Abstract
In this chapter we discuss the strengths, caveats and technical considerations of three approaches for reprogramming the chemical composition of selected amino acids within a membrane protein. In vivo nonsense suppression in the Xenopus laevis oocyte, evolved orthogonal tRNA and aminoacyl-tRNA synthetase pairs and protein ligation for biochemical production of semisynthetic proteins have been used successfully for ion channel and receptor studies. The level of difficulty for the application of each approach ranges from trivial to technically demanding, yet all have untapped potential in their application to membrane proteins.
Introduction
The ability to genetically alter the chemo-physical properties of an amino acid permits the detailed examination of the functional contributions of atomic characteristics of side- and main-chain chemistries. Further, emerging techniques are available to genetically encode non-canonical amino acids (ncAA) with fluorescent/spectral, photo-activated and bioorthogonal labeling properties. This chapter will focus on three established methods that have been successfully used for the site-directed incorporation of ncAAs into membrane proteins (Fig. 1). First, the in vivo nonsense suppression method in the context of the Xenopus laevis oocyte employs a chemo-enzymatically acylated orthogonal tRNA to incorporate the ncAA in the target protein encoded by the co-injected cRNA in the context of Xenopus laevis oocytes (Fig. 1A). This technique has been widely used for structure-function analysis and pharmacological characterizations of ligand, drug and toxin interactions with ligand- and voltage-gated ion channels. Second, orthogonal co-evolved tRNA and aminoacyl-tRNA synthetase (aa-RS) pairs, once generated, can simply be co-expressed with the target gene in the presence of the ncAA (Fig. 1B). This approach has been successfully applied in a broad spectrum of cell types, from E. coli and yeast to eukaryotic cell lines and even multicellular organisms. Third, ion channel semi-synthesis via chemical ligation is technically challenging but permits the use of amino acids that may be either toxic or not tolerated in a cellular context by bypassing ribosomal and translational quality control checks, limitations that have the potential to affect the use of truly unique amino acids (Fig. 1C). The technical aspects, considerations and limitations of each of these approaches will be discussed as well as their applications to the study of ion channels and membrane proteins.
Fig. 1.

Approaches for genetic code expansion with non-canonical amino acids. a In vivo non- sense suppression method: a non-canonical amino acid (ncAA, green star) is synthetically acylated to a suppressor tRNA (ncAA-tRNASUP) and microinjected into Xenopus laevis oocytes together with RNA coding for the protein of interest. To enable site-specific incorporation of the ncAA a codon is repurposed, most commonly it is the amber stop codon TAG (SUP, site of suppression). b Alternatively, tRNA can be misacylated inside the cell by aminoacyl-tRNA synthetases (aa-RSs). Orthogonal aa-RS and tRNA pairs can be co-evolved to be specific for certain ncAAs. For incorporation of ncAAs (green stars), the DNA encoding these pairs is transfected into the expression system of choice together with the gene of interest carrying a repurposed codon at the site of mutagenesis (SUP). The ncAA is taken up by the cell directly from the extracellular growth media. c Protein ligation through the use of bacterial intein sequences allows for chemical ligation of recombinant and synthetic peptide fragments.
Genetic code expansion in live cells comes with a variety of considerations. For one, because these techniques largely rely on the endogenous translation machinery, it is possible to simply ‘repurpose’ the codon at the incorporation site to encode for the new amino acid. There are 64 nucleotide codons – 61 that encode canonical amino acids and three, TAA (ochre), TGA (opal) and TAG (amber), that encode termination codons. Repurposing of such stop codons has proved successful for incorporation of ncAAs. The amber (TAG) stop codon is the rarest of the three stop codons and is therefore the one most often used for ‘nonsense suppression’ in order to minimize suppression of endogenous termination codons. However, the proportional usage of stop codons is variable between kingdoms and cell types and should be considered when choosing the suppressor codon. Four codon suppressor systems are also available for both nonsense suppression in oocytes (Rodriguez et al 2007a; Rodriguez et al 2007b) and evolved tRNA/aa-RS pairs (Neumann et al 2010b).
Not to be neglected are the prerequisites concerning the ncAA itself as it must be bioavailable, non-toxic and metabolically inert. Moreover, once acylated to the tRNA, the ncAA must be tolerated by cellular elongation factors Tu (EF-Tu) and the ribosome. Lastly, for any technique used, the imagined ncAA must first be synthesized at the mid (50-100 mg) to large (500 mg – 1 gram) scale, for in vivo nonsense suppression in oocytes or for evolved tRNA/aa-RS pair generation and application, respectively.
Many of these technical challenges may be bypassed through the application of protein ligation strategies that allow for the coupling of synthetic and recombinant expressed protein fragments to produce ‘semi-synthetic’ channels (Valiyaveetil et al 2002). Of note, unlike cell-based approaches, the amino acid is unrestricted by biological limitations. However, the technical challenges, such as protein refolding, which may be surmountable by some, represent a significant technical barrier to most investigators, and this task is especially onerous with membrane proteins.
1. Approaches of non-canonical amino acid incorporation
The technical options for designing new probes and altering the chemical properties of amino acids within membrane proteins are continually expanding, becoming more accessible to more laboratories and thus hold tremendous promise for a variety of applications. Herein, potential challenges and technical considerations of these methods are discussed in light of some examples of their application to ion channels and receptors.
1.1 In vivo nonsense suppression in Xenopus oocytes
In vivo nonsense suppression is a powerful approach for the incorporation of ncAAs in ion channel proteins in Xenopus oocytes that was built upon a multitude of incremental advances. Key amongst these breakthroughs were the demonstrations of tRNA chemical aminoacylation in vitro (Hecht et al 1978), and that these charged tRNAs could be used for the delivery specialized amino acids into a protein through the suppression of an introduced stop codon (Noren et al 1989). Subsequent adaption of the technique for microinjection of misacylated-tRNA with nicotinic acetylcholine receptor cRNA into Xenopus laevis oocytes (Nowak et al 1998) has since led to more than 60 published articles and the incorporation of over 100 non-natural amino acids in more than 25 channel and receptor types.
The general principles of in vitro amino-acylation of tRNA are shown in Figure 2 and have been described in depth elsewhere (Nowak et al 1998; Pless and Ahern 2013; Dougherty and Van Arnam 2014). Briefly, the ncAA is first chemically coupled to the dinucleotide pdCpA (Fig. 2A) which is then subsequently enzymatically ligated to a synthetic tRNA (Fig. 2B). The tRNA must be orthogonal to Xenopus laevis oocytes, such that the tRNA does not become edited or reacylated by endogenous aminoacyl-tRNA synthetases. Tetrahymena thermaphila has an irregular genetic code such that the glutamine is encoded by the UAG codon and thus the natural glutamyl-tRNA is ideal for nonsense suppression of introduced amber (TAG) stop codons (Saks et al 1996). The tRNA variant most often used for amber codon suppression in oocytes, THG73 (Tetrahymena thermophila G73), contains the U73G mutation at the acceptor stem to further obscure recognition of the tRNA from endogenous Gln synthetases (Fig. 2B). This THG73 tRNA is effective for nonsense suppression in Xenopus laevis oocytes, but is also orthogonal for in vitro translation with Escherichia coli (Cload et al 1996), rabbit reticulocyte (Rothman et al 2005) and wheat germ (England et al 1999) expression systems. Further, THG73 tRNA can suppress the opal (TGA) stop codon if the appropriate anti-codon is engineered into the tRNA (Rodriguez et al 2007a; Rodriguez et al 2007b). Additionally, E. coli tRNALeu (with the Leu anticodon mutated to CUA) is orthogonal in Xenopus (Kalstrup and Blunck 2013), as is E. coli tRNAAsn (Rodriguez et al 2007a; Rodriguez et al 2007b). Thus, with numerous viable tRNAs, it is possible to attempt to incorporate multiple ncAAs within the same protein, although the generally low incorporation rate may prove for this possibility to be especially challenging for the rescue of macroscopic current. Alternatively, multiple ncAAs have been incorporated via frameshift suppression in response to quadruplet codons CGGG and GGGU with yeast phenylalanine frameshift suppressor (YFFS) tRNAs (Rodriguez et al 2006; Rodriguez et al 2007a; Rodriguez et al 2007b). These YFFS tRNAs show lowered suppression efficiency compared to THG73, but the quadruplet codons are less likely to be ‘read-through’ at promiscuous sites, a spurious process described below in General Considerations. However, endogenous CGGG and GGGU sequences should be removed first, and their prevalence may be significant in the longer reading frames of some ion channel, receptor and transporter genes.
Fig. 2.
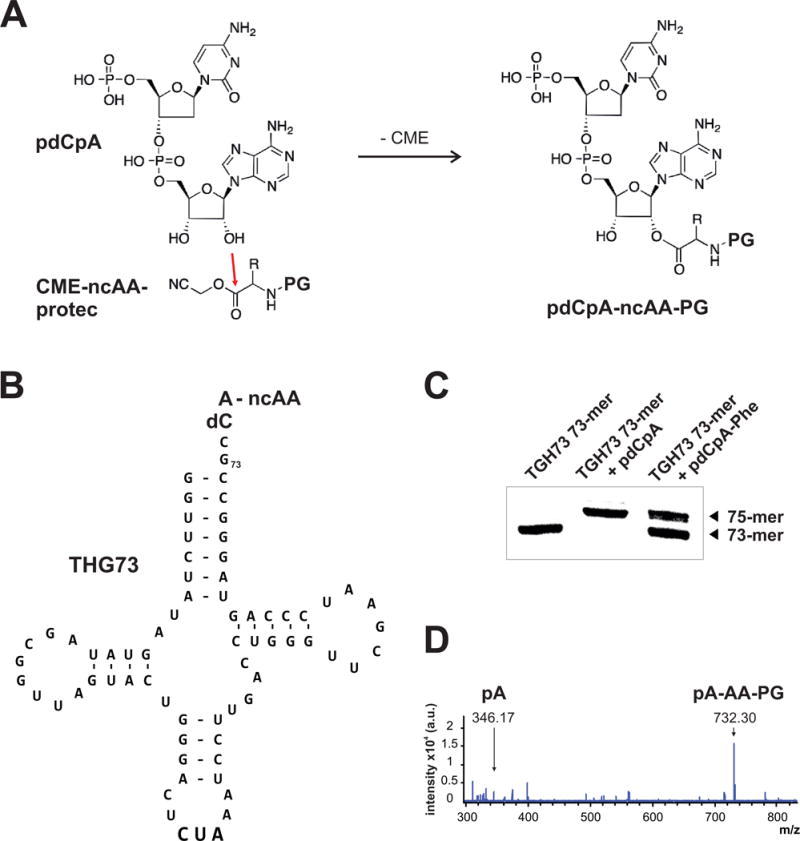
In vivo nonsense suppression. a The non-canonical amino acid (ncAA) is chemically cou- pled to the dinucleotide phosphodesoxy-cytosine phospho-adenosine (pdCpA) via ester bond for- mation between the free hydroxyl group (3′) of the ribose in the phospho-adenosine molecule and the carboxyl group of the ncAA. The starting product of the ncAA is a cyanomethyl ester (CME). A protecting group (PG) is shielding the alpha amino group from reactivity. b The ncAA-pdCpA conjugate is subsequently ligated to a refolded tRNA that lacks the 3′ dinucleotide CA. Here, the final product of the ligation is shown: a misacylated THG73 tRNA (Tetrahymena thermaphila glutamyl-tRNA U73G) carrying the anti-codon for TAG (CUA). c Efficiency of the ligation can be estimated via denaturing TBE/Urea polyacrylamide (15 %) gel electrophoresis: the ligated THG73 (75-mer) is separated from the non-reacted 73-mer. RNA was visualized with a commercial nucle- otide dye. d Qualitative proof of tRNA misacylation with the ncAA of interest can be achieved by mass spectrometry analysis. Spectrum shows fragments of misacylated tRNA after digest with S1 nuclease detected in reflector negative mode on a MALDI TOF instrument (CHCA matrix). Numbers correspond to measured mass that are in agreement with theoretical calculations. pA, 5′-phosphoryl-adenosine; pA-AA-PG, 5′-phosphoryl-adenosine esterified with an amino acid (AA) that carries a protecting group (PG) on the alpha amine (here: pA-Phe-NVOC).
