Abstract
The high mortality of pancreatic cancer demands that new therapeutic avenues be developed. The orally available small molecule inhibitor AT13148 potently inhibits ROCK1 and ROCK2 kinases that regulate the actomyosin cytoskeleton. We previously reported that ROCK kinase expression increases with human and mouse pancreatic cancer progression and that conditional ROCK activation accelerates mortality in a genetically modified LSL-KrasG12D; LSL-p53R172H; Pdx1-Cre; (KPC) mouse pancreatic cancer model. In this study, we show that treatment of KPC mouse and human TKCC5 patient-derived pancreatic tumor cells with AT13148, as well as the ROCK selective inhibitors Y27632 and H1152, act comparably in blocking ROCK substrate phosphorylation. AT13148, Y27632, and H1152 induced morphological changes and reduced cellular contractile force generation, motility on pliable discontinuous substrates, and three-dimensional collagen matrix invasion. AT13148 treatment reduced subcutaneous tumor growth and blocked invasion of healthy pancreatic tissue by KPC tumor cells in vivo without affecting proliferation, suggesting a role for local tissue invasion as a contributor to primary tumor growth. These results suggest that AT13148 has anti-tumor properties that may be beneficial in combination therapies or in the adjuvant setting to reduce pancreatic cancer cell invasion and slow primary tumor growth. AT13148 might also have the additional benefit of enabling tumor resection by maintaining separation between tumor and healthy tissue boundaries.
Introduction
Pancreatic cancer, and the predominant pancreatic ductal adenocarcinoma (PDAC) type in particular, is one of the deadliest of all major cancers with the lowest 5-year survival rate (1), and which has been projected to become the second leading cause of cancer deaths by 2030 in the United States (2). Despite intense efforts directed at improving patient outcomes, there has been little to no improvement in survival rates (3). Most patients are not treatable with surgical resection; chemotherapy is the most frequently used approach. One challenge for chemotherapy is that the mostly commonly mutated driver genes (KRAS, TP53, SMAD4 and CDKN2A (4,5)) are not targeted by current drugs. As a result, drugs that could be used in therapeutic combinations, in the adjuvant setting, or which might increase tumor resectability, could be beneficial and help to extend patient survival (6).
The RhoA and RhoC regulated ROCK1 and ROCK2 serine/threonine kinases play central and critical roles in the regulation of actomyosin cytoskeleton organization and dynamics, acting largely through the phosphorylation of substrates including regulatory myosin light chain 2 (MLC2), myosin-binding subunit of the MLC phosphatase (MYPT1), and LIM kinases 1&2 (LIMK) (7–9). Numerous lines of evidence indicate that the ROCK kinases contribute to tumor cell invasion and metastasis, by increasing cytoskeleton contractility and cellular tension to affect properties including adhesion and migration (7). Conditional genetic deletion of both Rock1 and Rock2 in mouse cells also revealed essential roles in cell cycle progression, although this effect only appears to be manifested following full or near complete loss of ROCK activity induced by gene deletion or high inhibitor concentrations (10).
The ROCK1 gene locus on human chromosome 18 is amplified in 15% of pancreatic tumors (11), an observation corroborated by a recent study in which ROCK1 gene amplification was observed in 12% of patient samples (12), and which was extended by the finding of concordancy between copy number and gene expression changes (4). We reported that there were significantly increased levels of ROCK1 and ROCK2 protein in human and mouse pancreatic tumors compared to healthy tissue, which were observed to increase in parallel with tumor progression (13). The observations of elevated ROCK1 protein in human pancreatic tumor tissues were also recently corroborated (12). Furthermore, siRNA-mediated knockdown of ROCK1/2 expression inhibited the proliferation and migration of pancreatic cancer cell lines in vitro (12). Importantly, elevated ROCK1 and/or ROCK2 expression was associated with reduced survival in human pancreatic patients, while conditional activation of ROCK2 in the genetically modified LSL-KrasG12D; LSL-p53R172H; Pdx1-Cre; (KPC) mouse pancreatic cancer model (14–16) also resulted in accelerated mortality (13). Conversely, treatment of KPC mice, or mice with orthotopically grown tumors of human TKCC5 patient-derived xenograft (PDX)-derived pancreatic cancer cells (17) with the selective ROCK inhibitor fasudil extended survival (13), consistent with ROCK inhibition having the potential to provide clinical benefit for pancreatic cancer patients. The physically stiff collagen-rich stroma associated with PDAC tumors was found to promote tumor growth via increased ROCK signaling (18), while in PDAC cells oncogenic KRAS drives increased transcription of the RhoA-activating ARHGEF2 guanine nucleotide exchange factor that promotes migration, invasion and colony formation (19). These findings are consistent with the additional scenario of a more general tumor promoting role for Rho-ROCK signaling in the absence of elevated ROCK1 or ROCK2 expression.
If ROCK inhibition were to be considered for clinical development as a pancreatic cancer chemotherapeutic, there are several key requirements that should be met, including high potency and good pharmacokinetic properties. Although fasudil extended the survival of KPC pancreatic cancer mice (13) and mice with human TKCC5 PDAC cell orthotopic tumors (17), and the compound is clinically used in Japan in an acute manner to treat cerebral vasospasm with very good safety profiles (20), the frequent dosing with high compound concentrations that are necessary to achieve in vivo responses make fasudil a sub-optimal choice for extended chemotherapy. The pyrazole-based AT13148 ((1S)-2-amino-1-(4-chlorophenyl)-1-[4-(1H-pyrazol-4-yl)phenyl]ethanol) AGC kinase inhibitor was discovered in a fragment-based screen and further developed to optimize its potency and pharmacokinetic properties (21). Although originally intended to be an AKT inhibitor, subsequent biological experiments revealed that it was more potent as an inhibitor of ROCK1 and ROCK2 (21). Through comparisons with non-ROCK targeting AKT inhibitors, it was determined that an inhibitory effect on melanoma cell motility was due to on-target ROCK inhibition (22). In addition, AT13148 was shown to slow the subcutaneous tumor growth of BT474 breast, PC3 prostate and MES-SA uterine human cancer cell lines, revealing its in vivo anti-tumor efficacy (21). Importantly, the therapeutic promise, satisfactory pharmacokinetic properties and selectivity profile of AT13148 contributed to its progression to phase 1 clinical trials (23).
To model PDAC, we used tumor cells isolated from the KPC mouse PDAC model that expresses mutant KRAS and p53 (16), and the human patient derived xenograft (PDX)-derived cell line TKCC5 (17). We now show that AT13148 phenocopied the selective ROCK inhibitors Y27632 and H1152 in blocking substrate phosphorylation in mouse and human PDAC cells. In addition, AT13148, Y27632 and H1152 induced morphological changes and reduced contractile force generation, cell motility on pliable discontinuous substrates and three-dimensional collagen matrix invasion. Treatment of mice with AT13148 or fasudil blocked invasion of healthy pancreatic tissue by KPC cells. Furthermore, AT13148 reduced KPC subcutaneous tumor growth, without affecting proliferation, suggesting a role for local tissue invasion as an important contributor to primary tumor growth. This study is the first to demonstrate that AT13148 inhibits ROCK activity in pancreatic cancer cells, and supports the conclusion that AT13148 has anti-tumor properties that make it a candidate for use in combination therapies, or in the adjuvant setting, to reduce tumor cell invasion and to slow primary tumor growth, which might have the additional benefit of enabling tumor resection by maintaining separation between tumor and healthy tissue boundaries.
Materials and Methods
Small molecule inhibitors
AT13148 (Astex Pharmaceuticals), Y27632 (Tocris 1254), H1152 (Tocris 2414), fasudil (LC Laboratoris F-4660), GM6001 (Millipore 142880-36-2).
Cell culture
The KPC PDAC tumor cell line was established from a Pdx1-Cre; LSL-KrasG12D/+; LSL-Trp53R172H/+ mouse at the Cancer Research UK Beatson Institute as described in reference (16). KPC cells were cultured in high glucose DMEM (Gibco 21969-035) supplemented with 10% fetal bovine serum (FBS: Gibco), 2 mmol/L L-glutamine (Gibco) and penicillin-streptomycin (Gibco). The TKCC5 human pancreatic cancer cell line was established from patient-derived xenograft (PDX)-derived tumor cells at The Kinghorn Cancer Centre, Garvan Institute of Medical Research, and their culture conditions were described in reference (17). Given that these pancreatic cancer cell lines were created at the institutions where the experiments described in this study were performed, there were no previous references against which their identities could be authenticated, and no methods of authentication were used. Cells were cultured only for up to ~2 months after recovery from vials of low passage number KPC or TKCC5 cells. All cell lines were routinely tested for mycoplasma by the Cancer Research UK Beatson Institute Molecular Services. For siRNA-mediated knockdown, 5 µl of 0.5 µmol/L siRNA mix was spotted into wells of 96 well black plates (Greiner 655090), then 15 µl of Optimem containing 0.15 µl Lipofectamine RNAiMAX (Invitrogen: 13778-030) was added and shaken 20-30 min at room temperature. Cells (15,000 in 80 µl DMEM with 10% FBS) were added and incubated for 24 h at 37°C.
Western blotting
KPC or TKCC5 cells (5x105) in complete media were plated into the wells of 6-well culture dishes and allowed to settle and grow overnight. Next day, cells were treated with inhibitors or DMSO vehicle in complete media for 1 h. Whole cell lysates were prepared with lysis buffer (1% SDS, 50 mM Tris pH 7.5), followed by protein concentration determination using the Bicinchoninic assay (Sigma). Standard protocols were used for immunoblotting (13). Membranes were incubated with primary antibodies: ROCK1 (BD-611136), ROCK2 (BD-610623), ROCK1/2 (Millipore 07-1458), total AKT (Cell Signaling 2920S), phospho-AKT (Cell Signaling 4060S), phospho-MYPT1 (Millipore 36003), phospho-MLC2 (Cell Signaling 3674), total MLC (MRCL3/MRLC2/MYL9; Santa Cruz sc-28329), phospho-PRAS40 (Cell Signaling 2997S), total PRAS40 (Cell Signaling 2691S), GAPDH (Millipore MAB374) and secondary antibodies: Alexa-Fluor 680 and DyLight 800 (Thermo Fisher Scientific)-conjugated antibodies. Infrared imaging (Li-Cor Odyssey) was used for detection and quantification of protein signals.
