Abstract
Background and aim
Some cancer-specific miRNAs are dysregulated in pancreatic adenocarcinoma (PAAD) and involved in cell autophagy, differentiation, proliferation, migration, invasion, and malignant transformation. The aim of our study was to determine a panel of new diagnostic and prognostic biomarkers for PAAD.
Methods
We conducted a comprehensive analysis of global miRNA-expression profiles and corresponding prognosis information of 168 PAAD patients from the Cancer Genome Atlas data set. A total of 16 differentially expressed miRNAs were identified as aberrantly expressed in PAAD, and six of these were evaluated for use as diagnostic markers for PAAD. Next, we confirmed a two-miRNA signature significantly associated with PAAD patient diagnosis and outcome prediction.
Results
The panel of two miRNAs showed outstanding diagnostic performance, with sensitivity of 100% and specificity of 87.5%. Finally, we divided the PAAD patients into high-risk and low-risk groups based on the expression profile of the two miRNAs. Kaplan–Meier analysis demonstrated that patients in the high-risk group had significantly worse prognosis than patients in the low-risk group. Univariate and multivariate Cox regression analysis showed that the two-miRNA signature was an independent prognostic factor for the overall survival of PAAD patients.
Conclusion
Taken together, the two-miRNA signature may serve as an accurate and sensitive biomarker for diagnosis and PAAD-outcome prediction, facilitating the diagnosis and potentially improving treatment outcome of PAAD.
Keywords: pancreatic adenocarcinoma, microRNA signature, TCGA, prognosis, diagnosis
Introduction
Pancreatic adenocarcinoma (PAAD) is one of the most lethal cancers and is associated with high morbidity and poor prognosis. The 5-year survival rate remains at only 7%.1 Due to the lack of effective early-diagnosis approaches or typical symptoms, most patients with PAAD are not diagnosed until reaching advanced stages.2 Although some progress has been made in recent years, the influence of environmental factors, genetic factors, epigenetic alterations, and chronic inflammation on PAAD development remains poorly understood, and therapeutic options for PAAD patients are still limited.3 Therefore, investigating the impact of multiple pathogenic factors on the clinical characteristics of PAAD is crucial for physiological assessment, treatment planning, and prognosis evaluation.4,5
miRNAs are small non-protein-coding RNAs that bind to complementary sequences on target-mRNA transcripts to modulate their expression and are considered efficient diagnostic and prognostic biomarkers in malignant disease.6 These small RNAs regulate the expression of up to 30% of all mammalian protein-coding genes. With advances in human genome-sequencing technology, an extremely large number of miRNAs have been discovered. Accumulating evidence has emerged to suggest that miRNAs participate in many fundamental biology processes, such as cell autophagy, apoptosis, differentiation, and proliferation, and may also participate in carcinogenesis.7 Several studies have demonstrated aberrant expression of some specific miRNAs in PAAD and other disease processes. By using murine orthotopic models, Mees et al found that miR224 and miR486 are involved in the progression of PAAD.8 Frampton et al suggested that specific miRNAs act as cooperative repressors of a network of tumor-suppressor genes.9 The underlying mechanisms by which cancer-specific miRNAs contribute to PAAD development are still unclear. Therefore, the identification of aberrantly expressed cancer-specific miRNAs may facilitate early PAAD diagnosis and survival prediction.
This study utilized miRNA-sequencing data and corresponding clinical information from the Cancer Genome Atlas (TCGA) database. The TCGA PAAD project includes the largest sequencing cohort currently available for PAAD. Here, we analyzed the differentially expressed miRNAs between PAAD tissue and matched normal tissue based on the miRNA-sequencing data. Based on our results, we identified a two-miRNA signature that may serve as an accurate diagnostic and prognostic biomarker for PAAD patients.
Materials and methods
miRNA-sequencing data processing
PAAD level 3 miRNA-sequencing data obtained from samples from 168 PAAD patients were downloaded from the TCGA database. The detailed clinical information included sex, age at diagnosis, tumor size, metastasis status, lymph-node status, and TNM stage (according to the seventh edition of the American Joint Committee on Cancer’s staging manual), and is shown in Table 1. The median diagnosis age was 64 years (range 35–88 years). The median follow-up time was 26.33 months (range 0–72.4 months).
Table 1.
