SUMMARY
Respiratory syncytial virus (RSV) is a leading cause of infant mortality, and there are currently no licensed vaccines to protect this vulnerable population. A comprehensive understanding of infant antibody responses to natural RSV infection would facilitate vaccine development. Here, we isolated over 450 RSV fusion glycoprotein (F)-specific antibodies from seven RSV-infected infants and found that half of the antibodies recognized only two antigenic sites. Antibodies targeting both sites showed convergent sequence features, and structural studies revealed the molecular basis for their recognition of RSV F. A subset of antibodies targeting one of these sites displayed potent neutralizing activity despite lacking somatic mutations, and similar antibodies were detected in RSV-naïve B cell repertoires, suggesting that expansion of these B cells in infants may be possible with suitably designed vaccine antigens. Collectively, our results provide fundamental insights into infant antibody responses and a framework for the rational design of age-specific RSV vaccines.
Keywords: pneumoviridae, germline, naïve B cells, crystal structures, VH3-21:VL1-40, VH3-11:VL1-40
INTRODUCTION
Respiratory syncytial virus (RSV) is a ubiquitous pathogen that causes bronchiolitis and pneumonia in infants and the elderly, resulting in nearly 60,000 deaths annually in children under the age of five (Shi et al., 2017). Currently, the only preventive measure available for RSV is passive prophylaxis with the monoclonal antibody Synagis® (The IMpact-RSV Study Group, 1998). Unfortunately, prophylaxis with Synagis® is costly and requires multiple doses per RSV season (Homaira et al., 2014; Kamal-Bahl et al., 2002), thereby restricting its use to high-risk infants in developed countries. Therefore, the development of an effective RSV vaccine and next-generation monoclonal antibodies is of great importance and ongoing clinical trials are evaluating numerous candidates (PATH, 2017).
An effective RSV vaccine has remained elusive due in part to the young age at which primary infection occurs (Glezen et al., 1986), a history of vaccine-enhanced disease in infants (Acosta et al., 2015), and a lack of long-lived immunity that results in frequent reinfections throughout life (Hall et al., 1991). Five target age groups for vaccination have been proposed—infants under six months of age, infants over six months of age, school-aged children, pregnant women, and adults over 65 years old—with the goal of either directly or indirectly protecting at-risk populations (Anderson et al., 2013). These target age groups have different immunological characteristics that may require different vaccination strategies for optimal protection. Although multiple modalities for an RSV vaccine are currently being pursued, most vaccination strategies seek to elicit neutralizing antibodies that recognize the RSV fusion glycoprotein (RSV F), which is targeted by the majority of RSV-neutralizing activity in human sera (Sastre et al., 2005).
RSV F is a class I fusion protein that mediates viral entry into host cells by converting from a metastable trimeric prefusion conformation (preF) to a highly stable postfusion conformation (postF). The antigenic topology of RSV F is substantially altered during this transition. Some groups of epitopes, referred to as antigenic sites, are generally conserved on both preF and postF, whereas others are found primarily on only one of the conformations (Graham, 2017). Molecules that prevent these structural changes can prevent viral fusion and have potential as therapeutics for RSV (Battles et al., 2016; Huang et al., 2010; Lambert et al., 1996; McLellan et al., 2013).
The first characterized RSV F-reactive antibodies bound to structural elements shared by both preF and postF and were therefore F-conformation-independent. These antibodies include Synagis®, which recognizes antigenic site II (Beeler and van Wyke Coelingh, 1989; McLellan et al., 2010b), and 101F, which recognizes antigenic site IV (McLellan et al., 2010a; Wu et al., 2007). Antibodies that preferentially bind to antigenic site I on postF were also readily isolated, but these were only weakly neutralizing (Anderson et al., 1986; Garcia-Barreno et al., 1989). The first preF-specific antibodies to be described recognized antigenic site Ø (“zero”), present at the apex of the preF trimer, and were extremely potent (McLellan et al., 2013). A second class of potently neutralizing antibodies, epitomized by MPE8, was later described and shown to recognize antigenic site III (Corti et al., 2013). Although the secondary structure elements that form site III are present on both preF and postF, they adopt a different spatial arrangement in postF that results in higher affinity binding to preF (Corti et al., 2013). Antigenic site V, located between sites Ø and III, was recently identified and shown to be the target of additional preF-specific antibodies that are potently neutralizing (Gilman et al., 2016; Mousa et al., 2017). A preF-specific antibody that targets site Ø and contains Fc modifications to extend serum half-life (MEDI8897) is currently in late-phase clinical trials (Griffin et al., 2017; Zhu et al., 2017).
An effective RSV vaccine will likely require the elicitation of potent neutralizing antibodies and a well-balanced T helper 1 (Th1) and Th2 response (Lambert et al., 2014; Legg et al., 2003). Infants present a number of unique challenges for vaccine development, including suppression of B cell responses by maternally derived antibody and immunological immaturity that results in reduced levels of T cell help, antibody class-switching, and somatic hypermutation (SHM) (Siegrist and Aspinall, 2009). Studies of convalescent infant sera have demonstrated that infants generally produce low titers of RSV-neutralizing antibodies after infection (Esposito et al., 2016; Murphy et al., 1986; Sande et al., 2014), although these titers are higher when levels of maternal antibody are low (Shinoff et al., 2008), suggesting that infants are capable of mounting neutralizing antibody responses to RSV. Serum studies have also suggested that different epitopes may be targeted as children age into adulthood (Fuentes et al., 2016), but little is known about how these changes are associated with antibody sequence or neutralization potency. An improved understanding of the specificities and neutralizing activities of antibodies induced by RSV infection in infants could facilitate the design of vaccine antigens that are less susceptible to interference by maternal antibody and that focus the response on epitopes associated with neutralizing activity.
Here, we sought to characterize the infant antibody response to mature, processed RSV F by isolating over 450 RSV F-specific antibodies from the peripheral B cells of seven RSV-infected infants. Infant responses were biased toward recognition of a limited number of antigenic sites that are distinct from those that dominate adult responses. Recognition of one of these sites was mediated by non-neutralizing antibodies that contained a convergent complementarity-determining region (CDR) H3 motif. A second site was recognized by antibodies that predominately utilized VH3-21:VL1-40 gene pairing, a subset of which displayed neutralizing activity despite lacking somatic mutations, suggesting that it may be possible to induce protective antibody responses in very young infants. Structural studies revealed the molecular basis for the conserved features of antibodies recognizing both sites, providing a blueprint for the rational design of vaccine immunogens capable of stimulating neutralizing antibody responses in young infants.
RESULTS
RSV F-specific antibodies isolated from young infants have low levels of SHM and biased V-gene usage
To analyze infant B cell responses to RSV F, we obtained blood samples from seven infants that were hospitalized due to complications associated with RSV infection. Of the seven infants, five were less than three months (< 3 mo.) and two were at least six months (≥ 6 mo.) of age at the time of hospitalization (Table S1). The youngest three infants were also ≤ 3 mo. at the time of blood draw. Six out of the seven infants were infected during the first RSV season of their life and were therefore likely experiencing a primary infection. One donor was 29.5 months old at the time of blood draw, but was also likely experiencing a primary infection because secondary RSV infections generally do not result in hospitalization (Glezen et al., 1986). To assess the magnitude of the B cell response to RSV F, peripheral blood mononuclear cells (PBMCs) were stained with fluorescently labeled tetramers of preF and postF trimers and analyzed by flow cytometry (Figures S1A and S1B). As expected, the frequency of class-switched B cells that were RSV F-specific was substantially lower in infants < 3 mo. compared with infants ≥ 6 mo. (Figure S1C). In infants < 3 mo., the frequency of RSV F-specific class-switched B cells ranged from 0.05–0.3%, whereas in infants ≥ 6 mo. the frequency ranged from 1.2–1.6%. Between 100 and 300 RSV F-reactive B cells from each donor were single-cell sorted and the antibody variable heavy (VH)- and variable light (VL)-chain sequences were rescued by single-cell PCR (Tiller et al., 2008). Due to the low frequency of RSV F-specific class-switched B cells in the five younger infants, all B cells that reacted with RSV F were single-cell sorted (Figure 1A). For the two infants that were ≥ 6 mo., only class-switched B cells were sorted (Figure 1B). Although all B cells that reacted with RSV F were sorted from infants < 3 mo., index sorting allowed for the determination of the B cell surface markers expressed on each sorted cell and revealed that 14–60% of the RSV F-specific B cells sorted from infants < 3 mo. were class-switched and/or CD27+, with the remaining B cells lacking classical memory markers (Figure 1C).
Figure 1. Anti-RSV F antibodies isolated from infant B cells display limited SHM and biased V-gene usage.
