (S) | >1 (R) | >1 (R) | >1 (R) | 1 (I) | ≤0.25 (S) |
| imipenem | 0.25 (S) | 0.25 (S) | >32 (R) | ND | >32 (R) | 0.25 (S) | 0.25 (S) |
| trimethoprim/sulfamethoxazole | ≤0.5/9.5 (S) | 1/19 (S) | >2/38 (R) | >2/38 (R) | >2/38 (R) | ≤0.5/9.5 (S) | ≤0.5/9.5 (S) |
| tetracycline | ≤2 (S) | 8 (I) | >8 (R) | 8 (I) | >8 (R) | ≤2 (S) | ≤2 (S) |
| gentamicin | ≤2 (S) | ≤2 (S) | >8 (R) | ≤2 (S) | >8 (R) | ≤2 (S) | ≤2 (S) |
| tobramycin | ≤2 (S) | ≤2 (S) | >8 (R) | ≤2 (S) | >8 (R) | ≤2 (S) | ≤2 (S) |
| amikacin | ≤8 (S) | ≤8 (S) | >128 (R) | ≤8 (S) | >128 (R) | ≤8 (S) | ≤8 (S) |
| ciprofloxacin | ≤0.5 (S) | 1 (S) | >2 (R) | 2 (I) | >2 (R) | ≤0.5 (S) | ≤0.5 (S) |
| tigecycline | 0.5 (S) | 2 (S) | 2 (S) | 2 (S) | 2 (S) | 0.125 (S) | 0.125 (S) |
| colistin | 0.5 (WT) | 0.5 (WT) | 0.25 (WT) | 0.25 (WT) | >16 (NWT) | 0.06 (WT) | 0.06 (WT) |
| chloramphenicol | 8 (S) | >256 (R) | >256 (R) | >256 (R) | >256 (R) | 16 (I) | 8 (S) |
| ceftazidime/avibactam | 0.125 (S) | 0.125 (S) | 2 (S) | 0.5 (S) | 1 (S) | 0.125 (S) | 0.125 (S) |
S, susceptible; I, intermediate; R, resistant; NWT, non-WT; NA, not applicable; ND, not determined.
Isolates 3, 4 and 8–12 were identified as XDR due to resistance to all antimicrobial categories tested with the exception of polymyxins (colistin; isolates 8, 9, 11 and 12 did acquire resistance and were defined as non-WT) and glycylcyclines (tigecycline). Ceftazidime/avibactam was not included in the definition in accordance with Magiorakos et al.8
Isolate 7 was identified as MDR as the isolate was resistant to at least one agent in nine categories (a minimum of three categories are required to meet the MDR definition).8 The AST profile at the time of obtaining the clinical culture was repeated due to the unusual profile (i.e. reduced susceptibility to certain cephalosporins but resistant to the carbapenems). The AST profile was confirmed upon repeat.
Figure 1.
Summary of the patient’s second hospital course with a timeline of events, fever curve, Gram-negative antibiotics and timing and location of isolates. *For isolate #7, from the kidney abscess drain (day 39), the AST profile was consistent with acquisition of the blaOXA-181-bearing ColKP3 plasmid by the hypervirulent ST23 strain. The AST profile at the time of obtaining the clinical culture was repeated due to the unusual profile (reduced susceptibility to certain extended-generation cephalosporins but resistant to the carbapenems). The AST profile was confirmed upon repeat and the isolate was found to be mCIM positive (positive for carbapenemase production). Unfortunately, by the time WGS was pursued the plasmid was lost in vitro and we were unable to demonstrate acquisition of the plasmid. AMP, ampicillin; CRO, ceftriaxone; CST, colistin; FEP, cefepime; MEM, meropenem; TOB, tobramycin; TGC, tigecycline; TZP, piperacillin/tazobactam.