It is worth briefly noting the unique nomenclature of tRNA. For one, the total number of nucleotides of a specific tRNA cannot be simply counted and extrapolated to a given site. In particular, stem nucleotides and loop positions have a fixed numbering regardless of their overall position within the tRNA oligonucleotide numbering sequence (Sprinzl et al 1996; Sprinzl and Vassilenko 2005). For example, “G73” in the THG73 tRNA is actually the 72nd nucleotide in the oligonucleotide sequence, and thus the full-length tRNA is a 75-mer oligonucleotide and not a 76-mer as often stated in the literature.
An established protocol for tRNA synthesis uses a cDNA template from a linearized plasmid containing the THG73 tRNA downstream of a T7 promoter and a 3′ FokI restriction digest site (Saks et al 1996; Nowak et al 1998). However, given recent improvements in commercial oligonucleotide synthesis, we have found that greater yields of tRNA can be simply obtained by using a synthetic DNA oligonucleotide template comprised of a 5′ T7 promoter followed by the sequence of the THG73 tRNA (Pless et al 2011a). This synthetic oligonucleotide or the linearized plasmid then serves as the template for any one of a number of commercially available T7 transcription kits, such as MEGAshortscript (Life Technologies, Grand Island, NY, USA) or T7-Scribe Standard RNA IVT (CELLSCRIPT, Madison, WI, USA). The in vitro translated tRNA is purified and folded (Nowak et al 1998), and is then ready for enzymatic ligation to the prepared amino acid-dinucleotide conjugate.
In parallel to tRNA synthesis and folding, the ncAA of interest is chemically coupled to the dinucleotide phosphodesoxy-cytosine phospho-adenosine (pdCpA) via attack of the pA ribose 3′ hydroxyl at the activated ester of the ncAA (Fig. 2A). Under typical reaction conditions both pA 2′ and 3′ hydroxyl groups may be esterified, depending on the structure of the ncAA, and such ‘di-coupled’ pdCpA-ncAA species have been reported to have enhanced expression properties (Duca et al 2008). The pdCpA dinucleotide can be synthesized or obtained commercially (GE Healthcare Dharmacon, Inc., Lafayette, CO, USA) and its reaction with activated ncAAs has been optimized (Robertson et al 1991). Protection of the ncAA a-amino group prior to coupling limits reactivity with pdCpA and the specific characteristics of the protecting group are worth considering. The majority of published studies of nonsense suppression in oocytes employ 4,5-Dimethoxy-2-nitrobenzyl chloroformate (NVOC) which can be photo-deprotected immediately prior to tRNA injection via UV exposure (Nowak et al 1998). Non-photolyzable protecting groups are also available, including the pentenoyl (Lodder et al 2005) and t-BOC (Hohsaka et al 1999) moieties which are chemically removed prior to tRNA acylation. These alternative chemical deprotection strategies may be preferable when expressing potentially photo-sensitive side chains. Lastly, the pdCpA-ncAA conjugate is then enzymatically coupled via RNA ligase to the folded THG73 tRNA or a similar orthogonal tRNA (Nowak et al 1998). The enzymatic tRNA acylation reaction can be followed by denaturing urea polyacrylamide gel electrophoresis (Fig. 2C) or by mass spectroscopy (Fig. 2D). The deprotected acylated tRNA is then micro-injected into a freshly isolated Xenopus laevis oocyte along with membrane protein cRNA containing a suppression codon at the site of interest, i.e. TAG, and can be subsequently analyzed by two-electrode voltage-clamp, with currents being detectable within 24-48 hours depending on the channel or receptor isoform.
In some instances, it is possible to see evidence (i.e. ionic current) of a cRNA-TAG clone in the absence of an acylated co-injected tRNA. This phenomenon, known as ‘read-through’ of an introduced stop site is highly variable from site to site and transcript to transcript and can be the source of much consternation. It is therefore imperative that the experimentalist performs rigorous controls consisting of cRNA containing a suppression codon that is co-injected with non-aminoacylated tRNA, i.e. pdCpA-tRNA. Such controls should be performed in parallel with aminoacylated tRNA at every site of incorporation and with each batch of oocytes and rounds of injections. In Xenopus oocytes, the sole expression of the cRNA containing the suppression codon alone is not an effective measure of read- through and does not confirm that measured function for rescued TAG sites with acylated tRNA is due to bona fide incorporation of the intended ncAA. The expression of ncAA containing proteins in the Xenopus oocyte does not generally produce biochemical scale amounts of rescued ion channel protein, however, advances in mass spectroscopy may soon facilitate the independent verification of site-specific ncAA incorporation.
1.2 Evolved orthogonal tRNA and aminoacyl-tRNA synthetase pairs
The use of evolved orthogonal tRNA and aminoacyl-tRNA synthetase (aa-RS) pairs provides a straightforward method for directed incorporation of the selected amino acid in either eukaryotic or prokaryotic expression systems. The strategy for expanding the genetic repertoire with orthogonal amino acid synthetase (aa-RS) and cognate tRNA pairs was first successfully demonstrated in E. coli by employing the yeast PheRS/tRNAPheCUA pair (Furter 1998a). Crucial to success of this approach is the ability to reprogram the specificity of aa-RSs for new amino acids. The evolution of aa-RS specificity has been accomplished first in E. coli in 2001 and soon after in eukaryotic expression systems (Wang et al 2001; Chin 2003). The strength of this approach is that, once an evolved tRNA/aa-RS pair is available for an ncAA, one needs only to transiently express the components (tRNA, RS and protein of interest) and supplement the ncAA to the cellular media. Therefore, this technique has potential to significantly level the playing field for the use of ncAAs for electrophysiological, biochemical and structural studies.
With this approach, the aa-RS and the tRNA must be specific for each other, compatible with the host translation machinery and orthogonal to it, i.e. the suppressor tRNA must not be a substrate for any endogenous aa-RS and the aa-RS must not aminoacylate any endogenous tRNA. The molecular determinants for tRNA-aa-RS recognition are conserved between archea and eukaryotes but are divergent from bacteria (Ibba and Soll 2000). Therefore, orthogonal aa-RS/tRNA pairs generally originate from a different kingdom of life than the host expression system. Further, the process of incorporation of a selected ncAA also requires the specificity of the enzyme for the desired amino acid substrate. And re-purposing an existing synthetase through targeted evolution of the ncAA binding pocket must leave tRNA recognition and orthogonality intact.
1.2.1 Genetically encoding non-canonical amino acids in prokaryotes
The archea Methanocaldococcus jannaschii tyrosyl-RS/tRNATyr pair was the first orthogonal aa-RS/tRNA pair imported into E. coli that was capable of site-specific, high fidelity and efficiency incorporation of ncAAs (Wang et al 2001). Here, the tRNATyr anticodon was mutated to CUA and the orthogonality was improved by screening a library of mutant tRNATyrCUA (Wang et al 2001; Wang and Schultz 2001). The tRNA evolution process is based on a double-sieve selection principle, whereby an initial negative selection in the absence of the cognate aa-RS removes mutant tRNAs from the library that are substrates for endogenous aa-RSs. Subsequent positive selection in the presence of the cognate aa-RS allows only orthogonal tRNAs with high affinity for the cognate enzyme to pass (Wang et al 2001; Wang and Schultz 2001).
The development of an approach for changing the substrate specificity of the orthogonal M. jannaschii Tyr-RS from tyrosine to a ncAA represented a significant breakthrough that benefitted greatly on structures of a Tyr-RS homologue from Bacillus steraothermophilus (Brick et al 1989). Specifically, access to the structural basis for amino acid recognition by the RS facilitated the rational design of libraries containing randomized residues in the amino acid binding site of the enzyme (Wang et al 2001). Later crystal structures for wild type and mutant Mj Tyr-RS (Kobayashi et al 2003; Zhang et al 2005; Liu et al 2007a; Young et al 2011) lead to libraries with up to 10 randomized residues in the active site of the enzyme (Peters et al 2009).
These RS libraries in the presence of the cognate tRNATyrCUA were subjected to a two-step selection process in E. coli (Fig. 3A). The positive selection relied upon successful suppression of an introduced TAG codon within the reading frame of the essential chloramphenicol acetyltransferase gene (Wang et al 2001; Fig. 3A). The growth in the presence of chloramphenicol and the ncAA of interest results in survival solely of clones containing functional aa-RSs that are able to aminoacylate the cognate tRNATyrCUA with either the desired ncAA or a canonical amino acid (Fig. 3A). Elimination of aa-RSs clones recognizing canonical amino acids (i.e. not specific for the ncAA) relies upon a negative selection, which is most commonly nonsense suppression of a cell-toxic gene (barnase) that contains introduced amber codons at permissive sites (Wang and Schultz 2001; Chin et al 2002a; Zhang et al 2002); Fig. 3A). As this selection is performed in the absence of the ncAA, all the aa-RSs that function with endogenous amino acids are removed from the library and only clones carrying ncAA-specific RSs survive (Fig. 3A). Multiple rounds of positive and negative selection are often required to identify an aa-RS that has both high incorporation efficiency and fidelity for the ncAA of interest. Simplified single plasmid versions of the screen are available as are alternatives for the negative selection (Santoro et al 2002; Melancon and Schultz 2009). To date, a vast variety of aa-RSs have been evolved from M. jannaschii Tyr-RS to charge its cognate tRNATyrCUA with more than 40 structurally different ncAAs (Liu and Schultz 2010) – highlighting the flexibility of the enzyme’s active site. Such pairs can exhibit excellent fidelity and capable yields (shake flask expression of soluble proteins: mg/l range; high-density fermentation expression of soluble proteins: g/l range; Liu and Schultz 2010). Despite these notable strengths, the evolved plasticity of the Mj Tyr-RS Tyr binding site is not limitless thus necessitating the construction of libraries based on other aa-RS/tRNA pairs. Indeed, several aa-RS/tRNA pairs from Saccharomyces cerevisiae have been shown to be orthogonal in E. coli (Furter 1998b; Ohno et al 1998; Liu and Schultz 2006), and some orthogonal pairs with hybrid or consensus components have been adapted for use in E. coli (Kowal et al 2001; Anderson and Schultz 2003; Santoro et al 2003; Anderson et al 2004). Further, orthogonal aa-RS/tRNA pairs have been identified in the methanogens Methanosarcina barkeri, Methanosarcina mazei and Desulfitobacterium hafniense that genetically encode Pyrrolysine, the so-called 21st amino acid (Srinivasan et al 2002; Krzycki 2005). These Pyl-RS/tRNAPylCUA pairs are discussed separately.
Fig. 3.