Cell viability assay
Cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega G7570). KPC or TKCC5 cells (2x104) in complete media were seeded into the wells of 96-well plates (Greiner 655083) and allowed to settle and grow overnight. Next day, medium was replaced by complete DMEM containing inhibitors or DMSO. After 18 h, cell viability was measured according to manufacturer’s instructions.
Cell cycle analysis
KPC cells (5x105) in complete DMEM were plated into the wells of 6-well culture dishes and allowed to settle and grow overnight. Cells were treated with inhibitors or DMSO vehicle in complete DMEM for 16 h. All cells were harvested by centrifugation for 3 min at 720 x g. Cell pellets were re-suspended in 300 µl PBS and 400 µl 70% EtOH/H2O added while vortexing. An additional 1.5 ml 70% EtOH/H2O was added to cells for fixation at 4°C overnight. Next day, cells were centrifuged for 3 min at 720 x g, washed with PBS, and centrifuged again for 3 min at 720 x g. Finally, cells were incubated with propidium iodide (50 µg/ml, Life Technologies P3566) and RNAse A (125 µg/ml, QIAGEN 1007885) in 500 µl PBS for at least 30 min prior to analysis by flow cytometry on the Attune Nxt Acoustic Focusing Cytometer (Life Technologies).
shRNA lentivirus production
HEK293 cells (106) were plated in 10 cm dishes and cultured in complete DMEM. The following day, medium was replaced with OPTIMEM for 1 h before transfections with 3.3 μg pLKO-Non-targeting control, 3.3 μg pLKO-ROCK1 shRNA (The RNAi Consortium, distributed by Dharmacon; TRCN0000121093) or 3.3 μg pLKO-ROCK2 shRNA (TRCN0000000978) with 3.3 μg psPAX2 and 3.3 μg VSVG plasmids using Fugene HD (Promega) according manufacturer’s recommendations. Medium was replaced with complete DMEM 6 h after transfection. Supernatants were collected 48 h after transfection, centrifuged at 1000 rpm for 5 minutes, collected and filtered through 0.45 μm pores. Polybrene was added at 8 μg/mL final concentration.
TKCC5 lentiviral transduction
TKCC5 cells were plated at 2 x 104 per well of a 6 well plate on the same day as HEK293 cells were plated for lentivirus production. After 4 days, medium was replaced twice in the same day with 4 mL of shRNA lentivirus supernatant. The next day, medium was replaced, which was supplemented ~7 h later with 1.25 µg/mL puromycin. Cells were selected with puromycin for 7 days with regular medium changes, then cell numbers were quantified using a CASY cell counter. Puromyosin resistant cells were further cultured in standard TKCC5 medium before western blot analysis and Operetta high content imaging analysis.
Immunofluorescence
KPC cells (6x104) in complete DMEM were seeded onto glass coverslips and allowed to settle and grow overnight. Next day, medium was replaced by complete DMEM containing inhibitors or DMSO vehicle. After 1 h of drug treatment, cells were fixed in 4% paraformaldehyde/PBS for 15 min and permeabilized with 0.5% Triton X-100/PBS for 5 min. F-actin was visualized by incubation with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). Cells were mounted with ProLong Diamond including DAPI (Molecular Probes). Z stack images were taken on a Zeiss 710 confocal microscope to generate maximum intensity projections using the ZEN 2010 software.
High content imaging
KPC cells (2x104) in complete DMEM were seeded into the wells of 96-well plates (Greiner 655090) and allowed to settle and grow overnight. Next day, medium was replaced by complete DMEM containing inhibitors or DMSO vehicle. After 1 h of drug treatment, cells were fixed in 4% paraformaldehyde/PBS for 15 min and permeabilized with 0.5% Triton X-100/PBS for 5 min. Cells were consecutively stained with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific), DAPI (0.15 µg/ml), and HCS CellMask Deep Red Stain (Thermo Fisher Scientific H32721). The Operetta High Content Imaging System (Perkin Elmer) and the Columbus Image Data Storage and Analysis System (Perkin Elmer) were used to determine cellular properties.
Metabolomics
KPC cells (5x105) in complete DMEM were seeded into the wells of 6-well culture dishes and allowed to settle and grow overnight. Next day, medium was replaced by complete DMEM containing 1 µM AT13148, 10 µM Y27632, 10 µM H1152, or DMSO vehicle. After 24 h of drug treatment, medium was removed, cells were washed 3 X with cold phosphate buffered saline (PBS), and then 600 µl of aqueous extraction solution (50% methanol, 30% acetonitrile) was added. After mixing at 4°C for 5 minutes, the extraction solution was removed to microcentrifuge tubes, shaken at 1400 rpm at 4°C for 10 minutes, and then centrifuged for 10 min at 16,100 x g, 4°C. Supernatents were transferred to glass HPLC vials and stored at -75°C prior to LC-MS. To determine protein concentration per well, plates were assayed using a modified Lowry method compared against a BSA protein standard curve. Metabolites were extracted and relative levels determined as described in (24).
An Exactive Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA, USA) was used together with a Thermo Scientific Accela HPLC system for LC-MS Analysis. The HPLC setup consisted of a ZIC-pHILIC column (SeQuant, 150 x 2.1mm, 5µm, Merck KGaA, Darmstadt, Germany), with a ZIC-pHILIC guard column (SeQuant, 20 x 2.1mm) and an initial mobile phase of 20% 20 mM ammonium carbonate, pH 9.4, and 80% acetonitrile. Cell and media extracts (5 µl) were injected and metabolites separated over a 15 minute mobile phase gradient, decreasing the acetonitrile content to 20%, at a flow rate of 200 μL/min and a column temperature of 45°C (25). The total analysis time was 23 minutes. All metabolites were detected across a mass range of 75-1000 m/z using the Exactive mass spectrometer at a resolution of 25,000 (at 200 m/z), with electrospray (ESI) ionization and polarity switching to enable both positive and negative ions to be determined in the same run. Lock masses were used and the mass accuracy obtained for all metabolites was below 5 ppm. Data were acquired with Thermo Xcalibur software.
The peak areas of different metabolites were determined using Thermo TraceFinder software where metabolites were identified by the exact mass of the singly charged ion and by known retention time on the HPLC column. Commercial standards of all metabolites detected had been analyzed previously on this LC-MS system with the pHILIC column.
Macropinocytosis assay
KPC cells in complete DMEM were seeded into the wells of 24-well culture dishes containing circular coverslips that had been washed with 95% ethanol, and allowed to settle and grow overnight. When the cells reached ~65% confluency, media was removed and cells were incubated with serum-free media for 2-4 h, then with serum-free media containing 0.1 mg/ml Tetramethylrhodamine (TMR)-Dextran and incubated at 37°C for 30 minutes. Plates were placed on ice and gently washed 5-7 times with ~2 ml/well cold PBS. Cells were fixed with 3.7% (v/v) formaldehyde solution in PBS at room temperature for 15 mins, then rinsed 3 times with PBS. Cells were consecutively stained with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific) and then DAPI (0.15 µg/ml). Coverslips were affixed to glass slides with DAKO mounting medium and stored on a flat surface in the dark overnight prior to imaging using an Olympus FV1000 microscope. The macropinocytic index was quantified as described in (26).
Collagen matrix invasion assay
The collagen matrix invasion assay was performed and analyzed as previously described (13). KPC or TKCC5 cells were each allowed to invade for 8 days. The area of invading KPC cells was divided by total area, and ratios were used to calculate the invasion index, relative to DMSO control. For TKCC5 cells, the invasive index was calculated using the following formula: (number of cells inside the collagen matrix)/(total number of cells), as described in (17).
Pliable discontinuous substrates for traction force microscopy
Pillar fabrication methods were adapted from Ghassemi et al. (27). Polydimethylsiloxane (PDMS; Sylgard 184, Dow Corning) was mixed at a 10:1 elastomer base:curing agent ratio, degassed and placed onto 35 mm glass bottom tissue culture dishes (Mattek). Silicon templates with hole structures were placed pattern side down onto the PDMS and allowed to cure at 70°C for 12 h, giving PDMS with 2 MPa Young’s modulus. Silicon templates were then removed from the cured PDMS while immersed in absolute ethanol. The resulting PDMS pillar dimensions were 1.8 µm height, 500 nm diameter, 1 µm center-to-center distance in a square array, and were calculated to have pillar bending stiffness of 3.16 nN/µm using the Euler-Bernoulli beam theory.
PDMS pillar substrates were coated with human fibronectin (R&D Systems) at 10 µg/ml final concentration for 1 h at 37°C. KPC cells were seeded at 5,000 cells/cm2 in complete DMEM and allowed to attach overnight. Before the start of timelapse microscopy and pillar tracking, 1 µM AT13148, 10 µM Y27632 or DMSO vehicle were added and incubated for 60 minutes.