Methods
Clinical cultures
Clinical specimens were processed following standard operating procedures based on specimen type and included inoculation onto tryptic soy agar with 5% sheep’s blood and MacConkey agar for recovery of Gram-negative organisms. Isolates were identified by MALDI-TOF MS (Bruker Daltonics Inc., Billerica, MA, USA) and AST performed using the BD Phoenix™ Automated Instrument (Becton Dickinson, Sparks, MD, USA). AST for tigecycline and ceftazidime/avibactam was performed using Etests (bioMérieux, Marcy-l’Étoile, France) and AST for colistin was performed using broth macrodilution and results were interpreted based on CLSI or FDA interpretive criteria.7 Isolates were considered MDR if non-susceptible to ≥1 agent in ≥3 antibiotic categories and XDR if non-susceptible to ≥1 agent in all but ≤2 categories.8
String test
The string test was considered indicative of an HMV K. pneumoniae isolate if the inoculation loop was able to generate a viscous string ≥5 mm in length while stretching bacterial colonies away from the agar plate (Figure S1).2
Detection of carbapenemase production
All carbapenem-resistant isolates were further tested using the phenotypic modified Carbapenem Inactivation Method (mCIM), to determine whether carbapenem resistance among the clinical isolates was due to carbapenemase production or non-carbapenemase mechanisms.7,9,10 To determine the genotypes of the carbapenemase-producing isolates, the Check-MDR CT103XL assay (Check-Points, Wageningen, The Netherlands) was performed on isolates positive by the mCIM.11
Infection control surveillance cultures
Additional case-finding strategies included a point prevalence survey of hospitalized patients who had epidemiological contact with the index patient. Rectal, wound, urine and/or sputum surveillance cultures were collected, as per the CDC recommendations, from 33 patients.12 Contact precautions were initiated pending negative surveillance culture results. Additionally, 10 high-touch surfaces from the patient room were sampled using a liquid Amies Elution Swab (Eswab, Copan) and cultured for carbapenem-resistant Enterobacteriaceae (CRE) using the CDC broth enrichment method.13
WGS
WGS was performed using both second-generation Illumina MiSeq (Illumina, San Diego, CA, USA) short-read sequencing and third-generation Oxford Nanopore MinION (Oxford, UK) long-read sequencing technologies. For details regarding the sequencing and analysis methods please see the Supplementary Methods and Tables S1 and S2.
Transformant studies
To determine the phenotype conveyed by acquisition of the blaOXA-181-bearing ColKP3, plasmid transformant studies were performed as previously described.14 Briefly, transformants were obtained by electroporation (Gene Pulser, 1.7 kV, 25 μF, 200 ohms) of 3 μL of total plasmid DNA with 50 μL of electrocompetent NEB 5-alpha Escherichia coli recipient and selected using agar plates containing 100 mg/L ampicillin. AST was performed on the recipient E. coli with and without the plasmid. Confirmation of acquisition of the blaOXA-181 gene was performed by the phenotypic mCIM and by the Check-MDR CT103XL assay, as described above.
Results
Characterization of the K. pneumoniae clinical and environmental isolates
Table 1 and Figure 1 summarize the timeline of the K. pneumoniae isolates acquired during the patient’s hospitalization. The K. pneumoniae isolates recovered from clinical cultures were highly mucoid and appeared phenotypically similar based on growth characteristics, whereas the AST profiles varied from highly susceptible to resistant to almost all antibiotics tested (Table 1). Isolates that demonstrated susceptibility to most antibiotics tested were also string test positive. It was unknown during the patient’s hospitalization whether there were one or multiple strains of K. pneumoniae causing the patient’s infection or whether the isolates were gaining or losing an MDR plasmid depending on selective pressure from the treatment regimens. Also, we were unable to recover both strains from the same source at the same time. It is unclear if this was due to the HMV and XDR strains causing concurrent but separate infections or whether the similar phenotypic appearance resulted in the isolation of one isolate per culture even though both may have been present. XDR isolates were determined to be positive for the carbapenemase blaOXA-48-like and the ESBL blaCTX-M-15 by the Check-MDR CT103XL assay. Of the 10 environmental sites sampled, the countertop of the patient’s room was positive for two XDR K. pneumoniae strains. Fortunately, no transmission events were identified.