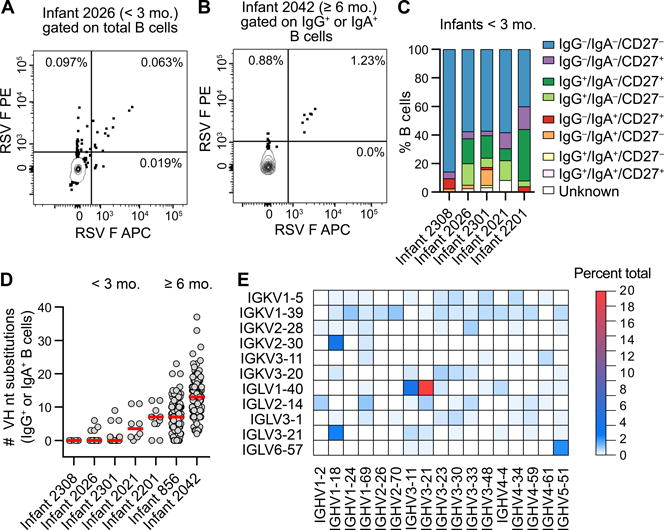
(A) Representative flow plot of the RSV F-specific B cell response in an infant < 3 mo. B cells were gated on CD14− CD8− CD3− CD19+ CD20+ before sorting on double-positive staining with dual-labeled RSV F probes. RSV F-specific B cells are in the upper-right quadrant. Results are representative of at least two independent experiments.
(B) Representative flow plot of the RSV F-specific B cell response in an infant ≥ 6 mo. B cells were gated on CD14− CD8− CD3− CD19+ CD20+ and IgG+ or IgA+ prior to sorting on double-positive staining with dual-labeled RSV F probes. RSV F-specific class-switched B cells are in the upper-right quadrant. Results are representative of at least two independent experiments.
(C) Index sort analysis of surface markers expressed on B cells from which RSV F-reactive antibodies were isolated. Infants are ordered from youngest to oldest at the time of hospitalization, left to right.
(D) Number of VH nucleotide substitutions for antibodies isolated from RSV F-specific class-switched B cells. Infants are ordered from youngest to oldest, left to right. Red bars indicate medians.
(E) Heat map of V-gene usage for all infants. Genes for which no VH:VL pairs were utilized in ≥ 0.5% of antibodies are omitted for clarity.
In total, over 450 cognate VH and VL pairs were cloned and expressed as full-length immunoglobulin Gs (IgGs). As expected, sequence analysis showed that the median level of SHM in antibodies isolated from class-switched B cells increased as a function of age (Figure 1D). Also, the majority of antibodies isolated from the youngest three infants lacked SHM, similar to what was observed previously in postF-reactive B cells (Williams et al., 2009). However, nearly 5% of antibodies isolated from these infants had VH genes containing at least five nucleotide substitutions, consistent with previous studies showing that SHM does occur in young infants, albeit at relatively low frequency (Rechavi et al., 2015; Ridings et al., 1998). The level of SHM in antibodies isolated from the two infants ≥ 6 mo. was relatively high, with a median of 7 and 13 VH nucleotide substitutions, respectively (Figure 1D). Analysis of V-gene usage showed that RSV F-reactive infant antibody responses were strongly biased toward either VH3-21:VL1-40 or the highly related VH3-11:VL1-40 gene pairing (Figure 1E). There was also a more modest preference for the VH1-18:VK2-30, VH1-18:VL3-21, and VH5-51:VL6-57 gene pairs (Figure 1E). The VH1-18:VK2-30 gene pair was present in 8.5% of RSV F-reactive antibodies isolated from adults and is associated with recognition of site V on preF (Gilman et al., 2016; Mousa et al., 2017). Overall, the results demonstrate that RSV infection induces B cell responses with higher levels of SHM in infants ≥ 6 mo. compared to < 3 mo., and that the responses in both age groups exhibit biased V-gene usage.
A subset of infant antibodies binds with high affinity to RSV F and potently neutralizes RSV
We next determined the apparent (IgG) binding affinity of each antibody for preF and postF. For the infants < 3 mo., 24–33% of the isolated antibodies bound to preF with affinities ≤ 5 nM compared with 45% and 91% for the two infants ≥ 6 mo. (Figure 2A, Medium and High). Although a total of 40 antibodies with preF affinities ≤ 5 nM were isolated from the youngest three infants, only two antibodies with ≤ 5 nM affinity for postF were isolated from these three infants (Figure 2B). In addition, the percentage of characterized antibodies from each infant that were preF-specific ranged from 41–100%, whereas substantially smaller percentages (0–18%) were postF-specific (Figure S2A). Antibodies recognizing both preF and postF comprised about 27–47% of the characterized antibody responses in the four oldest infants, but less than 10% of the response in the three youngest infants. Collectively, these results suggest that young infants generate a preF-biased antibody response that expands to include recognition of postF as infants age.
Figure 2. A subset of RSV F-specific infant antibodies binds with high affinity to RSV F and neutralizes RSV.
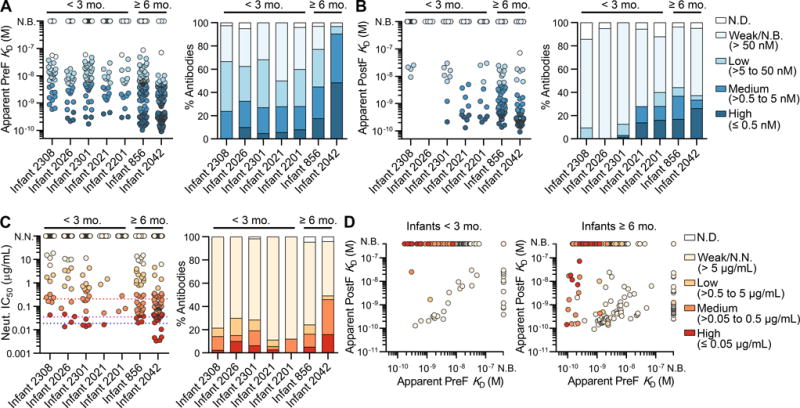
(A) Apparent (IgG) affinity of each antibody for preF (left). Percentage of antibodies with indicated affinities for preF (right). Infants are ordered from youngest to oldest, left to right. Coloring corresponds to the legend shown in (B). N.D., not determined; N.B., non-binding. Results are representative of at least two independent experiments.
(B) Same as (A), except that binding is to postF.
(C) Neutralization potency (IC50) of each antibody (left). Percentage of antibodies with indicated neutralization potencies (right). Red, purple, and blue dotted lines indicate the IC50 values for motavizumab, MPE8 and D25, respectively. N.N., non-neutralizing. Results are derived from a single experiment performed in duplicate.
(D) Affinities for postF plotted against affinities for preF, colored by neutralization potency, for antibodies isolated from infants < 3 mo. (left) and infants ≥ 6 mo. (right).
The antibodies were also tested for neutralizing activity using a high-throughput assay modified from a previously established flow-cytometry assay (Chen et al., 2010). This analysis revealed that 11–50% of the antibodies isolated from each infant had measurable neutralizing activity (IC50 < 100 μg/mL), and a subset of antibodies isolated from six out of the seven infants showed highly potent neutralizing activity (IC50 ≤ 0.05 μg/mL) (Figure 2C). Nearly 20% of the neutralizing antibodies lacked V-gene mutations (Table S2), suggesting that affinity maturation is not required for neutralization of RSV. However, the proportion of antibodies with medium-to-high neutralizing activity (IC50 ≤ 0.5 μg/mL) was about 3.5 times higher in the group of antibodies that contained SHM compared to those that lacked SHM, suggesting that affinity maturation does improve the overall response (Figure S2B). Analysis of the relationship between binding affinity and neutralization potency demonstrated that most highly potent neutralizing antibodies bound with high affinity to preF (KD ≤ 0.5 nM) and failed to bind to postF (Figure 2D). In addition, although approximately 20% of the neutralizing antibodies isolated from infants ≥ 6 mo. recognized both preF and postF, this type of neutralizing antibody was rare in infants < 3 mo. (Figures 2D and S2C), demonstrating that nearly all neutralizing antibodies in very young infants are preF-specific.
We next assessed the polyreactivity of the infant antibodies using a previously described assay (Jain et al., 2017). Although the fraction of medium-to-highly polyreactive antibodies was relatively low for all infants (≤ 15%), there was a higher proportion of polyreactive antibodies in the infants < 3 mo. compared to the infants ≥ 6 mo. (Figure S3A). This result could be related to differences in tolerance mechanisms in these two infant populations or to the higher frequency of antibodies lacking SHM in the younger infants. In support of the latter hypothesis, stratification of the antibodies based on their SHM levels showed that 12% of antibodies that lacked SHM displayed medium-to-high polyreactivity, compared with < 2% of antibodies that contained > 5 VH gene substitutions (Figure S3B). This is consistent with a prior study showing that the process of affinity maturation can result in decreased polyreactivity of human antibodies (Reed et al., 2016).
