Abstract
Autosomal recessive polycystic kidney disease (OMIM 263200) is a serious condition of the kidney and liver caused by mutations in a single gene, PKHD1. This gene encodes Fibrocystin/Polyductin (FPC, PD1), a large protein shown by in vitro studies to undergo Notch-like processing. Its cytoplasmic tail, reported to include a ciliary targeting sequence, a nuclear localization signal and a polycystin-2 binding domain, is thought to traffic to the nucleus after cleavage. We now report a novel mouse line with a triple HA-epitope “knocked-in” to the C-terminus along with lox P sites flanking exon 67, which encodes most of the C-terminus (Pkhd1Flox67HA). The triple HA-epitope has no functional effect as assayed by phenotype and allows in vivo tracking of Fibrocystin. We used the HA tag to identify previously predicted Fibrocystin cleavage products in tissue. In addition, we found that Polycystin-2 fails to co-precipitate with Fibrocystin in kidney samples. Immunofluorescence studies with anti-HA antibodies demonstrate that Fibrocystin is primarily present in a sub-apical location in kidney, biliary duct and pancreatic ducts, partially overlapping with the Golgi. In contrast to previous studies, the endogenous protein in the primary cilia was not detectable in mouse tissues. After Cre-mediated deletion, homozygous Pkhd1Δ67 mice are completely normal. Thus, Pkhd1Flox67HA is a valid model to track Pkhd1-derived products containing the C-terminus. Significantly, exon 67 containing the nuclear localization signal and the polycystin-2 binding domain is not essential for Fibrocystin function in our model.
Keywords: Autosomal Recessive Polycystic Kidney Disease, Fibrocystin, PKHD1 gene, protein cleavage, mouse model
INTRODUCTION
Autosomal recessive polycystic kidney disease (ARPKD) (OMIM 263200) is an often severe disorder that affects 1 in 20,000 live births1. It is a complex disease that presents with a wide range of clinical manifestations including enlarged echogenic kidneys primarily due to dilatation of collecting ducts, cystic proliferation of biliary ducts and congenital hepatic fibrosis1–3. Respiratory failure due to pulmonary hypoplasia is a leading cause of neonatal mortality, affecting 30–40%4. While congenital hepatic fibrosis is an invariant finding in ARPKD, renal manifestations are highly variable, being most severe in neonatal disease but milder in patients that reach adulthood2,5–9.
Mutations of a single gene, PKHD1, are responsible for all typical forms of the disease10,11. The gene extends over ~500kb, and its longest open reading frame is encoded by a 67-exon transcript that produces a 4074aa protein, fibrocystin/polyductin (FPC/PD1). The gene is reported to undergo a complex pattern of tissue-specific splicing with several alternative exons but the significance of this phenomenon is uncertain since mutations have thus far been exclusively found in the core 67 exons10,12,13. Mutations have been identified across the length of the gene, with two truncating mutations associated with a more severe phenotype than other mutational patterns2,9,13–15.
FPC is an approximately 500kDa protein with a long extracellular N-terminal region, a single transmembrane span and an intracellular cytoplasmic domain (ICD)10,11,16. It appears to be a ciliary protein, though it also has been localized to the basal body and the plasma membrane17–19. While FPC’s structure and its extracellular motifs suggest that it functions as a receptor, its ligand(s) remains unknown.
Previous in vitro studies of over-expressed recombinant, epitope-tagged human FPC showed that it undergoes Notch-like processing with multiple post-translational proteolytic steps20,21. The extracellular domain (PECD) is first cleaved by a likely pro-protein convertase and it then remains tethered to the stalk by disulfide bridges. The entire ectodomain undergoes regulated shedding, reportedly mediated by a metalloprotease, with gamma-secretase dependent release of an intracellular domain (ICD) that translocates to the nucleus where it may regulate transcriptional pathways20,21.
The ICD, encoded mostly by Pkhd1 exon 67, is reported to have several functional motifs including a ciliary targeting sequence (CTS), a nuclear localization signal (NLS) and a polycystin-2 (PC2) binding domain (PC2BD)21–26. PC2 is the protein encoded by the PKD2 gene, which is linked to the less common form of human autosomal dominant polycystic kidney disease27,28. The interaction between FPC and PC2 is reported to regulate the latter’s channel activity22. However, the functional relationship between PC2 and FPC has been questioned given the very different phenotypes associated with loss of either protein29. Previous studies, however, have reported a genetic interaction in mice between Pkhd1 and either Pkd1 or Pkd2 suggesting that FPC and the polycystins may cooperatively modulate signaling pathways25,30,31.
Whether Notch-like processing of FPC occurs in vivo is still uncertain. Bakeberg et al have reported detection of a 450kDa product in exosome-like vesicles that decreases to ~390kDa after de-glycosylation, which they conclude is likely the shed ectodomain32. However, biochemical analyses of samples from both wild type mice and a mouse line with a double V5-tag knocked into exon 3 suggest that the uncleaved 500kDa product is the predominant form present in vivo17,18,32. None of the studies report detection of the cleaved N-terminal fragment tethered to a C-terminal stalk in vivo or shorter C-terminal fragments.
Genetically modified mice are invaluable tools for elucidating the function of a gene and the protein it encodes in a physiological setting. To better track the C-terminal ICD fragment of FPC and to determine its physiological role “in vivo”, we targeted exon 67 and the 3’UTR of the Pkhd1 gene. We introduced Lox P sites into intron 66 and adjacent to the 3’UTR. In addition, we knocked a triple HA epitope into the C-terminus of FPC. Using this model, we find that the C-terminus of FPC can be detected in vivo. We also unexpectedly discovered that mice lacking exon 67, which encodes the NLS and PC2BD are completely normal.
RESULTS
Generation of a conditional Pkhd1Flox67HA knock-in mouse
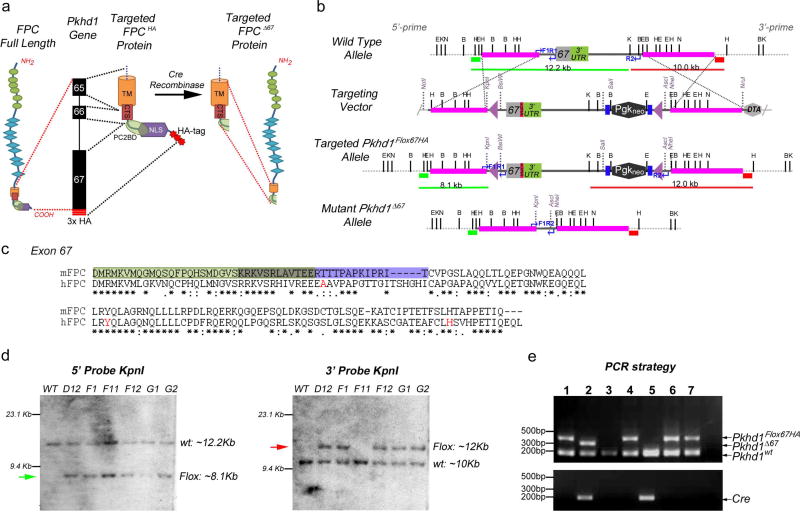
To investigate the functional role of the C-terminal tail of FPC we used a homologous recombination gene targeting strategy to flox exon 67 and to introduce a triple HA epitope-tag into the mouse Pkhd1 gene locus (Pkhd1Flox67HA) (Figure 1a–e). To make the targeting vector, three HA-tags were cloned in frame at the end of Pkhd1 exon 67, just prior to the stop codon (Figure 1a and b). The HA tags allow in vivo tracking of full length FPC (FPC-HA) and any cleavage fragments containing the C-terminus of the protein. A neomycin cassette, flanked by two FRT sites, was added after the 3′ UTR and LoxP sites were inserted into intron 66 and adjacent to the neomycin cassette (Figure 1b and d). This design allows Cre-mediated deletion of the C-terminal 137 amino acids of FPC (Figure 1a). This segment harbors the nuclear localization signal (NLS) and the PC2-binding site (PC2BD) and is a site for pathogenic mutations in humans (Figure 1a and c)9,33,34 and RWTH Aachen University PKHD1 mutation database, http://www.humgen.rwth-aachen.de/index.php).
Figure 1. Gene-targeting strategy used to generate a floxed HA-tagged allele for murine Pkhd1 (Pkhd1Flox67HA).