Timelapse light microscopy was performed using an EVOS FL Auto 1 microscope with an on-stage incubator (ThermoScientific) operating at standard cell culture conditions of 37°C and 5% CO2. Single cell images were captured every 2 minutes for 2 h using a 100X air objective with 0.95 numerical aperture (Olympus). Timecourse stacks of single cells were aligned using the MultiStackReg plugin for ImageJ. Using the ImageJ Nano Tracking plugin (28,29) subpixel movement of pillars were measured. The displacement of each pillar at one timepoint was calculated relative to the position of the same pillar in the timepoint immediately preceding it. Cellular traction forces at each timepoint were calculated by multiplying displacements with the pillar bending stiffness. Individual cell migration velocity and displacement were obtained by tracking single cell movement across time using the Manual Tracking plugin for ImageJ. A total of 4 independent experiments were performed with 4 cells per experiment and at least 10 pillars per cell were measured for each condition. Each data point per condition is the average measurement of ≥160 pillars from 16 cells at a single timepoint.
Cell motility
Individual cell motility was observed with the Livecyte label-free imaging system (phasefocus). KPC cells (1x105) in complete DMEM were seeded into the wells of 6-well plates (MatTek P06G-1.5.20-F) and allowed to settle and grow overnight. Wells were coated with 0.3 mg/ml rat tail collagen type I (Corning 354249) prior to plating of cells. Next day, medium was replaced by complete DMEM containing inhibitors or DMSO vehicle and, after a 30 min delay, cells were imaged every 5 min for 10 h, with a 10x objective lens. The image processing tool and the automated cell tracking function provided with the cell analysis toolbox was used to determine the instantaneous cell velocities.
Scratch wound migration assay
The IncuCyte Live Cell Analysis System (Essen Bioscience) was used for the measurement of cell migration in scratch wound assays. KPC cells (2x104) in complete DMEM were seeded into the wells of 96-well ImageLock plates (Essen Bioscience) and allowed to settle and grow overnight. Next day, a scratch wound was created in cell monolayers using the Woundmaker device (Essen Bioscience). Complete DMEM containing inhibitors or DMSO vehicle was added to cells and cells were imaged every 2 h for 48 h.
Animal models
All mouse experiments were approved by the University of Glasgow College of Medicine, Veterinary and Life Sciences Research Ethics Committee, and performed under a project license granted by the United Kingdom government Home Office, in line with the Animals (Scientific Procedures) Act 1986 and European Union Directive 2010/63/EU in a dedicated barriered facility. Immunodeficient CD-1 nude mice were purchased from Charles River. AT13148 was dissolved in 10% DMSO, 1% Tween and 89% water for oral administration of 40 mg/kg AT13148 on a schedule with three treatments per week. Fasudil was dissolved in water for oral administration and animals were treated daily with a fasudil dose of 2 mg/mouse. 250 µl bromodeoxyuridine (BrdU) was injected into the peritoneum 2 h before animals were sacrificed.
To analyze the effect of AT13148 on tumor cell invasion, KPC cells (1x106) in 100 µl PBS were injected into the peritoneum of CD-1 females. Similarly, KPC (1x106) cells in 100 µl PBS were IP injected into CD-1 males to assess the effect of fasudil. Animals were treated with AT13148 or fasudil respectively, and body weights were monitored. After 2 weeks, animals were sacrificed as per institutional guidelines.
To analyze the effect of AT13148 on KPC tumor cell growth, KPC cells (1x106) in 100 µl PBS were injected subcutaneously into the right flank of CD-1 females. Animals were treated with AT13148 till endpoint, which was typically tumor ulceration. Body weight was monitored over the whole time of the experiment. Tumor measurements were taken 3 times weekly, starting 11 days after KPC cell injection when tumors had reached about 50-100 mm3 volumes. Tumor volumes were calculated using a modified ellipsoid formula: volume = (length x width2/2) (30).
Histology
Tissues were fixed in 10% formalin at room temperature and standard histological methods were used for processing and staining. Paraffin-embedded sections were rehydrated and immersed in boiling 10 mM citric acid buffer at pH 6.0 for 20 min. After blocking with H2O2 and normal goat serum, tissues were incubated with primary anti-BrdU antibody (BD Bioscience, 347580), followed by incubation with Envision + System-HRP labeled Polymer (Dako), and Liquid DAB + Substrate (Dako).
The Leica SCN 400f scanner and Leica Slidepath Digital Image Hub software were used to image and analyze the tissues. KPC tumor cell invasion into the pancreas was determined on H&E stains. Regions that show KPC tumor cells in-between the normal pancreatic cells were measured and expressed as percentage of the total pancreas area. An algorithm was used to determine numbers of BrdU positive nuclei. Images were taken on an Olympus BX51 microscope.
Statistical analysis
Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test, or using two-way ANOVA and post-hoc Dunnett’s multiple comparison test as indicated in the figure legends. For comparisons between two conditions, either paired or unpaired Student’s t-tests were used, or exact p values calculated by Mann Whitney tests as indicated in the figure legends. IC50 values were determined by four parameter variable slope nonlinear curve fitting. Error bars indicate standard error of the mean (± SEM). *, p<0.05; **, p<0.01; ***, p<0.001.
Results
AT13148 efficiently inhibits ROCK-mediated substrate phosphorylation
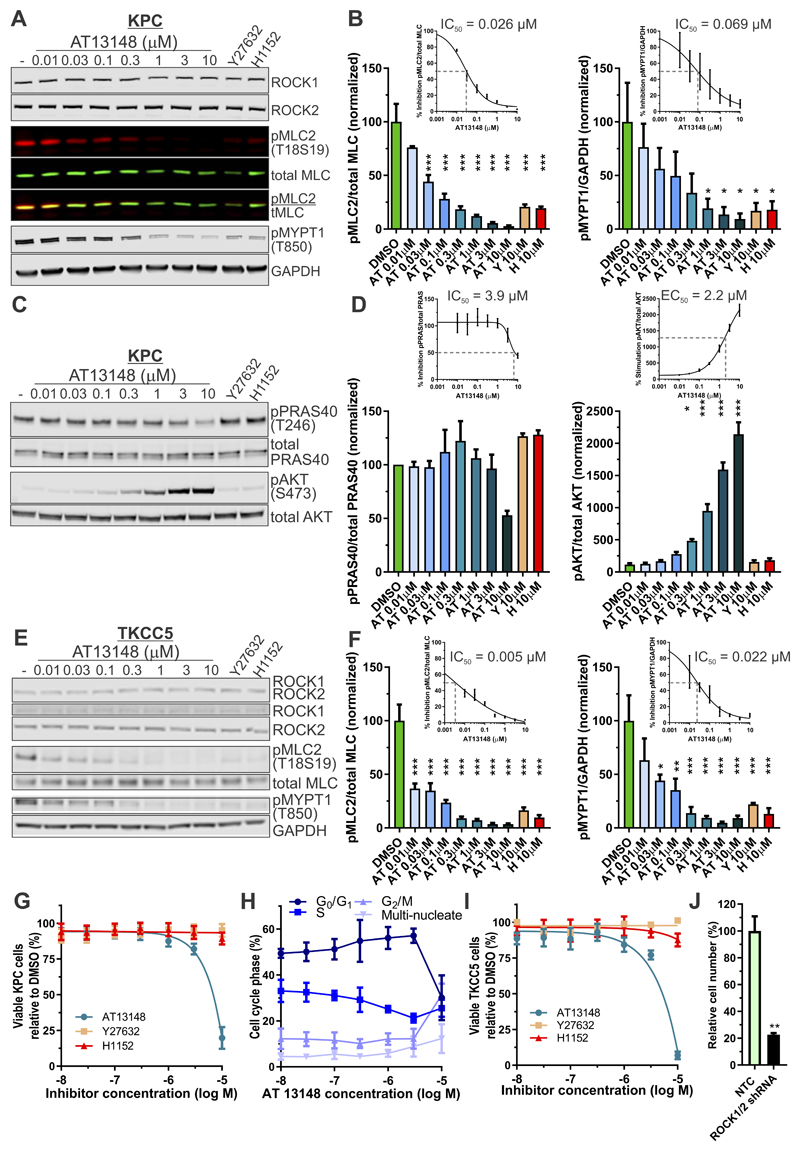
To determine whether AT13148 inhibited ROCK activity in PDAC cells, genetically defined mouse KPC (LSL-KrasG12D; LSL-p53R172H; Pdx1-Cre) tumor cells were treated with AT13148 ranging from 10 nM to 10 µM, or the ROCK selective inhibitors Y27632 (10 µM) (31) or H1152 (10 µM) (32) for 1 hour. Western blotting revealed significantly reduced phosphorylation of the ROCK substrate myosin light chain 2 (pMLC2) (33) relative to total MLC2 (tMLC) by AT13148 (AT) concentrations ≥ 0.03 µM, or by Y27632 (Y) and H1152 (H) compared to DMSO vehicle controls (Figs. 1A and 1B, left panel). Reduced phosphorylation was also observed for the ROCK substrate myosin-binding subunit of the MLC phosphatase (MYPT1) (34) relative to GAPDH by AT13148 concentrations ≥ 1 µM, or by Y27632 and H1152 (Figs. 1A and 1B, right panel). Expression of ROCK1 and ROCK2 were unaffected by any drug concentration (Fig. 1A, Supplemental Figs. S1A and S1B). Curve fitting gave IC50 values for the inhibition of MLC2 phosphorylation by AT13148 of 0.026 µM (Fig. 1B, left panel inset), and for the inhibition of MYPT1 phosphorylation of 0.069 µM (Fig. 1B, right panel inset).
Figure 1. Potent inhibition of substrate phosphorylation by AT13148.