Infant antibody responses are focused primarily on two antigenic sites that have different neutralization sensitivities
To define the epitopes recognized by the infant antibodies, each antibody was tested for competition with a panel of known RSV F-specific antibodies and assigned to an antigenic site based on the competition profile (Figures 3A and 3B). In the three youngest infants, responses were dominated by antibodies directed against site III, whereas in the other infants a larger proportion of the responses were directed against site I, and in some cases site IV (Figure 3B). The proportion of antibodies recognizing preF-specific sites Ø and V at the apex of the preF molecule was low, particularly in the three youngest infants. Interestingly, analysis of the V-gene usage for the site I-directed antibodies revealed that over 25% utilized the VK1-39 light chain gene (Figure S4A). Although these site I-directed antibodies utilized a variety of VH genes, many possessed a convergent CDR H3 motif, generated from recombination of the DH3-22 and JH-4 genes (Figure S4B). In contrast, nearly 85% of antibodies that recognized site III utilized either the VH3-21:VL1-40 or the related VH3-11:VL1-40 gene pairing (Figure S4C) and did not show evidence of a convergent CDR H3 sequence (Figure S4B).
Figure 3. Infant responses are focused toward two antigenic sites with different neutralization sensitivities.
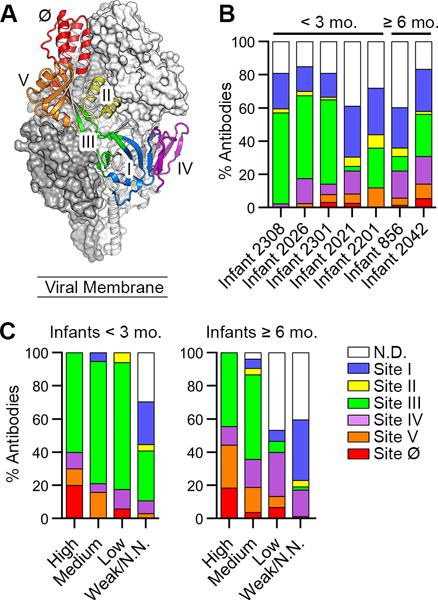
(A) The preF structure with two protomers displayed as gray molecular surfaces and one protomer displayed as ribbons colored according to antigenic site.
(B) Percentage of antibodies directed against each antigenic site, colored as in (A). Infants are ordered from youngest to oldest, left to right.
(C) Percentage of antibodies directed against each antigenic site—grouped according to neutralization potency—isolated from infants < 3 mo. (left) and infants ≥ 6 mo. (right). N.D., not determined; N.N., non-neutralizing.
The majority of site III-directed antibodies were preF-specific and nearly 50% had medium-to-high neutralizing activity, whereas antibodies that recognized site I preferentially bound to postF and tended to be weakly or non-neutralizing (Figures 3C, S4D and S4E). In infants < 3 mo., 60% of antibodies that displayed high neutralizing activity (IC50 ≤ 0.05 ug/mL) were directed against site III (Figure 3C). Therefore, although antibodies against both sites I and III are readily elicited during RSV infection in infants, site III-directed antibodies can potently neutralize RSV whereas site I-directed antibodies are typically non-neutralizing.
Site III-directed antibodies can neutralize RSV in the absence of SHM and are present in the naive B cell repertoire
We next analyzed the epitope specificities of the neutralizing antibodies that lacked SHM (Figure 4A). Of the 33 germline antibodies that displayed detectable RSV-neutralizing activity, 27 targeted antigenic site III. Analysis of the index sort data revealed that approximately half of the neutralizing germline antibodies originated from naïve B cells (IgG−IgA−CD27−) and the other half originated from memory B cells (IgG+, IgA+, and/or CD27+) (Figure 4B). The identification of site III-directed neutralizing antibodies from naïve B cells led us to investigate the occurrence of these antibody specificities in the naïve B cell repertoire. We therefore cloned and expressed 112 and 19 antibodies from RSV F-reactive cord blood B cells and healthy adult naïve B cells, respectively. Due to the low affinity of naïve B cell-derived antibodies, only 22/112 (20%) antibodies isolated from cord blood B cells and 9/19 (47%) antibodies isolated from adult naïve B cells bound with measurable affinity to RSV F as full-length IgGs. However, 11/22 (50%) and 7/9 (78%) of the RSV F-binding antibodies isolated from cord blood and adult naïve B cells, respectively, utilized VH3-21:VL1-40 or VH3-11:VL1-40 gene pairing (Figure 4C). Of these 18 antibodies, the 13 with binding affinities that allowed for analysis in a competition assay were all shown to recognize antigenic site III (Table S3). The affinities of these antibodies for preF were relatively high, ranging from 1.0–60 nM (Figure 4C). In addition, approximately 40% of these site-III directed antibodies were neutralizing, with IC50 values ranging from 1.5–4.4 μg/mL (Figure 4C). In contrast, none of the naïve B cell-derived antibodies that utilized other V-gene combinations showed detectable neutralizing activity (Figure 4C). Collectively, these results indicate that site III-directed antibodies can neutralize RSV in the absence of SHM and that these types of antibodies are present in the naïve B cell repertoire.
Figure 4. Germline antibodies that target antigenic site III can potently neutralize RSV and are present in the naïve B cell repertoire.
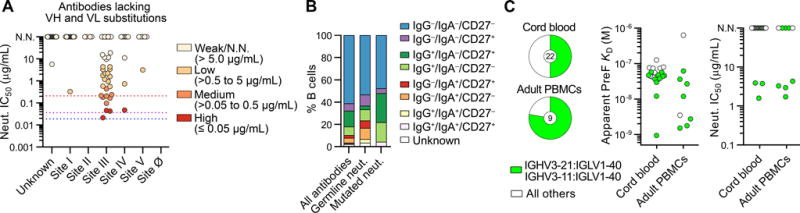
(A) Neutralization potency (IC50) of antibodies lacking V-gene nucleotide substitutions, grouped by antigenic site. IC50 values for motavizumab, MPE8, and D25 are shown with red, purple and blue dotted lines, respectively. N.N., non-neutralizing. No site Ø-directed antibodies lacking somatic hypermutation were isolated. Results are derived from a single experiment performed in duplicate.
(B) Index sort analysis of surface markers expressed on B cells from which RSV F-reactive antibodies were isolated for infants < 3 mo. Percentage of RSV F-specific antibodies derived from B cells with each surface phenotype is shown for all antibodies, neutralizing antibodies that lack somatic mutations (Germline neut.), and neutralizing antibodies that contain somatic mutations (Mutated neut.).
(C) Fraction of RSV F-reactive naïve B cells that utilized VH3-21:VL1-40 or VH3-11:VL1-40 gene pairing isolated from the cord blood of four donors (top) and peripheral blood of two adult donors (bottom). Naïve B cells were defined as CD14− CD8− CD3− CD19+ CD20+ IgM+ IgG− CD27− cells. The number in the center of the pie denotes the total number of antibodies with detectable binding to RSV F. Affinity of each of these antibodies for preF, colored according to V-gene usage (middle). Neutralization potency (IC50) for each antibody that displayed detectable binding (right). Results are derived from at least two independent experiments.
See also Figure S4, Table S2, and Table S3.
A site III-directed neutralizing antibody utilizes germline-encoded features of the VH3-21 and VL1-40 genes for high-affinity recognition of preF
To investigate the molecular basis of preferential V-gene pairing in antibodies targeting site III, we determined the 4.1-Å resolution crystal structure of ADI-19425 bound to a preF-stabilized variant of RSV F (PR-DM) (Figures 5A and S5A, Table S4) (Krarup et al., 2015). This antibody was potently neutralizing despite lacking SHM (Table S2, Figure S5B). The structure revealed that although the heavy chain accounts for roughly 65% of the buried surface area on preF, light chain residues Tyr31 (Kabat numbering) in CDR L1 and Tyr91 in CDR L3 are likely important, and contact the loop connecting α6 to α7 on preF (Figure 5B). Consistent with our structural analysis, substitution of Tyr31 or Tyr91 with alanine resulted in greater than 200- or 80-fold reductions in affinity, respectively, as measured by surface plasmon resonance (SPR) (Figure 5C).
Figure 5. Neutralizing antibody ADI-19425 uses germline-encoded features for high-affinity binding to antigenic site III on preF.