(a) Fibrocystin/Polyductin (FPC) Protein Targeting Strategy. The location of C-terminal protein domains within Pkhd1 exons 65–67 are indicated. The targeted FPC protein includes three HA-tags (red bars or stars) at its C-terminus. Cre recombinase mediated deletion results in loss of the targeted intracellular domain (ICD) containing the nuclear localization signal (NLS), the polycystin 2 binding domain (PC2BD) and the HA tags. The transmembrane domain (TM) and the ciliary targeting sequence (CTS) remain intact. (b) Pkhd1 Gene Targeting Strategy. Genomic maps of the wild-type Pkhd1 allele, the targeting vector, the Pkhd1Flox67HA allele after homologous recombination and following Cre-mediated deletion (Pkhd1Δ67) are indicated. Three HA-tags (red bar) were inserted in frame after exon 67 and before the stop codon. A neomycin cassette flanked by two FRT sites (blue bars) was cloned adjacent to the 3′ UTR and LoxP sites (purple triangles) were inserted into intron 66 and outside of the 3′ UTR. Neomycin was used for positive selection and DTA for negative selection of ES cells. The pink bars represent homology arms while the green and red bars indicate the 5’ and 3’ probes, respectively, that were used for Southern blot analysis. The blue arrows indicate the PCR primers developed for genotyping. Single letters identify the following restriction sites: B, BglII; E, EcoRV; H, HindIII; X, XbaI, K, Kpnl and N, Notl. (c) Alignment of murine FPC exon 67 (137 amino acids) with the homologous region of human FPC. The light green box indicates the putative PC2 binding domain that overlaps (olive box) with the nuclear localization signal (purple box). The amino acid residues in red are affected by pathogenic mutations in ARPKD patients. Asterisks indicate conserved residues. (d) Autoradiographs of Southern blots prepared from targeted ES cell clones (D12, F1, F11, F12, G1 and G2) digested with KpnI. Blots were hybridized to 5’ (green bar) and 3’ (red bar) probes in Panel B. Fragment sizes that correspond to those predicted for the wild type and targeted or floxed (green and red arrows) allele are shown. Clones D12, F1, F12, G1 and G2 yielded fragments of the correct sizes using both probes. (e) Three-primer PCR genotyping strategy that distinguishes 3 alleles Pkhd1wt, Pkhd1Flox67HA and Pkhd1Δ67. The location of primers and the size of expected bands are indicated in Panel b (Pkhd1wt 188bp, Pkhd1Flox67HA 320bp and Pkhd1Δ67 268bp). Cre recombinase specific PCR is shown in the bottom panel. Pkhd1Flox67HA heterozygotes were bred with Meox2tm1(Cre)Sor/J mice to delete Pkhd1 exon 67. Four animals (Lanes 1, 4, 6 and 7) were Pkhd1Flox67HA/WT compound heterozygotes. The Pkhd1Δ67 band is not seen since the animals are Cre negative. Two animals (Lanes 3 and 5) were Pkhd1wt homozygotes. The animal in lane 2, which is Cre positive, has the genotype Pkhd1Δ67/WT after deletion of exon 67.
Once we obtained germ line transmission of the targeted allele, we deleted PKHD1 exon 67 by breeding Pkhd1Flox67HA mice with a Meox2-Cre transgenic line (Meox2tm1(cre)Sor/J) to produce Pkhd1Δ67 offspring35. Cre-recombinase mediated deletion also results in loss of the HA tag. We devised a single PCR genotyping strategy that allowed us to distinguish 3 products corresponding to the untagged, wild type Pkhd1 (188bp, Pkd1wt), HA-tagged Pkhd1 (320bp, Pkhd1Flox67HA) and exon 67 deleted Pkhd1 (268bp, Pkhd1Δ67) (Figure 1e).
Pkhd1Flox67HA mice can be used to study native FPC
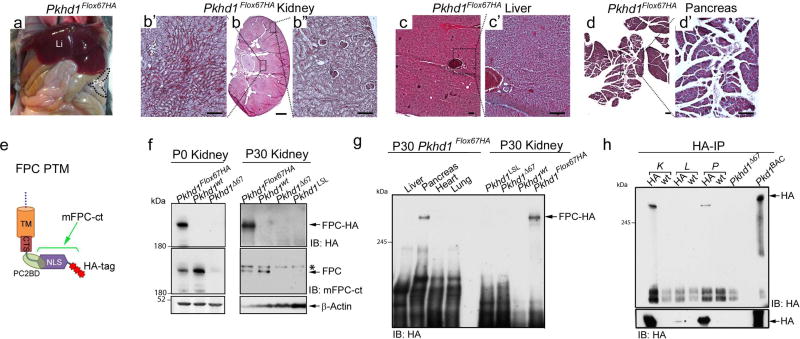
Pkhd1Flox67HA homozygous mice were viable and fertile with normal behavior throughout postnatal and adult life (Supplementary Table 1). Renal, hepatic and pancreatic histology from adult Pkhd1Flox67HA mice was indistinguishable from that of Pkd1wt control littermates (Figure 2a–d) suggesting that epitope tagging of FPC at its C-terminus does not have overt functional consequences.
Figure 2. Characterization of Pkhd1Flox67HA mice.
(a) Gross anatomy of a 6-month old Pkhd1Flox67HA mouse showing normal liver and pancreas. Li: liver, Pa: pancreas. (b–d) Representative histologic sections from kidney (b, b’, b”), liver (c, c’) and pancreas (d, d’) derived from a 6-month old Pkhd1Flox67HA mouse. The black dotted lines and squares indicate corresponding enlarged areas. Scale bars represent 1mm in panel b (magnification 1×) and 200µm in panels b’, b”, c, c’, d and d’. Magnification: c, d 4 ×, Magnification b, b’, b”, c’, d’ 10 ×. (e) Schematic representation of the FPC C-terminal transmembrane fragment (FPC PTM) including the transmembrane domain (TM), the ciliary targeting sequence (CTS), the polycystin-2 binding domain (PC2BD) the nuclear localization signal (NLS) and 3 HA tags. The epitope recognized by the mFPC-ct antibody is contained within the bracketed region. (f) Western blot analysis of total lysates prepared from murine kidneys of the indicated genotypes at post-natal day 0 (P0) and 30 (P30). Western blots were probed with antibodies against HA (upper panel), mFPC-ct (middle panel) and actin as a loading control (lower panel). The asterisk indicates a non-specific band in this blot recognized by the mFPC-ct antibody. (g) Western blot survey of total lysates prepared from adult Pkhd1Flox67HA tissues probed with anti-HA. Total lysates from Pkhd1LSL, Pkhd1Δ67 and Pkhd1wt kidneys serve as negative controls. (h) Anti-HA Affinity Matrix was used to Immunoprecipitate (IP) HA tagged FPC from adult kidney (K), liver (L), and pancreas (P) (upper panel). Western blots were prepared and probed with anti-HA antibodies. IP from Pkd1BAC-HA lungs and Pkhd1Δ67 kidneys serve as positive and negative controls, respectively. Overnight exposure of the blot (lower panel) demonstrates liver expression of FPC.
We took advantage of the C-terminal HA tag to survey FPC expression. We prepared total lysates from murine tissues and probed Western blots with anti-HA antibody. In neonatal and adult kidney as well as in adult pancreas, we detected a band of ~500kDa, compatible with the predicted size of full-length tagged FPC (FPC-HA) as well as a few fainter, less consistently detected high molecular weight bands ranging in size from 180kDa to just under 500kDa in P0 kidney (Figure 2e–g). Anti-HA did not detect tagged FPC in negative controls lacking HA, including wild type (Pkhd1wt) and Pkhd1Δ67 mice where Cre recombinase deleted the HA tag. A new rat monoclonal antibody generated against the C-terminal region of FPC (mFPC-ct) recognized a ~500kDa protein, expressed at comparable levels, in Pkhd1wt and Pkhd1Flox67HA kidneys (Figure 2e and f). The mFPC-ct antibody did not detect FPC in either Pkhd1Δ67 or Pkhd1LSL kidneys indicating that this antibody is specific (Figure 2f). Pkhd1LSL is a previously described null allele that produces no Pkhd1 mRNA32.
We used immunoprecipitation (IP) with anti-HA affinity agarose beads followed by immunoblotting with anti-HA to enhance detection of FPC-HA. Using this enrichment strategy, we not only identified FPC-HA in pancreas and kidney but also in the liver (Figure 2h and Supplementary Figure S1a and b). Low hepatic FPC levels are likely due to restricted expression of FPC in cholangiocytes, which make up a small fraction of the cell types found in the liver. FPC-HA could not be detected in adult heart or lung, indicating that FPC is either absent or present in very low abundance (Supplementary Figure S1a). In some gels FPC resolved as a doublet, as has been previously reported (Figure 2h and Supplementary Figure S1a and b)32. Immunoprecipitation with anti-HA was negative in tissues from all mice lacking FPC-HA (Figure 2h and Supplementary Figure S1a and b).
FPC undergoes Proteolytic Cleavage in vivo
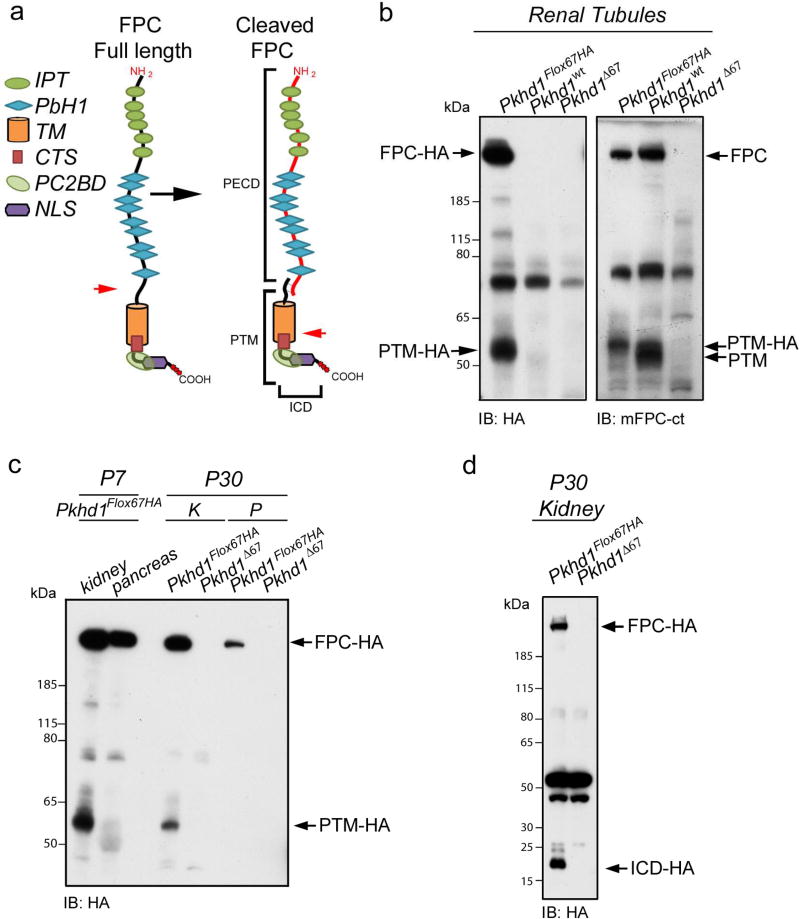
Several studies have demonstrated that FPC undergoes proteolytic cleavage when over expressed in cell culture (Figure 3a)20,21. However, FPC protein processing has never been confirmed in vivo due to a dearth of antibodies targeting the C-terminus of the protein. We prepared lysates from freshly isolated renal tubules and total kidney using a sucrose containing lysis buffer. Western blots were probed with anti-HA or mFPC-ct antisera (Figure 3b–d). Both antibodies detected a specific band of ~55kDa in Pkhd1Flox67HA renal tissue that was absent in the appropriate negative control samples (Figure 3b and c). This band corresponds to the predicted PTM (PD1 transmembrane fragment) that was reported to result from cleavage at a pro-protein convertase site. The PTM detected by mFPC-ct in Pkhd1wt tubules is slightly smaller (~51KDa) due to the absence of the 3X-HA tag. We did not observe a PTM fragment in the pancreas.