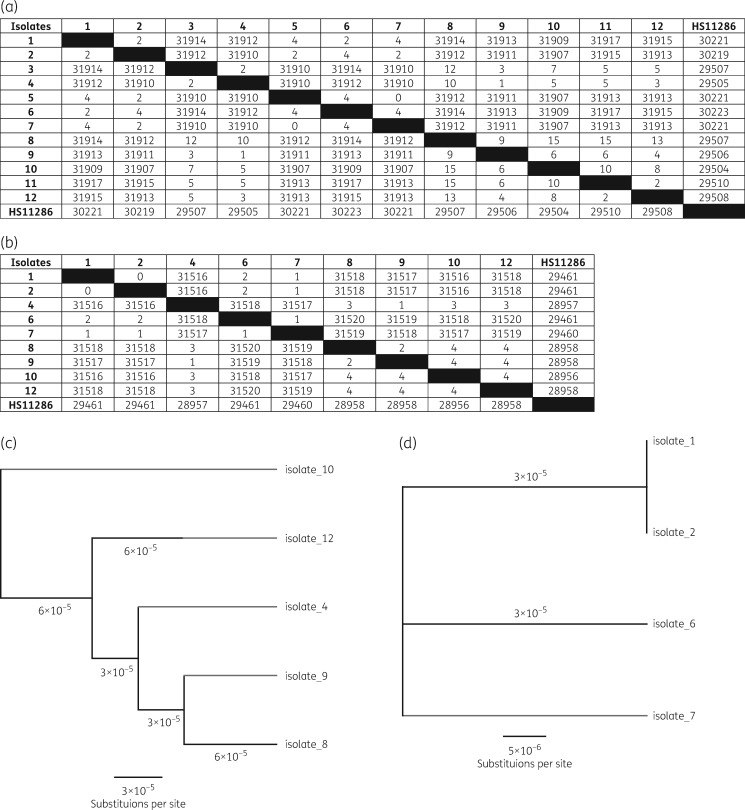
Phylogenetic analysis of strains (MLST and SNP analysis)
WGS analysis revealed that there were two unrelated K. pneumoniae strains isolated from the patient (Figure 2): one strain correlating with the HMV K. pneumoniae ST23 and one strain correlating with the XDR K. pneumoniae ST147. The complete genomes of the HMV K. pneumoniae and the XDR K. pneumoniae were ∼5.4 Mb in size with >99% identity to the HS11286 K. pneumoniae reference genome. The XDR K. pneumoniae was most closely related to K. pneumoniae MS6671, which was isolated from a patient that was hospitalized in the United Arab Emirates and a K. pneumoniae SKGH01 posted to the National Center for Biotechnology Information (NCBI) database by Sheikh Khalifa General Hospital in the United Arab Emirates.15 The HMV K. pneumoniae was most closely related to ED23, an ST23 HMV K. pneumoniae, which was isolated from patients with primary liver abscess and metastatic meningitis in Taiwan (Figure S2).16
Figure 2.
Assessing the genetic relatedness of the K. pneumoniae strains by WGS, including SNP analysis using the SPAdes assembled MiSeq short read data (a; number of bp substitutions); SNP analysis using Pilon MiSeq corrected long read Nanopore data (b) with corresponding scaled phylogenetic trees of the core-genome alignment of the XDR OXA-181-producing K. pneumoniae (c) and the HMV K. pneumoniae (d) strains.
SNP analysis using the assembled short-read MiSeq data only or the hybrid MiSeq corrected long-read Nanopore data for all strains revealed that the HMV ST23 strains evolved by up to 4 SNPs/genome, while the XDR ST147 strains evolved by up to 15 SNPs/genome over the patient’s hospitalization (Figure 2 and Figure S3). Differences in SNP results were observed between MiSeq and Nanopore assemblies as highlighted in Figure 2.