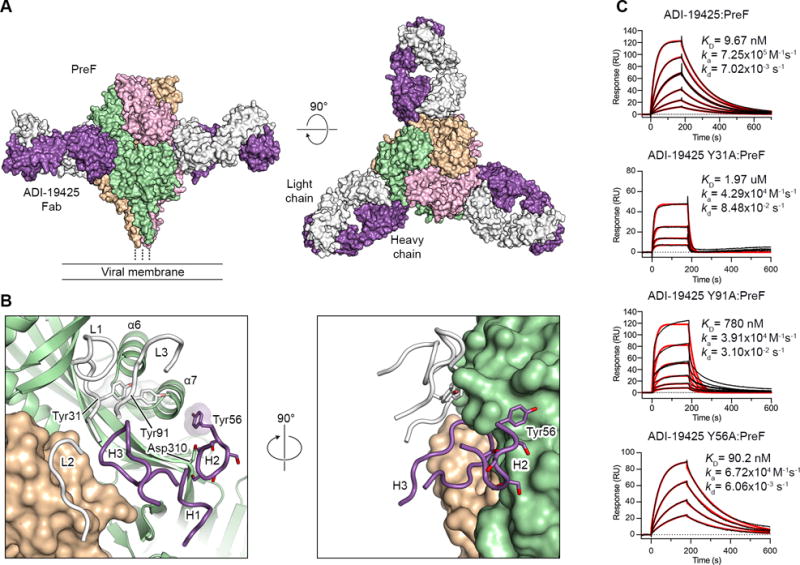
(A) Crystal structure of ADI-19425 Fab in complex with preF shown as molecular surfaces viewed along (left) and above (right) the viral membrane. The preF protomers are colored tan, pink and green, and the ADI-19425 heavy and light chains are colored purple and white, respectively.
(B) Magnified view of the antibody interface (left) and a 90° rotation about the vertical axis (right), colored as in (A). CDRs are shown as tubes and one RSV F protomer is shown as ribbons. Germline-encoded tyrosine and serine residues are shown as sticks with oxygen atoms colored red. Transparent molecular surfaces are shown for the three labeled tyrosine residues.
(C) Sensorgrams for the binding of ADI-19425 and the Y31A, Y91A and Y56A variants to preF measured using SPR. The data were double-reference subtracted (black) and fit to a 1:1 binding model (red). Results are representative of a single experiment.
In addition to the contacts formed by the light chain, the CDR H2 encoded by VH3-21 contains a stretch of five consecutive serine residues, three of which are near Asp310 on β6 of preF and could form hydrogen bonds with this residue. The VH3-11 and VH3-48 genes, which share a high degree of sequence identity with VH3-21, were utilized by site III-directed antibodies at a much lower frequency than VH3-21. One explanation for this could be the presence of a tyrosine residue (Tyr56) directly following the polyserine motif in VH3-21. Our structure shows that Tyr56 of ADI-19425 is buried in a small groove neighboring antigenic site II on preF (Figure 5B). This tyrosine is not present in VH3-48 and is only present in some alleles of VH3-11. Although we isolated three site III-directed antibodies that utilized either VH3-48 or VH3-11 and contained a threonine at position 56 (ADI-19461, ADI-22776 and ADI-19440), these had low affinity for preF and were non-neutralizing (Table S2). Consistent with these observations, substitution of Tyr56 with alanine resulted in a more than 9-fold decrease in affinity to preF (Figure 5C).
In contrast to the V-gene-specific features highlighted above, there were fewer restrictions on the sequences of the CDR H3s of site III antibodies that utilized VH3-21:VL1-40 or VH3-11:VL1-40 gene pairing. The CDR H3s varied from 10 to 20 amino acids in length, with some preference toward usage of glycine, serine and tyrosine residues at positions 96–100c (Figure S4B). Our structure reveals that although the ADI-19425 CDR H3 buries approximately 250 Å2 on preF, it does not form hydrogen bonds or salt bridges with either protomer (Figure 5B). Notably, the neutralizing antibody MPE8, which has recently been structurally characterized (Wen et al., 2017), also utilizes the VH3-21 and VL1-40 gene pair to recognize site III with a binding mode nearly identical to that of ADI-19425, despite a substantially different CDR H3 (Figures S5C–D). This is consistent with our observation that multiple CDR H3 sequences can be utilized by this family of antibodies to recognize preF.
A site I-directed non-neutralizing antibody recognizes postF using a convergent CDR H3 motif and germline-encoded regions of the VK1-39 light chain
We determined the 3.0-Å resolution structure of a site I-directed antibody, ADI-14359, in complex with postF, to define the molecular determinants of the convergent antibody features (Figure 6A, Table S4). The structure revealed that the 18-amino-acid long CDR H3, generated from the convergent usage of DH3-22/JH-4, is partially inserted into a small groove near the top of the postF trimer (Figure 6B) and makes a number of hydrogen bonds with postF residues in and around this groove (Figure 6C). CDR H3 residues Tyr100c and Tyr100d, which are uncommon in D genes other than DH3-22, form hydrogen bonds with postF residues Glu31 and Glu378, respectively, which are located on the ridge surrounding the groove. The tip of the CDR H3 loop is composed of three small amino acids that allow it to to stack against Trp314, which is positioned at the floor of the groove. These small residues also make hydrogen bonds with residues Asp344 and Asn380 of postF. In addition, heavy chain residue Tyr100g, which is unique to the JH-4 gene, stacks against Tyr49 in the light chain, which may help properly orient the CDR H3.
Figure 6. Non-neutralizing antibody ADI-14359 uses a convergent CDR H3 motif and germline features of the VK1-39 light chain for binding to antigenic site I on postF.
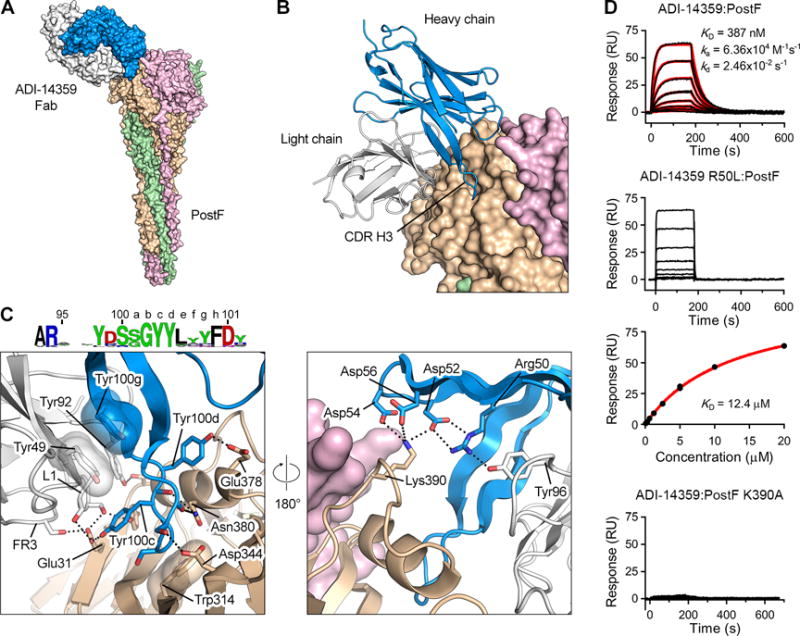
(A) Crystal structure of ADI-14359 Fab (VH2-70:VK1-39) in complex with postF shown as molecular surfaces with the three postF protomers colored in tan, pink and green, and the ADI-14359 heavy and light chains colored blue and white, respectively.
(B) The variable domain of ADI-14359 is shown as ribbons and postF is shown as molecular surfaces.
(C) Magnified view of the antibody interface (left) and a 180° rotation about the vertical axis (right), colored as in (A). The variable domain of ADI-14359 and one postF protomer are shown as ribbons. Selected residues are shown as sticks with oxygen atoms colored red and nitrogen atoms colored blue. Hydrogen bonds and salt bridges are depicted as black dotted lines. The residues in the sequence logo of the convergent CDR H3 are colored according to chemical property (Crooks et al., 2004). FR3, framework region 3.
(D) Sensorgram for the binding of ADI-14359 to post F measured using SPR (top). The data were double-reference subtracted (black) and fit to a 1:1 binding model (red). Rate constants for the R50L variant binding to postF were too fast to be accurately determined (second from top) and therefore the equilibrium responses were plotted against the concentration of Fab and fit to a steady-state affinity model (red line) (third from top). Binding of ADI-14359 to the K390A variant of postF was too weak to determine an affinity (bottom). Results are representative of a single experiment.
Antibodies that utilized the convergent CDR H3 were biased toward pairing with the VK1-39 light chain gene. Several germline-encoded residues within CDR L1 and the framework region 3 (FR3) of VK1-39 form hydrogen bonds with Glu31 on postF (Figure 6C). The light chain of ADI-14359 is predicted to clash substantially with β22 of preF, which rearranges during the transition to postF (Figure S6). Therefore, preferential binding to postF by this type of antibody appears to be mediated by the light chain. In addition, Tyr92 at the start of the CDR L3 is a unique feature of VK1-39, and forms a hydrogen bond with Ser35 on the F2 subunit of postF. These features, combined with the interaction of Tyr49 of VK1-39 with Tyr100g of the CDR H3, may help to explain the preferential usage of VK1-39 by this group of antibodies.