Figure 3. FPC is cleaved in vivo.
(a) Schematic representation of full length FPC (left) and cleaved FPC (right) with proposed cleavage sites indicated by red arrows. Cleavage of full length FPC at a probable proprotein convertase site produces a large extracellular domain (PECD) that is tethered to a C-terminal fragment (PTM) by disulfide bridges. The PTM is further cleaved by a γ-secretase to release an intracellular C-terminal fragment (ICD) containing the putative nuclear localization signal (NLS). IPT: Ig like, plexins, transcripton factor domain PbH1: Parallel beta-helix repeat, TM: transmembrane segment (b) Western blots were prepared from total lysates derived from renal tubules isolated at P7. Blots were probed with anti-HA (left panel) or mFPC-ct (right panel). Anti-HA detects full-length FPC (FPC-HA, ~500KDa) and another FPC related band at ~55KDa that corresponds with the predicted PTM (PTM-HA). These bands were present only in Pkhd1Flox67HA but not in Pkhd1wt or Pkhd1Δ67 tubules. The mFPC-ct antibody also detected full-length FPC and the PTM in Pkhd1Flox67HA and Pkhd1wt tubules but not in Pkhd1Δ67 tubules. (c) Western blots of total lysates prepared from Pkhd1Flox67HA and Pkhd1Δ67 adult kidney (K) and pancreas (P), probed with anti-HA. Anti-HA detects FPC-HA and PTM-HA only in Pkhd1Flox67HA tissues. The PTM is absent in pancreas. (d) Western blots prepared from total adult kidney lysates run on 4–12% Bis-Tris acetate gels to enhance detection of small proteins. Anti-HA detects FPC-HA and a small 17KDa fragment harboring that corresponds with the size of the cleaved C-terminal ICD product (ICD-HA). The PTM isn’t visualized because it is presumably obscured by a non-specific band just above 50KDa
To resolve small fragments, we ran total lysates on 4–12% Bis-Tris acetate gels. Anti-HA detected a fragment of about 17 kDa in total kidney and in DBA positive cells isolated from Pkhd1Flox67HA kidneys (Figure 3d and Supplementary Figure S1c). This band was not found in either Pkhd1wt or Pkhd1Δ67 tissues lacking FPC-HA. This band is similar in size to the ICD fragment that has been previously observed in over expression systems and which is thought to be the result of γ-secretase dependent cleavage20,21. We found that detection of PTM and ICD was highly dependent on the conditions that were used for tissue preparation, including the use of freshly isolated tissue and the composition of lysis buffer. The PTM was best visualized when tissues were lysed in a sucrose based buffer whereas the ICD could best be resolved when a triton based buffer was employed.
FPC Fragments are Shed in Urine
It has been previously reported that the FPC extracellular PECD fragment is present in whole urine and urinary exosome-like vesicles (ELVs)32. To better define which FPC fragments are found in urine, we prepared urinary Western blots from several FPC mouse models (Supplementary Figure S1d and e). We confirmed that FPC PECD was detected by anti-V5 in unprocessed urine as well as in ELVs from mice bearing a 5’-V5 tag (Pkhd1Pk(+)). In contrast anti-HA detected a clear PTM band only in ELVs but not in unprocessed urine from Pkhd1Flox67HA mice (Supplementary Figure S1d and e). These results suggest that there are two different classes of FPC molecules in urine, a highly abundant free PECD and a second less abundant PECD that is either tethered to or shed with the PTM in ELVs.
Cell type-specific expression of Endogenous FPC
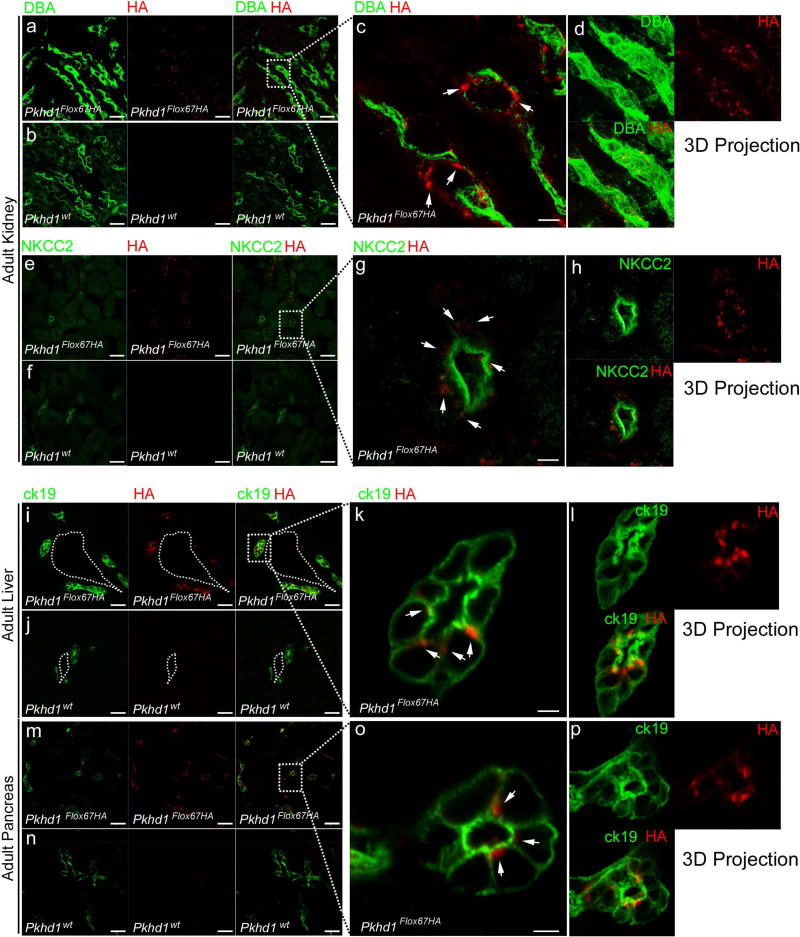
To localize endogenous FPC we co-stained frozen kidney sections from neonatal and adult Pkhd1Flox67HA mice and controls with anti-HA antibody and segment-specific markers (Figure 4a–h, Supplementary Figure S2 and Supplementary Figure S3). In neonatal mice (P0), we observed robust anti-HA staining in Dolichos biflorus agglutinin (DBA) positive tubules (Supplementary Figure S3a–d) but also faint staining in some LTL positive renal segments (Supplementary Figure S2e–h). In adult mice, however we detected anti-HA staining only in DBA and Na-K-Cl cotransporter 2 (NKCC2) positive segments (Figure 4a–h) but not in LTL expressing tubules (Supplementary Figure S2a–d). Taken together, our data show that FPC is expressed primarily in the distal tubule and thick ascending limb of the loop of Henle but there appears to be low-level patchy expression in the proximal tubule before renal development is complete at P0.
Figure 4. Immunolocalization of endogenous FPC-HA in adult kidney, liver and pancreas.
(a–h) Adult kidney cryosections from Pkhd1Flox67HA and Pkhd1wt mice stained with anti-HA (red), Dolichos biflorus agglutinin (DBA) (green in panels a–d) and anti-Na-K-Cl cotransporter 2 (NKCC2), (green in panels e–h). (i–p) Cryosections from Pkhd1Flox67HA and Pkhd1wt Adult liver (i–l) and pancreas (m–p) stained with anti-HA (red) and ck19 (cytokeratin 19) (green). Arrows in c, g, k and o show accumulation of FPC-HA in sub-apical vesicle-like structures. Original magnification × 40 (scale bar 20 µm) in a, b, e, f, i, j, m, n. Original magnification × 63 (scale bar 2 µm) in c, g, k, o. The dotted lines and squares indicate corresponding enlarged areas. Dotted lines in i and j outline the portal vein.
In the liver (Figure 4i–l and Supplementary Figure S3e–h) and the pancreas (Figure 4m–p and Supplementary Figure S3i–k), FPC-HA was identified only in cells expressing cytokeratin 19 (ck19), which is a ductule-specific marker36. The distribution of FPC within cells appeared to be primarily in cystoplasmic sub-apical vesicle-like structures (Figure 4g, h, k, l, o, p and Supplementary Movie S1). Anti-HA staining was negative in all control (Pkhd1wt and Pkhd1Δ67) tissues, indicating that antibody staining is specific (Figure 4b, f, j and n and Supplementary Movie S1).