Clinical characteristics of PAAD patients
| Variables | n (%) |
|---|---|
| Age at diagnosis | |
| <60 years | 50 (29.8%) |
| ≥60 years | 118 (70.2%) |
| Gender | |
| Female | 75 (44.6%) |
| Male | 93 (55.4%) |
| Metastasis | |
| M0 | 78 (46.4%) |
| M1 | 4 (2.4%) |
| MX | 86 (51.2%) |
| Lymph-node status | |
| N0 | 23 (13.7%) |
| N1 | 62 (36.9%) |
| NX | 1 (0.6%) |
| Clinical stage | |
| I | 15 (8.9%) |
| II | 144 (85.7%) |
| III | 3 (1.8%) |
| IV | 4 (2.4%) |
| T stage | |
| T1 | 6 (3.6%) |
| T2 | 19 (11.3%) |
| T3 | 139 (82.7%) |
| T4 | 3 (1.8%) |
| Neoplasm histological grade | |
| G1 | 24 (14.3%) |
| G2 | 94 (56.0%) |
| G3 | 47 (28.0%) |
| G4 | 1 (0.6%) |
| GX | 2 (1.2%) |
| Surgical margin resection status | |
| R0 | 98 (58.3%) |
| R1 | 51 (29.8%) |
| R2 | 5 (3.0%) |
| RX | 4 (2.4%) |
| Tumor size | |
| ≤4 cm | 105 (62.5%) |
| >4 cm | 50 (29.8%) |
| Race | |
| Asian | 11 (6.5%) |
| Black/African-American | 6 (3.6%) |
| White | 147 (87.5%) |
Abbreviation: PAAD, pancreatic adenocarcinoma.
Raw-count miRNA-sequencing data were processed with EdgeR, a Bioconductor package based on the R language, to screen differentially expressed miRNAs between PAAD tissue and adjacent normal tissue using the unpaired t-test. For all P-values, a false-discovery rate (FDR) was applied to correct the statistical significance of multiple testing. Fold changes (FCs) in the expression of individual miRNA were calculated, and differentially expressed miR-NAs with FDR values <0.05 and FCs >1 were considered significant.
Diagnostic performance of differently expressed miRNAs
Receiver-operating characteristic (ROC) analysis was performed to evaluate the diagnostic performance of differently expressed miRNAs (DEmiRNAs). To compare the superiority or inferiority of DEmiRNAs for diagnostic performance, ROC curves were drawn. Area under the ROC curve (AUROC) was used to calculate the diagnostic sensitivity and specificity. P<0.05 was considered significant.
Association between DEmiRNAs and patient prognosis
All patients were classified into high or low miRNA-expression groups according to the median value. Overall survival (OS) and disease-free survival were used in survival analysis. OS and disease-free survival data were downloaded from TCGA. Kaplan–Meier and log-rank methods were used to test differences between the two groups. P<0.05 was considered significant.
Target-gene prediction of two miRNAs and functional analysis
The mRNAs targeted by the two miRNAs were predicted using two online analysis tools: miRDB (www.mirdb.org) and TargetScan (www.targetscan.org). Next, overlapping target genes were analyzed using a Venn diagram. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed using the Database for Annotation, Visualization, and Integrated Discovery bioinformatics tool.
Statistical analysis
Data are presented as mean ± SD. The two-miRNA signature was determined based on significant miRNAs in OS and diagnostic performance. Univariate and multivariate Cox proportional-hazard regression models were used to evaluate the prognostic significance of the two-miRNA signature. All statistical analysis was conducted using SPSS 22.0 (IBM, Armonk, NY, USA). All tests were two-sided, and P<0.05 was considered significant.
Results
Identification of DEmiRNAs
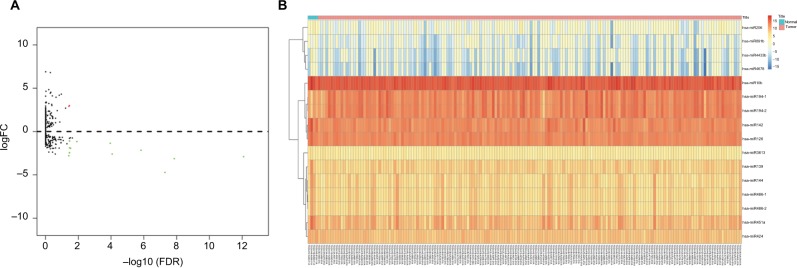
We identified a total of 16 DEmiRNAs from the dataset of TCGA, of which two were upregulated and 14 downregulated, with cutoffs of FDR <0.05 and FCs >1 (Table 2). A volcano plot was drawn to show the relationships between FDR and FCs of each DEmiRNA (Figure 1A). Then, we organized the 16 miRNA data points as a heat map to visualize the distinct expression profile between PAAD and normal tissue (Figure 1B).
Table 2.