A, KPC mouse PDAC cells were treated with DMSO vehicle or indicated concentrations of AT13148, Y27632 or H1152 for 1 hour, then immunoblotted for ROCK1, ROCK2, phosphorylation of MLC2 (pMLC2; T18S19), total MLC (tMLC), phosphorylation of MYPT (pMYPT1; T850) and GAPDH B, Left panel Quantifications represent the ratio of pMLC2 to total MLC as percentage relative to DMSO vehicle. Means ± SEM are shown from n = 3 independent experiments. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. ***, p<0.001. Inset Four parameter variable slope non-linear curve fitting of pMLC2/total MLC percentages was used to determine IC50 value. Right panel Relative pMYPT1 to GAPDH signal as percentage of DMSO vehicle. Means ± SEM are shown from n = 3 independent experiments. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. *, p<0.05. Inset Four parameter variable slope non-linear curve fitting of pMPYT1/GAPDH percentages was used to determine IC50 value. C, KPC cells were treated with DMSO or indicated concentrations of AT13148, Y27632 or H1152 for 1 hour, then immunoblotted for phosphorylation of PRAS40 (pPRAS40; T246), total PRAS40, phosphorylation of AKT (pAKT; S473), or total AKT. D, Left panel Ratio of pPRAS40 to PRAS40 as percentage of DMSO vehicle. Means ± SEM are shown from n = 3 independent experiments. Inset Four parameter variable slope non-linear curve fitting of pRAS40/PRAS percentages was used to determine IC50 value. Right panel Ratio of pAKT to AKT as percentage of DMSO vehicle. Means ± SEM are shown from n = 3 independent experiments. Inset Four parameter variable slope non-linear curve fitting of pAKT/AKT percentages was used to determine IC50 value. E, TKCC5 human PDAC cells were treated with DMSO vehicle or indicated concentrations of AT13148, Y27632 or H1152 for 1 hour, then immunoblotted for ROCK1, ROCK2, pMLC2, total MLC, pMYPT1, and GAPDH. F, Left panel Ratio of pMLC2 to total MLC as percentage of DMSO vehicle. Means ± SEM are shown from n = 3 independent experiments. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. ***, p<0.001. Inset Four parameter variable slope non-linear curve fitting of pMLC2/total MLC percentages was used to determine IC50 value. Right panel Relative pMYPT1 to GAPDH signal as percentage of DMSO vehicle. Means ± SEM are shown from n = 3 independent experiments. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. *, p<0.05. **, p<0.01. ***, p<0.001. Inset Four parameter variable slope non-linear curve fitting of pMPYT1/GAPDH percentages was used to determine IC50 value. G, Effects of AT13148, Y27632 or H1152 on KPC cell viability after 18 h drug treatment. Means ± SEM are shown from n = 4 independent experiments. H, Cell cycle analysis of KPC cells treated with a concentration range of AT13148 for 16 h. Means ± SEM are shown from n = 3 independent experiments. I, Effects of AT13148, Y27632 or H1152 on TKCC5 cell viability after 18 h drug treatment. Means ± SEM are shown from n = 3 independent experiments. J, Relative TKCC5 cell numbers 7 days after transduction with non-targeting control (NTC) or combined ROCK1/2 shRNA lentivirus. Means ± SEM are shown from n = 3 independent experiments. Statistical significance was determined for the comparisons of NTC with ROCK1/2 knockdown by unpaired Student’s t-test. **, p<0.01.
Given that AT13148 also inhibits AKT in vitro kinase activity significantly, albeit less potently than ROCK1 (IC50 = 6 nM) or ROCK2 (IC50 = 4 nM), with IC50 values for AKT1 (38 nM), AKT2 (402 nM) and AKT3 (50 nM) (21), KPC cells were treated with varying AT13148 concentrations from 10 nM to 10 µM, as well as 10 µM Y27632 and 10 µM H1152. Western blotting revealed reduced phosphorylation of the AKT substrate PRAS40 (Proline-Rich AKT Substrate) (35) relative to total PRAS40 by AT13148 only at 10 µM, with a calculated IC50 = 3.9 µM (Figs. 1C and 1D, left panel and inset). It has previously been determined that AKT inhibitors can lead to hyperphosphorylation of AKT on regulatory sites (including S473), leading to AKT activation that can counteract the effects of small molecule inhibitors (36). Western blotting of AT13148 treated cells indeed revealed profound AKT S473 phosphorylation (Figs. 1C and 1D, right panel and inset), providing an explanation for the relatively low potency of AT13148 as an AKT inhibitor in pancreatic cancer cells. Both Y27632 and H1152 did not affect PRAS40 or AKT phosphorylation (Figs. 1C and 1D).
To determine the potency of AT13148 in human pancreatic cancer cells, PDX-derived TKCC5 cells were treated with AT13148 ranging from 10 nM to 10 µM, or the ROCK selective inhibitors Y27632 (10 µM) or H1152 (10 µM) for 1 hour. Western blotting revealed significantly reduced phosphorylation of the ROCK substrate pMLC2 relative to total MLC2 by AT13148 concentrations ≥ 0.01 µM, or by Y27632 and H1152 compared to DMSO vehicle controls (Figs. 1E and 1F, left panel). Reduced phosphorylation was also observed for the ROCK substrate MYPT1 relative to GAPDH by AT13148 concentrations ≥ 0.03 µM, or by Y27632 and H1152 (Figs. 1E and 1F, right panel). Expression of ROCK1 and ROCK2 were unaffected by any drug concentration (Fig. 1E). Curve fitting gave IC50 values for the inhibition of MLC2 phosphorylation by AT13148 of 0.005 µM (Fig. 1F, left panel inset), and for inhibition of MYPT1 phosphorylation of 0.022 µM (Fig. 1F, right panel inset), revealing ROCK sensitivity to AT13148 inhibition in human tumor cells 3-5 times greater than in mouse PDAC cells.
The effects of AT13148 at concentrations from 10 nM to 10 µM, as well as 10 µM Y27632 and 10 µM H1152, on viable KPC cell numbers were examined after 18 hours of treatment. While Y27632 and H1152 were without effect, AT13148 reduced viable cell numbers at the highest 10 µM treatment (Fig. 1G), which was associated with a reduced proportion of cells in G0/G1 and increased proportion in G2/M cell cycle phase (Fig. 1H), consistent with the reported effect of complete genetic deletion of Rock1 and Rock2 on cell cycle progression in mouse embryonic fibroblasts (10). Comparable effects of AT13148 on viability were also observed for human TKCC5 PDAC cells (Fig. 1I). Consistent with the reported inhibition of proliferation of pancreatic cancer cell lines in vitro following siRNA-mediated knockdown of ROCK1/2 (12) and supporting the effect of AT13148 on proliferation being the result of ROCK inhibition, shRNA-mediated ROCK1/2 knockdown (Supplemental Figure S2A) significantly reduced TKCC5 cell numbers (Fig. 1J). Subsequent experiments used non-toxic AT13148 concentrations up to 3 µM. These results indicate that AT13148 is a potent ROCK inhibitor, but ineffective AKT inhibitor due to compensatory mechanisms, in mouse and human PDAC cells.
AT13148 effects on cell morphology
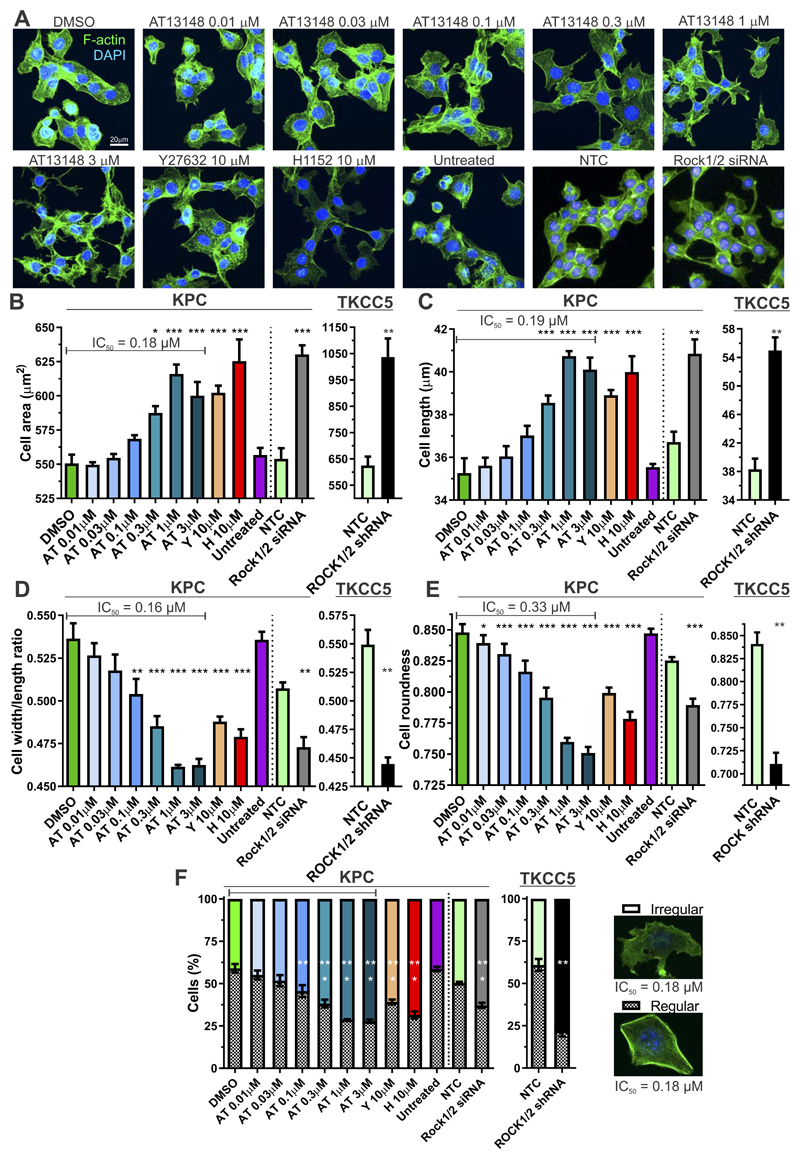
Given the central and critical role of ROCK kinases in actomyosin cytoskeleton and cell morphology regulation (7), the effects on KPC cells of a range of AT13148 concentrations were compared to the ROCK selective inhibitors Y27632 and H1152, or to shRNA-mediated knockdown of ROCK1 and ROCK2 in TKCC5 cells (Supplementary Fig. S2A) or siRNA-mediated knockdown of Rock1 and Rock2 in KPC cells (Supplemental Fig. S2B). Cells were fixed and stained with fluorescent phalloidin to visualize filamentous actin (F-actin) structures one hour after drug treatment (Fig. 2A). High content analysis (HCA) revealed dose dependent increases in total cell area (Fig. 2B and Supplemental Fig. S2C; IC50 = 0.18 µM), and cell length (Fig. 2C and Supplemental Fig. S2D; IC50 = 0.19 µM) induced by AT13148 that were significant at doses ≥ 0.3 µM, and for 10 µM Y27632 and H1152. Accompanying the increased cell area and length, the width/length ratios were significantly decreased at AT13148 concentrations ≥ 0.1 µM, and for 10 µM Y27632 and H1152 (Fig. 2D and Supplemental Fig. S2E; IC50 = 0.16 µM), while overall cell roundness also significantly decreased at AT13148 concentrations ≥ 0.01 µM, and for 10 µM Y27632 and H1152 (Fig. 2E and Supplemental Fig. S2F; IC50 = 0.33 µM). Consistent with the effect of the small molecular inhibitors being the result of ROCK inhibition, siRNA-mediated Rock1 and Rock2 knockdown in KPC cells, or shRNA-mediated ROCK1 and ROCK2 knockdown in TKCC5 cells, induced significant changes in each morphological parameter relative to non-targeting control (NTC) cells, in the same direction as AT13148, Y27632 or H1152 treatment (Figs. 2B-E). Using multiparametric analysis and applying a machine-learning algorithm (37) to classify cells either into “regular” or “irregular” morphologies, it was determined that AT13148 significantly shifted the distribution of cell morphologies from regular towards irregular at concentrations ≥ 0.1 µM, as did 10 µM Y27632, 10 µM H1152, or siRNA-mediated Rock1 and Rock2 knockdown in KPC cells or shRNA-mediated ROCK1 and ROCK2 knockdown in TKCC5 cells (Fig. 2F, Supplemental Figs. S2G and S2H; IC50 = 0.18 µM). In addition, there were subtle but significant effects on nuclear morphology in KPC cells induced by AT13148, Y27632 and H1152 treatments (Supplemental Figs. S3A and S3B). Taken together, these observations reveal potent effects of AT13148 on cell morphology that phenocopy the actions of the ROCK selective inhibitors Y27632 and H1152, or the effect of ROCK knockdown induced by siRNA or shRNA.