Subcellular Localization of Endogenous FPC
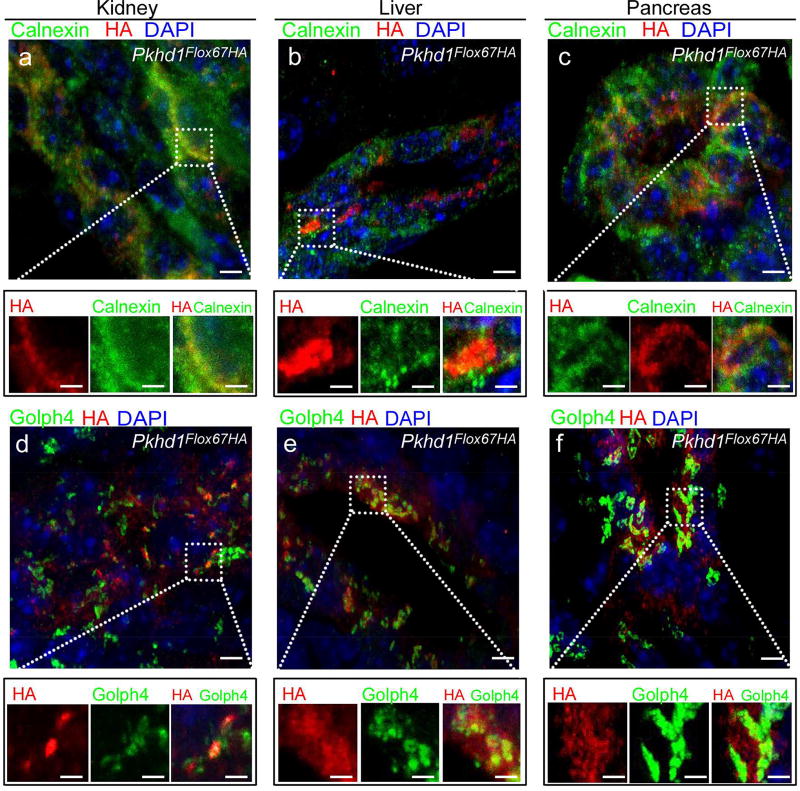
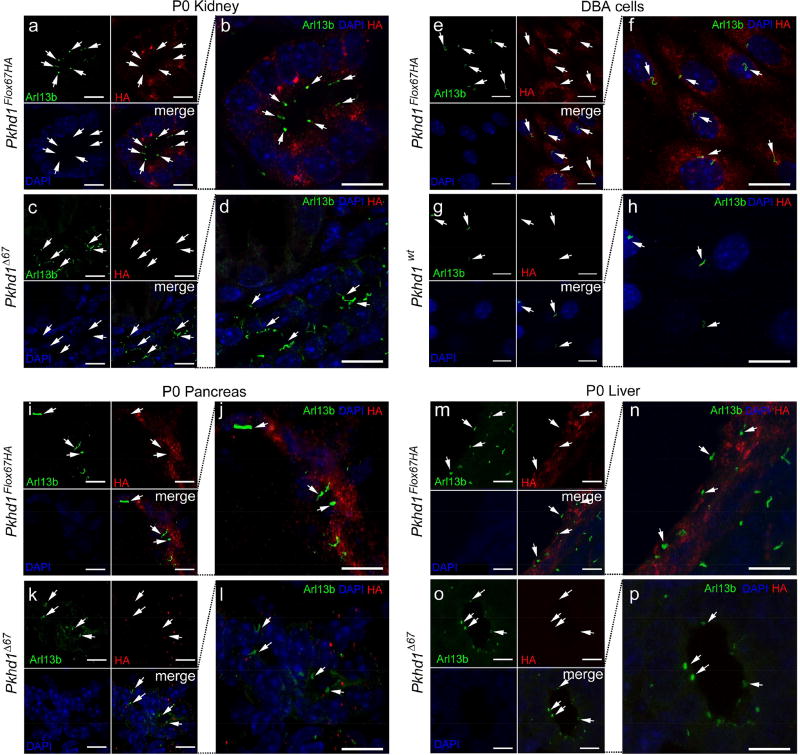
Next, we examined the subcellular localization of FPC-HA in adult tissues from Pkhd1Flox67HA mice (Figure 5a–f and Supplementary Figure 4 Sa–d). We found that FPC-HA co-localized with the endoplasmic reticulum (ER) marker calnexin and with the golgi marker Golph4 in renal tubular epithelial cells (Figure 5a and d and Supplementary Figure S4a–d) and in hepatic cholangiocytes and pancreatic ducts (Figure 5b and c). We were unable to detect FPC-HA in the cilia of any cell types that we tested including renal tubule epithelial cells (Figure 6a–d and Supplementary Figure S5), DBA positive cells isolated from Pkhd1Flox67HA kidneys (Figure 6e–h) or in duct lining cells of the pancreas (Figure 6i–l) and liver (Figure 6m–p).
Figure 5. Subcellular localization of endogenous FPC-HA in adult kidney, liver and pancreas.
(a–f) Cryosections from Pkhd1Flox67HA distal tubules (a, d) and hepatic biliary (b, e) and pancreatic (c, f) ducts stained with anti-HA (red). Cryosections were also labeled with the endoplasmic reticulum (ER) marker calnexin (green in a–c) and with the cis-medial Golgi cisternae marker Golph4 (green in d–f). There is a small amount of co-localization of FPC-HA with calnexin but much more with Golph 4. Original magnification × 63 (scale bar 2 µm) in (a–f). The dotted lines and squares indicate corresponding enlarged areas (scale bar 0.5µm). All panels are 3D projections of 10–15 Z-stacks.
Figure 6. FPC-HA is not detected in cilia.
(a–d) Cryosections from Pkhd1Flox67HA and Pkhd1Δ67 P0 kidneys were stained with anti-HA (red), Arl13b (green) and DAPI (blue). The merged images are magnified in panels b and d. (e–h) Immunostaining of DBA-positive cells isolated from Pkhd1Flox67HA and Pkhd1wt kidneys. The cells were stained with anti-HA (red), DAPI (blue) and a ciliary marker anti-Arl13 (green). Merged images are magnified in panels (f) and (h). (i–p) Cryosections from P0 pancreas (i–l) and P0 liver (m–p) from Pkhd1Flox67HA and Pkhd1Δ67 mice were stained with anti-HA (red) DAPI (blue) and anti-Arl13 (green). Merged images are magnified in panels (j, l, n, p). White arrows indicate cilia that lack FPC-HA staining. Original magnification × 63 (scale bar 5 µm).
FPC is a highly glycosylated protein in vivo
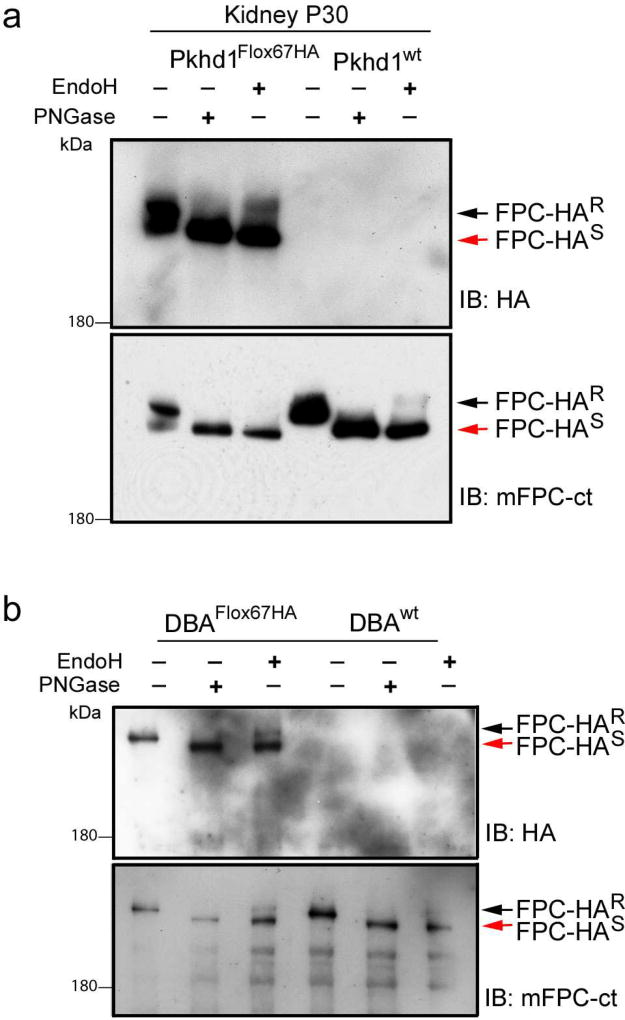
To further characterize endogenous FPC trafficking, we performed glycosylation studies using total lysates from Pkhd1Flox67HA adult kidneys (Figure 7a) and from DBA positive Pkhd1Flox67HA renal epithelial cells (Figure 7b). We treated total lysates with PNGase and found that the predominant 500kDa band shifted down to ~450kDa indicating that FPC is highly glycosylated as has been previously reported32. Treatment with EndoH revealed that only about 5–10% of FPC was EndoH resistant. This suggests that the largest pool of FPC remains in the ER with a small fraction trafficking beyond the golgi. We repeated these experiments using the mFPC-ct antibody and confirmed that untagged FPC is highly glycosylated with a similar fraction of the protein remaining EndoH resistant (Lower panels in Figure 7a and b). We obtained similar results using pancreas lysates (Supplementary Figure S6a) and total lysates from an MDCK cell line transfected with V5-tagged FPC (Supplementary Figure S6b).
Figure 7. Endogenous FPC is an N-glycosylated protein.
(a) Total lysates from P30 Pkhd1Flox67HA and Pkhd1wt kidneys were treated with PNGaseF or EndoH as indicated. Western blots were probed with anti-HA antibody (upper panel) or mFPC-ct (lower panel). Mature FPC-HA is resolved as a doublet but the predominant 500kDa band shifts down to ~450kDa upon treatment with PNGaseF. A small fraction of FPC is EndoH resistant (black arrow, FPC-HAR). The red arrow indicates the EndoH sensitive fragment (FPC-HAS). (b) Total lysates were prepared from DBA-positive renal epithelial cells isolated from Pkhd1Flox67HA and Pkhd1wt kidneys. Lysates were treated with PNGaseF or EndoH and Western blots were probed with anti-HA (upper panel) or mFPC-ct (lower panel). In these cells a small fraction of FPC is EndoH resistant (black arrow, FPC-HAR).