DEmiRNAs between PAAD and normal controls
| miRNAs | LogFC | P-value | FDR | |
|---|---|---|---|---|
| Upregulated | hsa-miR194-1 | 2.995 | 3.92−4 | 3.54−2 |
| hsa-miR194-2 | 2.9078 | 4.75−4 | 3.82−2 | |
| Downregulated | hsa-miR139 | −2.9103 | 5.72−16 | 8.28−13 |
| hsa-miR451a | −3.1417 | 1.97−11 | 1.42−8 | |
| hsa-miR4433b | −4.7319 | 1.09−1 | 5.28−8 | |
| hsa-miR142 | −2.1612 | 4.18−9 | 1.51−6 | |
| hsa-miR144 | −2.6036 | 3.00−7 | 8.68−5 | |
| hsa-miR424 | −1.3554 | 4.69−7 | 1.13−4 | |
| hsa-miR126 | −1.1608 | 5.99−5 | 1.24−2 | |
| hsa-miR486-1 | −1.9358 | 1.87−4 | 3.01−2 | |
| hsa-miR486-2 | −1.8879 | 2.80−4 | 3.46−2 | |
| hsa-miR891b | −2.4688 | 3.07−4 | 3.46−2 | |
| hsa-miR10b | −1.107 | 3.21−4 | 3.46−2 | |
| hsa-miR3613 | −1.0856 | 3.35−4 | 3.46−2 | |
| hsa-miR206 | −2.4482 | 3.82−4 | 3.54−2 | |
| hsa-miR4678 | −2.8038 | 5.07−4 | 3.86−2 |
Abbreviations: DEmiRNAs, differently expressed miRNAs; FC, fold change; FDR, false-discovery rate; PAAD, pancreatic adenocarcinoma.
Figure 1.
Volcano plot and heat map of DEmiRNAs.
Notes: (A) Plots of log2FC vs –log10 (FDR) for DEmiRNAs. Each red dot represents a significantly upregulated miRNA and each green dot a significantly downregulated miRNA. (B) Sixteen differentially expressed miRNAs were visualized by heat map. Each column represents one sample and each row one miRNA.
Abbreviations: DEmiRNAs, differently expressed miRNAs; FC, fold change; FDR, false-discovery rate.
Diagnostic performance of DEmiRNAs
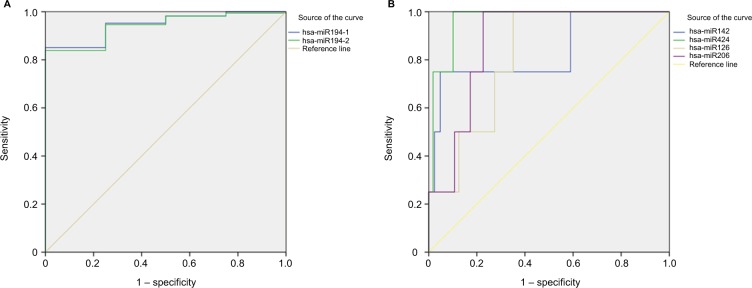
ROC curves and AUROC were used to evaluate the ability of DEmiRNAs to distinguish PAAD tissue from normal tissue. AUROC >0.8 can discriminate tumor from normal tissue. The upregulated miRNAs miR194-1 and miR194-2 had good diagnostic performance, with AUROC values of 0.946 and 0.940, respectively (Figure 2A). The downregulated miRNAs miR142, miR424, miR126, and miR206 also showed good diagnostic performance, with AUROC values of 0.835, 0.966, 0.812, and 0.874 respectively (Figure 2B). Sensitivity and specificity are listed in Table 3.
Figure 2.
ROC curves of DEmiRNAs in distinguishing PAAD tissues from normal tissue.
Notes: (A) Upregulated DEmiRNAs (AUROC >0.8); (B) downregulated DEmiRNAs (AUROC >0.8).
Abbreviations: AUROC, area under receiver-operating characteristic; DEmiRNAs, differently expressed miRNAs; PAAD, pancreatic adenocarcinoma; ROC, receiver-operating characteristic.
Table 3.
Diagnostic performance of specific miRNAs (AUROC >0.8)
| Variable(s) | AUROC | P-value | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|
| Upregulated | has-miR194-1 | 0.946 | 0.002 | 95.2 | 75 |
| has-miR194-2 | 0.94 | 0.003 | 94.6 | 75 | |
| Downregulated | has-miR142 | 0.835 | 0.022 | 75 | 95.2 |
| has-miR424 | 0.966 | 0.001 | 100 | 89.9 | |
| has-miR126 | 0.812 | 0.033 | 100 | 64.9 | |
| has-miR206 | 0.874 | 0.011 | 100 | 77.4 |
Abbreviation: AUROC, area under receiver-operating characteristic.