Figure 2. AT13148 affects cell morphology.
A, Representative confocal microscope maximum intensity projection images of F-actin (green) and DAPI (blue) stained KPC cells treated with indicated concentrations of AT13148, Y27632 or H1152 for 1 h. Alternatively, cells were transfected with non-targeting control (NTC) or a mixture of Rock1 + Rock2 (Rock1/2) siRNAs and imaged on an Operetta High Content Imaging System. High content imaging of KPC cells after 1 h of AT13148, Y27632 or H1152 drug treatment, or 24 h post siRNA transfection (left graphs), or >7 days post lentiviral transduction of TKCC5 cells with NTC or ROCK1 and ROCK2 targeted shRNAs (right graphs), was used to determine B, cell area; C, cell length; D, cell width/length ratios; E, cell roundness. Means ± SEM are shown from n = 3 independent experiments for drug treatments, n = 4 independent experiments for siRNA transfections, or n = 3 independent experiments with shRNA knockdown cells. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test for AT13148, Y276732 or H1152 treatments. Statistical significance was determined for the comparisons of corresponding NTC with Rock1/2 or ROCK1/2 knockdown by unpaired Student’s t-test. *, p<0.05; **, p<0.01; ***, p<0.001. F, Multiparametric analysis was used to classify cells as irregular (white bar) or regular (hatched bar), as typified by the indicated cells. Means ± SEM are shown from n = 3 independent experiments, n = 4 independent experiments for siRNA transfections, or n = 3 independent experiments with shRNA knockdown cells. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test for AT13148, Y276732 or H1152 treatments. Statistical significance was determined for the comparisons of corresponding NTC with Rock1/2 or ROCK1/2 knockdown by unpaired Student’s t-test. **, p<0.01; ***, p<0.001.
Effect of ROCK inhibitors on tumor cell metabolism
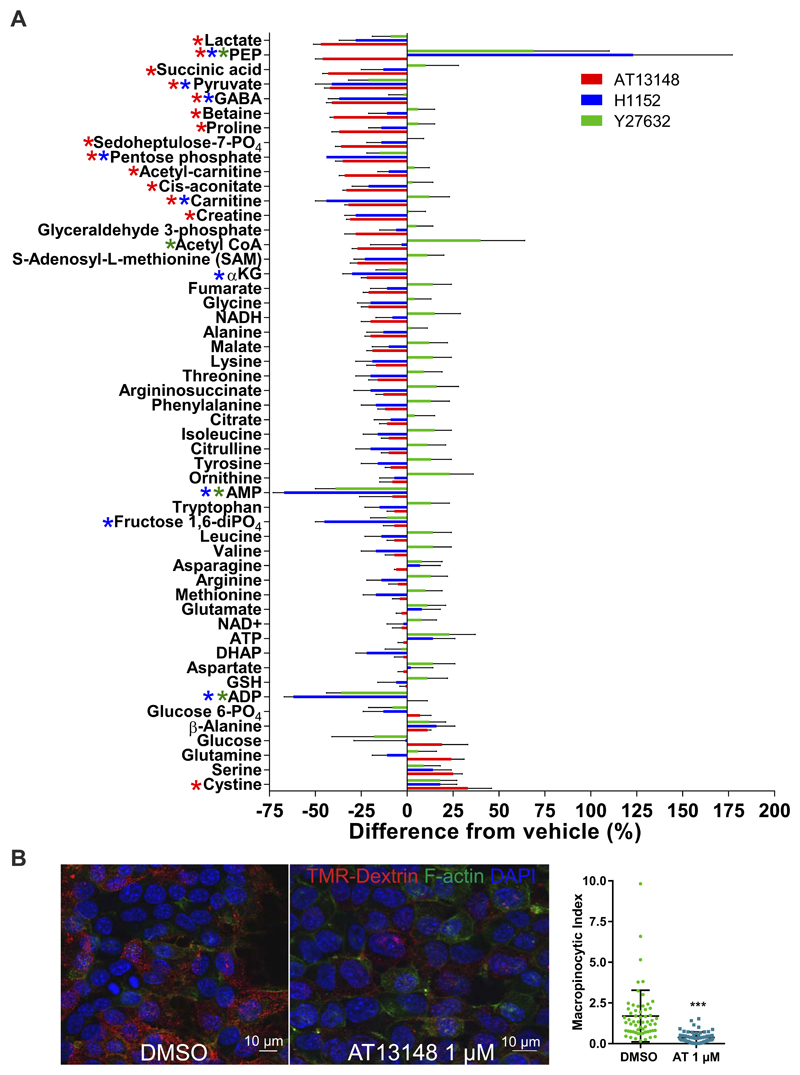
To characterize the metabolic alterations induced by 1 µM AT1318, 10 µM Y27632 or 10 µM H1152 treatment, KPC cell extracts were prepared and metabolites analyzed as described in Tardito et al. (24). Results in Figure 3A are plotted at the negative or positive percentage change relative to DMSO vehicle treatment (100%), and two-way ANOVA and post-hoc Dunnett’s multiple comparison tests were used to identify significant effects relative to DMSO treatment. Given that the concentrations of these three inhibitors potently inhibited ROCK substrate phosphorylation, and that there were no significant effects in the same direction induced by all three inhibitors, ROCK activity likely does not play a major role in regulating metabolism in PDAC cells. There were some significant responses to 1 µM AT13148, including decreased lactate, succinic acid and pyruvate, which suggest that this compound may have ROCK and AKT independent effects on metabolism. Interestingly, H1152 and Y27632 significantly increased phosphoenolpyruvate (PEP) levels, while AT13148 significantly decreased PEP (Fig. 3A). In addition, both H1152 and Y27632 significantly decreased AMP and ADP levels, which were unaffected by AT13148. Taken together, these results are consistent with all three small molecules having spectra of off-target effects on metabolic pathways, which have not previously been considered either as part of their apparent mechanisms of action, or as potential therapeutic liabilities.
Figure 3. Effect of ROCK inhibitors on PDAC cell metabolism.
A, Normalized levels of intracellular metabolites in cells treated with 1 µM AT13148, 10 µM H1152 or 10 µM Y27632 for 24 hours as negative or positive relative differences from DMSO treated controls. Means ± SEM are shown from n = 4 independent experiments. Colored * indicates p<0.05 for corresponding drug treatment using two-way ANOVA and post-hoc Dunnett’s multiple comparison test compared to DMSO. B, Macropinocytic index was measured as described in (26) by examining the uptake of tetramethylrhodamine (TMR)-Dextran over 30 minutes by confocal microsopy. Means ± SEM are shown for 80 DMSO and 80 AT13148 treated cells. Statistical significance was determined by unpaired Student’s t-test. ***, p<0.001.
PDAC cells have also been shown to obtain nutrients via scavenging of extracellular proteins acquired through macropinocytosis (38,39). When KPC cells were treated with 1 µM AT1318, macropinocytic activity measured by tetramethylrhodamine (TMR) dextran uptake was significantly inhibited (Fig. 3B). Therefore, under conditions of nutrient deprivation, AT13148 would be expected to limit access to this alternative source.