Pkhd1Δ67 mice do not exhibit a kidney or liver phenotype
To determine if the C-terminal ICD of FPC plays a critical role in vivo we generated Pkhd1Δ67 mice lacking Pkhd1 exon 67. We found that these mice were viable and were born in appropriate Mendelian ratios (Supplementary Table 2). We compared the renal phenotype of Pkhd1Δ67 mice with that of a previously reported Pkhd1 allele lacking exons 3 and 4 (Pkhd1Δ3–4), both in a C57BL/6 background30. At 6 months of age we were unable to detect histological differences between the kidneys of Pkhd1Δ67 homozygotes (N=24), Pkhd1wt littermates (N=8) or Pkhd1Δ3–4 homozygotes (N=6) (Supplementary Figure S7a–c). Overall, both tubular and glomerular structures were normal with no evidence of dilatation or cyst formation. This was true regardless of sex.
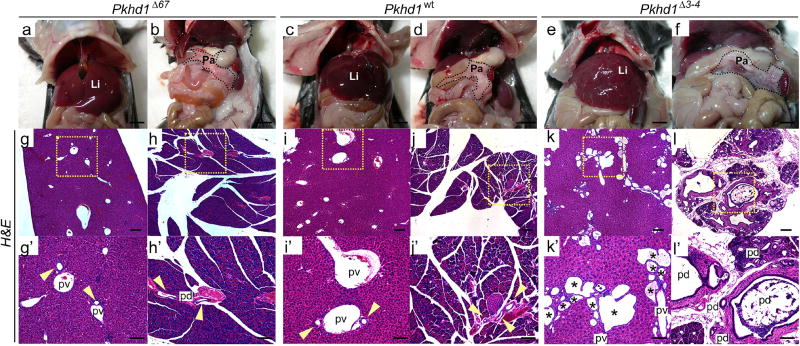
In contrast to what has been described for all other Pkhd1 targeted alleles reported to date, homozygous Pkhd1Δ67 mice did not develop an obvious liver phenotype (Figure 8a–l and Supplementary Figure S8a–c)25,30,32,37–40. At 6 months of age Pkhd1Δ67 homozygotes (N=24) exhibited no evidence of ductal plate malformation and their livers and pancreas were histologically indistinguishable from those of their Pkhd1wt littermates (Figure 8a–d and g–j and Supplementary Figure S8a, b, d and e). Pkhd1Δ3–4 mice on the other hand displayed dilated bile ducts as early as post-natal day 15 (data not shown) and all the mice analyzed (N=6) had liver cysts and pancreatic cysts by 6 months of age with ck19 staining showing evidence of bile duct proliferation (Figure 8 e, f, k and l and Supplementary Figure S8c and f).
Figure 8. Pkhd1Δ67 mice exhibit normal biliary tract histopathology.
(a–f) Representative gross anatomical appearance of liver (a, c, e) and pancreas (b, d, f) in a ~6-month old Pkhd1Δ67 mouse (a, b), a control Pkhd1wt littermate (c, d) and a ~6-month old Pkhd1Δ3–4 mouse (e, f). Scale bars: 0.5mm (g–l) H&E staining of representative liver (g, i, k) and pancreas (h, j, l) sections from a ~6-month old Pkhd1Δ67 mouse (g, h), a control Pkhd1wt littermate (I, j) and a ~6-month old Pkhd1Δ3–4 mutant mouse (k, l). Black squares in (g–l) indicate magnified areas in (g’–l’). Yellow arrows indicate biliary ducts in Pkhd1Δ67 and Pkhd1wt livers. Asterisks mark cysts in the Pkhd1Δ3–4 mouse. Li: liver, P: pancreas, pv: portal vein, pd: pancreatic duct. Scale bars: 200 µm. Magnification 4× in g–l, 10× in g’–l’.
FPC does not interact with PC2 in vivo
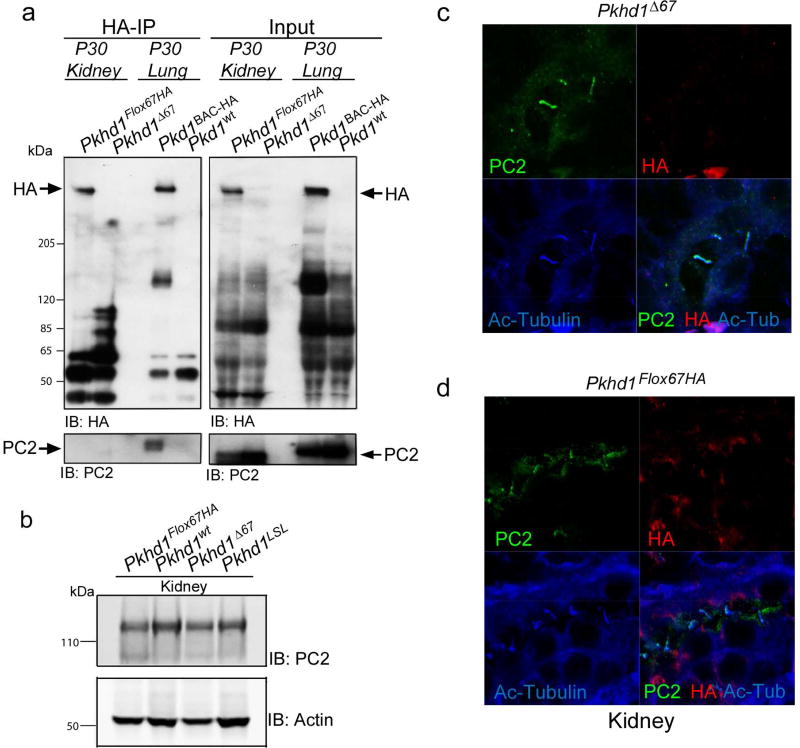
Next, we took advantage of HA-tagged FPC to test for a physical interaction between FPC and PC2 in vivo (Figure 9). We used anti-HA affinity agarose beads to IP FPC-HA from kidney. Although PC2 was present in total kidney lysates, it was undetectable in the IP products (Figure 9a). In contrast, we could co-IP PC1-HA and PC2 from lung lysates derived from a BAC transgenic mouse model harboring HA tagged PC131. We found no difference in total PC2 levels by Western blot in Pkhd1Δ67 kidneys, despite deletion of the PC2BD (Figure 9b). In addition, PC2 could be detected by immunohistochemistry in the cilia of Pkhd1Δ67 renal epithelial cells (Figure 9c and d).
Figure 9. FPC does not interact with PC2 in vivo.
(a) Co-Immunoprecipitation of FPC and PC2. Anti-HA Affinity Matrix was used to IP proteins from Pkhd1Flox67HA and Pkhd1Δ67 adult kidneys and from Pkd1BAC-HA and Pkd1WT lungs as positive and negative controls, respectively (Left). Blots were probed with anti-HA (Top) and anti-PC2 antisera (bottom). Input is shown in the panels on the right. Anti-HA can co-IP PC1-HA and PC2 but not FPC-HA and PC2. (b) Total kidney lysates from adult kidneys of the indicated genotypes were used to prepare Western blots probed with anti-PC2. Actin was used as a loading control. (c,d) Cryosections from Pkhd1Δ67 (c) and Pkhd1Flox67HA (d) P5 kidneys were stained with anti-PC2 (green), anti-HA (red) and Acetylated Tubulin (blue). PC2 can be detected in cilia of both genotypes.
DISCUSSION
Relatively little has been learned about FPC since the initial discovery of PKHD1. There are no robust cell culture read-outs that can be used to assess function, and all previously described genetic modifications of mouse Pkhd1 have disrupted the gene in its extracellular domains25,30,32,37–40. The renal phenotype has been highly variable both with respect to its severity and to the nephron segments that are affected. Even our Pkhd1Δ3–4 model that was previously described as having characteristic disease in a mixed genetic background lost its renal phenotype after the mutation was backcrossed into the 129/Sv and C57BL/6 strains30. Pancreatic cysts also have been a highly variable finding. In contrast, all reported mouse models have fibrocystic liver disease as an invariant finding. The lack of a clear phenotype-genotype correlation has made it difficult to attribute specific functional properties to any domain or exon.
Tracking endogenous protein in an unambiguous way is often a challenge for modestly expressed genes. For FPC, Bakeberg et al knocked a double V5 tag into the N-terminus, which they used to track the protein in vivo32. In membrane preparations, they detected a 500kDa uncleaved protein in tissues and a shorter version shed into urine and urinary exosomes. They concluded that the shed version likely represents the cleaved ectodomain previously described by Kaimori et al and that cleavage occurs upon shedding but provided no direct evidence for this20,21. Unfortunately, antibodies for the V5 epitope generally are poor for immunolocalization studies and none were reported in the Bakeberg study.