Plasmid analysis—WGS and transformant studies
WGS revealed up to seven circular plasmids carried by the individual strains. The HMV K. pneumoniae strains harboured an IncH1B plasmid (∼230 000 bp) with no associated resistance genes but harboured many of the virulence factor genes (as described below; Table S3). The XDR strains harboured up to seven plasmids including blaOXA-181-bearing ColKP3 (∼13 000 bp), ColpvC (78 000 bp), Col (∼3000 bp), IncF1B (∼118 000 bp), blaCTX-M-15-bearing IncR (∼82 000 bp), IncN2 (55 000 bp) and IncFII (∼100 000 bp) plasmids. The IncR and IncFII plasmids were identified as MDR plasmids and had variable copy numbers of certain resistance genes among the XDR isolates [aac(6ʹ)Ib-cr, rmtF, dfrA14; Table S4]. Furthermore, plasmid diversification was observed by the fusion of the IncR and IncFII plasmids (∼180 000 bp) in isolate #9 where long reads from Nanopore sequencing spanned the fusion areas.
Transformant studies support the likelihood that the HMV strain acquired the ColKP3 plasmid harbouring blaOXA-181 from the XDR K. pneumoniae strain providing a similar resistance profile to the transformant (isolate #7 and transformant; Table 1). The ColKP3 plasmid bearing blaOXA-181 provided elevated MICs of ampicillin/sulbactam, piperacillin/tazobactam, cefazolin and ertapenem.
Detection of antibiotic resistance and virulence genes by WGS
The associated antibiotic resistance genes and their location (i.e. chromosomal versus plasmid-mediated) are summarized in Table S4. Plasmid-mediated antimicrobial resistance genes identified among the XDR strains included the β-lactamase genes blaTEM-1, blaCTX-M-15 and blaOXA-181 and those encoding aminoglycoside [aac(6ʹ)Ib-cr, APH(3ʹʹ)Ib, APH(6)Id, rmtF], rifampicin (arr-2), chloramphenicol (cat, catII), trimethoprim (dfrA12, dfrA14) and fluoroquinolone [aac(6ʹ)Ib-cr, qnrB1] resistance among others. Chromosomally encoded blaSHV11 (narrow-spectrum β-lactamase in K. pneumoniae), oqxA/oqxB (efflux pumps) and fosA5 (fosfomycin resistance) genes were identified among all HMV and XDR isolates. Chromosomal mutations in gyrA and parC encoding fluoroquinolone resistance were observed among XDR strains. Mobilization of blaCTX-M-15 by ISEcp1 resulted in disruption and inactivation of ompK35 in the XDR strains leading to further carbapenem resistance. Mutations in ompK36 were observed among both the HMV and XDR strains. Lastly, while under the selective pressure of colistin, the XDR strains developed colistin resistance via a 2 bp insertion of adenine and guanine at position 120 of the mgrB gene, a negative regulator of the phoPQ two-component regulatory system. Mutations in mgrB result in constitutive expression of LPS-modifying genes yielding colistin resistance.
Virulence factor genes are summarized in Table S3. The HMV strains harboured multiple virulence genes for iron-scavenging (aerobactin, salmochelin and yersiniabactin), for the K1 capsular type and for the HMV phenotype (rmpA/A2) where the majority were harboured on the H1B plasmid. In contrast, the XDR isolates harboured few virulence factors (yersiniabactin and a type III fimbrial adhesion) that were chromosomally encoded.
Discussion
We report a case of a patient with a concurrent disseminated infection with a highly virulent HMV Klebsiella pneumoniae and an OXA-181-producing XDR K. pneumoniae with recent hospitalization in India. The co-acquisition of the HMV and XDR strains most likely occurred while the patient was hospitalized in India. This is concerning as HMV isolates have a propensity to cause abscesses and metastatic infections among both healthy and immunocompromised individuals,2 and isolates harbouring blaOXA-181 are often resistant to most available antibiotics.17 The combination of these two traits is a significant public health concern.