Evolving substrate specificity of aminoacyl-tRNA synthetases (aa-RSs) for ncAAs. a In prokaryotic expression systems, a two-step evolution procedure for aa-RSs is well established. After choosing an aa-RS/tRNA pair orthogonal to the host, a library containing randomized resi- dues in the amino acid binding site of the enzyme is constructed. This population of aa-RS variants (grey and red spheres) is subjected together with its cognate tRNA (black cross) to a round of positive selection and screened for activity with either canonical (grey circle) or non-canonical (red star) amino acids, by virtue of their ability to suppress an introduced stop codon and so allow to complete translation of a gene that is essential for survival. To eliminate aa-RSs that recognize natural amino acids (grey spheres) a negative selection in the absence of the ncAA is performed. Here, nonsense suppression of a cell-toxic gene takes place, thus allowing for survival of clones that carry aa-RSs specific for the ncAA only (red spheres). Scheme was adapted from Davis and Chin 2012. b In eukaryotic expression systems, evolution of aa-RSs is more complicated but follows the same principles. The main difference is that the TAG stop codons are introduced not directly into genes that are responsible for survival or death of clones during the selection but into the gene of a transcription factor (GAL4) that drives the expression of reporter genes (HIS3, URA3) causing growth or death of the cells. For details of the procedure please refer to the main text.
1.2.2 Genetically encoding non-canonical amino acids in eukaryotes
Genetic code expansion in eukaryotes holds tremendous promise for the advanced study of membrane proteins in native cellular environments and for ultimately revealing molecular mechanisms of cell biology and physiology. Unfortunately, directed ncAA-RS evolution in mammalian cells is unfeasible due to low transformation efficiencies, slow generation times and comparably low efficiency of survival-death selection. However, the translation mechanism of the lower eukaryote Saccharomyces cerevisiae is conserved with higher eukaryotes, genetically well characterized and is susceptible to manipulation, qualities that have made it a go-to host organism for directed evolution of orthogonal ncAA-RS/tRNA pairs for eukaryotic expression. To date, several E. coli pairs have been proven to be orthogonal in eukaryotes: E. coli Leu-RS/E. coli tRNALeu, E. coli Glu-RS/human initiator tRNALeu and E. coli Tyr-RS/E. coli tRNATyr, the last of which has been broadly employed (Edwards and Schimmel 1990; Chin 2003; Wu et al 2004). The Pyl-RS/tRNAPylCUA pairs exhibit dual orthogonality (Blight et al 2004; Mukai et al 2008; Chen et al 2009), and thus have been applied in prokaryotes as well as eukaryotes.
The evolution procedure in S. cerevisiae is similar to the two-step selection developed in E. coli, differing in that the TAG codons are encoded into a transcription factor that drives the expression of reporter genes that result in growth or death of the cells (Fig. 3B; Chin 2003; Chin et al 2003; Cropp et al 2007). The first library design employed the Ec Tyr-RS and relied on the randomization of five residues in the Tyr binding pocket that were identified in the crystal structure of the homologue Tyr-RS from Bacillus steraothermophilus (Brick et al 1989; Chin 2003). This library contained ~107 variants and was transformed along with the cognate Ec tRNATyrCUA into a yeast strain that is auxotrophic for Histidine and Uracil and harbors the GAL4 transcription factor gene that contained TAG codons for nonsense suppression at permissive sites (Chin 2003; Chin et al 2003; Cropp et al 2007). The successful suppression of the GAL4 TAG sites promotes transcriptional activation of GAL4-responsive HIS3 and URA3 reporter genes that are enlisted for positive and negative selections (Fig. 3B). The positive selection is performed in the presence of the ncAA of interest. First, on Histidine-deficient medium, only those clones will grow that carry aa-RSs capable to aminoacylate the tRNATyrCUA with either at least one canonical or the non-canonical amino acid (or a combination of both) thereby allowing transcriptional activation of HIS3 and thus Histidine biosynthesis that is essential for survival (Fig. 3B). Cells with non-functional aa-RSs would die. The second positive selection on Uracil-deficient medium follows the same principle and results in clones that are capable to compensate for the Uracil auxotrophy of the yeast strain (Fig. 3B). Aminoacyl-tRNA synthetases that recognize endogenous amino acids are removed in a negative selection performed in absence of the ncAA and in presence of 5-fluoroorotic acid (5-FOA), a substrate for the URA3 gene product. Upon successful translation of GAL4 and subsequent transcriptional activation of URA3, 5-FOA is converted into a cell-toxic product resulting in cell death (Fig. 3B). Hence, only clones survive which carry active aa-RS/tRNA pairs with a substrate specificity reprogrammed for the ncAA and those that utilize canonical amino acids are unviable (Fig. 3B). This directed evolution is carried out in at least three rounds of consecutive positive and negative selection until the desired fidelity and efficiency of ncAA incorporation are achieved. This approach has been used successfully to evolve aa-RS/tRNA pairs that encode about 40 ncAAs in eukaryotes (Liu and Schultz 2010). After the initial directed evolution in S. cerevisiae the orthogonal ncAA-RS/tRNA pairs can be shuttled to vectors for higher eukaryotic expression systems (Sakamoto et al 2002; Wang et al 2007b; Liu et al 2007b; Mukai et al 2008; Chen et al 2009). Such pairs have been used for genetic code expansion in a variety of proteins in primary and secondary cell cultures, as well as whole multicellular organisms such as Caenorhabditis elegans and Drosophila melanogaster (Liu and Schultz 2010; Greiss and Chin 2011; Bianco et al 2012; Chang et al 2013).
A significant challenge for efficient incorporation of ncAAs into eukaryotic proteins in vivo arises from intrinsic differences in the transcription and processing of tRNAs in prokaryotes and eukaryotes. For one, bacterial tRNAs are transcribed through promoters upstream of the tRNA gene, whereas eukaryotic tRNAs are transcribed through promoter elements within the tRNA genes (Galli et al 1981; Wang et al 2009b). These promoter elements, termed A- and B-boxes, are absent in most prokaryotic tRNAs. Moreover, in prokaryotes the full tRNA sequence is encoded, yet in eukaryotes the 3′-CCA trinucleotide is added enzymatically after transcription of the tRNA. And while the Ec tRNATyrCUA contains an B-box but not the A-box, a homologue tRNATyr from the B. stearothermophilus has been identified that contains both promoter elements naturally and is orthogonal in mammalian cells as well as compatible with the Ec Tyr-RS. Replacing the Ec tRNATyrCUA with the Bs tRNATyrCUA flanked by a 5′ sequence of the human tRNATyr and a 3′ termination sequence enabled efficient nonsense suppression in mammalian cells (Sakamoto et al 2002). Alternatively, to increase the transcription level of prokaryotic tRNA and to facilitate its processing in eukaryotes an external RNA-polymerase III promoter that contains the consensus eukaryotic A- and B-box sequences can be placed upstream of the bacterial tRNA gene lacking the 3′ CCA tail. This strategy has been demonstrated successfully in yeast with the promoters RPR1 or SNR52 (Wang and Wang 2008; Lee et al 2009; Majmudar et al 2009) as well as in mammalian cells using U6 or H1 promoters (Wang et al 2007b; Mukai et al 2008; Gautier 2010). To further increase tRNA expression, the expression cassette can be repeated multiple times (Sakamoto et al 2002; Mukai et al 2008; Gautier 2010). However, caution is advised especially in stable cell lines, as repeating sequences can be associated with recombination during amplification as well as gene silencing (Hsieh and Fire 2000).
1.2.3 Achaeal pyrrolysyl-tRNA-synthetase/tRNA pairs display orthogonality in prokaryotes as well as eukaryotes
The identification of pyrrolysine as a natural expansion of the genetic code lead to the discovery of pyrrolysyl-tRNA-synthetase/tRNA pairs from different methanogenic species, including Methanosarcina barkeri and Methanosarcina mazei (Hao et al 2002; Srinivasan et al 2002; Polycarpo et al 2004). Several features of the Pyl-RS/tRNA pairs make them exceptionally useful for expanding the methodology of ncAA incorporation. Most importantly, the Pyl-RS/tRNAPylCUA pair exhibits orthogonality in prokaryotes as well as eukaryotes (Blight et al 2004; Mukai et al 2008; Chen et al 2009). This unique property allows the evolution of new synthetase specificities to be performed in E. coli, where the selection procedure allows for large synthetase libraries to be screened quickly. The newly developed pair can then be employed in any prokaryotic or eukaryotic expression system for ncAA incorporation. Moreover, the Pyl-RS does not recognize any of the 20 canonical amino acids, hence there is no need to destroy the natural synthetase activity before creating a new one. Indeed, several ncAAs have been incorporated using the wild type Pyl-RS (Polycarpo et al 2006; Li et al 2009). Furthermore, Pyrrolysine is encoded naturally by the amber stop codon TAG, thus eliminating the need of tRNA anticodon mutagenesis (James et al 2001; Srinivasan et al 2002).
At present, over 30 ncAAs have been incorporated into various proteins using the Pyl-RS/tRNA pairs (Fekner et al 2010; Davis and Chin 2012; Neumann 2012). Some of the incorporated pyrrolysine analogues mimic posttranslational modifications (Anderson et al 2004; Farrell et al 2005; Wang et al 2009a; Huang et al 2010; Isaacs et al 2011; Johnson et al 2011), others are photocaged canonical amino acids (Mohibullah and Hahn 2008; Tamura et al 2009; Yamano et al 2010; Mukai et al 2010; Shiota et al 2011). Yet a growing majority of pyrrolysine analogues are being used for bioorthogonal chemical reactions allowing site-specific labeling of proteins with a variety of probes, like fluorophores and biotin (Chin et al 2002c; Chin et al 2002b; Rackham and Chin 2005; Wang et al 2007a; Neumann et al 2010b; Neumann et al 2010a; Barrett and Chin 2010; Ai et al 2011). Remarkably, even whole multicellular organisms, like C. elegans and D. melanogaster, have been generated to incorporate several ncAAs for bona fide in vivo biological studies (Greiss and Chin 2011; Bianco et al 2012; Chang et al 2013). However to our knowledge, a systematic comparison between Mb Pyl-RS/tRNAPyl and Mm Pyl-RS/tRNAPyl has not been reported to speak to inherent differences in enzyme stability, their amenability to directed evolution, nonsense suppression efficiency and tRNA stability in different organisms.
1.2.4 Optimization of ncAA incorporation efficiencies and protein yields
In the last decade, the methodology of ncAA incorporation has advanced greatly and has steadily allowed for an increasing number of labs starting to using these tools to tackle various biological questions. However, the efficiencies of ncAA suppression of target genes and ultimately, the expression of an ncAA into a desired protein, can be highly variable and are thus the subject of numerous optimization efforts. Some improvements in incorporation efficiency have stemmed from advances in engineering of translational components such as increasing the efficiency and fidelity of the tRNA/aa-RS pairs themselves or the interaction of acylated-tRNA with the elongation factor EF-Tu (Cooley et al 2014). Additionally improved protein yield can result from expression conditions and expression plasmid constructs. For instance, ncAA incorporation efficiencies are enhanced significantly after integrating the orthogonal aa-RS/tRNA pairs into a single vector and increasing their promoter strengths as well as the plasmid copy numbers (Ryu and Schultz 2006; Chen et al 2007; Hammill et al 2007; Cellitti 2008; Liu et al 2009; Liu and Schultz 2010; Peeler and Mehl 2012; Chatterjee et al 2013a). And as mentioned previously, the efficient transcription of the orthogonal tRNA is key in determining the overall yield of ncAA-containing proteins in eukaryotes.