We have generated a novel mouse model that is a complement to the V5-tagged Pkhd1 mouse line. Our new line includes a triple HA tag knocked into the extreme C-terminus of Pkhd1 to allow in vivo tracking of both the full-length uncleaved protein and the cleaved C-terminus. The model also includes LoxP sites flanking the last exon, which encodes several domains previously thought essential for FPC function. We show that introducing these changes into the Pkhd1 locus has no ill effects as the mice are phenotypically normal. Using both anti-HA antibodies and a newly generated antibody that recognizes the C-terminus, we too found that full length FPC is abundant in neonatal and adult kidneys and pancreas. In contrast to prior work, however, we also detected 55kDa and 17kDa products in kidneys that likely represent the PTM and ICD, respectively. This is the first evidence indicating that Notch-like processing likely also occurs in vivo. Interestingly, these cleavage products were not detected in pancreas, suggesting either tissue-specific differences in FPC processing or tissue-specific differences in the half-life of cleaved FPC. Levels of expression in total liver lysates were too low to determine whether the smaller products also are present in that organ. A small number of minor bands ranging in size from ~75kDa up to just under 500kDa were inconsistently seen and it is not clear whether these represent products of alternative Pkhd1 splicing, regulated proteolytic processing or non-specific degradation
While one would not have expected Bakeberg et al to have detected either PTM or ICD with their V5 tag, it is surprising that they did not detect the cleaved N-terminus (PECD) in their membrane preparations as one would have expected it to remain tethered to the PTM after cleavage20,21,32. One possibility is that the PECD is quickly shed into the urine after cleavage whereas the PTM remains behind in a stable form. Our studies of urinary FPC are consistent with this model as we see abundant PECD and no PTM in unprocessed urine. This result would be somewhat unexpected, however, if FPC follows the Notch receptor model whereby activation of the receptor results in ectodomain shedding and gamma-secretase dependent release of its intracellular domain. Selective retention of the PTM would suggest that it has intrinsic functional properties independent of the PECD, or that an additional signal is required for triggered release of its ICD. An alternative explanation is that the lysis buffers used to isolate the V5 tagged products included reducing agents prior to membrane purification. We had previously found that addition of DTT to the lysis buffer disrupted PECD-PTM tethering20.
The presence of two classes of FPC molecules in the urine likely has functional implications. The majority of urinary FPC is in the form of the free, shed ectodomain (PECD). If FPC follows the Notch schema, the free pool of PECD results from receptor activation, indicating that a substantial fraction of FPC is constitutively activated under normal conditions. The stimuli that trigger shedding of free PECD (i.e. a ligand and/or a mechanical process like flow) remain to be determined. It also is unknown whether the primary effector pathways reside in the cells that release PECD, with the shed product simply representing a byproduct of the process, or whether the PECD itself is being sent as a signal to targets in other cells. We predict the latter but further study is required to distinguish between these possibilities. A second pool of urinary FPC consists of ELVs harboring the mature, processed FPC (PECD) along with the PTM. In vitro studies suggest that these two fragments remain tethered after cleavage by pro-protein convertase. The ELVs could deliver this FPC complex to target cells to produce bi-directional signaling, including release of the ICD. The cellular targets and processes regulated by these two distinct classes of FPC are unresolved mysteries but further investigation into this issue should yield novel insights into the protein’s function.
Our data also raise new questions about the reported FPC-PC2 interaction22,25. We were not able to co-precipitate PC2 with FPC from tissue using robust and specific methods, and we found that deletion of the PC2 binding domain had no obvious pathologic effect on mice (discussed later). Moreover, deletion of this domain did not affect the total levels of PC2 in kidney or the ability of PC2 to reach the cilium. These observations, coupled with concerns raised in a prior commentary, suggest that an FPC-PC2 interaction, if it occurs in vivo, is not essential for either gene’s function29.
An advantage of the HA tag over the V5 tag and others is that anti-HA antibodies work well for immunolocalization studies. Using an anti-HA antibody, we unambiguously detected the tagged protein in the collecting ducts, the thick ascending limb of Henle (TAL) and in the epithelial cells lining biliary ductules and pancreatic ducts. HA-tagged FPC was distributed in vesicle-like structures, in the ER and the Golgi. While we did not detect FPC-HA in the adult proximal tubule, we did observe patchy low-level anti-HA staining in proximal tubules at P0. This is consistent with reports that human ARPKD is associated with a phase of transient proximal tubule cyst formation during fetal development41.
In contrast to the published literature, we were not able to detect the HA tag in primary cilia or in the nucleus. One possible explanation for these discrepant findings is that our methods were not sensitive enough to detect the ciliary or nuclear products in vivo. This is quite likely given that we had to use amplification methods to detect them in our earlier in vitro studies20. Our de-glycosylation studies also suggest that ≥80% of full length FPC is Endo-H sensitive, indicating that the vast majority of it likely localizes to the ER or cis-Golgi. Our immunoblot studies show that at least half of all HA-signal is associated with the uncleaved full-length protein, with the remainder split between the PTM and NLS products. Therefore, the pool of HA signal that localizes to the nucleus or is associated with the PECD is quite low and unless highly clustered could be minimally higher than background and thus easily missed. An alternative explanation is that only the cleaved N-terminus (PECD) dissociated from the PTM traffics to the cilia. This would be consistent with the Bakeberg study that found only an ~450kDa form of FPC, which they presumed was PECD, in the urine and urinary exosome-like vesicles32. We would have been unable to detect PECD alone using either anti-HA or mFPC-ct antisera. Confirmation of this model will require antibodies that can unambiguously recognize PECD by immunostaining.
The most surprising finding of this study is that deletion of exon 67 had no apparent effect. Even with aging, homozygous mutant mice had normal kidney, pancreas and liver histology. Remarkably, this is the first rodent model where manipulation of Pkhd1 has not resulted in liver disease25,30,32,37–40. This deletion removes both the putative PC2 binding site and the nuclear localization signal while leaving intact the transmembrane segment and putative ciliary targeting sequence. We targeted this exon because of these alleged functional domains and because at least two human truncating mutations have been reported to disrupt exon 67 (RWTH Aachen University PKHD1 mutation database, http://www.humgen.rwth-aachen.de/index.php).
So how does one interpret these findings? One possibility is that exon 67 in mice and humans serve different functions. In fact, the mouse C-terminus is one of the least conserved parts of the protein compared to the human peptide sequence16. In addition, there is abundant evidence to suggest that murine and human FPC have somewhat different properties. While there is considerable variability in the phenotype of humans with bi-allelic mutations of PKHD1, most mutations are thought to result in kidney disease, and a substantial fraction of affected individuals have severe perinatal presentations4,15,42. In contrast, kidney disease, when present in the rodent models, is generally mild--none of the Pkhd1 mouse models reported have severe neonatal PKD-- and greatly influenced by other modifiers25,30–32,37–40. These observations suggest there must be some redundancy in mice for FPC function at least in the kidney, which is influenced by genetic strain differences. Finally, a much less likely explanation for the discordant human/mouse data is that the two human truncating mutations that have been reported (RWTH Aachen University PKHD1 mutation database, http://www.humgen.rwth-aachen.de/index.php) to disrupt exon 67 in fact are not pathogenic. In this schema, exon 67 is not essential for either species. There are no data to support this interpretation, however.
The lack of an essential function for the ICD domains in the mouse does raise the question of why this region of FPC has been evolutionarily retained. While somewhat surprising given the conserved functional domains, there are numerous examples of orthologous genes that are essential in humans but where inactivation in mice results in no apparent phenotype43,44. In many cases, it was subsequently discovered that the gene had previously unknown functions that were missed because the investigators did not know to screen for them43,44. In other examples, the gene’s redundant but important functional properties were unmasked by either environmental or other genetic stresses44. What if any factors can elicit a phenotype in the Pkhd1Δ67 mouse line remains to be determined.
In sum, we have generated a novel mouse model of Pkhd1 that includes an epitope tag suitable for biochemical analyses, immunopurification and immunolocalization studies. This feature will be useful in characterizing the protein’s binding partners in vivo. The model also has been used to analyze the functional properties of several FPC domains, revealing hidden complexity that will require further study to understand better.
MATERIAL AND METHODS
Animal Care and Use
Mouse work was performed at the University of Maryland and at the National Institutes of Health (NIH). All animal studies were reviewed and approved by the IACUC and conducted in accordance with the University of Maryland and NIH Animal Care and Use Committee guidelines and procedures. The PkhdFlox67HA, Pkhd1Δ67, Pkhd1Δ3–4 and Pkhd1LSL mice described in this study were maintained on an inbred C57BL/6 background. Pkhd1LSL mice were obtained from Jackson Laboratories (Stock #019423). Mice of both sexes were used in this study.
Generation of the Epitope-Tagged Pkhd1 Knock-in Mouse
The targeting construct was prepared using genomic PCR and standard cloning strategies. Thirty-µg of the final construct was linearized with SwaI and electroporated into ~107 C57BL/6 ES cells (Taconic) and selected with 200 µg/mL of G418. Seven plates of G418 resistant ES clones (~672) were selected for screening. The primary ES screening was performed by Southern blot using 5’ end probes and by PCR testing for the distal LoxP (Taconic). Six potential targeted clones (D12, F1, F11, F12, G1 and G2) were identified and expanded for further analysis. After expansion, additional Southern and PCR confirmation analysis were performed. Based on this analysis, five out of the six expanded clones (D12, F1, F12, G1 and G2) were confirmed to have incorporated the construct via homologous recombination. The proper insertion of the distal LoxP site and the HA tag in these clones was also confirmed by PCR using the following primer pairs: 1) the 5’ LoxP site was confirmed using Primer LoxPF: 5’-GTGGAATTAGTATGGAACATGTGTTGACTTT-3’ and Primer LoxPR: 5’-GAAATCCATATCACAGCCAAGGCC-3’ (Amplicon size: 180bp for the wild type allele and 320bp for the inserted LoxP site). 2) The HA-tag sequence in the targeted allele was confirmed using the primers: HA-F: 5’-GCTGGACAAAGGGAGTGACTGCACT-3’ and HA-R: 5’-GAGTTCACATTCTCCTTATTTGTCCAAAC-3’ (Amplicon size: 310bp for the inserted HA sequence and 220bp for the wild type allele).
Clone D12 was selected for co-isogenic B6 blastocyst (Tyr) injection. Highly chimeric males were bred with C57BL/6 females and germ line transmission of the targeted Pkhd1Flox67HA allele was obtained. Heterozygous Pkhd1Flox67HA mice were subsequently intercrossed to generate the homozygous progeny. Excision of the floxed fragment was achieved by crossing the homozygous Pkhd1Flox67HA with the Meox2 tm1(Cre)Sor/J (Jackson Laboratories, Stock #03755 in B6) line, which induces Cre-mediated recombination at a high rate in somatic tissues.