Although site I-directed antibodies did not show convergent VH-gene usage, the heavy chain utilized by ADI-14359 also makes critical interactions with postF (Figure 6C). Arg50 forms a salt bridge with Asp52 in the CDR H2 and appears to aid in coordinating an electrostatic interaction between Asp52, Asp54 and Asp56 of the CDR H2 with Lys390 on postF. Arg50 also forms a hydrogen bond with light-chain residue Tyr96, which is unique to the IGK-J2 gene utilized by ADI-14359. To investigate the contribution of these VH-gene-mediated interactions to binding, we generated an R50L variant of ADI-14359, since a leucine is present at position 50 in other alleles of VH2-70. The affinity of the R50L variant for postF was reduced by more than 30-fold as measured by SPR (Figure 6D). In addition, substitution of Lys390 with alanine (K390A) on postF almost entirely ablated ADI-14359 binding (Figure 6D). The presence of three acidic residues in the CDR H2 therefore appears to be critical for the interaction of ADI-14359 with postF. However, the contribution of the CDR H2 to binding may vary among other members of this group, since many of the VH germlines utilized lack this acidic motif.
DISCUSSION
Our results demonstrate that infant antibody responses to RSV F differ substantially from those of healthy adults, not only in affinity and neutralization potency, but also in the patterns of epitope recognition. The infant responses were focused on two major regions of the RSV F trimer—antigenic sites I and III—neither of which are dominant in adult responses (Gilman et al., 2016). These differences were the most extreme in infants under three months of age, with infants older than six months exhibiting responses that began to resemble healthy adults. This observation is consistent with previous studies showing that the infant immune system begins to mature at around six months of age (Ridings et al., 1998).
The majority of antibodies that recognized antigenic site III utilized the same V-gene pairing, but were not restricted in D- and J-gene usage. A subset of these antibodies showed potent neutralizing activity despite lacking SHM. Recent work has shown that polyclonal IgM antibodies purified from RSV-naïve infant sera are capable of neutralizing RSV, and it was suggested that these antibodies may represent natural anti-RSV antibodies that react with the N- and O-linked glycans present on the RSV surface glycoproteins (Jans et al., 2017). However, unlike natural IgM antibodies—which rely on avidity, typically recognize common surface antigens, and exhibit some degree of polyreactivity (Panda and Ding, 2015)—the site III-directed antibodies described here bound with high affinity in an IgG backbone, specifically recognized an epitope on RSV F that lacks N-linked glycans, and generally showed little to no polyreactivity, suggesting that they are distinct from previously described natural IgM antibodies. Similar germline-mediated recognition in the adaptive immune response has also been described for other viral pathogens, including hepatitis C virus (Bailey et al., 2017) and human cytomegalovirus (Thomson et al., 2008), and for bacterial pathogens such as Staphylococcus aureus (Yeung et al., 2016). The presence of functional germline antibodies in the human antibody repertoire has been proposed to serve as a type of innate humoral response to life-threatening pathogens that are likely to be encountered early in life (Lerner, 2011). The isolation of this class of antibodies from all seven infants studied here, as well as from cord blood B cells, adult naïve B cells, and memory B cells from previously characterized adult donors (Gilman et al., 2016), suggests that the naïve B cell precursors encoding these antibody specificities are likely present in most individuals and that these B cells can be activated in response to antigen exposure and undergo affinity maturation. This is in contrast to HIV broadly neutralizing antibodies, which only develop in a subset of infected individuals and whose inferred germline precursors display limited reactivity with native HIV envelope antigens (Burton and Hangartner, 2016). Although further studies will be required to determine the levels and function of these antibodies in infant sera and elucidate their role in protection from severe RSV disease, our results suggest that expansion of these cells may be a feasible goal for infant vaccination strategies.
Neutralizing antibodies that react with both preF and postF were identified in healthy adults and infants over 6 months old, but were almost entirely absent in the youngest infants analyzed here. Although postF antigens are capable of eliciting neutralizing antibodies that also bind to preF, their inability to elicit preF-specific antibodies would likely prove problematic for use in a young infant population. In addition, our results show that a relatively large fraction of the infant antibody response is directed against antigenic site I, which is well-exposed on postF. Antibodies targeting this site generally lacked neutralizing activity, suggesting that vaccination with postF antigens could drive infant antibody responses toward ineffective recognition of RSV F. Formalin-inactivated RSV (FI-RSV), the preparation that resulted in vaccine-enhanced disease in infants in the 1960s, displays an abundance of postF on the surface of the virus (Killikelly et al., 2016). Although many factors contribute to the development of vaccine-enhanced disease (Acosta et al., 2015), our data suggest that the high abundance of postF on FI-RSV could result in the induction of high levels of site I-directed antibodies and a low fraction of neutralizing antibodies, which is a property associated with the formation of immune complexes that contribute to lung pathology in vaccine-enhanced illness (Murphy and Walsh, 1988; Polack et al., 2002).
We observed an age-dependent increase in the response against antigenic sites Ø and V, which are both present near the apex of the preF trimer. Although infant antibodies that targeted these epitopes tended to be potently neutralizing, they were present at low abundance, particularly in infants under three months of age. These data suggest that although the presence of site Ø is likely important for generating neutralizing antibody responses later in life, eliciting a neutralizing response in young infants may depend on the presentation of antigenic site III. These differences in the dominant epitopes targeted by infant and adult responses provide a unique opportunity for prevention strategies that seek to combine passive and active immunization. For example, vaccines could be designed to preferentially elicit site III antibodies, which would not compete for binding with second-generation prophylactic antibodies that target antigenic site Ø, such as MEDI8897 (Griffin et al., 2017; Zhu et al., 2017). In addition, antibodies elicited by a site-III-specific vaccine would not block access to the apex of the preF trimer on infectious virions, allowing the development of neutralizing antibodies directed against antigenic sites in this region to occur during RSV infection.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Laura Walker (laura.walker@adimab.com).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Families of infants were approached at the time of hospitalization for documented RSV infection. At that point, a Dartmouth Committee for the Protection of Human Subjects approved consent was signed to obtain 5–10 cc of blood approximately 1–10 months after discharge from the Children’s Hospital at Dartmouth (CHaD). Families were contacted at the planned time for phlebotomy and arrangements were made for blood to be drawn either at CHaD or at a medical facility closer to their home. All infants were between 0.35-26 months of age at the time of hospitalization. Three of the infants were male and four were female.
METHOD DETAILS
Production of RSV F sorting probes
To generate sorting probes with high avidity, and uniformly oriented F proteins, and reduced propensity for “daisy-chaining”, we produced preF (DS-Cav1) and postF (F ΔFP) trimers with a single biotinylated C-terminal AviTag before coupling to streptavidin-PE or -APC (Gilman et al., 2016). For each variant, expression vectors containing either a C-terminal 6x His-tag–AviTag or a C-terminal Strep-tag II were co-transfected into FreeStyle 293-F cells at a 1:2 ratio. The protein was purified from the cell supernatant using Ni–nitrilotriacetic acid (NTA) resin to remove trimers lacking the 6x His-tag–AviTag. The elution from the Ni-NTA column was then purified over StrepTactin resin, which was washed thoroughly to remove trimers containing only one StrepTagII. The elution from the StrepTactin resin therefore contained trimers with two StrepTagII trimers and one 6x His-tag–AviTag trimer. These proteins were then biotinylated using birA (Avidity) and then separated from excess biotin by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) in PBS.
Single B cell sorting
PBMCs from RSV-infected infants were stained using anti-human IgG (BV605), IgA (FITC), CD27 (BV421), CD8 (PerCP-Cy5.5), CD14 (PerCP-Cy5.5), CD3 (PerCP-Cy5.5), CD19 (PECy7), CD20 (PECy7) and a mixture of dual-labeled preF and postF tetramers (50 nM each). For infants ≥ 6 months of age, class-switched B cells binding to RSV F tetramers were single-cell sorted. For infants < 3 months of age, total B cells binding to RSV F tetramers were single-cell sorted. For naïve B cell sorting, cord blood or PBMCs from healthy adult donors were stained with anti-human IgG (BV605), IgM (FITC), CD27 (BV421), CD8 (PerCP-Cy5.5), CD14 (PerCP-Cy5.5), CD19 (PECy7), CD20 (PECy7), CD21 (APC-Cy7) and a mixture of dual-labeled preF and postF tetramers (50 nM each). CD8–CD14–CD3–CD19+CD20+IgG–IgM+CD27– B cells binding to RSV F tetramers were single-cell sorted. Tetramers were prepared fresh for each experiment. Single cells were sorted on a fluorescence-activated cell sorter (FACS) Aria II (BD Biosciences) into 96-well PCR plates (BioRad) containing 20 μL/well of lysis buffer [5 μL of 5X first strand cDNA buffer (Invitrogen), 0.25 μL RNaseOUT (Invitrogen), 1.25 μL dithiothreitol (Invitrogen), 0.625 μL NP-40 (New England Biolabs), and 12.6 μL dH2O]. Plates were immediately stored at −80 °C.