Prognostic performance of DEmiRNAs
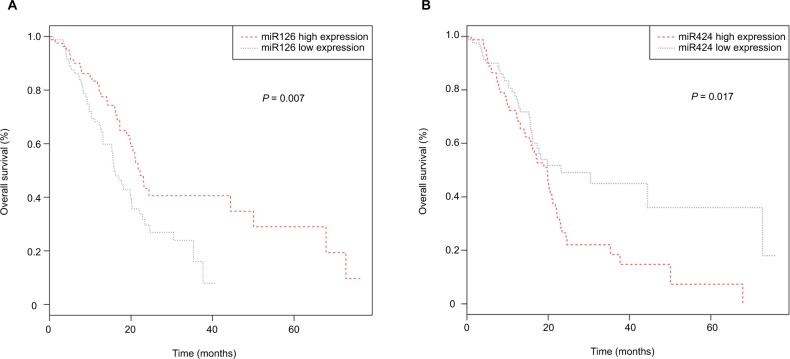
To reveal the relationship between DEmiRNAs and prognosis, we performed Kaplan–Meier analysis. Our results revealed that miR126 (P=0.007) and miR424 (P=0.017) were significantly associated with PAAD patients’ OS (Figure 3 A and B). To discover potential prognostic biomarkers for recurrence, we assessed the influence of expression of all DEmiRNAs on disease-free survival in PAAD patients. The results revealed that no DEmiRNA expression was significantly correlated with disease-free survival. We next evaluated prognostic values of differentially expressed miRNAs using univariate Cox regression analysis. The results showed that miR424 and miR126 were independent prognostic factors for PAAD patients (Table 4).
Figure 3.
Kaplan–Meier survival curves for two miRNAs associated with overall survival: (A) miR126; (B) miR424.
Table 4.
Univariate Cox regression analysis of DEmiRNAs associated with overall survival
| HR | Z | P-value | |
|---|---|---|---|
| hsa-miR424 | 1.512 | 2.468 | 0.014 |
| hsa-miR126 | 0.646 | −2.324 | 0.020 |
| hsa-miR3613 | 0.717 | −2.030 | 0.042 |
| hsa-miR206 | 0.835 | −1.509 | 0.131 |
| hsa-miR142 | 0.852 | −1.391 | 0.164 |
| hsa-miR486-1 | 0.897 | −1.315 | 0.189 |
| hsa-miR4433b | 1.453 | 1.205 | 0.228 |
| hsa-miR139 | 0.882 | −1.148 | 0.251 |
| hsa-miR451a | 0.924 | −1.092 | 0.275 |
| hsa-miR486-2 | 0.923 | −0.995 | 0.320 |
| hsa-miR194-2 | 0.945 | −0.706 | 0.480 |
| hsa-miR194-1 | 0.948 | −0.678 | 0.498 |
| hsa-miR10b | 0.910 | −0.602 | 0.547 |
| hsa-miR144 | 0.958 | −0.595 | 0.552 |
| hsa-miR4678 | 0.742 | −0.562 | 0.574 |
| hsa-miR891b | 0.975 | −0.132 | 0.895 |
Abbreviations: DEmiRNAs, differently expressed miRNAs; HR, hazard ratio.
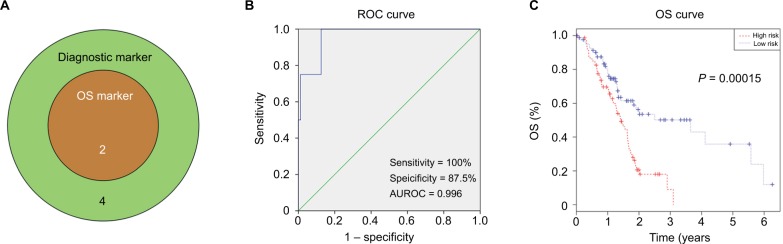
We used a Venn diagram to find DEmiRNAs with both excellent diagnostic and prognostic performance, and miR126 and miR424 met the criteria (Figure 4A). ROC analysis was preformed to assess the diagnostic performance of the two-miRNA signature. The AUROC of the two-miRNA signature was 0.966 (95% CI 0.911–1.000), with sensitivity of 100% and specificity of 87.5% (Figure 4B). Next, we evaluated the prognostic performance of the two-miRNA signature using multivariate Cox regression analysis and calculated a risk score for each patient. PAAD patients were divided into a high-risk group and a low-risk group based on the median risk score. The results demonstrated that patients with high-risk scores had significantly worse prognoses (P=0.0015) than PAAD patients with low-risk scores (Figure 4C). Univariate and multivariate Cox regression analyses indicated that the two-miRNA signature was an independent prognostic factor in OS (hazard ratio [HR] 2.318, 95% CI 1.482–3.626; P <0.001; Table 5).
Figure 4.
Two-miRNA signature in diagnosis and prognosis of PAAD.
Notes: (A) Venn analysis of overlapping statistically significant miRNAs among OS markers and diagnostic markers; (B) ROC curves of two-miRNA signature in differentiating PAAD tissues from normal tissue; (C) Kaplan–Meier survival curves of two-miRNA signature in overall survival prediction.