Endogenous gene regulation pathways have the potential to interfere or assist with nonsense suppression strategies. For instance, the mRNA surveillance mechanism – nonsense-mediated mRNA decay (NMD) – identifies mRNAs that contain pre-mature stop codons and targets those for rapid degradation and thus the stability of mRNAs containing nonsense codons may be a consideration in eukaryotic cell types (Maquat 2004; Amrani et al 2006). The mechanism is most efficient when the stop codon is located closer to the 5′ than the 3′ end of the mRNA, consequently NMD-deficient yeast strains exhibit increased yields for proteins that carried ncAAs in the N-terminal two thirds of the sequence (Wang and Wang 2008; Wang et al 2009b).
Also to be considered is the competition between endogenous release factors, such as RF-1, and the supplied suppressor tRNA for binding to TAG. This becomes particularly obvious when attempting ncAA incorporation at multiple sites of the same protein. A simple deletion of RF-1 in E. coli is lethal (Rydén and Isaksson 1984) which has led to a variety of strategies to overcome this phenotype (Wang et al 2009a; Huang et al 2010; Mukai et al 2010; Isaacs et al 2011; Johnson et al 2011; Lajoie et al 2013). An elegant example of one such strategy can be found with a genomically recoded organism (GRO) that was generated with an in vivo genome-editing approach (Isaacs et al 2011). This genomic approach allowed for the replacement all known TAG codons in an E. coli strain with TAA, as well as deletion of RF-1 without disturbing prototrophy or morphology of the cells (Lajoie et al 2013). Thus, in the resulting GRO the TAG codon has been reassigned fully for the first time to a sense codon for robust ncAA incorporation.
Low protein yields in cellular environments may also result from impaired bioavailability and/or internalization of the ncAA, and this may be especially prevalent for permanently charged ncAAs. To overcome this drawback, the ncAA of interest can be incorporated into dipeptides which becomes cleaved once internalized or the ncAA can be modified to more hydrophobic and metabolically labile acetoxymethyl (AM) esters (Takimoto et al 2010). Alternatively, a non-specific amino acid transporter can be overexpressed in the plasma membrane. Thus, a variety of strategies are available to enhance ncAA uptake and incorporation.
Lastly, the incorporation of multiple or different ncAAs into the same protein presents a particular challenge. However, the successful incorporation of two distinct ncAAs into the same protein has been achieved by employing complementary amber- and ochre-suppressor pairs (Wan et al 2010; Chatterjee et al 2013a). Reassignment of triplet nonsense codons limits the number of chemically distinct ncAAs that can be incorporated into one protein but quadruple codons, which theoretically offers access to 256 new blank codons, has been used for coding two distinct ncAAs in the same protein (Anderson et al 2004; Wang et al 2014). Further, Jason Chin and coworkers engineered an orthogonal ribosome (Rackham and Chin 2005), termed ribo-X, which translates only mRNAs containing artificial 5′ sequences and opens the door for entirely new ribosomal functionalities. For one, subsequent efforts have identified mutations in the orthogonal 16S-rRNA that decrease the interaction with RF-1 and other mutations that enhance decoding of quadruplet codons (Wang et al 2007a; Neumann et al 2010b; Barrett and Chin 2010). Hence, such orthogonal ribosomes enable not only a high efficiency translation with ncAAs in prokaryotes but also offer the possibility to produce proteins with multiple ncAAs or even proteins entirely composed of ncAAs. The insertion of several different ncAAs into the same protein also requires the availability of a sufficient number of evolved orthogonal tRNA/RS pairs. Currently, such pairs are limiting and therefore attempts are underway to identify new or alternatively designed new pairs based on existing ones (Neumann et al 2010a; Chatterjee et al 2012; Chatterjee et al 2013b). In one such example, a heterologous archaeal Pro-RS/tRNAPro pair (Pyrococcus horikoshii Pro-RS /Archaeoglobus fulgidus tRNAPro) was developed for ncAA mutagenesis in E. coli (Chatterjee et al 2012). Interestingly, by reprogramming the anticodon binding pocket of the RS, the authors succeeded in generating Proline-RS variants that recognize specifically engineered tRNAPro with three different anticodons, forming mutually orthogonal pairs (Chatterjee et al 2012). Also, the Mj Tyr-RS/tRNACUA pair has been duplicated through several rounds of mutagenesis and selection to create a new pair that decodes four-base codons and is orthogonal to the parent pair (Neumann et al 2010a).
1.3 Semi-synthetic approaches
Chemical synthesis is a very powerful method for protein modification as it enables the incorporation of a large number of unnatural amino acids as well as for allowing for changes to the protein backbone. A key advantage of chemical synthesis over the cell-based nonsense suppression approaches is that it is not dependent on the ability of the ribosome to incorporate the modification. Therefore, a wider variety of unnatural side chains and peptide backbone modifications can be introduced using chemical synthesis compared to nonsense suppression. Further, issues with the fidelity of incorporation i.e. ‘read-through’ in not a factor when using chemical synthesis for protein modification.
A major consideration in using of chemical synthesis is the size of the protein. Chemical synthesis is carried out using solid phase peptide synthesis protocols (SPPS), which is currently limited to peptides 50-60 amino acids in length. The synthesis of peptides longer than ~60 amino acids is not efficient and in most cases results in very low yields. A key advance in the field of peptide synthesis was the development of the native chemical ligation reaction (NCL). NCL is a reaction between a peptide with a C-terminal thioester and a peptide with an N-terminal Cys that links the peptides together with a native peptide bond at the ligation site (Fig. 1). The NCL reaction can therefore be used to synthesize a protein from a number of component peptides. The NCL reaction can also be used for protein semisynthesis in which the protein is assembled from a synthetic peptide and protein segment or segments obtained by recombinant means (Muir et al 1998). The advantage of a semisynthetic approach is that it allows us to use chemical synthesis to modify the region of interest, while the use of recombinant means for obtaining the remainder of the protein has the advantage that recombinant expression, unlike SPPS, in not limited to peptides of a certain length. The use of NCL and semisynthetic strategies have greatly extended the size limits of proteins that can be modified using chemical synthesis (Muir 2003).
Semisynthetic strategies are particularly useful in the chemical synthesis of integral membrane proteins. The synthesis and purification of the transmembrane segments in a membrane protein is technically challenging therefore a “total synthesis approach” in which all the peptide components required are generated using SPPS is practical only for very small membrane proteins (< 150 amino acids). Membrane proteins of interest such as voltage gated K+ channels fall well outside the purview of total synthesis but can be modified using a semisynthetic approach.
The semisynthetic approach can involve a two fragment or a three fragment approach depending upon the position of the region of interest in the protein under investigation. If the region of interest is within 50-60 amino acids (the size limits of SPPS) of the N or the C-terminus then a two part ligation strategy is used while a three part strategy is used if the region of interest is greater than 60 amino acids from either terminus. In a three part semisynthesis, the central peptide corresponds to the region of interest and is obtained by chemical synthesis while the flanking segments are obtained using recombinant expression. Figure 4 B demonstrates the strategy used for the 3-part semisynthesis of the KcsA channel used for modification of the selectivity filter.
Fig. 4.
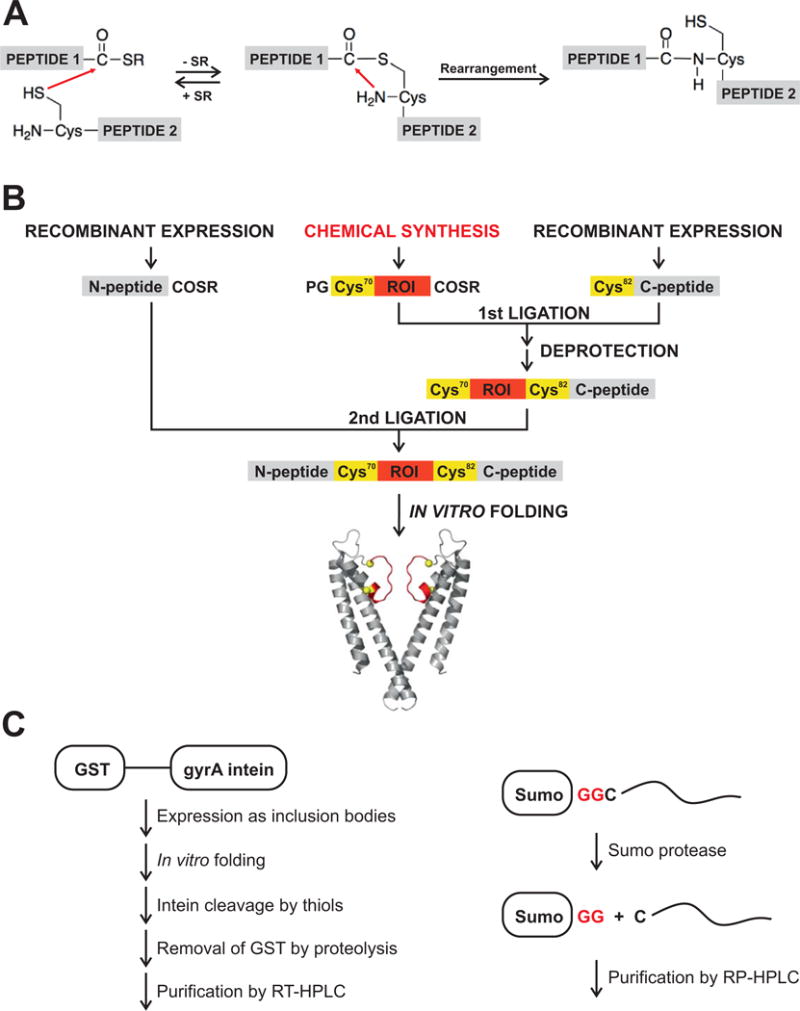
Incorporation of ncAAs via native chemical ligation and semisynthesis of proteins. a In the native chemical ligation reaction a peptide with a C-terminal thioester reacts with a peptide with an N-terminal Cys to link the peptides together with a native peptide bond. b Modular semisynthe- sis of the KcsA channel. The KcsA polypeptide is synthesized by two sequential native chemical ligation reactions. The first ligation reaction between a recombinantly expressed C-peptide (residues 82–165) and a synthetic peptide (residues 70–81) peptide yields the intermediate peptide. The protecting group (PG) on the N-terminal Cys of the intermediate peptide is removed and the deprotected intermediate peptide is ligated to a recombinantly expressed N-peptide thioester (1–69) to yield the KcsA polypeptide. The KcsA polypeptide is folded in vitro to the native state. The protein segments obtained by chemical synthesis are colored red (ROI), while the protein seg- ments obtained by recombinant means are colored grey (N-peptide, C-peptide). The ligation sites (Cys70 and Cys82) are represented in yellow. c The recombinant expression of protein segments for semisynthesis is shown: left, the sandwich fusion strategy for obtaining membrane spanning thioester peptides and, right, the Sumo fusion/proteolysis strategy for obtaining membrane spanning N-Cys peptides.