Single B cell cloning
Antibody variable genes (IgH, Igλ and Igκ) were amplified by RT-PCR and nested PCR reactions using cocktails of IgG-, IgM-, and IgA-specific primers (Tiller et al., 2008). The primers used in the second round of PCR contained 40 base pairs of 5ʹ and 3ʹ homology to the digested expression vectors to allow for cloning by homologous recombination into Saccharomyces cerevisiae (Swers et al., 2004). PCR products were cloned into S. cerevisiae using the lithium acetate method for chemical transformation (Gietz and Schiestl, 2007). Each transformation reaction contained 10 μL of unpurified heavy chain and light chain PCR product and 200 ng of cut heavy and light chain plasmids. Following transformation, individual yeast colonies were picked for sequencing and characterization.
Expression and purification of IgGs and Fab fragments
RSV F-specific IgGs were expressed in S. cerevisiae cultures grown in 24-well plates. After 7 days, the supernatant was harvested and IgGs were purified with Protein A. IgGs were eluted with 200 mM acetic acid, 50 mM NaCl pH 3.5 into 1/8th volume 2 M HEPES pH 8.0. IgGs were then buffer exchanged into PBS pH 7.0. Fab fragments used for competition assays were generated by digesting the IgGs with papain for 2 h at 30 °C. The digestion was terminated by the addition of iodoacetamide, and the Fab and Fc mixtures were passed over Protein A agarose to remove Fc fragments and undigested IgG. The flowthrough of the Protein A resin was then passed over CaptureSelect™ IgG-CH1 affinity resin (ThermoFischer Scientific), and eluted with 200 mM acetic acid, 50 mM NaCl pH 3.5 into 1/8th volume 2 M HEPES pH 8.0. Fab fragments were then buffer exchanged into PBS pH 7.0.
Biolayer interferometry binding analysis
IgG binding affinities for DS-Cav1 and RSV F ΔFP were measured by BLI using a FortéBio Octet HTX instrument (Pall Life Sciences). For high-throughput KD determination, IgGs were immobilized on AHQ sensors (Pall Life Sciences) and exposed to 100 nM antigen in PBS containing 0.1% BSA (PBSF) for an association step, followed by a dissociation step in PBSF buffer. Data were analyzed using the FortéBio Data Analysis Software 7. IgG KDs were calculated for antibodies with BLI responses >0.1 nm. The data were fit to a 1:1 binding model to calculate association and dissociation rate constants, and KD was calculated using the ratio kd/ka.
Antibody competition assays
Antibody competition was measured by the ability of a control anti-RSV F Fab to inhibit binding of yeast surface-expressed anti-RSV F IgGs to either DS-Cav1 or F ΔFP. 50 nM biotinylated DS-Cav1 or F ΔFP was pre-incubated with 1 μM competitor Fab for 30 min at room temperature and then added to a suspension of yeast expressing anti-RSV F IgG. Unbound antigen was removed by washing with PBS containing 0.1% BSA (PBSF). After washing, bound antigen was detected using streptavidin Alexa Fluor 633 at a 1:500 dilution (Life Technologies) and analyzed by flow cytometry using a FACSCanto II (BD Biosciences). The degree of competition was analyzed by measuring the fold reduction in antigen binding in the presence of competitor Fab relative to an antigen-only control. Antibodies that showed a greater than five-fold reduction in binding in the presence of competitor Fab were considered competitors.
Polyreactivity assay
Antibody polyreactivity assay was performed essentially as described previously (Jain et al., 2017). IgGs presented on the surface of yeast were incubated with biotinylated CHO cell membrane preparations and incubated on ice for 20 minutes. Cells were then washed and re-suspended in secondary antibody mix (Extravidin-R-PE, anti-human LC-FITC, and propidium iodide). The mixture was incubated on ice for 20 minutes and then washed twice with PBSF. Cells were then re-suspended in PBSF and run on a FACSCanto II (BD Biosciences). The mean fluorescence intensities of binding were normalized using control antibodies that display high, medium, or low polyreactivity to assess non-specific binding.
High-throughput fluorescence plate reader neutralization assay
A total of 2.4 × 104 HEp-2 cells/well in 30 μL culture medium were seeded in 384-well black optical-bottom plates (Nunc®384-well plates, Thermo Scientific). Antibodies were diluted four-fold starting at 100 μg/mL. An equal volume of recombinant mKate-RSV A2 or mKate-RSV B 18537 was then added and incubated at 37 °C for 1hr. After incubation, 50 μL of the antibody–virus mixture was added to the HEp-2 cells and incubated at 37 °C for 22–24 hours. After incubation, the fluorescence intensity of each well was measured using a microplate reader at an excitation of 588 nm and an emission of 635 nm (SpectraMax Paradigm, Molecular Devices). Neutralization IC50s were calculated using GraphPad Prism (GraphPad Software Inc.).
Production of ADI-14359, ADI-19425 and AM22 Fabs and variants
Plasmids encoding the heavy and light chains of ADI-14359, ADI-19425 or AM22 were co-transfected at a 1:1 ratio into Expi293F cells. Point mutants were generated using MegaPrimer PCR and were expressed in FreeStyle 293-F cells. Fabs were purified using CaptureSelect IgG-CH1 affinity matrix (Life Technologies) and were further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare).
Production of protein complexes for crystallization
Mammalian expression vectors encoding either PR-DM (preF) (Krarup et al., 2015) or RSV F ΔFP (postF) with a C-terminal HRV 3C cleavage site, 8x His-tag and StrepTagII were transfected into FreeStyle 293-F cells and 5 μM kifunensine was added approximately 4 hours after transfection. The secreted proteins were purified using Strep-Tactin resin (IBA), then treated with 10% (wt/wt) EndoH to remove N-linked glycans, followed by 10 U/mg of HRV 3C to remove tags. The proteins were then purified by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) in buffer containing 2 mM Tris pH 8, 200 mM NaCl and 0.02% NaN3.
To produce the ADI-14359 Fab–postF complex, purified postF was combined with a 1.5-fold molar excess of ADI-14359 Fab relative to each postF protomer and incubated at room temperature for approximately 30 minutes. Excess Fab was separated from the complex by size-exclusion chromatography using a Superose 6 column (GE Healthcare) in buffer containing 2 mM Tris pH 8, 200 mM NaCl and 0.02% NaN3. The complex eluted with a retention volume indicative of a complex with 1–2 Fabs bound per postF trimer, suggesting that ADI-14359 Fab may bind sub-stoichiometrically to postF.
To produce the ADI-19425–AM22–preF ternary complex, purified PR-DM was combined with a 1.5-fold molar excess of both ADI-19425 Fab and AM22 Fab relative to each preF protomer. Binding took place at room temperature for roughly 30 minutes before the ternary complex and excess Fab were separated by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) in 2 mM Tris pH 8, 200 mM NaCl and 0.02% NaN3.
Crystallization and data collection
The ADI-14359 Fab–postF complex was crystallized by the hanging-drop vapor-diffusion method by mixing 1.3 μL of protein at a concentration of 4.45 mg/mL with 0.7 μL of reservoir solution composed of 13% (w/v) polyethylene glycol (PEG) 8000 and 0.43 M ammonium citrate pH 8.5. Cryo-preservation was performed by hanging the looped crystal over a 1 M sodium chloride solution for approximately 2 minutes prior to plunge freezing in liquid nitrogen. Data were collected to 3.0 Å resolution at SSRL (Stanford Synchrotron Radiation Lightsource, National Accelerator Laboratory).
The unbound ADI-19425 Fab was initially crystallized using the sitting-drop vapor-diffusion method by combining 50 nL protein at 8.78 mg/mL with 100 nL reservoir solution containing 2.0 M ammonium sulfate and 0.1 M HEPES pH 7.5. These crystals were vortexed in the presence of a polytetrafluoroethylene bead to generate a seed solution used for microseed matrix screening. The final crystals were obtained using 150 nL protein at 8.78 mg/ml, 50 nL seed solution and 100 nL reservoir solution containing 1.5 M ammonium sulfate, 0.1 M sodium chloride, and 0.1 M Bis-Tris pH 6.5. Crystals were soaked in a solution of reservoir containing a final concentration of 2.5 M ammonium sulfate before being frozen in liquid nitrogen. Data were collected to 1.7 Å resolution at the SBC beamline 19-BM (Advanced Photon Source, Argonne National Laboratory).
The ADI-19425–AM22–preF ternary complex was crystallized by the sitting-drop vapor-diffusion method by combining 100 nL of protein solution at a concentration of 4.80 mg/mL with 100 nL of reservoir solution containing 0.1 M sodium citrate pH 5.5, 10% isopropanol and 10% (w/v) PEG 4000. Crystals were soaked in a cryoprotectant solution containing reservoir solution plus 15% 2R,3R-butanediol before being frozen in liquid nitrogen. Data were collected to 4.1-Å resolution at the SBC beamline 19-ID (Advanced Photon Source, Argonne National Laboratory).