Abbreviations: AUROC, area under receiver-operating characteristic; OS, overall survival; PAAD, pancreatic adenocarcinoma; ROC, receiver-operating characteristic.
Table 5.
Univariate and multivariate Cox regression analysis in pancreatic adenocarcinoma patients
| Univariate
|
P-value | Multivariate
|
P-value | |||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |||
| Sex | 0.752 | 0.498–1.135 | 0.175 | |||
| Age (continuous variant) | 1.021 | 1.000–1.042 | 0.046 | 1.019 | 0.998–1.041 | 0.074 |
| Metastasis (M1 vs M0) | 1.066 | 0.255–4.462 | 0.93 | |||
| Lymph-node status (N1 vs N0) | 1.76 | 1.076–2.879 | 0.024 | 1.624 | 0.990–2.663 | 0.055 |
| Clinical stage (III + IV vs I + II) | 0.745 | 0.235–2.363 | 0.617 | |||
| T stage (T3 + T4 vs T1 + T2) | 1.185 | 0.629–2.234 | 0.599 | |||
| Two-miRNA signature (high risk vs low risk) | 2.318 | 1.482–3.626 | <0.001 | 2.136 | 1.352–3.375 | 0.001 |
Abbreviation: HR, hazard ratio.
Target-gene prediction and functional enrichment analysis
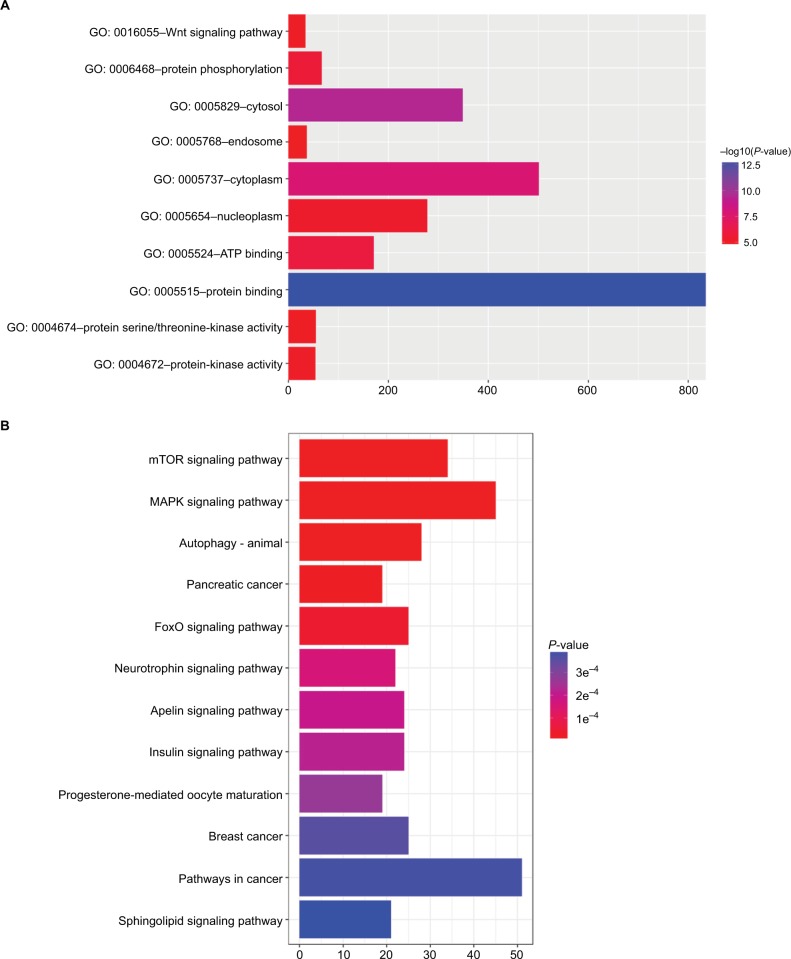
To elucidate the biological functions of DEmiRNAs, we performed Gene Ontology and KEGG pathway-enrichment analysis to identify target genes of both miR126 and miR424, as predicted by miRDB and Target Scan. The Gene Oncology annotation included protein binding, ATP binding, protein phosphorylation, protein-kinase activity, the Wnt-signaling pathway, and other functional classes (Figure 5A). KEGG enrichment pathways were mainly associated with the mTOR-signaling pathway, MAPK-signaling pathway, autophagy, FoxO-signaling pathway, and insulin-signaling pathway (Figure 5B).
Figure 5.
GO and KEGG pathway-enrichment analysis of DEmiRNA-targeted genes.
Notes: (A) Enriched gene-ontology annotation of target genes; (B) enriched KEGG pathway of target genes.