PDAC invasion is inhibited by AT13148
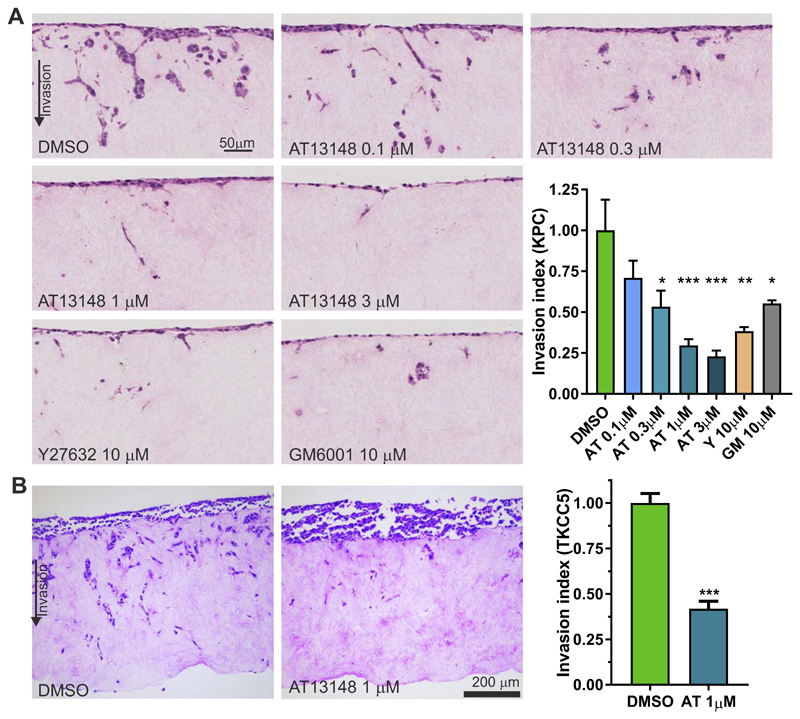
Pancreatic tumor masses have significant collagen content that cells move through to enable primary tumor growth, invade local tissue and to metastasize. A 3-dimensional (3D) collagen matrix invasion assay, in which cells invade through fibroblast-remodeled collagen matrix towards serum-containing medium, was used previously to demonstrate ROCK dependence of PDAC invasive behavior (13,40). Invasion of KPC mouse PDAC cells was significantly inhibited by AT13148 at concentrations ≥ 0.3 µM and by 10 µM Y27632 (Fig. 4A). In addition, invasion was significantly inhibited by the broad specificity matrix metalloproteinase inhibitor GM6001 (10 µM) (41). Consistent with these observations, treatment of human PDX-derived TKCC5 tumor cells with 1 µM AT13148 significantly inhibited 3D collagen invasion (Fig. 4B). Interestingly, the inhibition of invasion resulting from GM6001 was less than the inhibition induced by 1 µM or 3 µM AT13148, or by 10 µM Y27632, suggesting that ROCK activity was required for an activity additional to promoting collagen degradation, or an activity more important than MMP activity for invasion, such as cell motility.
Figure 4. Collagen matrix invasion is efficiently inhibited by AT13148.
A, H&E-stained sections of KPC cell invasion into fibroblast-conditioned collagen matrix after 8 days, which included treatment with AT13148, Y27632 or GM6001 as indicated. The invasion index was normalized to DMSO control conditions, with means ± SEM shown from n = 3 independent experiments. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. *, p<0.05; **, p<0.01; ***, p<0.001. B, H&E-stained sections of TKCC5 cell invasion into fibroblast-conditioned collagen matrix after 8 days, which included treatment with AT13148. The invasion index was normalized to DMSO control conditions, with means ± SEM shown from n = 3 independent experiments. Statistical significance was determined using unpaired Student’s t-test. ***, p<0.001.
Cell migration and contractile force generation
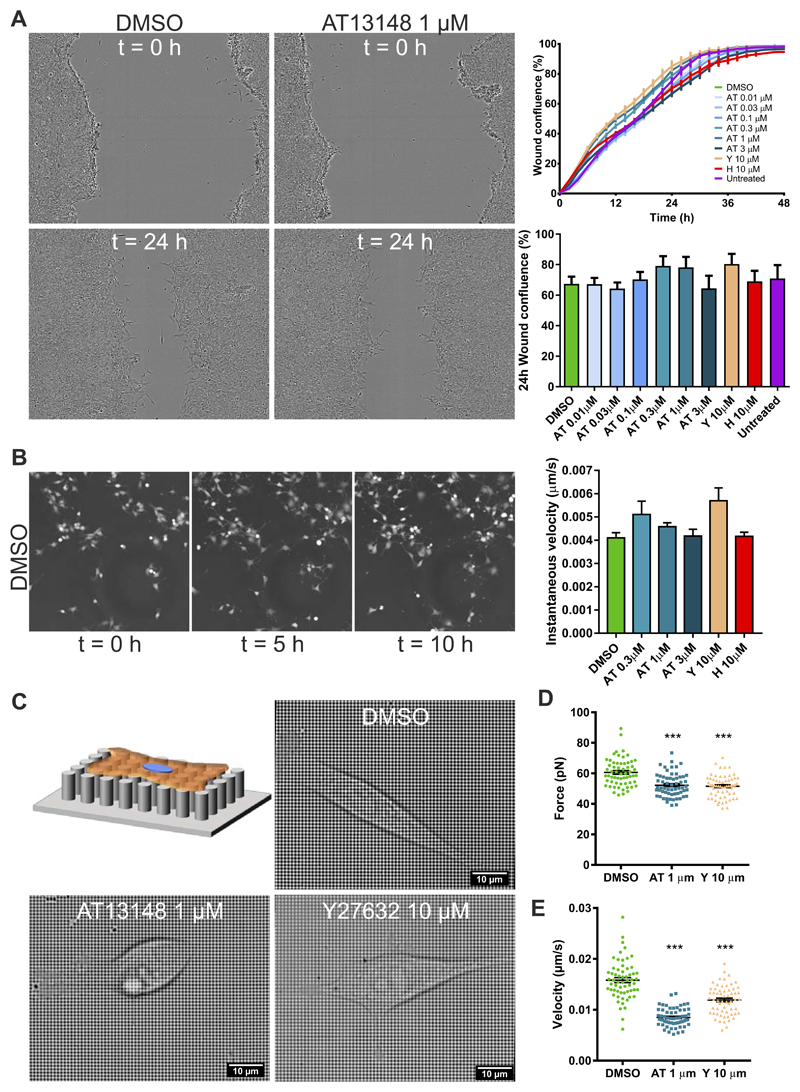
A possible explanation for the significant effect of ROCK inhibition on collagen invasion was decreased cell motility. To determine whether ROCK activity was required for migration across two-dimensional (2D) surfaces, confluent KPC monolayers were scratched and the migration of cells to close the wound was monitored in real time over 48 hours. Concentrations of AT13148 from 0.01 µM to 3 µM, as well as 10 µM Y27632 or 10 µM H1152, did not significantly affect wound closure over all time points or at 24 hours when wound closure was ~50% (Fig. 5A). Furthermore, instantaneous random cell migration velocities were not affected when KPC cells were monitored for 10 hours in the presence of AT13148 from 0.3 µM to 3 µM, 10 µM Y27632 or 10 µM H1152 (Fig. 5B). These results indicate that migration across planar stiff surfaces, akin to “mesenchymal” migration that is largely dependent on actin polymerization to push cells forward, was not sensitive to ROCK inhibition (42).
Figure 5. AT13148 reduces physical force and migration on pliable substrates.
A, Directed scratch wound migration of AT13148, Y27632 or H1152 drug-treated cells was monitored over 48 hours. Left: Representative images at 0 and 24 hours for DMSO and 1 µm AT13148 treated cells. Upper right graph shows mean ± SEM percentage wound confluence acquired every 2 hours over 48 hours for n=6 replicates. Lower right graph shows mean ± SEM percentage wound confluence at 24 hours for n=3 independent experiments. B, Random migration of cells treated with AT13148, Y27632 or H1152 was monitored over 10 hours. Left panels show representative images at 0, 5 and 10 hours. Means ± SEM are shown from n = 3 independent experiments. C, Schematic diagram and representative images of cells on nanopillars with DMSO, AT13148 or Y27632 treatments as indicated. D, Forces leading to nanopillar deflection were determined for ≥10 pillars under 16 cells for AT13148 and Y27632 treatments, and for 32 cells for DMSO. Mean force values determined at each 2 minute interval over 120 minutes are plotted for each condition, with indicated means ± SEM. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. ***, p<0.001. E, Velocities of 16 cells for AT13148 and Y27632 treatments, and of 32 cells for DMSO were determined from time lapse images acquired every 2 minutes over 2 hours. Mean velocities determined at each 2 minute interval over 120 minutes are plotted for each condition, with indicated means ± SEM. Statistical significance was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. ***, p<0.001.
To determine whether ROCK inhibition by AT13148 or Y27632 would affect forces applied to the substrate on which cells were attached, KPC cells were plated on grids of flexible nanopillars (27) and pillar deflection distances collected over 2 hours was used to calculate absolute forces exerted (Fig. 5C). Consistent with the role of ROCK in the generation of actomyosin contractile force, treatment with 1 µM AT13148 or 10 µM Y27632 significantly decreased the force exerted on nanopillars (Fig. 5D). The imaging of single cells on nanopillars was also used to determine cell velocity; 1 µM AT13148 or 10 µM Y27632 significantly decreased cell movement on the flexible substrate (Fig. 5E). Unlike migration on planar stiff surfaces, movement on flexible discontinuous substrates apparently does require contractile force, consistent with previous studies that modelled migration modes on varying substrate geometries (43).