To date, there are sporadic reports in the USA of OXA-48-like carbapenemase-producing Enterobacteriaceae and most have been associated with exposure to the healthcare setting outside the USA, in particular the Middle East or South Asia, as seen in this case.17 Reports of HMV strains have generally been isolated to Asia but are increasingly reported worldwide.2,3 Until recently, the majority of HMV K. pneumoniae strains have been described as susceptible to most antibiotics, with reports of ESBL-positive and occasional carbapenem-resistant HMV K. pneumoniae strains (mostly KPC producers) localized to China with a few cases from India and South America.3,18–22 Recently, there was a fatal outbreak of ST11, hypervirulent KPC-2-producing K. pneumoniae reported from a Chinese hospital.4 To date, there is only one previous report of two HMV isolates acquiring a blaOXA-48-like gene.23 To the best of our knowledge, this is the first report of a carbapenem-resistant HMV K. pneumoniae in the USA.
In the case of our patient, their highly susceptible HMV K. pneumoniae strain became resistant to carbapenems while sparing most of the extended-generation cephalosporins, providing an AST profile consistent with the acquisition of the blaOXA-181 gene harboured on the ColKP3 plasmid. Zhang et al.22 published a similar phenomenon of HMV K. pneumoniae strains acquiring blaKPC-2 under the selective pressure of imipenem exposure. Unfortunately, we were unable to demonstrate acquisition of the plasmid by WGS as the plasmid was lost on further subculture and had reverted to the susceptible phenotype by the time follow-up studies were completed. Transformant studies did support the likelihood that the strain acquired the ColKP3 plasmid harbouring blaOXA-181 from the concurrent XDR K. pneumoniae strain providing a similar resistance profile to the transformant. Potron et al.24 previously demonstrated the association of blaOXA-181 with the small ColKP3 plasmid scaffold and demonstrated the broad host-range specificity of the plasmid through successful transformant studies with Pseudomonas aeruginosa.
Importantly, the HMV strain in this study was unable to preserve the resistance phenotype without the selective pressure of meropenem and it lost the plasmid on further subcultures. Similarly, Huang et al.25 demonstrated the in vitro loss of blaNDM-1 by K. pneumoniae with the removal of selective pressures, which occurred through reduced copy number of the blaNDM-bearing pKPX-1 plasmid or the loss of blaNDM-1 via directed repeat-mediated slippage. Another study demonstrated the possible in vivo loss of blaNDM in K. pneumoniae through a 5 kb deletion on the blaNDM-7-harbouring IncX3 plasmid.26 These studies suggest that there can be a negative impact on the fitness of K. pneumoniae if they harbour certain carbapenemase genes and/or carbapenemase-encoding plasmids and that acquisition of these genes can be relatively unstable in the absence of selective pressure. The instability may be linked to many factors including the K. pneumoniae strains, plasmid incompatibility groups and/or specific carbapenemase genes. Nevertheless, environments allowing for prolonged selective pressures may result in the further evolution of these strains to become fit to harbour carbapenemase genes in the absence of selective pressures. This case provides a strong argument for the judicious use of antibiotics in order to reduce the development and spread of antibiotic resistance. This is further supported by the remarkable recovery of the patient while off antibiotics as they were discharged to hospice care due to the poor prognosis given repeated bloodstream infections.
The XDR phenotype among strains was linked to two MDR plasmids (IncR and IncFII), the blaOXA-181-bearing plasmid (ColKP3) and chromosomal mutations (ompK35, ompK36, gyrA, parC, ±mgrB). The hypervirulent HMV phenotype was linked to an IncH1B plasmid harbouring many of the associated virulence factor genes including the capsular polysaccharide regulator genes responsible for the HMV phenotype (rmpA/A2) and multiple iron-scavenging systems (aerobactin, salmochelin and yersiniabactin) among other chromosomally located virulence factors. In light of the multiple virulence factors associated with this patient’s HMV K. pneumoniae isolate, it is not surprising that they had an aggressive infection with multi-organ involvement. The concurrent infection with both an HMV isolate and an XDR K. pneumoniae can make for a highly virulent combination of epidemiological concern, as was observed in our patient.