Abbreviations: DEmiRNA, differently expressed miRNA; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
miRNAs can modulate the expression of multiple genes, playing essential roles in many cellular processes in human malignant disease, and thus miRNAs can directly influence the development and progression of cancer.10–12 Recent studies have revealed distinct miRNA-expression profiles in PAAD, indicating key roles of miRNAs in the development and progression of PAAD.13–20 In this study, we screened miRNA-sequencing data and corresponding clinical information obtained from the TCGA PAAD project. Sixteen miRNAs were identified as aberrantly expressed in PAAD tissue: two were upregulated and 14 downregulated. A previous study suggested that combinations of different miRNAs may be more sensitive and specific than the currently individual miRNA markers.21 Therefore, based on our results, we constructed a two-miRNA signature, which had outstanding diagnostic performance and independent prognostic significance for PAAD.
The distinct expression profiles of cancer-specific miR-NAs in PAAD have attracted attention for use as diagnostic and prognostic markers. Several recent studies have revealed that some cancer-specific miRNAs are aberrantly expressed in PAAD and could be used as diagnostic and prognostic markers.22–24 However, these studies had several limitations. There was clinical and molecular heterogeneity in different studies. There were methodological differences, limiting reproducibility and normalization among these studies. The number of patients enrolled in each study was generally small. The TCGA PAAD project includes gene-sequencing data and corresponding clinical information in PAAD tissue and adjacent normal tissue. Translating “big data” information into a better understanding of underlying biological mechanisms can help identify potential diagnostic and prognostic markers for PAAD.
Recently, one study used univariate Cox regression to analyze TCGA data and found that ten highly expressed miRNAs were associated with worse prognosis and three highly expressed miRNAs associated with better prognosis.25 Here, we analyzed the TCGA data using EdgeR, a Bioconductor package based on the R language, and identified 16 DEmiRNAs with FDR <0.05 and FCs >1. Additional studies have demonstrated that some of these miRNAs are aberrantly expressed in PAAD, which lends credibility to our study. Jiao et al reported that expression profiling revealed down-regulation of 21 miRNAs, including miR126 and miR451 in pancreatic ductal adenocarcinoma.26 Conversely, Nakata et al performed microdissection analysis and showed that miR10b exhibited higher expression levels in pancreatic cancer cells than in normal pancreatic ductal cells.27
To establish an accurate and efficient biomarker in diagnosis and prognosis, we identified a two-miRNA signature of miR126 and miR424 based on a Venn diagram of significant miRNAs, with excellent diagnostic and prognostic performance. These two miRNAs are generally considered related to malignant disease and patient OS. Located in 9q34.3, miR126 is the most commonly dysregulated miRNA in tumors. Emerging evidence has demonstrated that miR126 acts as a tumor-suppressor gene by targeting many oncogenes associated with metastasis, proliferation, migration, and invasion. Liu et al showed that deactivation of miR126 induced a pseudohypoxia state due to increased HIFα expression, which further enhanced therapeutic resistance and cell motility mediated by SLC7A5 and SERPINE1, respectively.28 Yuan et al showed that miR126 suppressed the migration, proliferation, and invasion of colon cancer by inactivating RHOA signaling via CXCR4.29 Chen et al showed that downregulation of miR126 expression clearly upregulated expression of VEGFA and its downstream signaling pathways.30 Jia et al demonstrated that miR126 inhibited invasion in bladder cancer by targeting ADAM9.31 In invasive ductal PAAD, Hamada et al showed reduced expression of miR126.32 In addition, there have been conflicting reports of the roles of miR424 in the progression and chemotherapy resistance of PAAD. Zhang et al reported that miR424 may contribute to non-small-cell lung cancer progression and metastasis by affecting migration, invasion, and proliferation via inhibition of.33 Rodriguez-Barrueco et al observed that loss of miR424 promoted chemotherapy resistance due to upregulation of BCL2 and IGF1R.34 In this study, we observed that miR424 was downregulated in PAAD tissue and high miR424 expression associated with worse outcome. Therefore, the conflicting function of miR424 should be investigated further. Here, we constructed ROC curves to verify that the two-miRNA signature was an efficient diagnostic marker in distinguishing PAAD tissue from adjacent normal tissue, with sensitivity of 100% and specificity of 87.5%. More importantly, the two-miRNA signature showed excellent performance in predicting OS in PAAD, indicating the two-miRNA signature may be a prognostic biomarker.
Although our results did not support use of miR10b and miR206 as prognosis biomarkers, these miRNAs have also been implicated in PAAD. Ouyang et al demonstrated that miR10b stimulated pancreatic cancer-cell invasion by decreasing TIP30 expression and enhancing EGFR signaling, suggesting miR10b may serve as a diagnostic marker in ductal PAAD.35 Nakata et al found that miR10b was overexpressed in pancreatic cancer and may participate in invasiveness in pancreatic cancer cells, thereby leading to poor prognosis.27 In addition, Duell et al revealed that the plasma level of miR10b may serve as a noninvasive biomarker for early ductal PAAD.36 Feng et al reported that simultaneous overexpression of miR206 and miR34a enhanced antitumor efficacy, affecting cancer cell viability, invasion, and apoptosis in PAAD cells.37 Considering the influence of miR10b and miR206 on PAAD progression, it remains unclear whether these two miRNAs would be useful for prognosis evaluation in PAAD patients.