The native chemical ligation requires a Cys residue and therefore the first step in a semisynthesis is to identify residues flanking the region of interest at which a Cys substitution is well tolerated and can serve as the ligation site. The location of the ligation sites determines the recombinant and synthetic peptides that are required for protein assembly. To satisfy the requirements of the NCL reaction the N-terminal recombinant fragment has a C-terminal thioester, the synthetic fragment has a N-terminal Cys and a C-terminal thioester while the C-terminal recombinant fragment bears an N-terminal Cys and the recombinant thioester peptides are obtained using inteins. To generate a thioester, an intein is fused to the C-terminal of the protein segment, the intein fusion protein is overexpressed, purified and the thioester is then generated by cleaving the intein fusion with a thiol like mercaptoethanesulfonic acid or ethanethiol. The presence of hydrophobic membrane spanning segments in the protein segment complicates the overexpression of an intein fusion. This is because the overexpression of hydrophobic segments is toxic to E. coli. Further, the fusion of an intein to a hydrophobic segment results in targeting the intein to the protein secretory pathway which is also lethal to E. coli. To overcome these problems, a sandwich fusion approach has been used in which the polypeptide of interest is sandwiched between glutathione-S-transferase (GST) at the N-terminus and the gyrA intein at the C-terminus (Figure 4C). The presence of the GST at the N-terminus drives the expression of the protein to inclusion bodies and thereby avoids any cell lethality that can be caused by the overexpression of a membrane spanning polypeptide or the targeting of the intein to the secretory system. Following expression, the fusion protein which is in inclusion bodies is purified and refolded in vitro. The GST protein at the N-terminus is removed by proteolysis, following which the intein is cleaved with a thiol to generate the desired peptide thioester that is then purified using RP-HPLC.
For the purpose of generating recombinant peptides with a N-terminal Cysteine, proteolysis strategies using factor Xa, Tobacco Etch Virus (TEV) protease, thrombin or sumo-protease have been used for soluble proteins. Of these approaches, the sumo-fusion and proteolysis strategy has been successfully applied to membrane spanning peptides (Fig. 4C). In this strategy, the sumo protein is appended to the N-terminus of the peptide of interest. Following expression and purification of the fusion protein, the sumo tag is removed by the use of the sumo protease to release the peptide with a Cys at the N-terminus, which is purified using RP-HPLC.
The ligation reactions between the thioester peptide and the N-Cys peptide is carried out in the presence of detergents to keep the peptides soluble during the course of the reaction. The ligation reaction is initiated by the addition of a thiol catalyst. Thiophenol has been mainly used as the catalyst though other thiols such as mercaptoethanesulfonic acid or mercaptophenylacetic acid can also be used. The ligation reaction is easily monitored by SDS-PAGE. In the three part ligation, the middle synthetic peptide bears both a C-terminal thioester and a N-terminal Cys. It is necessary to protect the N-Cys of the central peptide to prevent cyclization and/or polymerization. A thiazolidine (Thz) group is commonly used for this purpose due to the ease of removal of the Thz group following the first ligation reaction. Following the ligation steps to assemble the polypeptide, it is folded in vitro to the native state. A key requirement for using protein synthesis or semisynthesis for protein modification is the ability to fold the membrane protein in vitro. In vitro folding is necessary as the synthetic steps only provide the unfolded polypeptide which has to be folded to the native state for functional and structural characterization. In the family of ion channels, in vitro folding has been demonstrated for the K+ channels, KcsA and KvAP, the non-selective channel NaK and the mechanosensitive channel MscL. Following folding, the semisynthetic channel is purified similar to the recombinant channel and then reconstituted into lipid bilayers for functional activity or crystallized for structural studies.
The ligation chemistry requires a Cys residue and it is important to ensure that the Cys substitution does not adversely affect the structure/function of the protein. If the Cys substitution is disruptive, then one solution is to use a Cys residue for the ligation reaction and to convert the Cys residue to another residue after the ligation reaction. Reaction conditions have been published that allow facile conversion of a Cys to either an Ala (Pentelute and Kent 2007) or a pseudo-glutamine (addition of a S atom to the Gln side chain). Thus by carrying out the ligation at a site that has either an Ala or a Gln in the native channel, we can eliminate the disruptive effect of the Cys substitution by transmutation of the Cys residue after the ligation reaction.
2. Applications of genetic code expansion to ion channels and receptors
2.1 Applications of in vivo nonsense suppression
A strength of the application of nonsense suppression with orthogonal tRNAs in the Xenopus laeivs oocyte is that a single tRNA, often THG73, can be used to efficiently deliver a wide variety of chemically distinct side-chains. Indeed, the overall approach, and its many applications to the study of ligand and voltage-gated channels has been reviewed extensively (Nowak et al 1998)(Pless and Ahern 2013; Dougherty and Van Arnam 2014). This powerful technique has been used extensively to divine subatomic (i.e electron densities) insights into the vast biological and pharmacological manifestations of the so-called cation-pi interaction: a sterically privileged, non-covalent, electrostatic interaction between an organic cation (neurotransmitter, drug or toxin) and the quadrupole moment generated the pi electrons of an aromatic Phe, Tyr or Trp side-chains (Dougherty 1996; Ma and Dougherty 1997; Gallivan and Dougherty 2000; Zacharias and Dougherty 2002). Further, electrostatic contributions of the pi-electrons of aromatic side-chains in a structural setting have been predicted to be common (Gallivan and Dougherty 1999) but few have been thus far identified and characterized (Pless et al 2011b). None the less, cation-pi interactions have been experimentally characterized between ligands and a multitude of receptor subtypes, including nACHRs (Zhong et al 1998; Xiu et al 2009; Puskar et al 2011), GABARs (Beene et al 2004; Lummis et al 2005; Padgett et al 2007; Lummis et al 2011), GlyRs (Pless et al 2008; Pless et al 2011c) and 5-HT3Rs (Beene et al 2002). And these interactions have been shown to support the extracellular blockade of block of Shaker potassium channels by tetraethyl-ammonium (TEA), and voltage-gated sodium channels by tetrodotoxin (TTX) (Santarelli et al 2007) as well playing a central role in their use-dependent, therapeutic inhibition by local anesthetics and class Ib anti-arrhythmic compounds (Ahern et al 2008; Pless et al 2011a). Nonsense suppression has also revealed novel roles for back-bone chemistries in structure function relationships in ligand gated channels and with interactions between permeant potassium ions and main-chain selectivity filter carbonyls (Lu et al 2001; Valiyaveetil et al 2006; Devaraneni et al 2013; Matulef et al 2013).
Further, isosteric manipulations that neutralize side-chains have been crucial in determining the roles of putative salt-bridges in pentameric subunit composition and voltage-sensor energetic (Cashin et al 2007; Pless et al 2011b; Pless et al 2011d; Pless et al 2014). Recently a subtle tryptophan derivative that lacks the ability to hydrogen bond (H-bond) at the indole nitrogen (Ind) was used to characterize an intricate H-bond network in the vicinity of the potassium channel selectivity filter that acts as a molecular timing mechanism to control ionic conductance during prolonged exposure to activating voltages (Pless et al 2013). The mechanisms, functionalities and energetics revealed in these studies results from the incorporation of amino acid derivatives with very subtle atomic, or subatomic (electron), chemical manipulations. Such approaches allow for the precise functional determination of amino acids on the small scale, a need which is becoming more important as the list of high resolution crystal structures of ion channels, receptor and transporters beg functional description. While these modest chemical and atomic alterations have been a rich source of mechanistic detail, there is also tremendous promise for the genetic incorporation probes with spectroscopic properties for macroscopic fluorescence (Kalstrup and Blunck 2013) as well as single molecule imaging (Pantoja et al 2009). Thus, there is a bright future for the application of misacylated orthogonal tRNAs in tandem with nonsense suppression to study ion channels and receptors.
2.2 Applications of orthogonal ncAA-RS/tRNA pairs
One obvious advantage of the use of ncAA-RS/tRNA pairs is that, once generated, one needs only transiently express the RS, tRNA and target membrane protein clones and supplement the ncAA to the growth media. Given that the acylation and incorporation chemistry is functionally “outsourced” to the cell and the ncAA-RS/tRNA pair, this approach is by far the most within reach of the general investigator.
2.2.1 Early adaptation and application of ncAA-RS/tRNA pairs
To date, most work associated with membrane proteins and with ncAA-RS/tRNA pairs has been done in the field of G-protein coupled receptors (GPCRs) Daggett and Sakmar 2011). This vast membrane protein gene family supports a diverse array of physiologically important cellular signaling pathways and GPCRs are established targets of more than a quarter of all therapeutic drugs. However, the details of their complex functioning in biological membranes remain to be fully understood. To dissect local conformational changes during activation of a GPCR, p-azido-L-phenylalanine (AzF) has been used as a genetically encoded infrared probe (Ye et al 2009; Ye 2010). This azido-functionalized ncAA can also serve as a unique chemical handle to attach probes, e.g. fluorophores, through the Staudinger-Bertozzi ligation with triarylphosphine derivatives (Okuda and Tokuda 2009) or through 1,3-cycloaddition between the azido and cyclooctynes (Kaiser 2006; Lakshmipathy 2007). Also, the keto group of the p-acetyl-L-phenylalanine (AcF) side-chain allows for site-specific labeling with hydrazide or hydroxylamine derivatives (Braig et al 2009). These approaches and their applications to the study of GPCRs have been reviewed in detail by Daggett and Sakmar, 2011.
2.2.2 Promise and potential caveats with the applications of cross-linkers
The ability to genetically encode side-chains with cross-linking abilities in response to exposure to light (often UV) has tremendous promise for the characterization of the multitude of non-covalent cellular interactions. Many such interactions are transient and their low affinity prohibits study by conventional approaches, and moreover, genetic encoding allows for specific placement of cross-linking side-chains into proteinaceous nooks and interaction surfaces that would otherwise be inaccessible to solvent accessible probes. There are a number of available encodable cross-linkers for expression in a variety if systems for biochemical and functional analyses. The photo-crosslinking amino acids p-benzoyl-L-phenylalanine (Bpa) and p-trifluoromethyl-diazirinyl-L-phenylalanine (tmdF) have been encoded separately with evolved M. jannaschii and E. coli. tyrosyl-RS/tRNATyr for bacterial and eukaryotic expression and both are capable of forming covalent bonds with an interacting partner upon UV illumination (Tippmann et al 2007; Chou et al 2011). TmdF has been reported to have higher degree of incorporation and higher crosslinking efficiency than Bpa when exposed to 365 nm light (Hino et al 2011). Alternatively, the diarerine crosslinkers ABK (3′-azibutyl-N-carbamoyl-lysine) and DiZPK ((3-(3-methyl-3H-diazirine-3-yl)-propaminocarbonyl-Nε–L-lysine) are encoded by the M. barkeri pyrrolysyl-tRNA-synthetase/tRNA and have been reported to have improved crosslinking efficiency over Bpa. Moreover, Ffact (p-fluoroacetyl-phenylalanine) forms covalent bonds with nearby cysteine residues (Coin et al 2013). As a photo-crosslinker, Bpa has several potential advantages over AzF, ABK and DiZPK. Upon UV illumination, Bpa forms a biradical that under certain geometries can abstract an H atom from C-H bonds in a distance of about 3-4 Å to form a covalent adduct (Sato et al 2011). However, n the absence of a reacting partner, Bpa returns to the ground state from which it can be re-excited (Dormán and Prestwich 1994). In contrast, the photo-activation of AzF, DiZPK and ABK is irreversible. However, DiZPK has been shown to crosslink with higher efficiency compared to Bpa when exposed to 365 nm light (Hino et al 2011). Alternatively, AzF has a range of potential targets and its photo-activation is also triggered by ambient light (Grunbeck et al 2011) and it is worth noting that non-specific crosslinking or photo-damage may occur when using AzF as it is excited at shorter wavelengths (Dormán and Prestwich 1994; Chin et al 2002c). Thus while each cross-linker has inherent benefits and caveats in their specific application to the study membrane proteins, they have been used previously (and successfully) to reveal ligand-receptor interactions, as well as inter-subunit and -domain contacts (Schlieker 2004; Haslberger 2007; Boos et al 2008; Umanah et al 2009; Ye et al 2009; Liu 2010; Wu et al 2010; Grunbeck et al 2011; Coin et al 2011; Grunbeck et al 2012; Coin et al 2013).