WGS was applied to all isolates recovered throughout the 3 month hospital course to demonstrate the changes that occurred in the resistome over time. WGS revealed plasmid diversification (fusion of the IncR and IncFII plasmids), mobilization of resistance elements (ompK35 inactivation by ISEcp1-blaCTX-M-15 mobilization, varying numbers of resistance genes on plasmid scaffolds) and chromosomal mutations (mutations in mgrB leading to colistin resistance) leading to further antibiotic resistance. For the most part, these antibiotic resistance changes could be linked to selective antibiotic pressure (i.e. the likely acquisition of the blaOXA-181 by the HMV strain during exposure to meropenem and chromosomal mutations in mgrB during exposure to colistin). This case demonstrates the evolution of the resistome in these strains under selective antibiotic pressure.
Tracing the mutational profile revealed that the isolates varied by up to 15 SNPs in the genome between related strains, while differences in results were observed when interpreting MiSeq versus Nanopore data. As previously suggested by Yang et al.,27 these differences in results and interpretation of SNPs support the need for the development of standards for utilizing SNP analysis for epidemiological and diversification assessments of strains. Furthermore, SNP analysis revealed the potential source of isolates, linking clinical isolates recovered from this patient as the likely source of environmental contamination [i.e. the sputum isolate was most closely related (two SNP differences) to the environmental isolate]. WGS has been demonstrated as a powerful tool in outbreak investigations allowing linking of patients and mapping transmission events.27,28 We further demonstrate the power of WGS both as an epidemiological tool but also as a tool to track evolution of antibiotic resistance among strains undergoing selective antibiotic pressure.
In summary, we present a complex case of a concurrent infection with an HMV K. pneumoniae and an XDR OXA-181-producing K. pneumoniae from a patient with prior hospitalization in India. Through WGS, we demonstrate the changes that occurred in the resistome of the isolates were linked to the selective pressures of treatment. Ultimately, this case demonstrates the role of antibiotic pressure in the acquisition and loss of important genetic elements leading to XDR bacteria. Furthermore, we demonstrate the power of WGS as a tool to track antibiotic resistance among strains undergoing selective antibiotic pressure.
Supplementary Material
Acknowledgements
We would like to acknowledge: Tracy Howard, Ambinbola Thompson, Krizia Chambers, Ava Roberts, Christos Galanis and Gyanu Lamichhane for their assistance with characterization of the isolates; Meklit Workneh for assistance with the clinical management of the patient; and Melanie Curless and Lisa Maragakis for their assistance with infection prevention management.
Funding
The work was supported by funding from the Sherrilyn and Ken Fisher Center for Environmental Diseases (P. J. S.), the National Institutes of Health (R21AI130608 awarded to P. J. S., 1K23AI127935 awarded to P. D. T. and R01-HG006677 awarded to M. C. S.) and the National Science Foundation (DBI-1350041 awarded to M. C. S.).
Transparency declarations
P. J. S. and W. T. have received travel funds from Oxford Nanopore to speak at their user group meetings. In addition, W. T. has two patents (US20110226623 A1 and US20120040343 A1) that are licensed to Oxford Nanopore. All other authors: none to declare.
Supplementary data
Figures S1 to S3, Supplementary Methods and Tables S1 to S4 are available as Supplementary data at JAC Online.