In order to confirm our results, we conducted independent validation using the Pancreatic Expression Database, which brings together the largest collection of pancreatic data from the literature, including transcriptomic, proteomic, miRNA, and genomic profiles.38 We searched for miR424 and miR126 expression in all studies included in the database. Bauer et al examined miRNA expression in 129 tissue samples, including normal pancreas, chronic pancreatitis, and ductal PAAD using Geniom Biochip, and found that expression of miR424 in ductal PAAD was lower than that of chronic pancreatitis, but higher than that of normal pancreas, suggesting that miR424 may play an important role in inflammatory-to-cancer transformation.39 Szafranska et al investigated miRNA expression in 25 samples containing normal pancreas, chronic pancreatitis, and ductal PAAD, as well as ductal PAAD-derived cell lines using miRNA arrays. The study showed that expression of miR424 in ductal PAAD cell lines was significantly lower than chronic pancreatitis and normal pancreas, and a similar expression pattern was found in miR126.40 Piepoli et al investigated expression profiling of 866 human miRNAs in 17 pancreatic cancers and matched adjacent normal tissue, and found that miR126 was upregulated in pancreatic cancers.41 Some of the results in these studies were similar to ours and some different. On one hand, these studies were based on different miRNA-array platforms. On the other hand, sample sizes for these studies varies widely: only 17 samples for Piepoli et al and 25 for Szafranska et al. However, our study had some limitations. We do not have our own PAAD-patient cohort to validate our results. In our future work, we will collect enough specimens and clinical data to verify our findings.
In this study, we analyzed miRNA-sequencing data in 168 samples from the TCGA database. We not only found differentially expressed miRNAs but also investigated their diagnostic and prognostic performance using bioinformatics analysis. We found that the two-signature miRNA was an excellent diagnostic biomarker and independent prognostic biomarker in PAAD patients. Additionally, the biological function and underlying mechanisms of candidate miRNAs in the development and progression of PAAD need to be elucidated.
Conclusion
Aberrations in miRNA expression are associated with PAAD progression. We conducted a comprehensive analysis of miRNA-expression profiles in PAAD tissue and adjacent matched tissue. Our data indicated that two-signature miRNA was an accurate and efficient diagnostic biomarker in PAAD and an independent prognostic biomarker in PAAD patients. However, further functional studies are needed to reveal the underlying molecular mechanisms for these miRNAs in PAAD progression.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Klein AP. Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer. 2013;13(1):66–74. doi: 10.1038/nrc3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63(5):318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiaravalli M, Reni M, O’Reilly EM. Pancreatic ductal adenocarcinoma: state-of-the-art 2017 and new therapeutic strategies. Cancer Treat Rev. 2017;60:32–43. doi: 10.1016/j.ctrv.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Kota J, Hancock J, Kwon J, Korc M. Pancreatic cancer: stroma and its current and emerging targeted therapies. Cancer Lett. 2017;391:38–49. doi: 10.1016/j.canlet.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Giovannetti E, van der Borden CL, Frampton AE, Ali A, Firuzi O, Peters GJ. Never let it go: stopping key mechanisms underlying metastasis to fight pancreatic cancer. Seminars in cancer biology. 2017;44:43–59. doi: 10.1016/j.semcancer.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 8.Mees ST, Mardin WA, Sielker S, et al. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol. 2009;16(8):2339–2350. doi: 10.1245/s10434-009-0531-4. [DOI] [PubMed] [Google Scholar]
- 9.Frampton AE, Castellano L, Colombo T, et al. Integrated molecular analysis to investigate the role of microRNAs in pancreatic tumour growth and progression. Lancet. 2015;385(Suppl 1):S37. doi: 10.1016/S0140-6736(15)60352-X. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57(2):840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 13.Rachagani S, Macha MA, Heimann N, et al. Clinical implications of miRNAs in the pathogenesis, diagnosis and therapy of pancreatic cancer. Adv Drug Deliv Rev. 2015;81:16–33. doi: 10.1016/j.addr.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang HL, Zhou R, Liu J, et al. MicroRNA-196b inhibits late apoptosis of pancreatic cancer cells by targeting CADM1. Sci Rep. 2017;7(1):11467. doi: 10.1038/s41598-017-11248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duell EJ, Lujan-Barroso L, Sala N, et al. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141(5):905–915. doi: 10.1002/ijc.30790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quattrochi B, Gulvady A, Driscoll DR, et al. MicroRNAs of the mir-17~92 cluster regulate multiple aspects of pancreatic tumor development and progression. Oncotarget. 2017;8(22):35902–35918. doi: 10.18632/oncotarget.16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong G, Feng M, Yang G, et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017;397:94–102. doi: 10.1016/j.canlet.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 18.van Kampen JG, van Hooij O, Jansen CF, et al. miRNA-520f reverses epithelial-to-mesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res. 2017;77(8):2008–2017. doi: 10.1158/0008-5472.CAN-16-2609. [DOI] [PubMed] [Google Scholar]