In addition to characterizing transient protein-protein interactions, encoded cross-linkers also have the potential for the external photo-control of a specific ion channel or receptor. An elegant example of such an application can be found in the use of RS/tRNA pairs to homomeric GluA2 or heteromeric GluA2:GluA1 AMPA receptors in HEK 293 cells (Klippenstein et al 2014). AMPA receptors are essential in physiology for fast, glutamate-activated, post-synaptic currents and are thus of great interest in optogenetic studies. The photo cross-linkers AzF and Bpa were incorporated at different positions of the ligand-binding domain D2 which allowed for the identification of positions with state-dependent and state-independent photo-crosslinking behavior. The state-dependent cross-linking sites allowed for the remote and rapid ‘photo-inactivation’ of Bpa expressing receptors. While several methods have been used to inactivate glutamate receptors in vivo previously (Adesnik et al 2005; Tracy et al 2011), these approaches lack cell specificity or develop only very slowly, thus the use of Bpa here represents a significant advancement and it will be exciting to see this approach applied in a more native context.
The AzF and Bpa Rs/tRNA pairs have also been expressed in N-methyl-D-aspartate receptors (NMDRs) and in the context of Xenopus laevis oocytes (Ye et al 2013). Here, the microinjected DNA plasmids encoding the Bs tRNATyrCUA and Ec Tyr-RS AzF and Bpa variants were optimized and the ncAAs were supplemented to the culture media, suggesting that the ncAAs can be internalized into the oocyte. Moreover, the Bpa-RS was demonstrated to exhibit a higher translational fidelity than AzF-RS. The photo-cross-linkers were introduced into the N-terminal domain of GluN2 at an inter-lobe position where UV photo-treatment locked the receptor in a discrete functional state, reinforcing the importance of this region in controlling NMDAR gating and pharmacology (Ye et al 2013).
2.2.3 Recoding voltage-gated enzymes and potassium channels: optogenetics, fluorophores and structure-function insights
Studies that apply orthogonal ncAA-RS/tRNA pairs for investigating ion channels and other ion conducting membrane proteins are beginning to emerge. Utilizing evolved Ec Tyr-RS and Ec Leu-RS derivatives in combination with optimized transcription and processing of Ec tRNACUA in different eukaryotic expression systems Wang and colleagues incorporated O-methyl-L-tyrosine (OmeTyr; Chin 2003) and 2-amino-3-(5-(dimethylamino)-naphthalene-1-sulfonamido)propanoic acid (DanAla; Summerer et al 2006) into the N-terminus of the neuronal voltage-gated K+ channel Kv1.4 to gauge steric limitations in fast-inactivation (Wang et al 2007b). More importantly, this work paved the way for their later development of a light-activated K+ channel as an optogenetic, neuronal silencing tool (Kang et al 2013). Here, the photoreactive 4,5-dimethoxy-2-nitrobenzyl-cysteine (Cmn) was genetically incorporated into the pore of the inwardly rectifying K+ channel Kir2.1, using a specifically evolved Ec Leu-RS variant paired with its cognate tRNALeuCUA (Kang et al 2013). The presence of Cmn in the pore of Kir2.1 rendered the channel non-conducting, however, UV photolysing the dimethoxynitrobenzyl group leaves behind a cysteine side-chain (Rhee et al 2008). Thus, brief illumination with UV light irreversibly releases the channel block and permits K+ current to flow, resulting in suppression of neuronal firing, as demonstrated in isolated rat hippocampal neurons as well as embryonic mouse cortex (Kang et al 2013). Further, a lentivirus-based gene delivery method was used to express DanAla into the voltage-sensing domain (VSD) of the Ciona intestinalis voltage-sensing phosphotase CiVSP to report membrane depolarization in differentiating neuronal cells (Shen et al 2011).
The Blunck group has utilized a tRNA/RS pair that encodes the fluorescent ncAA Anap, 3-(6-acetylnaphthalen-2-ylamino)-2-aminopropionic acid to investigate dynamics of Shaker potassium channel pore conformations (Lee et al 2009)(Kalstrup and Blunck 2013). Compared to dansyl- or coumarin containing ncAAs, Anap is smaller in size and displays enhanced environmental sensitivity with comparable or increased brightness (Lee et al 2009). At present, the most powerful way to associate structural and dynamic (functional) information for electrogenic membrane proteins is voltage-clamp fluorometry (VCF), described in Chapter 4, (Mannuzzu et al 1996; Cha and Bezanilla 1997). However, traditional VCF is restricted to extracellular surface rearrangements due to the cysteine attachment chemistry required for covalent attachment of fluorophores. By encoding Anap into cytoplasmic regions of the Shaker channel, the authors found that pore opening occurs in two sequential transitions (Kalstrup and Blunck 2013). This work also further demonstrates that orthogonal RS/tRNA pairs may be used to incorporate ncAAs in X. laevis oocytes by simply microinjecting the DNA encoding the orthogonal pair into the oocyte’s nucleus prior cytosolic injection of the target cRNA together with the ncAA.
2.3 Applications of protein semisynthesis
Protein semisynthesis has been reported for the K+ channels, KcsA and KvAP, and the non-selective channel NaK. In the K+ channels, semisynthesis has been used to investigate mechanisms of selectivity, ion permeation and C-type inactivation while in the NaK channel, semisynthesis has been used to investigate the mechanism of block by divalent ions. Here, we discuss the use of semisynthesis to investigate C-type inactivation in K+ channels.
C-type (or slow) inactivation is a conformational change that takes place at the selectivity filter, the ion binding sites in a K+ channel and converts it from a conductive state through which permeation of K+ can take place through to a non-conductive state. C-type inactivation is widespread in K+ channels and plays an important physiological role in regulating neuronal firing and in pacing cardiac action potentials. C-type inactivation is also observed in the bacterial K+ channel KcsA and the archaeal K+ channel KvAP and these channels have been used as model systems to investigate the process of C-type inactivation. The following two modifications have been introduced using semisynthesis in studies of C-type inactivation.
2.3.1. D-Ala substitution in the K+ selectivity filter
Structural studies on the KcsA channel under conditions that favored C-type inactivation showed a non-conductive conformation of the selectivity filter which was referred to as the collapsed state. It was proposed that the collapsed state of the selectivity filter represents the C-type inactivated state. To determine if the collapsed state does indeed correspond to the C-type inactivated state, semisynthesis was used to generate a channel in which collapse of the selectivity filter was blocked. This was accomplished by replacing the first conserved Gly in the selectivity filter, Gly77 with D-Ala. Using crystallography and functional studies, it was demonstrated structure of the selectivity filter of the D-Ala mutant is similar to the wild type KcsA channel (at 150 mM K+) and that the ion permeation and selectivity properties of the D-Ala mutant are similar to the wild type channel. However, the presence of the additional methyl group due to the D-Ala substitution prevents the selectivity filter from attaining the collapsed conformation. This was demonstrated by structural studies at low K+. It was observed that under conditions of low K+ (at 1 mM K+) at which the selectivity filter of the wild type channel is in the collapsed state, the selectivity filter of the D-Ala mutant stays in the conductive state. An investigation of C-type inactivation in the D-Ala mutant did not indicate any significant changes in C-type inactivation or in the recovery from the inactivated state. Similar results were also observed for the KvAP channel in which semisynthesis was used to replace the equivalent glycine residue in the selectivity filter, G198 with D-Ala. These results show that blocking the collapsed state of the selectivity filter does not affect C-type inactivation which indicates that the collapsed conformation of the selectivity filter does not correspond to the C-type inactivated state. The use of semisynthesis is critical in these experiments as it enabled the incorporation of D-Ala into the channel.
2.3.2 Ester substitutions in the K+ selectivity filter
One of the characteristics of C-type inactivation is a dependence on the permeant ion. The rate of C-type inactivation decreases when the K+ concentration is increased or when the permeant ion is changed from K+ to Rb+. The mechanism by which these changes in the concentration or the nature of the ion influence C-type inactivation is not known. It has been proposed that C-type inactivation is linked to ion occupancy at specific sites in the selectivity filter. To investigate the role of ion occupancy at the selectivity filter on C-type inactivation, semisynthesis used was to alter ion binding at specific sites in the selectivity filter. The S1-3 sites in the selectivity filter are constructed using the backbone carbonyl oxygen atoms. Semisynthesis of the KcsA channel was used to generate amide to ester substitutions in the protein backbone of the selectivity filter. An ester linkage is iso-steric to an amide bond but shows reduced electronegativity at the carbonyl oxygen, which reduces ion binding at the selectivity filter site. The ester substitutions thereby provide a means to investigate the effect of ion occupancy at the selectivity filter on C-type inactivation. Ester substitutions were introduced at the S1-3 sites in the selectivity filter of the KcsA channel using protein semisynthesis. It was observed that the ester substitution at the S1 site did not affect inactivation, while the ester substitutions at the S2 and the S3 sites dramatically reduced inactivation. The ester substitution at the S2 site in the KcsA channel was also introduced using the nonsense suppression approach. Structural studies on S2 ester mutant of the KcsA channel showed that the ester substitution resulted in a decrease in ion occupancy at the S2 site. The nonsense suppression approach was also used to introduce an ester substitution at the S2 site in the KvAP channel and demonstrated to cause a decrease in C-type inactivation as observed for the KcsA channel. These observations led to the conclusion that C-type inactivation in a K+ channel requires ion occupancy at the S2 site.
Outlook and future directions
The future is bright for the application of genetic code expansion for the study of membrane proteins. The sophistication and functionality of the approaches described herein are improving with each technological iteration and application to membrane proteins. While much effort has been made in genetic code expansion in prokaryotic and model mammalian cells lines, techniques to all for the genetic encoding of new amino acids into native cell-types of neuroendocrine, muscle, neurons and live animals will be especially important as they will permit the use of encoded reporters for optogenetic applications as well as having the promise to yield transformative insights into molecular physiological and the basis for disease. However, such complex cell-types may hold unique challenges in terms of RNA stability as well as efficient and stable expression of tRNA and synthetases. Further, adding new amino acids to cell-free protein reactions has the ability to reveal fundamental new insights into ion channel function. For one, such systems lack inclusion bodies that can rob membrane protein yields needed for biochemical and structural studies and thus can produce large-scale preparations of pure membrane protein, as well as allowing for a wider scale of expression conditions. Further, combining cell free systems with novel encoded amino acids with spin label or gadolinium chelates would be transformative for structure-function studies and for structural refinement. And the continued development of robust approaches for the efficient expression of multiple amino acids with single molecule fluorescent or spin label properties will open many new doors for biochemical and cellular applications. Thus current and future approaches of ncAA encoding, and overcoming the challenges of their implementation, will ultimately have widespread impacts on the molecular and physiological understanding of ion channels and receptors, from atoms to animals.