References
- 1. Weiner LM, Webb AK, Limbago B. et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol 2016; 37: 1288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shon AS, Bajwa RP, Russo TA.. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 2013; 4: 107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arena F, Henrici De Angelis L, D'Andrea MM. et al. Infections caused by carbapenem-resistant Klebsiella pneumoniae with hypermucoviscous phenotype: a case report and literature review. Virulence 2017; 8: 1900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gu D, Dong N, Zheng Z. et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 2017; 18: 37–46. [DOI] [PubMed] [Google Scholar]
- 5. Rock C, Curless MS, Cantara M. et al. Resolution of carbapenemase-producing Klebsiella pneumoniae outbreak in a tertiary cancer center; the role of active surveillance. Infect Control Hosp Epidemiol 2017; 38: 1117–9. [DOI] [PubMed] [Google Scholar]
- 6. Snitkin ES, Zelazny AM, Thomas PJ. et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012; 4: 148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27. CLSI, Wayne, PA, USA, 2017. [Google Scholar]
- 8. Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 9. Simner PJ, Gilmour MW, DeGagne P. et al. Evaluation of five chromogenic agar media and the Rosco Rapid Carb screen kit for detection and confirmation of carbapenemase production in Gram-negative bacilli. J Clin Microbiol 2015; 53: 105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasoo S, Cunningham SA, Kohner PC. et al. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol 2013; 51: 3097–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuzon G, Naas T, Bogaerts P. et al. Evaluation of a DNA microarray for the rapid detection of extended-spectrum β-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J Antimicrob Chemother 2012; 67: 1865–9. [DOI] [PubMed] [Google Scholar]
- 12. Sehulster L, Chinn RY.. Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). MMWR Recomm Rep 2003; 52: 1–42. [PubMed] [Google Scholar]
- 13. CDC. Laboratory Protocol for Detection of Carbapenem-Resistant or Carbapenemase-Producing, Klebsiella spp. and E. coli from Rectal Swabs.https://www.cdc.gov/HAI/pdfs/labSettings/Klebsiella_or_Ecoli.pdf.
- 14. Simner PJ, Zhanel GG, Pitout J. et al. Prevalence and characterization of extended-spectrum β-lactamase- and AmpC β-lactamase-producing Escherichia coli: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011; 69: 326–34. [DOI] [PubMed] [Google Scholar]
- 15. Zowawi HM, Forde BM, Alfaresi M. et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 2015; 5: 15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin HH, Chen YS, Hsiao HW. et al. Two genome sequences of Klebsiella pneumoniae strains with sequence type 23 and capsular serotype K1. Genome Announc 2016; 4: e01097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lyman M, Walters M, Lonsway D. et al. Notes from the field: carbapenem-resistant Enterobacteriaceae producing OXA-48-like carbapenemases—United States, 2010-2015. MMWR Morb Mortal Wkly Rep 2015; 64: 1315–6. [DOI] [PubMed] [Google Scholar]
- 18. Andrade LN, Vitali L, Gaspar GG. et al. Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol 2014; 52: 2530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cejas D, Fernández Canigia L, Rincόn Cruz G. et al. First isolate of KPC-2-producing Klebsiella pneumoniae sequence type 23 from the Americas. J Clin Microbiol 2014; 52: 3483–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Li XY, Wan LG. et al. Virulence and transferability of resistance determinants in a novel Klebsiella pneumoniae sequence type 1137 in China. Microb Drug Resist 2014; 20: 150–5. [DOI] [PubMed] [Google Scholar]
- 21. Yao B, Xiao X, Wang F. et al. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis 2015; 37: 107–12. [DOI] [PubMed] [Google Scholar]
- 22. Zhang R, Lin D, Chan EW. et al. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 2016; 60: 709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shankar C, Nabarro LE, Devanga Ragupathi NK. et al. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announc 2016; 4: e01081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Potron A, Nordmann P, Lafeuille E. et al. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 2011; 55: 4896–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang TW, Chen TL, Chen YT. et al. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One 2013; 8: e62774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lynch T, Chen L, Peirano G. et al. Molecular evolution of a Klebsiella pneumoniae ST278 isolate harboring blaNDM-7 and involved in nosocomial transmission. J Infect Dis 2016; 214: 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang S, Hemarajata P, Hindler J. et al. Evolution and transmission of carbapenem-resistant Klebsiella pneumoniae expressing the blaOXA-232 gene during an institutional outbreak associated with endoscopic retrograde cholangiopancreatography. Clin Infect Dis 2017; 64: 894–901. [DOI] [PubMed] [Google Scholar]
- 28. Sievert DM, Ricks P, Edwards JR. et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol 2013; 34: 1–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.