Structure determination, model building and refinement
Diffraction data were indexed and integrated using iMOSFLM (Battye et al., 2011) and merged and scaled with AIMLESS (Evans and Murshudov, 2013). Molecular replacement solutions were obtained with PHASER (McCoy et al., 2007) and the structures were refined using PHENIX (Adams et al., 2002) and built manually using Coot (Emsley and Cowtan, 2004). Software used for processing and visualization of X-ray diffraction data was curated by SBGrid and accessed using the CCP4i interface (Collaborative Computational Project, 1994; Morin et al., 2013; Potterton et al., 2003). Data collection and refinement statistics for the three crystal structures are presented in Table S4.
The ADI-14359–postF complex formed crystals in space group P212121 and a molecular replacement solution was found using the previously solved postF structure (PDB ID: 3RRT), the heavy chain from 2D1 Fab (PDB ID: 3QHZ), and the light-chain from 5-51/O12 Fab (PDB ID: 4KMT) as search models. The asymmetric unit contained one postF trimer with only one ADI-14359 Fab bound per trimer. The model was built manually in Coot and refined in PHENIX using non-crystallographic symmetry (NCS) restraints and reference model restraints to an Rwork/Rfree of 22.2/25.5%.
The unbound ADI-19425 Fab also formed crystals in P212121, and the heavy chain from MJ5 Fab (PDB ID: 3EYQ) and the light chain from LDLR-competitive Fab (PDB ID: 3H42) were used as search models in molecular replacement. The structure was manually built in Coot and refined in PHENIX to an Rwork/Rfree of 17.4/20.4%. The ADI-19425–AM22–preF complex formed crystals in space group P41212 and a molecular replacement solution was found using the refined structures of the unbound ADI-19425 Fab and the complex of preF bound to AM22 Fab as search ensembles. The asymmetric unit contained a single preF trimer bound by three molecules of AM22 Fab and three molecules of ADI-19425 Fab. The model was built manually in Coot and refined in PHENIX using NCS restraints and reference model restraints to an Rwork/Rfree of 20.4/25.6%.
Fab affinity measurements for ADI-14359, ADI-19425 and variants
The affinity of ADI-14359 Fab for postF was measured using SPR. Purified postF (RSV F ΔFP) with a C-terminal HRV 3C cleavage site, 8x His-tag and StrepTagII was captured on the sample flow cell of an NTA sensor chip to approximately 115 RU per cycle using a Biacore X100 (GE Healthcare). The NTA chip was regenerated between each cycle with 0.35 M EDTA followed by 0.5 mM NiCl2. A buffer-only reference sample (HBS-P+ pH 8) was injected over both flow cells, followed by a 2-fold serial dilution of ADI-14359 Fab from 800 nM to 6.25 nM, starting with the lowest concentration, with a duplication of the 100 nM sample. The data were double-reference subtracted, then fit to a 1:1 binding model using Scrubber. Binding of ADI-14359 Fab to the postF K390A variant was measured in a similar manner, with capture of approximately 100 RU per cycle and injection of a buffer-only reference, followed by a 2-fold serial dilution of Fab from 1600 nM to 6.25 nM with a duplication of the 100 nM concentration. The data were double-reference subtracted, but the total response was too low to allow an affinity to be calculated. For the germline variant of ADI-14359 (R50L), approximately 115 RU of postF was captured on the NTA chip before injection of a buffer-only reference, followed by a 2-fold serial dilution of ADI-14359 R50L Fab from 20 μM to 78 nM. The data were double reference subtracted and fit using a steady-state affinity model in Scrubber.
Similar SPR experiments were performed to measure the binding between ADI-19425 Fab and preF. Purified preF (DS-Cav1) with a C-terminal 8x His-tag and AviTag was captured on the sample flow cell of an NTA sensor chip to approximately 150 RU. A buffer-only reference sample (HBS-P+ pH 8.0) was injected over both the sample and reference flow cells, followed by a 2-fold serial dilution of ADI-19425 Fab from 40 nM to 1.25 nM, with a duplication of the 10 nM concentration. For the ADI-19425 Fab variants (heavy chain Y56A, light chain Y31A, and light chain Y91A), roughly 150 RU of preF was captured on the NTA chip before the injection of a buffer-only reference, followed by a 2-fold serial dilution of ADI-19425 Fab variant from 250 nM to 31.25 nM, with a duplication of the 250 nM concentration. The data were double reference subtracted and fit using a 1:1 binding model in Scrubber.
QUANTIFICATION AND STATISTICAL ANALYSIS
Neutralization curves were fit by nonlinear regression using GraphPad prism software.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Antibody sequences were deposited in GenBank. GenBank accession numbers are listed in Table S2. Atomic coordinates and structure factors for the ADI-14359–postF complex structure, the unbound ADI-19425 Fab, and the ADI-19425–AM22–preF complex structure have been deposited with the Protein Data Bank under accession codes 6APB, 6APC, and 6APD.
Supplementary Material
Acknowledgments
We thank Emilie Shipman for assistance with protein production, members of the McLellan laboratory for critical reading of the manuscript, and Cody Williams for assistance preparing the graphical abstract. PBMC processing was carried out in DartLab, the Immune Monitoring and Flow Cytometry Shared Resource, supported by a National Cancer Institute Cancer Center Support Grant to the Norris Cotton Cancer Center (P30CA023108-37) and an Immunology COBRE Grant (P30GM103415-15) from the National Institute of General Medical Sciences. Results shown in this report are derived in part from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, as well as the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract No. DE-AC02-06CH11357. The Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). Support for this work was provided by the National Institute of General Medical Sciences of the NIH awards T32GM008704 (M.S.A.G.) and P20GM113132 (J.S.M.) and by intramural funding from the National Institute of Allergy and Infectious Diseases to support work at the Vaccine Research Center (B.S.G.).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, P.F.W, L.M.W, B.S.G, and J.S.M.; Investigation, M.G., E.G., R.I.C., M.C., S.M.M., J.O.N., A.S., and D.W.; Writing – Original Draft, M.G.; Writing – Reviewing & Editing, M.G., D.W., L.M.W, E.G., R.I.C, M.C., and J.S.M.; Visualization, M.G. and D.W.; Supervision, L.M.W., B.S.G. and J.S.M.
DECLARATION OF INTERESTS
L.M.W. is an inventor on pending patent applications describing the RSV antibodies (“Anti-respiratory syncytial virus antibodies, and methods of their generation and use,” USSN 62/411,500, USSN 62/411,508, and USSN 62/411,510). M.C., J.S.M., and B.S.G. are inventors on a patent entitled “Prefusion RSV F proteins and their use,” (US patent No. 9,738,689). L.M.W., A.S., and E.G. have an equity position in Adimab LLC.