Genotype analysis
Genomic DNA was isolated from mouse tails and genotyped using a 3 primer PCR strategy that allow us to distinguish the floxed, wild type and mutant Pkhd1Δ67 alleles. The PCR products were resolved on 2% NuSieve agarose gels, yielding products of ~320, ~268 and ~188 base pairs, respectively (Fig 1e). Primer sequences: Primer 1F: 5’-GTGGAATTAGTATGGAA CATGTGTTG-3’, 1R: 5’-GAAATCCATATCACAGCCAAGGCC-3’ and 2R: 5’-CATTTGCCTAGGAATTTCGTG-3’ with 56 °C annealing temperature and 35 PCR cycles. The Cre transgene was detected using the following primers: Cre 200F: 5’-ATTGCTGTCACTTGGTCGTGGC-3’ and Cre 200R: 5’-GGAAAATGCTTCTGTCCGTTTGC-3’ with 59 °C annealing and 35 PCR cycles. The PCR product was resolved on 2% NuSieve agarose gels, yielding a product of ~200 base pairs.
Immunohistochemistry
Mice were euthanized and tissues were fixed with 4% paraformaldehyde and embedded in paraffin, sectioned and stained with hematoxylin-eosin (H&E) using standard protocols.
Immunofluorescence
Tissues were fixed in 4% paraformaldehyde, buffered in 30% cold sucrose and frozen-embedded in OCT. Five to ten µm sections of kidney, liver, and pancreas were incubated for 1 h in blocking solution (1%BSA, 5%FBS and 0.1% Triton X-100). Primary antibodies were diluted in blocking solution and incubated overnight at 4°C. Samples were washed three times using PBT (0.5%Triton in PBS) and incubated with the appropriate secondary antibody (Alexa Fuor®, Life Technologies) for 1h at room temperature. After washing in PBT, sections were mounted in Fluoromount-G® (SouthernBiotech) and analyzed using a confocal multichannel point scanning microscope LSM 510 Duo (Zeiss).
Primary cell isolation
DBA or LTL positive epithelial cells were isolated from kidneys using the CELLection™ Biotin binder Dynabeads (Invitrogen). Briefly, kidneys were minced and digested in digestion buffer (0.2% collagenase and 2U/mL DNase I in F12/DMEM) for 45 minutes at 37°C. The digested tubules were incubated for 30 minutes at 4°C with biotin binder Dynabeads (Vector Laboratories Inc.) conjugated to DBA or LTL. DBA or LTL-bound cells were selected using a magnet, plated and then cultured in growth media (5%FBS, 5ug/mL Insulin, 5nM Dexamethasone, 5ng/mL Na Selenite, 5ug/mL transferrin, 1nM Triiodothyronine, 4ng/mL EGF, 50U/mL Pen/Strep).
Immunocytochemistry
Cells were washed with PBS and fixed using 4% Formaldehyde (Thermo Scientific) for 10 minutes, washed thoroughly with PBS, and incubated with blocking buffer (5%FBS, 2%BSA and 0.1% Triton-X100 in PBS) for 1 hour. Cells were incubated overnight with primary antibodies, washed with 0.5% TritonX-100 in PBS and incubated with the appropriate fluorescence-conjugated secondary antibodies (Alexa Fluor®, Life Technologies). Cells were washed three times for 5 minutes each with 0.5% TritonX-100 in PBS and stained with DAPI. Samples were mounted in Fluoromount-G® (SouthernBiotech) and analyzed using a LSM 510 Duo confocal multichannel point scanning microscope (Zeiss).
Western Blot analysis and immunoprecipitation
Total lysates were prepared from primary epithelial cells and freshly isolated organs including kidneys, pancreas, liver, lung, and heart. 70µg of protein was loaded per gel lane. Equal loading was confirmed using β-actin.
To isolate renal tubules, kidney fragments (~1mm3) were incubated in Collagenase solution (5%FBS, 250µg/µL, 50 µg/µL Insulin, 20 mg/mL Collagenase A, 20 mg/mL Trypsin and 2U/mL DNase I in D-MEM/F12) for 20 minutes at 37°C and then centrifuged at 1500 rpm for 10 minutes. The supernatant was aspirated and the pellet containing tubules was washed and re-suspended in 10mL D-MEM/F12. The preparation was centrifuged for 3 seconds at 1500 rpm and the wash was repeated 3 more times.
The PTM band was detected using fresh tissue or renal tubules that had been lysed using Buffer A [0.25M sucrose, 10mM TrisCl pH 7.4, 1mM EDTA pH 7.4, 1mM PMSF, and 1:50 protease cocktail inhibitor (Sigma)]. The ICD band was detected using fresh tissue lysed using buffer B [20 mM Na-P (pH 7.2), 150 mM NaCl, 1mM EDTA, 10% Glycerol, 1% Triton-X100 and a cocktail of protease inhibitors (Sigma). The lysates were homogenized with a motor-driven Polytron. The samples were rotated in lysis buffer for 40 minutes and centrifuged at 14000rpm for 20 minutes at 4°C. The supernatant was transferred to a fresh tube and the protein concentration measured using the BCA protein assay kit (Pierce).
Fresh urine was collected from mice and precleared by centrifugation at 4000g for 15 minutes at 4°C as previously described32. To enrich for exosome-like vesicles (ELVs), precleared urine was centrifuged at 100,000g for 1 hour at 4°C. The supernatant was carefully removed and pellets re-suspended in SDS buffer for Western analysis.
For immunoprecipitation studies, total lysates (~5 mg total) were incubated overnight with mouse monoclonal Anti-HA antibody conjugated to agarose (Sigma). Samples were resolved on a 3–8% Tris-Acetate (PTM or full length) or 4–12% BT NuPage gel (ICD) (Invitrogen) and transferred to a PDVF membrane. Blots were probed with primary antibodies and resolved using a chemoluminiscence kit ECL™ Prime Western Blotting Detection Reagents (GE Healthcare).
Antibodies
The following primary antibodies were used in these studies: rat antisera directed against HA 1:500 (Roche, 3F10), human PC2 1:1000 (Boletta et al., 2000), anti-GM130 EP892Y 1:250 from (Novus Biologicals), fluorescein labeled Dolichos Biflorus Agglutinin (DBA) 1:500 and fluorescein labeled Lotus Tetragonolobus Lectin (LTL) 1:500 both from Vector Laboratories, anti-rabbit Arl13b 1:500, anti-mouse Alix (clone 3A9) 1:1000 from Cell Signaling and anti-Acetylated Tubulin antibody produced in mouse 1:2000 (Sigma-Aldrich), rabbit anti-cytokeratin 19 (ck19) 1:1000 (kindly provided by Dr. Ben Stanger), mouse anti-Actin 1:5000 (Sigma-Aldrich), rabbit anti-NKCC2 (kindly provided by Dr. Pedro Jose), rabbit anti-Calnexin 1:500 (Enzo Life science), Golph4 1:500 (Abcam).
The rat monoclonal FPC antibody (mFPC-ct) was amplified by PCR using a 549-base pair cDNA fragment of mouse Pkhd1 (11858–12406 nt, NM_153179) corresponding to the entire C-terminal (183 amino acid residues of FPC) and cloned into pET21 (Novagen). After expression in E.coli, the Histidine-tagged FPC fragment was affinity-purified with Ni-agarose beads and then used to raise monoclonal antibodies in rats. Hybridoma clones were initially screened by an ELISA assay against the purified FPC fragment and positives tested for western detection of recombinant full-length mouse FPC and mouse tissues.
Enzymatic De-glycosylation Studies
Total Lysates (~150µg) from kidneys, primary cells and pancreas were quickly denatured at 95°C using the Glycoprotein Denature Buffer (New England Biolabs), cooled and mixed with 1/10 concentrated EndoH (0.5 M sodium citrate, pH 5.5) or PNGaseF [0.5 M sodium phosphate, pH 7.5 containing 10% (v/v) Nonidet P40]. Samples were digested at 37°C for 1½ hours and separated using NuPAGE® Novex® 3–8% Tris-Acetate Gels (Life Technologies).
Mouse full-length Pkhd1 expression vector
To generate the mouse full-length Pkhd1 expression vector, we used RT-PCR to amplify three overlapping fragments from mouse mRNA. The RT-PCR products spanning 67 Pkhd1 exons were found to be 100% identical to the NCBI Reference Sequence (NM_153179.3). The fragments were assembled into a single full-length cDNA using unique restriction enzyme sites common to the overlapping segments. The final product was cloned into a mammalian expression vector (pcDNA5, Invitrogen). A VSV-G epitope tag was added to the N-terminus immediately C-terminal to the signal-peptide cleavage site and V5-epitope tag was added to the C-terminus. This construct was used to establish inducible stable MDCK II cell lines as previously described45.
Supplementary Material
Acknowledgments
Sources of support: This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK): R01-DK051259 and 1 ZIA-DK075042 to GGG, R01-DK095036 and R01-DK076017 to TW, R01-DK62199 to FQ. This work was also supported by a mini-grant from The National Kidney Foundation of Maryland (POG) and a research grant from the Polycystic Kidney Disease Foundation (FQ). These studies utilized resources provided by the NIDDK-sponsored Baltimore Polycystic Kidney Disease Research and Clinical Core Center (P30DK090868) and also The George M. O’Brien Kidney Center at Yale (P30 DK079310).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Part of this work was presented at the Annual Meeting of the American Society of Nephrology held October 30–November 4, 2012 in San Diego, California.
DISCLOSURES
The authors have no relevant conflicts to disclose.