In vivo inhibition of PDAC invasion and tumor growth by AT13148
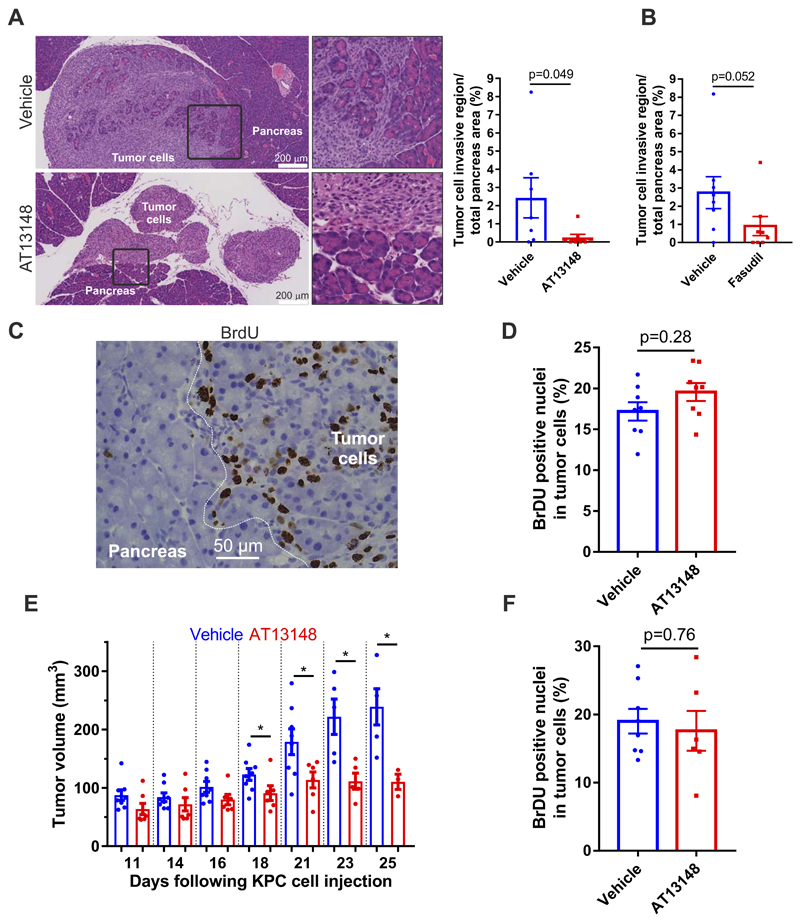
Although the 3D collagen invasion assay recapitulates aspects of the in vivo environment, only an in vivo model would resemble the complex environments encountered by invasive tumor cells. KPC cells (106) were injected into the peritoneal cavity of CD-1 immunocompromised mice, which were then treated for 2 weeks with 40 mg/kg AT13148 three times per week, daily treatment with 2 mg/mouse (~80 mg/kg) of the ROCK selective inhibitor fasudil, or corresponding vehicle solutions with matched volumes. Over the 2 weeks, there were no significant effects of either compound on mouse weights (Supplemental Figs. S4A and S4B), indicating that both were well tolerated. Histopathology revealed that tumor cells had homed to the pancreas, where they grew onto and invaded into healthy tissue (Fig. 6A upper panels; note the pale tumor cells mixed with darker healthy pancreas). In contrast, in AT13148 treated mice, tumor cells homed and grew on the pancreas, but a clearly demarcated border was consistently observed between tumor and normal tissue (Fig. 6A lower panels). When the area of the invasive region, in which tumor cells were intermixed with normal pancreas cells, was expressed as a percentage of the total pancreas for each condition, there was a significant decrease in the invasive region for AT13148 treated mice (Fig. 6A), which approached significance for the less potent ROCK inhibitor fasudil treated mice (Fig. 6B). To determine whether the anti-invasive effect of AT13148 treatment was due to decreased proliferation, mice were injected with bromodeoxyuridine (BrdU) 2 h before sacrifice, and then tissues were stained with anti-BrdU antibody (Fig. 6C). Counting the percentage of BrdU positive tumor cells indicated that there was no effect of AT13148 on proliferation (Fig. 6D), indicating that reduced invasion of pancreatic tissue was a direct effect on invasive behaviors.
Figure 6. AT13148 blocks PDAC invasion and tumor growth in vivo.
A, Mouse PDAC cells were injected into the peritoneum of CD-1 mice and animals were treated with vehicle or AT13148 (40 mg/kg, 3 x per week) for 14 days. Left: Representative H&E-stained sections of pancreas (dark staining) and invading tumor cells (light staining). Right: Quantification of pancreas area invaded by KPC tumor cells. Means ± SEM are shown for n=7 for vehicle-treated and n=8 for AT13148-treated mice. Exact P value by Mann-Whitney test. B, Quantification of pancreas area invaded by KPC tumor cells. Means ± SEM are shown for n=8 for vehicle-treated and n=8 for mice treated daily with 2 mg/mouse fasudil. Exact P value by Mann-Whitney test. C, Representative image of BrdU staining of normal pancreas and tumor cells, with white line indicating normal pancreas from region with tumor cells. D, Quantification of percentage BrdU staining in KPC tumor cells. Means ± SEM shown for n=8 for vehicle-treated and n=8 for AT13148-treated mice. Exact P value by Mann-Whitney test. E, Volumes of subcutaneous KPC cell tumors in CD-1 mice treated with AT13148 (40 mg/kg, 3 x per week) or vehicle control for up to 25 days (dots represent individual mice). Statistical significance was determined for vehicle versus AT13148 for each day by unpaired Student’s t-test. *, p<0.05. F, Quantification of percentage BrdU staining in KPC tumor cells. Means ± SEM shown for n=8 for vehicle-treated and n=6 for AT13148-treated mice. Exact P value by Mann-Whitney test.
AT13148 was previously reported to reduce the growth of subcutaneous xenograft tumors of BT474 breast, PC3 prostate and MES-SA uterine cancer cell lines (21). However, the mechanism of tumor growth inhibition was not determined, and proliferation was not directly examined. Given that it has been proposed that even short range cell dispersal leads to faster tumor growth (44), we wished to determine whether PDAC xenograft tumor growth would be affected by the anti-invasion effect of AT13148. KPC cells (106) in 100 µl PBS were injected subcutaneously into the right flank of CD-1 mice, and were treated with 40 mg/kg AT13148 or vehicle control until endpoint, which was typically tumor ulceration. Body weights again showed no difference induced by AT13148 treatment (Supplemental Fig. S4C). Tumor measurements were taken 3 times weekly, starting 11 days after KPC cell injection when tumors had reached 50-100 mm3 volumes (Supplemental Figs. S5A and S5B). Pairwise comparisons revealed significantly reduced tumor growth at each day between days 18 to 25 (Fig. 6E). As before, mice were treated with BrdU 2 hours prior to sacrifice and BrdU positive cells quantified for each condition. As before, AT13148 did not significantly affect proliferation as indicated by no difference in BrdU positivity between conditions (Fig. 6F), consistent with the anti-invasive actions of AT13148 being responsible for reducing tumor growth, rather than an anti-proliferative effect.
Discussion
The lack of effective therapies for pancreatic cancer has led to considerable effort being directed at discovering new therapeutic strategies (6). Difficulties with drugging the major oncogenic drivers mean that alternative approaches are required. We found that the potent inhibitor AT13148 has anti-tumor properties that support its development for PDAC treatment as part of a chemotherapeutic combination or in an adjuvant setting.
Originally discovered on the basis of being an AKT inhibitor, it was subsequently determined that AT13148 is a multi-AGC kinase inhibitor with significant potency for ROCK1 and ROCK2 (21). We found that AT13148 induced comparable, if not greater, effects compared to the selective ROCK inhibitors Y27632 and H1152 on substrate phosphorylation (Fig. 1), cell morphology (Fig. 2), migration on discontinuous pliable substrates (Fig. 5) and invasion into three-dimensional collagen matrix (Fig. 4) in mouse and human PDAC cells, consistent with ROCK inhibition being a key mechanism of action. Interestingly, AT13148 was determined to be a relatively poor AKT inhibitor (Figs. 1C and 1D), likely due a previously described compensatory mechanism that leads to AKT phosphorylation on the activating S473 site (36). We also determined that AT13148 significantly reduced macropinocytosis (Fig. 3B), which allows mutant KRAS expressing PDAC cells to obtain nutrients via scavenging of extracellular proteins. Previous studies revealed that Kaposi's Sarcoma-Associated Herpesvirus entry via macropinocytosis was sensitive to ROCK inhibition (45), consistent with ROCK on-target actions of AT13148 being responsible for the reduction in macropinocytic activity.
Invasion of three dimensional collagen matrices by KPC mouse PDAC tumor cells was previously found to be sensitive to Y27632 (40), which we confirmed and extended by showing dose-dependent inhibition by AT13148 (Fig. 4A). In addition, collagen invasion by the human PDX-derived TKCC5 PDAC cell line was significantly inhibited by AT13148 (Fig. 4B). Interestingly, collagen invasion was more sensitive to ROCK inhibition by Y27632 or AT13148 than to the general MMP inhibitor GM6001, suggesting that ROCK activity was required for an activity in addition to MMP-mediated collagen degradation, or that ROCK contributed to a function that was more important for collagen invasion than matrix degradation (Fig. 4A). Given that invasion is dependent on the ability of cells to move, we examined motility on stiff planar surfaces as well as on discontinuous pliable substrates. Interestingly, ROCK inhibition did not affect migration on two dimensional surfaces as determined for monolayer wound healing (Fig. 5A) or individual random cell migration (Fig. 5B). However, movement on pliable surfaces was affected by AT13148 and Y27632 (Fig. 5E), which was paralleled by significantly reduced force being exerted on underlying substrates (Fig. 5D). These results reveal different requirements for ROCK activity and contractile force for movement on varying surfaces, suggesting that ROCK activity was dispensable for “mesenchymal” migration on stiff planar substrates, but necessary for migration on discontinuous and pliable surfaces that more closely reflect the in vivo environment.