- 19.Amponsah PS, Fan P, Bauer N, et al. MicroRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017;388:107–117. doi: 10.1016/j.canlet.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 20.Yuan P, He XH, Rong YF, et al. KRAS/NF-κB/YY1/miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Res. 2017;77(1):100–111. doi: 10.1158/0008-5472.CAN-16-1898. [DOI] [PubMed] [Google Scholar]
- 21.Zampini M, Bisio V, Leszl A, et al. A three-miRNA-based expression signature at diagnosis can predict occurrence of relapse in children with t(8;21) RUNX1-RUNX1T 1 acute myeloid leukaemia. Br J Haematol. 2017 Sep 29; doi: 10.1111/bjh.14950. Epub. [DOI] [PubMed] [Google Scholar]
- 22.Yuan W, Tang W, Xie Y, et al. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7(48):80033–80045. doi: 10.18632/oncotarget.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Zhu CF, Ma MZ, et al. Micro-RNA-155 is induced by K-Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotarget. 2015;6(25):21148–21158. doi: 10.18632/oncotarget.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia X, Zhang K, Cen G, et al. MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4. Oncotarget. 2015;6(25):21046–21063. doi: 10.18632/oncotarget.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Huang Z, Xu L, et al. A panel of 13-miRNA signature as a potential biomarker for predicting survival in pancreatic cancer. Onco-target. 2016;7(43):69616–69624. doi: 10.18632/oncotarget.11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao LR, Frampton AE, Jacob J, et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One. 2012;7(2):e32068. doi: 10.1371/journal.pone.0032068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakata K, Ohuchida K, Mizumoto K, et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150(5):916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, Chen H, Wong N, Haynes W, Baker CM, Wang X. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett. 2017;394:65–75. doi: 10.1016/j.canlet.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan W, Guo YQ, Li XY, et al. MicroRNA-126 inhibits colon cancer cell proliferation and invasion by targeting the chemokine (C-X-C motif) receptor 4 and Ras homolog gene family, member A, signaling pathway. Oncotarget. 2016;7(37):60230–60244. doi: 10.18632/oncotarget.11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Li L, Wang S, et al. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget. 2014;5(23):11873–11885. doi: 10.18632/oncotarget.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia AY, Castillo-Martin M, Bonal DM, Sanchez-Carbayo M, Silva JM, Cordon-Cardo C. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer. 2014;110(12):2945–2954. doi: 10.1038/bjc.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamada S, Satoh K, Fujibuchi W, et al. miR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10(1):3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Gao C, Yang Y, et al. miR-424 promotes non-small cell lung cancer progression and metastasis through regulating the tumor suppressor gene TNFAIP1. Cell Physiol Biochem. 2017;42(1):211–221. doi: 10.1159/000477314. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Barrueco R, Nekritz EA, Bertucci F, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553–566. doi: 10.1101/gad.292318.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang H, Gore J, Deitz S, Korc M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene. 2014;33(38):4664–4674. doi: 10.1038/onc.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duell EJ, Lujan-Barroso L, Sala N, et al. Plasma microRNAs as bio-markers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141(5):905–915. doi: 10.1002/ijc.30790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng SD, Mao Z, Liu C, et al. Simultaneous overexpression of miR-126 and miR-34a induces a superior antitumor efficacy in pancreatic adenocarcinoma. Onco Targets Ther. 2017;10:5591–5604. doi: 10.2147/OTT.S149632. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Chelala C, Hahn SA, Whiteman HJ, et al. Pancreatic expression database: a generic model for the organization, integration and mining of complex cancer datasets. BMC Genomics. 2007;8:439. doi: 10.1186/1471-2164-8-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer AS, Keller A, Costello E, et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS One. 2012;7(4):e34151. doi: 10.1371/journal.pone.0034151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 41.Piepoli A, Tavano F, Copetti M, et al. miRNA expression profiles identify drivers in colorectal and pancreatic cancers. PLoS One. 2012;7(3):e33663. doi: 10.1371/journal.pone.0033663. [DOI] [PMC free article] [PubMed] [Google Scholar]