References
- Acosta PL, Caballero MT, Polack FP. Brief History and Characterization of Enhanced Respiratory Syncytial Virus Disease. Clin Vaccine Immunol. 2015;23:189–195. doi: 10.1128/CVI.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Anderson LJ, Dormitzer PR, Nokes DJ, Rappuoli R, Roca A, Graham BS. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine. 2013;31(Suppl 2):B209–215. doi: 10.1016/j.vaccine.2012.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LJ, Hierholzer JC, Stone YO, Tsou C, Fernie BF. Identification of epitopes on respiratory syncytial virus proteins by competitive binding immunoassay. J Clin Microbiol. 1986;23:475–480. doi: 10.1128/jcm.23.3.475-480.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, Flyak AI, Cohen VJ, Li H, Wasilewski LN, Snider AE, Wang S, Learn GH, Kose N, Loerinc L, et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. 2017;2:92872. doi: 10.1172/jci.insight.92872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battles MB, Langedijk JP, Furmanova-Hollenstein P, Chaiwatpongsakorn S, Costello HM, Kwanten L, Vranckx L, Vink P, Jaensch S, Jonckers TH, et al. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat Chem Biol. 2016;12:87–93. doi: 10.1038/nchembio.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol. 1989;63:2941–2950. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chang JS, Nason M, Rangel D, Gall JG, Graham BS, Ledgerwood JE. A flow cytometry-based assay to assess RSV-specific neutralizing antibody is reproducible, efficient and accurate. J Immunol Methods. 2010;362:180–184. doi: 10.1016/j.jim.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Esposito S, Scarselli E, Lelii M, Scala A, Vitelli A, Capone S, Fornili M, Biganzoli E, Orenti A, Nicosia A, et al. Antibody response to respiratory syncytial virus infection in children <18 months old. Hum Vaccin Immunother. 2016;12:1700–1706. doi: 10.1080/21645515.2016.1145847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S, Coyle EM, Beeler J, Golding H, Khurana S. Antigenic Fingerprinting following Primary RSV Infection in Young Children Identifies Novel Antigenic Sites and Reveals Unlinked Evolution of Human Antibody Repertoires to Fusion and Attachment Glycoproteins. PLoS Pathog. 2016;12:e1005554. doi: 10.1371/journal.ppat.1005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barreno B, Palomo C, Penas C, Delgado T, Perez-Brena P, Melero JA. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J Virol. 1989;63:925–932. doi: 10.1128/jvi.63.2.925-932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol. 2016;1:eaaj1879. doi: 10.1126/sciimmunol.aaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- Graham BS. Vaccine development for respiratory syncytial virus. Curr Opin Virol. 2017;23:107–112. doi: 10.1016/j.coviro.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin MP, Khan AA, Esser MT, Jensen K, Takas T, Kankam MK, Villafana T, Dubovsky F. Safety, Tolerability, and Pharmacokinetics of MEDI8897, the Respiratory Syncytial Virus Prefusion F-Targeting Monoclonal Antibody with an Extended Half-Life, in Healthy Adults. Antimicrob Agents Chemother. 2017;61:e01714–01716. doi: 10.1128/AAC.01714-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis. 1991;163:693–698. doi: 10.1093/infdis/163.4.693. [DOI] [PubMed] [Google Scholar]
- Homaira N, Rawlinson W, Snelling TL, Jaffe A. Effectiveness of Palivizumab in Preventing RSV Hospitalization in High Risk Children: A Real-World Perspective. Int J Pediatr. 2014;2014:571609. doi: 10.1155/2014/571609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Incognito L, Cheng X, Ulbrandt ND, Wu H. Respiratory syncytial virus-neutralizing monoclonal antibodies motavizumab and palivizumab inhibit fusion. J Virol. 2010;84:8132–8140. doi: 10.1128/JVI.02699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci U S A. 2017;114:944–949. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Pettengill M, Kim D, van der Made C, de Groot R, Henriet S, de Jonge MI, Ferwerda G, Levy O. Human newborn B cells mount an interferon-α/β receptor-dependent humoral response to respiratory syncytial virus. J Allergy Clin Immunol. 2017;139:1997–2000 e1994. doi: 10.1016/j.jaci.2016.10.032. [DOI] [PubMed] [Google Scholar]
- Kamal-Bahl S, Doshi J, Campbell J. Economic analyses of respiratory syncytial virus immunoprophylaxis in high-risk infants: a systematic review. Arch Pediatr Adolesc Med. 2002;156:1034–1041. doi: 10.1001/archpedi.156.10.1034. [DOI] [PubMed] [Google Scholar]
- Killikelly AM, Kanekiyo M, Graham BS. Pre-fusion F is absent on the surface of formalin-inactivated respiratory syncytial virus. Sci Rep. 2016;6:34108. doi: 10.1038/srep34108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJ, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR., Jr Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert L, Sagfors AM, Openshaw PJ, Culley FJ. Immunity to RSV in Early-Life. Front Immunol. 2014;5:466. doi: 10.3389/fimmu.2014.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg JP, Hussain IR, Warner JA, Johnston SL, Warner JO. Type 1 and type 2 cytokine imbalance in acute respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med. 2003;168:633–639. doi: 10.1164/rccm.200210-1148OC. [DOI] [PubMed] [Google Scholar]
- Lerner RA. Rare antibodies from combinatorial libraries suggests an S.O.S. component of the human immunological repertoire. Mol Biosyst. 2011;7:1004–1012. doi: 10.1039/c0mb00310g. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Chang JS, Yang Y, Kim A, Graham BS, Kwong PD. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol. 2010a;84:12236–12244. doi: 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Kim A, Yang Y, Graham BS, Kwong PD. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010b;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa JJ, Kose N, Matta P, Gilchuk P, Crowe JE., Jr A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol. 2017;2:16271. doi: 10.1038/nmicrobiol.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Alling DW, Snyder MH, Walsh EE, Prince GA, Chanock RM, Hemming VG, Rodriguez WJ, Kim HW, Graham BS, et al. Effect of age and preexisting antibody on serum antibody response of infants and children to the F and G glycoproteins during respiratory syncytial virus infection. J Clin Microbiol. 1986;24:894–898. doi: 10.1128/jcm.24.5.894-898.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Walsh EE. Formalin-inactivated respiratory syncytial virus vaccine induces antibodies to the fusion glycoprotein that are deficient in fusion-inhibiting activity. J Clin Microbiol. 1988;26:1595–1597. doi: 10.1128/jcm.26.8.1595-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Ding JL. Natural antibodies bridge innate and adaptive immunity. J Immunol. 2015;194:13–20. doi: 10.4049/jimmunol.1400844. [DOI] [PubMed] [Google Scholar]
- PATH. RSV Vaccine and mAb Snapshot. 2017 http://www.path.org/publications/files/CVIA_rsv_snapshot_final.pdf.
- Polack FP, Teng MN, Collins PL, Prince GA, Exner M, Regele H, Lirman DD, Rabold R, Hoffman SJ, Karp CL, et al. A role for immune complexes in enhanced respiratory syncytial virus disease. J Exp Med. 2002;196:859–865. doi: 10.1084/jem.20020781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Rechavi E, Lev A, Lee YN, Simon AJ, Yinon Y, Lipitz S, Amariglio N, Weisz B, Notarangelo LD, Somech R. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci Transl Med. 2015;7:276ra225. doi: 10.1126/scitranslmed.aaa0072. [DOI] [PubMed] [Google Scholar]
- Reed JH, Jackson J, Christ D, Goodnow CC. Clonal redemption of autoantibodies by somatic hypermutation away from self-reactivity during human immunization. J Exp Med. 2016;213:1255–1265. doi: 10.1084/jem.20151978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridings J, Dinan L, Williams R, Roberton D, Zola H. Somatic mutation of immunoglobulin V(H)6 genes in human infants. Clin Exp Immunol. 1998;114:33–39. doi: 10.1046/j.1365-2249.1998.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande CJ, Cane PA, Nokes DJ. The association between age and the development of respiratory syncytial virus neutralising antibody responses following natural infection in infants. Vaccine. 2014;32:4726–4729. doi: 10.1016/j.vaccine.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre P, Melero JA, Garcia-Barreno B, Palomo C. Comparison of affinity chromatography and adsorption to vaccinia virus recombinant infected cells for depletion of antibodies directed against respiratory syncytial virus glycoproteins present in a human immunoglobulin preparation. J Med Virol. 2005;76:248–255. doi: 10.1002/jmv.20349. [DOI] [PubMed] [Google Scholar]
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoff JJ, O’Brien KL, Thumar B, Shaw JB, Reid R, Hua W, Santosham M, Karron RA. Young infants can develop protective levels of neutralizing antibody after infection with respiratory syncytial virus. J Infect Dis. 2008;198:1007–1015. doi: 10.1086/591460. [DOI] [PubMed] [Google Scholar]
- Siegrist CA, Aspinall R. B-cell responses to vaccination at the extremes of age. Nat Rev Immunol. 2009;9:185–194. doi: 10.1038/nri2508. [DOI] [PubMed] [Google Scholar]
- Swers JS, Kellogg BA, Wittrup KD. Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The IMpact-RSV Study Group. Palivizumab, a Humanized Respiratory Syncytial Virus Monoclonal Antibody, Reduces Hospitalization From Respiratory Syncytial Virus Infection in High-risk Infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- Thomson CA, Bryson S, McLean GR, Creagh AL, Pai EF, Schrader JW. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27:2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE, Jr, Jardetzky TS. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol. 2017;2:16272. doi: 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Weitkamp JH, Blum DL, LaFleur BJ, Crowe JE., Jr The human neonatal B cell response to respiratory syncytial virus uses a biased antibody variable gene repertoire that lacks somatic mutations. Mol Immunol. 2009;47:407–414. doi: 10.1016/j.molimm.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Schmidt A, Beil EJ, Day ND, Branigan PJ, Liu C, Gutshall LL, Palomo C, Furze J, Taylor G, et al. Characterization of the epitope for anti-human respiratory syncytial virus F protein monoclonal antibody 101F using synthetic peptides and genetic approaches. J Gen Virol. 2007;88:2719–2723. doi: 10.1099/vir.0.82753-0. [DOI] [PubMed] [Google Scholar]
- Yeung YA, Foletti D, Deng X, Abdiche Y, Strop P, Glanville J, Pitts S, Lindquist K, Sundar PD, Sirota M, et al. Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire. Nat Commun. 2016;7:13376. doi: 10.1038/ncomms13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, McLellan JS, Kallewaard NL, Ulbrandt ND, Palaszynski S, Zhang J, Moldt B, Khan A, Svabek C, McAuliffe JM, et al. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci Transl Med. 2017;9:aaj1928. doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