References
- 1.Zerres K, Rudnik-Schoneborn S, Steinkamm C, Becker J, Mucher G. Autosomal recessive polycystic kidney disease. J Mol Med (Berl) 1998;76(5):303–309. doi: 10.1007/s001090050221. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann C, Senderek J, Windelen E, et al. Clinical consequences of PKHD1 mutations in 164 patients with autosomal-recessive polycystic kidney disease (ARPKD) Kidney Int. 2005;67(3):829–848. doi: 10.1111/j.1523-1755.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 3.Guay-Woodford LM, Desmond RA. Autosomal recessive polycystic kidney disease: the clinical experience in North America. Pediatrics. 2003;111(5 Pt 1):1072–1080. doi: 10.1542/peds.111.5.1072. [DOI] [PubMed] [Google Scholar]
- 4.Guay-Woodford LM, Muecher G, Hopkins SD, et al. The severe perinatal form of autosomal recessive polycystic kidney disease maps to chromosome 6p21.1-p12: implications for genetic counseling. Am J Hum Genet. 1995;56(5):1101–1107. [PMC free article] [PubMed] [Google Scholar]
- 5.Guay-Woodford LM. Autosomal recessive polycystic kidney disease: the prototype of the hepato-renal fibrocystic diseases. Journal of pediatric genetics. 2014;3(2):89–101. doi: 10.3233/PGE-14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunay-Aygun M, Turkbey IB, Bryant J, et al. Hepatorenal Findings in Obligate Heterozygotes for Autosomal Recessive Polycystic Kidney Disease. Molecular genetics and metabolism. 2011;104(4):677–681. doi: 10.1016/j.ymgme.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denamur E, Delezoide AL, Alberti C, et al. Genotype-phenotype correlations in fetuses and neonates with autosomal recessive polycystic kidney disease. Kidney Int. 2010;77(4):350–358. doi: 10.1038/ki.2009.440. [DOI] [PubMed] [Google Scholar]
- 8.Adeva M, El-Youssef M, Rossetti S, et al. Clinical and molecular characterization defines a broadened spectrum of autosomal recessive polycystic kidney disease (ARPKD) Medicine (Baltimore) 2006;85(1):1–21. doi: 10.1097/01.md.0000200165.90373.9a. [DOI] [PubMed] [Google Scholar]
- 9.Bergmann C, Senderek J, Kupper F, et al. PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD) Hum Mutat. 2004;23(5):453–463. doi: 10.1002/humu.20029. [DOI] [PubMed] [Google Scholar]
- 10.Onuchic LF, Furu L, Nagasawa Y, et al. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet. 2002;70(5):1305–1317. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward CJ, Hogan MC, Rossetti S, et al. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet. 2002;30(3):259–269. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- 12.Boddu R, Yang C, O'Connor AK, et al. Intragenic motifs regulate the transcriptional complexity of Pkhd1/PKHD1. J Mol Med (Berl) 2014;92(10):1045–1056. doi: 10.1007/s00109-014-1185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp AM, Messiaen LM, Page G, et al. Comprehensive genomic analysis of PKHD1 mutations in ARPKD cohorts. J Med Genet. 2005;42(4):336–349. doi: 10.1136/jmg.2004.024489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furu L, Onuchic LF, Gharavi A, et al. Milder presentation of recessive polycystic kidney disease requires presence of amino acid substitution mutations. J Am Soc Nephrol. 2003;14(8):2004–2014. doi: 10.1097/01.asn.0000078805.87038.05. [DOI] [PubMed] [Google Scholar]
- 15.Gunay-Aygun M, Tuchman M, Font-Montgomery E, et al. PKHD1 sequence variations in 78 children and adults with autosomal recessive polycystic kidney disease and congenital hepatic fibrosis. Mol Genet Metab. 2010;99(2):160–173. doi: 10.1016/j.ymgme.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagasawa Y, Matthiesen S, Onuchic LF, et al. Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J Am Soc Nephrol. 2002;13(9):2246–2258. doi: 10.1097/01.asn.0000030392.19694.9d. [DOI] [PubMed] [Google Scholar]
- 17.Ward CJ, Yuan D, Masyuk TV, et al. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet. 2003;12(20):2703–2710. doi: 10.1093/hmg/ddg274. [DOI] [PubMed] [Google Scholar]
- 18.Menezes LF, Cai Y, Nagasawa Y, et al. Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int. 2004;66(4):1345–1355. doi: 10.1111/j.1523-1755.2004.00844.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Luo Y, Wilson PD, Witman GB, Zhou J. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol. 2004;15(3):592–602. doi: 10.1097/01.asn.0000113793.12558.1d. [DOI] [PubMed] [Google Scholar]
- 20.Kaimori JY, Nagasawa Y, Menezes LF, et al. Polyductin undergoes notch-like processing and regulated release from primary cilia. Hum Mol Genet. 2007;16(8):942–956. doi: 10.1093/hmg/ddm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiesberger T, Gourley E, Erickson A, et al. Proteolytic cleavage and nuclear translocation of fibrocystin is regulated by intracellular Ca2+ and activation of protein kinase C. J Biol Chem. 2006;281(45):34357–34364. doi: 10.1074/jbc.M606740200. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Dai XQ, Li Q, et al. Kinesin-2 mediates physical and functional interactions between polycystin-2 and fibrocystin. Hum Mol Genet. 2006;15(22):3280–3292. doi: 10.1093/hmg/ddl404. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Zhang J, Nauli SM, et al. Fibrocystin/polyductin, found in the same protein complex with polycystin-2, regulates calcium responses in kidney epithelia. Mol Cell Biol. 2007;27(8):3241–3252. doi: 10.1128/MCB.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Follit JA, Li L, Vucica Y, Pazour GJ. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol. 2010;188(1):21–28. doi: 10.1083/jcb.200910096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim I, Fu Y, Hui K, et al. Fibrocystin/polyductin modulates renal tubular formation by regulating polycystin-2 expression and function. J Am Soc Nephrol. 2008;19(3):455–468. doi: 10.1681/ASN.2007070770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim I, Li C, Liang D, et al. Polycystin-2 expression is regulated by a PC2-binding domain in the intracellular portion of fibrocystin. J Biol Chem. 2008;283(46):31559–31566. doi: 10.1074/jbc.M805452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mochizuki T, Wu G, Hayashi T, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272(5266):1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 28.Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 2009;76(2):149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaimori JY, Germino GG. ARPKD and ADPKD: first cousins or more distant relatives? J Am Soc Nephrol. 2008;19(3):416–418. doi: 10.1681/ASN.2008010033. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Gonzalez MA, Menezes LF, Piontek KB, et al. Genetic interaction studies link autosomal dominant and recessive polycystic kidney disease in a common pathway. Hum Mol Genet. 2007;16(16):1940–1950. doi: 10.1093/hmg/ddm141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedeles SV, Tian X, Gallagher AR, et al. A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet. 2011;43(7):639–647. doi: 10.1038/ng.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakeberg JL, Tammachote R, Woollard JR, et al. Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J Am Soc Nephrol. 2011;22(12):2266–2277. doi: 10.1681/ASN.2010111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denamur E, Delezoide AL, Alberti C, et al. Genotype-phenotype correlations in fetuses and neonates with autosomal recessive polycystic kidney disease. Kidney Int. 2010;77(4):350–358. doi: 10.1038/ki.2009.440. [DOI] [PubMed] [Google Scholar]
- 34.Bergmann C, Senderek J, Sedlacek B, et al. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1) J Am Soc Nephrol. 2003;14(1):76–89. doi: 10.1097/01.asn.0000039578.55705.6e. [DOI] [PubMed] [Google Scholar]
- 35.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26(2):113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Zong Y, Panikkar A, Xu J, et al. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136(10):1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser M, Matthiesen S, Kirfel J, et al. A mouse model for cystic biliary dysgenesis in autosomal recessive polycystic kidney disease (ARPKD) Hepatology. 2005;41(5):1113–1121. doi: 10.1002/hep.20655. [DOI] [PubMed] [Google Scholar]
- 38.Woollard JR, Punyashtiti R, Richardson S, et al. A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int. 2007;72(3):328–336. doi: 10.1038/sj.ki.5002294. [DOI] [PubMed] [Google Scholar]
- 39.Gallagher AR, Esquivel EL, Briere TS, et al. Biliary and pancreatic dysgenesis in mice harboring a mutation in Pkhd1. Am J Pathol. 2008;172(2):417–429. doi: 10.2353/ajpath.2008.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams SS, Cobo-Stark P, James LR, Somlo S, Igarashi P. Kidney cysts, pancreatic cysts, and biliary disease in a mouse model of autosomal recessive polycystic kidney disease. Pediatr Nephrol. 2008;23(5):733–741. doi: 10.1007/s00467-007-0735-4. [DOI] [PubMed] [Google Scholar]
- 41.Nakanishi K, Sweeney WE, Jr, Zerres K, Guay-Woodford LM, Avner ED. Proximal tubular cysts in fetal human autosomal recessive polycystic kidney disease. J Am Soc Nephrol. 2000;11(4):760–763. doi: 10.1681/ASN.V114760. [DOI] [PubMed] [Google Scholar]
- 42.Guay-Woodford LM. Autosomal recessive polycystic kidney disease: the prototype of the hepato-renal fibrocystic diseases. J Pediatr Genet. 2014;3(2):89–101. doi: 10.3233/PGE-14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105(19):6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbaric I, Miller G, Dear TN. Appearances can be deceiving: phenotypes of knockout mice. Brief Funct Genomic Proteomic. 2007;6(2):91–103. doi: 10.1093/bfgp/elm008. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, Xu H, Yao Q, et al. Ciliary membrane proteins traffic through the Golgi via a Rabep1/GGA1/Arl3-dependent mechanism. Nat Commun. 2014;5:5482. doi: 10.1038/ncomms6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.