References
- Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. doi: 10.1016/j.neuron.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Ahern CA, Eastwood AL, Dougherty DA, Horn R. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circ Res. 2008;102:86–94. doi: 10.1161/CIRCRESAHA.107.160663. [DOI] [PubMed] [Google Scholar]
- Ai H-W, Shen W, Sagi A, et al. Probing protein–protein interactions with a genetically encoded photo-crosslinking amino acid. Chembiochem. 2011;12:1854–1857. doi: 10.1002/cbic.201100194. [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Schultz PG. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry (Mosc) 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wu N, Santoro SW, et al. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett OPT, Chin JW. Evolved orthogonal ribosome purification for in vitro characterization. Nucleic Acids Res. 2010;38:2682–2691. doi: 10.1093/nar/gkq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beene DL, Brandt GS, Zhong W, et al. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry (Mosc) 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- Beene DL, Price KL, Lester HA, et al. Tyrosine residues that control binding and gating in the 5-hydroxytryptamine3 receptor revealed by unnatural amino acid mutagenesis. J Neurosci Off J Soc Neurosci. 2004;24:9097–9104. doi: 10.1523/JNEUROSCI.2429-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Townsley FM, Greiss S, et al. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- Blight SK, Larue RC, Mahapatra A, et al. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- Boos D, Kuffer C, Lenobel R, et al. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J Biol Chem. 2008;283:816–823. doi: 10.1074/jbc.M706748200. [DOI] [PubMed] [Google Scholar]
- Braig D, Bar C, Thumfart JO, Koch HG. Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J Mol Biol. 2009;390:401–413. doi: 10.1016/j.jmb.2009.04.061. [DOI] [PubMed] [Google Scholar]
- Brick P, Bhat TN, Blow DM. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Cashin AL, Torrice MM, McMenimen KA, et al. Chemical-scale studies on the role of a conserved aspartate in preorganizing the agonist binding site of the nicotinic acetylcholine receptor. Biochemistry (Mosc) 2007;46:630–639. doi: 10.1021/bi061638b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellitti SE. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9268–9281. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Chang H, Han M, Huang W, et al. Light-induced protein translocation by genetically encoded unnatural amino acid in Caenorhabditis elegans. Protein Cell. 2013;4:883–886. doi: 10.1007/s13238-013-3118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Sun SB, Furman JL, et al. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry (Mosc) 2013a;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Schultz PG. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:14841–14846. doi: 10.1073/pnas.1212454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Yang P-Y, et al. A tryptophanyl-tRNA synthetase/tRNA pair for unnatural amino acid mutagenesis in E. coli. Angew Chem Int Ed Engl. 2013b;52:5106–5109. doi: 10.1002/anie.201301094. [DOI] [PubMed] [Google Scholar]
- Chen PR, Groff D, Guo J, et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew Chem Int Ed Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Schultz P, Brock A. J Mol Biol. 2007;371:112. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Chin JW. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Chu S, et al. Progress toward an expanded eukaryotic genetic code. Chem Biol. 2003;10:511–519. doi: 10.1016/s1074-5521(03)00123-6. [DOI] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, et al. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2002a;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, et al. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci U S A. 2002b;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Santoro SW, Martin AB, et al. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002c;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- Chou C, Uprety R, Davis L, et al. Genetically encoding an aliphatic diazirine for protein photocrosslinking. Chem Sci. 2011;2:480–483. [Google Scholar]
- Cload ST, Liu DR, Froland WA, Schultz PG. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem Biol. 1996;3:1033–1038. doi: 10.1016/s1074-5521(96)90169-6. [DOI] [PubMed] [Google Scholar]
- Coin I, Katritch V, Sun T, et al. Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell. 2013;155:1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin I, Perrin MH, Vale WW, Wang L. Photo-cross-linkers incorporated into G-protein-coupled receptors in mammalian cells: a ligand comparison. Angew Chem Int Ed Engl. 2011;50:8077–8081. doi: 10.1002/anie.201102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley RB, Karplus PA, Mehl RA. Gleaning unexpected fruits from hard-won synthetases: probing principles of permissivity in non-canonical amino acid-tRNA synthetases. Chembiochem Eur J Chem Biol. 2014;15:1810–1819. doi: 10.1002/cbic.201402180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropp TA, Anderson JC, Chin JW. Reprogramming the amino-acid substrate specificity of orthogonal aminoacyl-tRNA synthetases to expand the genetic code of eukaryotic cells. Nat Protoc. 2007;2:2590–2600. doi: 10.1038/nprot.2007.378. [DOI] [PubMed] [Google Scholar]
- Daggett KA, Sakmar TP. Site-specific in vitro and in vivo incorporation of molecular probes to study G-protein-coupled receptors. Curr Opin Chem Biol. 2011;15:392–398. doi: 10.1016/j.cbpa.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- Devaraneni PK, Komarov AG, Costantino CA, et al. Semisynthetic K+ channels show that the constricted conformation of the selectivity filter is not the C-type inactivated state. Proc Natl Acad Sci. 2013;110:15698–15703. doi: 10.1073/pnas.1308699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormán G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry (Mosc) 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- Dougherty DA. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- Dougherty DA, Van Arnam EB. In vivo incorporation of non-canonical amino acids by using the chemical aminoacylation strategy: a broadly applicable mechanistic tool. Chembiochem Eur J Chem Biol. 2014;15:1710–1720. doi: 10.1002/cbic.201402080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca M, Chen S, Hecht SM. Aminoacylation of transfer RNAs with one and two amino acids. Methods San Diego Calif. 2008;44:87–99. doi: 10.1016/j.ymeth.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Edwards H, Schimmel P. Mol Cell Biol. 1990;10:1633. doi: 10.1128/mcb.10.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England P, Zhang Y, Dougherty D, Lester H. Cell. 1999;96:89. doi: 10.1016/s0092-8674(00)80962-9. [DOI] [PubMed] [Google Scholar]
- Farrell IS, Toroney R, Hazen JL, et al. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- Fekner T, Li X, Chan MK. Pyrrolysine Analogs for Translational Incorporation into Proteins. Eur J Org Chem. 2010;2010:4171–4179. doi: 10.1002/ejoc.201000204. [DOI] [Google Scholar]
- Furter R. Protein Sci. 1998a;7:419. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter R. Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci Publ Protein Soc. 1998b;7:419–426. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. A Computational Study of Cation−π Interactions vs Salt Bridges in Aqueous Media: Implications for Protein Engineering. J Am Chem Soc. 2000;122:870–874. doi: 10.1021/ja991755c. [DOI] [Google Scholar]
- Gautier A. Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- Greiss S, Chin JW. Expanding the genetic code of an animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunbeck A, Huber T, Abrol R, et al. Genetically encoded photo-cross-linkers map the binding site of an allosteric drug on a G protein-coupled receptor. ACS Chem Biol. 2012;7:967–972. doi: 10.1021/cb300059z. [DOI] [PubMed] [Google Scholar]
- Grunbeck A, Huber T, Sachdev P, Sakmar TP. Mapping the ligand-binding site on a G protein-coupled receptor (GPCR) using genetically encoded photocrosslinkers. Biochemistry (Mosc) 2011;50:3411–3413. doi: 10.1021/bi200214r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammill JT, Miyake-Stoner S, Hazen JL, et al. Preparation of site-specifically labeled fluorinated proteins for 19F-NMR structural characterization. Nat Protoc. 2007;2:2601–2607. doi: 10.1038/nprot.2007.379. [DOI] [PubMed] [Google Scholar]
- Hao B, Gong W, Ferguson TK, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- Haslberger T. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell. 2007;25:247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Hecht S, Alford B, Kuroda Y, Kitano S. J Biol Chem. 1978;253:4517. [PubMed] [Google Scholar]
- Hino N, Oyama M, Sato A, et al. Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells. J Mol Biol. 2011;406:343–353. doi: 10.1016/j.jmb.2010.12.022. [DOI] [PubMed] [Google Scholar]
- Hohsaka T, Ashizuka Y, Sisido M. Incorporation of two nonnatural amino acids into proteins through extension of the genetic code. Nucleic Acids Symp Ser. 1999:79–80. doi: 10.1093/nass/42.1.79. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Fire A. Recognition and silencing of repeated DNA. Annu Rev Genet. 2000;34:187–204. doi: 10.1146/annurev.genet.34.1.187. [DOI] [PubMed] [Google Scholar]
- Huang Y, Russell WK, Wan W, et al. A convenient method for genetic incorporation of multiple noncanonical amino acids into one protein in Escherichia coli. Mol Biosyst. 2010;6:683–686. doi: 10.1039/b920120c. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Carr PA, Wang HH, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CM, Ferguson TK, Leykam JF, Krzycki JA. The amber codon in the gene encoding the monomethylamine methyltransferase isolated from Methanosarcina barkeri is translated as a sense codon. J Biol Chem. 2001;276:34252–34258. doi: 10.1074/jbc.M102929200. [DOI] [PubMed] [Google Scholar]
- Johnson DBF, Xu J, Shen Z, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- Kalstrup T, Blunck R. Dynamics of internal pore opening in K(V) channels probed by a fluorescent unnatural amino acid. Proc Natl Acad Sci U S A. 2013;110:8272–8277. doi: 10.1073/pnas.1220398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-Y, Kawaguchi D, Coin I, et al. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80:358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippenstein V, Ghisi V, Wietstruk M, Plested AJR. Photoinactivation of glutamate receptors by genetically encoded unnatural amino acids. J Neurosci Off J Soc Neurosci. 2014;34:980–991. doi: 10.1523/JNEUROSCI.3725-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nureki O, Ishitani R, et al. Nat Struct Biol. 2003;10:425. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- Kowal AK, Kohrer C, RajBhandary UL. Twenty-first aminoacyl-tRNA synthetase-suppressor tRNA pairs for possible use in site-specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc Natl Acad Sci U S A. 2001;98:2268–2273. doi: 10.1073/pnas.031488298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzycki J. Curr Opin Microbiol. 2005;8:706. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Lajoie MJ, Kosuri S, Mosberg JA, et al. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy SK. Identification of nascent chain interaction sites on trigger factor. J Biol Chem. 2007;282:12186–12193. doi: 10.1074/jbc.M609871200. [DOI] [PubMed] [Google Scholar]
- Lee HS, Guo J, Lemke EA, et al. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J Am Chem Soc. 2009;131:12921–12923. doi: 10.1021/ja904896s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-T, Mahapatra A, Longstaff DG, et al. Specificity of pyrrolysyl-tRNA synthetase for pyrrolysine and pyrrolysine analogs. J Mol Biol. 2009;385:1156–1164. doi: 10.1016/j.jmb.2008.11.032. [DOI] [PubMed] [Google Scholar]
- Liu C. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature. 2010;463:197–202. doi: 10.1038/nature08651. [DOI] [PubMed] [Google Scholar]
- Liu C, Cellitti S, Geierstanger B, Schultz P. Nat Protoc. 2009;4:1784. doi: 10.1038/nprot.2009.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotech. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. Adding New Chemistries to the Genetic Code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- Liu W, Alfonta L, Mack AV, Schultz PG. Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases. Angew Chem Int Ed Engl. 2007a;46:6073–6075. doi: 10.1002/anie.200701990. [DOI] [PubMed] [Google Scholar]
- Liu W, Brock A, Chen S, et al. Nat Methods. 2007b;4:239. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- Lodder M, Wang B, Hecht SM. The N-pentenoyl protecting group for aminoacyl-tRNAs. Methods San Diego Calif. 2005;36:245–251. doi: 10.1016/j.ymeth.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lu T, Ting AY, Mainland J, et al. Probing ion permeation and gating in a K+ channel with backbone mutations in the selectivity filter. Nat Neurosci. 2001;4:239–246. doi: 10.1038/85080. [DOI] [PubMed] [Google Scholar]