When KPC mouse PDAC cells were injected into the peritoneal cavity of CD-1 immunocompromised mice, a proportion of cells homed to the pancreas where they formed tumors and invaded healthy tissue (Fig. 6A). By treating mice with AT13148, invasion into pancreatic tissue was effectively inhibited, resulting in clear separation between tumor and pancreas (Fig. 6A), an observation largely replicated in fasudil treated mice (Fig. 6B). Interestingly, BrdU labelling of tumor cells was not different between control and AT13148 treated mice (Fig. 6D), indicating that the anti-invasive effect was independent of anti-proliferative effects. Although we did observe inhibition of cell cycle progression at the highest 10 µM AT13148 concentration in vitro, which was consistent with the cell cycle arrest induced by Rock1/Rock2 deletion in mouse cells (10), it seems likely that the concentrations attained in pancreatic tissue in vivo were below levels that would result in attenuated proliferation. When tumor cells were injected subcutaneously, AT13148 significantly slowed increases in tumor volume (Fig. 6E and Supplemental Fig. S5), which also was not associated with differences in BrdU labelling of tumor cells (Fig. 6F). These results are consistent with the effect of AT13148 on tumor growth is not due to inhibition of proliferation, but is more likely a reflection of reduced movement of cells within the tumor mass and invasion of local adjacent tissue. It was recently proposed that even short range cell dispersal leads to faster tumor growth (44), which suggests that inhibition of invasion and cell motility would have broad positive consequences on slowing primary tumor growth, in addition to the potential benefits from reducing the invasion of local healthy tissue (Fig. 6A) and from inhibiting distant metastasis (22).
The results from this study support the potential for further development of AT13148 for pancreatic cancer therapy. Its clinical value would arise from its ability to restrict tumor cell invasion, which would slow tumor growth and possibly enable surgical resection by helping to maintain separation between tumor and healthy tissue. In addition to patients with amplified or elevated ROCK expression (13), activation of ROCK activity by stiff desmoplastic stroma (18,46) suggests that a broad range of patients would likely benefit from ROCK inhibitor treatment. A recent study found that treatment of mice bearing PDX-derived xenograft PDAC tumors with fasudil increased therapeutic responses to gemcitabine and nab-paclitaxel, increased the sensitivity of tumor cells to shear stress, and decreased metastasis to the liver (17). It was proposed that transient ROCK inhibition with fasudil affected the tumor-associated extracellular matrix and vasculature, thus “priming” tumors to be more responsive to standard chemotherapies. Consistent with these conclusions, the use of AT13148 would likely be restricted to combination therapy or in an adjuvant setting, given that its actions as a single agent significantly slowed tumor growth, but have not been seen to result in overt tumor regression in this or prior in vivo studies (21,47).
Supplementary Material
Acknowledgements
Funding was provided from the Engineering and Physical Sciences Research Council SoftMech Centre for Mathematical Sciences in Healthcare (EP/N014642/1), and from Cancer Research UK to MFO (A18276), JJK (C50242/A17728) and to the Beatson Institute (A17196), as well as a Len Ainsworth Pancreatic Cancer Fellowship from NHMRC. AT13148 was a gift from Astex Pharmaceuticals. Silicon templates were a gift from Prof. J Hone, Columbia University. NG acknowledges partial support from ERC funding through FAKIR 648892 Consolidator Award.
Footnotes
Conflict of interest statement: The authors have no potential conflicts of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 4.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 5.Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vennin C, Rath N, Pajic M, Olson MF, Timpson P. Targeting ROCK activity to disrupt and prime pancreatic cancer for chemotherapy. Small GTPases. 2017 doi: 10.1080/21541248.2017.1345712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK) Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–8. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott RW, Olson MF. LIM kinases: function, regulation and association with human disease. J Mol Med. 2007;85:555–68. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 10.Kümper S, Mardakheh FK, McCarthy A, Yeo M, Stamp GW, Paul A, et al. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. eLife. 2016;5:e12203. doi: 10.7554/eLife.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whatcott CJ, Ng S, Barrett MT, Hostetter G, Von Hoff DD, Han H. Inhibition of ROCK1 kinase modulates both tumor cells and stromal fibroblasts in pancreatic cancer. PLoS One. 2017;12:e0183871. doi: 10.1371/journal.pone.0183871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rath N, Morton JP, Julian L, Helbig L, Kadir S, McGhee EJ, et al. ROCK signaling promotes collagen remodeling to facilitate invasive pancreatic ductal adenocarcinoma tumor cell growth. EMBO Mol Med. 2017;9:198–218. doi: 10.15252/emmm.201606743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 16.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–51. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vennin C, Chin VT, Warren SC, Lucas MC, Herrmann D, Magenau A, et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8504. eaai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laklai H, Miroshnikova YA, Pickup MW, Collisson EA, Kim GE, Barrett AS, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent OA, Sandí M-J, Burston HE, Brown KR, Rottapel R. An oncogenic KRAS transcription program activates the RHOGEF ARHGEF2 to mediate transformed phenotypes in pancreatic cancer. Oncotarget. 2016;8 doi: 10.18632/oncotarget.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson MF. Applications for ROCK kinase inhibition. Curr Opin Cell Biol. 2008;20:242–8. doi: 10.1016/j.ceb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yap TA, Walton MI, Grimshaw KM, te Poele RH, Eve PD, Valenti MR, et al. AT13148 Is a Novel, Oral Multi-AGC Kinase Inhibitor with Potent Pharmacodynamic and Antitumor Activity. Clini Cancer Res. 2012;18:3912–23. doi: 10.1158/1078-0432.CCR-11-3313. [DOI] [PubMed] [Google Scholar]
- 22.Sadok A, McCarthy A, Caldwell J, Collins I, Garrett MD, Yeo M, et al. Rho kinase inhibitors block melanoma cell migration and inhibit metastasis. Cancer Res. 2015;75:2272–84. doi: 10.1158/0008-5472.CAN-14-2156. [DOI] [PubMed] [Google Scholar]
- 23.Kumar R, Mateo J, Smith AD, Khan KH, Ruddle R, Swales KE, et al. First-in-human, first-in-class phase 1 study of a novel oral multi-AGC kinase inhibitor AT13148 in patients (pts) with advanced solid tumors. J Clin Oncol. 2014;32:2554. [Google Scholar]
- 24.Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17:1556–68. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karvela M, Baquero P, Kuntz EM, Mukhopadhyay A, Mitchell R, Allan EK, et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy. 2016;12:936–48. doi: 10.1080/15548627.2016.1162359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Commisso C, Flinn RJ, Bar-Sagi D. Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protocols. 2014;9:182–92. doi: 10.1038/nprot.2014.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc Natl Acad Sci USA. 2012;109:5328–33. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelles J, Schnapp BJ, Sheetz MP. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331:450–3. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 29.Wolfenson H, Meacci G, Liu S, Stachowiak MR, Iskratsch T, Ghassemi S, et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat Cell Biol. 2016;18:33–42. doi: 10.1038/ncb3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Euhus DM, Hudd C, Laregina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–34. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 31.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–4. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 32.Ikenoya M, Hidaka H, Hosoya T, Suzuki M, Yamamoto N, Sasaki Y. Inhibition of rho-kinase-induced myristoylated alanine-rich C kinase substrate (MARCKS) phosphorylation in human neuronal cells by H-1152, a novel and specific Rho-kinase inhibitor. J Neurochem. 2002;81:9–16. doi: 10.1046/j.1471-4159.2002.00801.x. [DOI] [PubMed] [Google Scholar]
- 33.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, et al. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–9. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 34.Ichikawa K, Ito M, Hartshorne DJ. Phosphorylation of the Large Subunit of Myosin Phosphatase and Inhibition of Phosphatase Activity. J Biol Chem. 1996;271:4733–40. doi: 10.1074/jbc.271.9.4733. [DOI] [PubMed] [Google Scholar]
- 35.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, et al. Identification of a Proline-rich Akt Substrate as a 14-3-3 Binding Partner. J Biol Chem. 2003;278:10189–94. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 36.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, et al. Inhibitor hijacking of Akt activation. Nat Chem Biol. 2009;5:484–93. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massey AJ. Multiparametric Cell Cycle Analysis Using the Operetta High-Content Imager and Harmony Software with PhenoLOGIC. PLoS One. 2015;10:e0134306. doi: 10.1371/journal.pone.0134306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human Pancreatic Cancer Tumors Are Nutrient Poor and Tumor Cells Actively Scavenge Extracellular Protein. Cancer Res. 2015;75:544–53. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2017;23:235–41. doi: 10.1038/nm.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, et al. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res. 2011;71:747–57. doi: 10.1158/0008-5472.CAN-10-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grobelny D, Poncz L, Galardy RE. Inhibition of human skin fibroblast collagenase, thermolysin, and Pseudomonas aeruginosa elastase by peptide hydroxamic acids. Biochemistry. 1992;31:7152–4. doi: 10.1021/bi00146a017. [DOI] [PubMed] [Google Scholar]
- 42.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–83. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tozluoglu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat Cell Biol. 2013;15:751–62. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 44.Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak MA. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–4. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veettil MV, Kumar B, Ansari MA, Dutta D, Iqbal J, Gjyshi O, et al. ESCRT-0 Component Hrs Promotes Macropinocytosis of Kaposi's Sarcoma-Associated Herpesvirus in Human Dermal Microvascular Endothelial Cells. J Virol. 2016;90:3860–72. doi: 10.1128/JVI.02704-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rath N, Olson MF. Regulation of pancreatic cancer aggressiveness by stromal stiffening. Nat Med. 2016;22:462–3. doi: 10.1038/nm.4099. [DOI] [PubMed] [Google Scholar]
- 47.Xi Y, Niu J, Shen Y, Li D, Peng X, Wu X. AT13148, a first-in-class multi-AGC kinase inhibitor, potently inhibits gastric cancer cells both in vitro and in vivo. Biochem Biophys Res Commun. 2016;478:330–6. doi: 10.1016/j.bbrc.2016.01.167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.