- Lummis SCR, L Beene D, Harrison NJ, et al. A cation-pi binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem Biol. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Lummis SCR, McGonigle I, Ashby JA, Dougherty DA. Two amino acid residues contribute to a cation-π binding interaction in the binding site of an insect GABA receptor. J Neurosci Off J Soc Neurosci. 2011;31:12371–12376. doi: 10.1523/JNEUROSCI.1610-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JC, Dougherty DA. The Cationminus signpi Interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- Majmudar CY, Lee LW, Lancia JK, et al. Impact of nonnatural amino acid mutagenesis on the in vivo function and binding modes of a transcriptional activator. J Am Chem Soc. 2009;131:14240–14242. doi: 10.1021/ja904378z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Matulef K, Komarov AG, Costantino CA, Valiyaveetil FI. Using protein backbone mutagenesis to dissect the link between ion occupancy and C-type inactivation in K+ channels. Proc Natl Acad Sci. 2013;110:17886–17891. doi: 10.1073/pnas.1314356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon C, Schultz P. Bioorg Med Chem Lett. 2009;19:3845. doi: 10.1016/j.bmcl.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir T. Annu Rev Biochem. 2003;72:249. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Hayashi A, Iraha F, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Kobayashi T, Hino N, et al. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- Neumann H. Rewiring translation - Genetic code expansion and its applications. FEBS Lett. 2012;586:2057–2064. doi: 10.1016/j.febslet.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Neumann H, Slusarczyk AL, Chin JW. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J Am Chem Soc. 2010a;132:2142–2144. doi: 10.1021/ja9068722. [DOI] [PubMed] [Google Scholar]
- Neumann H, Wang K, Davis L, et al. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010b;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- Noren C, Anthony-Cahill S, Griffith M, Schultz P. Science. 1989;244:182. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- Nowak MW, Gallivan JP, Silverman SK, et al. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- Ohno S, Yokogawa T, Fujii I, et al. Co-expression of yeast amber suppressor tRNATyr and tyrosyl-tRNA synthetase in Escherichia coli: possibility to expand the genetic code. J Biochem (Tokyo) 1998;124:1065–1068. doi: 10.1093/oxfordjournals.jbchem.a022221. [DOI] [PubMed] [Google Scholar]
- Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA. 2009;106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett CL, Hanek AP, Lester HA, et al. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation-pi interaction between GABA and beta 2Tyr97. J Neurosci Off J Soc Neurosci. 2007;27:886–892. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja R, Rodriguez EA, Dibas MI, et al. Single-molecule imaging of a fluorescent unnatural amino acid incorporated into nicotinic receptors. Biophys J. 2009;96:226–237. doi: 10.1016/j.bpj.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeler JC, Mehl RA. Site-specific incorporation of unnatural amino acids as probes for protein conformational changes. Methods Mol Biol Clifton NJ. 2012;794:125–134. doi: 10.1007/978-1-61779-331-8_8. [DOI] [PubMed] [Google Scholar]
- Pentelute BL, Kent SBH. Selective desulfurization of cysteine in the presence of Cys(Acm) in polypeptides obtained by native chemical ligation. Org Lett. 2007;9:687–690. doi: 10.1021/ol0630144. [DOI] [PubMed] [Google Scholar]
- Peters F, Brock A, Wang J, Schultz P. Chem Biol. 2009;16:148. doi: 10.1016/j.chembiol.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Ahern CA. Unnatural amino acids as probes of ligand-receptor interactions and their conformational consequences. Annu Rev Pharmacol Toxicol. 2013;53:211–229. doi: 10.1146/annurev-pharmtox-011112-140343. [DOI] [PubMed] [Google Scholar]
- Pless SA, Elstone FD, Niciforovic AP, et al. Asymmetric functional contributions of acidic and aromatic side chains in sodium channel voltage-sensor domains. J Gen Physiol. 2014;143:645–656. doi: 10.1085/jgp.201311036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Galpin JD, Frankel A, Ahern CA. Molecular basis for class Ib anti-arrhythmic inhibition of cardiac sodium channels. Nat Commun. 2011a;2:351. doi: 10.1038/ncomms1351. [DOI] [PubMed] [Google Scholar]
- Pless SA, Galpin JD, Niciforovic AP, et al. Hydrogen bonds as molecular timers for slow inactivation in voltage-gated potassium channels. eLife. 2013;2:e01289. doi: 10.7554/eLife.01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat Chem Biol. 2011b;7:617–623. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Hanek AP, Price KL, et al. A cation-π interaction at a phenylalanine residue in the glycine receptor binding site is conserved for different agonists. Mol Pharmacol. 2011c;79:742–748. doi: 10.1124/mol.110.069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Leung AWY, Galpin JD, Ahern CA. Contributions of conserved residues at the gating interface of glycine receptors. J Biol Chem. 2011d;286:35129–35136. doi: 10.1074/jbc.M111.269027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Millen KS, Hanek AP, et al. A cation-pi interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J Neurosci Off J Soc Neurosci. 2008;28:10937–10942. doi: 10.1523/JNEUROSCI.2540-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpo C, Ambrogelly A, Bérubé A, et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci U S A. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpo C, Herring S, Berube A, et al. FEBS Lett. 2006;580:6695. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskar NL, Xiu X, Lester HA, Dougherty DA. Two neuronal nicotinic acetylcholine receptors, alpha4beta4 and alpha7, show differential agonist binding modes. J Biol Chem. 2011;286:14618–14627. doi: 10.1074/jbc.M110.206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- Rhee H, Lee J-S, Lee J, et al. Photolytic control and infrared probing of amide I mode in the dipeptide backbone-caged with the 4,5-dimethoxy-2-nitrobenzyl group. J Phys Chem B. 2008;112:2128–2135. doi: 10.1021/jp074776z. [DOI] [PubMed] [Google Scholar]
- Robertson SA, Ellman JA, Schultz PG. A general and efficient route for chemical aminoacylation of transfer RNAs. J Am Chem Soc. 1991;113:2722–2729. doi: 10.1021/ja00007a055. [DOI] [Google Scholar]
- Rodriguez E, Lester H, Dougherty D. Proc Natl Acad Sci USA. 2006;103:8650. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez EA, Lester HA, Dougherty DA. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: minimizing misacylation. RNA N Y N. 2007a;13:1703–1714. doi: 10.1261/rna.666807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez EA, Lester HA, Dougherty DA. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: evaluating suppression efficiency. RNA N Y N. 2007b;13:1715–1722. doi: 10.1261/rna.667607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman DM, Petersson EJ, Vázquez ME, et al. Caged phosphoproteins. J Am Chem Soc. 2005;127:846–847. doi: 10.1021/ja043875c. [DOI] [PubMed] [Google Scholar]
- Rydén SM, Isaksson LA. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet MGG. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Hayashi A, Sakamoto A, et al. Nucleic Acids Res. 2002;30:4692. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks M, Sampson J, Nowak M, et al. J Biol Chem. 1996;271:23169. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- Santarelli VP, Eastwood AL, Dougherty DA, et al. Calcium block of single sodium channels: role of a pore-lining aromatic residue. Biophys J. 2007;93:2341–2349. doi: 10.1529/biophysj.107.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S, Wang L, Herberich B, et al. Nat Biotechnol. 2002;20:1044. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Anderson JC, Lakshman V, Schultz PG. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 2003;31:6700–6709. doi: 10.1093/nar/gkg903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Mimasu S, Sato A, et al. Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry (Mosc) 2011;50:250–257. doi: 10.1021/bi1016183. [DOI] [PubMed] [Google Scholar]
- Schlieker C. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- Shen B, Xiang Z, Miller B, et al. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cells Dayt Ohio. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Mabuchi H, Tanaka-Yamano S, et al. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci U S A. 2011;108:15179–15183. doi: 10.1073/pnas.1105921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Steegborn C, Hübel F, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1996;24:68–72. doi: 10.1093/nar/24.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- Summerer D, Chen S, Wu N, et al. Proc Natl Acad Sci USA. 2006;103:9785. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto JK, Xiang Z, Kang J-Y, Wang L. Esterification of an unnatural amino acid structurally deviating from canonical amino acids promotes its uptake and incorporation into proteins in mammalian cells. Chembiochem Eur J Chem Biol. 2010;11:2268–2272. doi: 10.1002/cbic.201000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, et al. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol. 2009;184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippmann EM, Liu W, Summerer D, et al. A genetically encoded diazirine photocrosslinker in Escherichia coli. Chembiochem. 2007;8:2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- Tracy TE, Yan JJ, Chen L. Acute knockdown of AMPA receptors reveals a trans-synaptic signal for presynaptic maturation. EMBO J. 2011;30:1577–1592. doi: 10.1038/emboj.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umanah G, Huang L-Y, Schultz PG, et al. Incorporation of the unnatural amino acid p-benzoyl-L-phenylalanine (Bpa) into a G protein-coupled receptor in its native context. Adv Exp Med Biol. 2009;611:333–335. doi: 10.1007/978-0-387-73657-0_149. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, Leonetti M, Muir TW, Mackinnon R. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 2006;314:1004–1007. doi: 10.1126/science.1133415. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, MacKinnon R, Muir TW. Semisynthesis and folding of the potassium channel KcsA. J Am Chem Soc. 2002;124:9113–9120. doi: 10.1021/ja0266722. [DOI] [PubMed] [Google Scholar]
- Wan W, Huang Y, Wang Z, et al. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed Engl. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009a;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007a;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- Wang K, Sachdeva A, Cox DJ, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brock A, Herberich B, Schultz P. Science. 2001;292:498. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- Wang L, Schultz P. Chem Biol. 2001;8:883. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009b;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- Wang W, Takimoto J, Louie G, et al. Nat Neurosci. 2007b;10:1063. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EYT, Mol CD, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Deiters A, Cropp TA, et al. A genetically encoded photocaged amino acid. J Am Chem Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- Xiu X, Puskar NL, Shanata JAP, et al. Nicotine binding to brain receptors requires a strong cation-pi interaction. Nature. 2009;458:534–537. doi: 10.1038/nature07768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem. 2010;285:41222–41231. doi: 10.1074/jbc.M110.163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Riou M, Carvalho S, Paoletti P. Expanding the genetic code in Xenopus laevis oocytes. Chembiochem Eur J Chem Biol. 2013;14:230–235. doi: 10.1002/cbic.201200515. [DOI] [PubMed] [Google Scholar]
- Ye SX. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- Ye SX, Huber T, Vogel R, Sakmar TP. FTIR analysis of GPCR activation using azido probes. Nat Chem Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DD, Young TS, Jahnz M, et al. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry (Mosc) 2011;50:1894–1900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias N, Dougherty DA. Cation-pi interactions in ligand recognition and catalysis. Trends Pharmacol Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Schultz PG, Wilson IA. Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine. Protein Sci Publ Protein Soc. 2005;14:1340–1349. doi: 10.1110/ps.041239305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang L, Brock A, Schultz P. Angew Chem Int Ed Engl. 2002;41:2840. doi: 10.1002/1521-3773(20020802)41:15<2840::AID-ANIE2840>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Zhong W, Gallivan JP, Zhang Y, et al. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci U S A. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]


