Abstract
Diseases are characterized by distinct changes in tissue molecular distribution. Molecular analysis of intact tissues traditionally requires preexisting knowledge of, and reagents for, the targets of interest. Conversely, label-free discovery of disease-associated tissue analytes requires destructive processing for downstream identification platforms. Tissue-based analyses therefore sacrifice discovery to gain spatial distribution of known targets or sacrifice tissue architecture for discovery of unknown targets. To overcome these obstacles, we developed a multimodality imaging platform for discovery-based molecular histology. We apply this platform to a model of disseminated infection triggered by the pathogen Staphylococcus aureus, leading to the discovery of infection-associated alterations in the distribution and abundance of proteins and elements in tissue in mice. These data provide an unbiased, three-dimensional analysis of how disease affects the molecular architecture of complex tissues, enable culture-free diagnosis of infection through imaging-based detection of bacterial and host analytes, and reveal molecular heterogeneity at the host-pathogen interface.
INTRODUCTION
All diseases result in characteristic changes in the abundance and localization of proteins, small molecules, and other analytes within tissues. However, detection of these disease-associated molecular events in intact tissues is subject to two limitations. First, spatial resolution of analytes in tissue samples normally requires a priori knowledge of the target so that appropriate detection reagents can be applied. This is true for common assays such as immunohistochemistry, in which target detection depends on the availability of reagents, as well as predictions regarding localization and temporal expression. Conversely, discovery-based methods aimed at unbiased detection of macromolecules typically require destructive processing of the tissue specimen, thereby negating the ability to define analyte localization. These shortcomings are particularly problematic for diseases characterized by well-circumscribed lesions involving only a portion of an otherwise healthy tissue.
The inflammatory lesions that occur during invasive bacterial infection typify tissue pathology for which traditional probe-based and tissue-disruptive protocols pose challenges for delineating macromolecular changes. Abscesses are the hallmark lesions of the innate response to bacterial pathogens and consist of a bacterial “microcolony” surrounded by neutrophils (1). Because these lesions are inherently heterogeneous and bacterial pathogens occupy only a small fraction of the abscess, it has been difficult to determine the bacterial and host proteins that are localized to the host-pathogen interface.
The competition for metal between host and pathogen is one of the most important factors dictating the outcome of infection. Because metals are cofactors for essential biochemical reactions, they are vital nutrients for all forms of life. This requirement for nutrient metals is exploited by innate immune factors that sequester metals as a defense against infection in a process termed nutritional immunity (2, 3). Because metal acquisition is crucial for pathogen survival, a considerable amount of research has focused on the mechanisms by which microbes obtain, and hosts withhold, essential metals (4). However, it has been technically difficult to delineate infection-associated shifts in nutrient metal, particularly in heterogeneous lesions such as abscesses. Analysis of the bacterial-host interface in abscessed tissues, and the mechanisms by which pathogens obtain nutrients in this environment, therefore requires new approaches to drive discovery of fundamental processes in microbial pathogenesis.
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) and laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) are emerging technologies for imaging the distribution of proteins and elements in intact tissues, respectively (5–8). These technologies mitigate the limitations of both probe-based and discovery-based technologies in that they obtain data at discrete x,y coordinates throughout the tissue, thereby delineating the spatial distribution of analytes. When performed serially on adjacent tissue sections, these imaging modalities can generate a map of molecular abundance throughout a whole organ or animal (9). To successfully harness the power of MALDI IMS and LA-ICP-MS for molecular analysis of diseased tissues, the modalities must be precisely integrated in three dimensions with one another and with more traditional imaging of tissue architecture. Moreover, high resolution and sensitivity are needed to enable the detection of molecular alterations that occur in rare cell populations or that are present only in small portions of a complex tissue, such as abscesses. Finally, incorporation of bioluminescent imaging (BLI) permits any molecular alterations associated with disease to be analyzed in the context of gene expression, enabling conclusions to be drawn regarding how molecular heterogeneity affects bacterial or host responses.
Here, we describe the development of a high-resolution multimodality imaging platform for defining the impact of infection on the abundance and spatial distribution of host and bacterial molecules in intact infected tissues, and for delineating how these alterations affect bacterial gene expression in vivo. We focus specifically on the competition for metals between host and pathogen as a paradigm for innate immune responses to invading bacteria. By combining the imaging platform with proteomics, we identify host proteins involved in the innate responses to Staphylococcus aureus, as well as bacterial proteins that specifically mark the bacterial nidus within abscesses. In addition to providing a powerful new tool to examine host-pathogen interactions in intact tissue, this platform may ultimately have utility for the culture-free pathologic diagnosis of invasive infectious diseases. Moreover, this integration of powerful imaging modalities will enable study of many disease processes characterized by alterations in molecular abundance or distribution. Accordingly, molecular imaging has the potential to elucidate fundamental pathophysiologic processes during diverse disease states.
RESULTS
3D BLI reveals nutrient stress responses in tissue abscesses
To characterize bacterial-host interactions in abscessed tissue, we created a multimodal imaging platform and applied it to a murine model of disseminated S. aureus infection. We focused on the competition for iron as a paradigm for critical host-pathogen interactions. Iron is one of the most important factors determining the outcome of infection, and iron abundance serves as a powerful signal to pathogens heralding the invasion of host tissues (10). However, iron concentrations vary across host tissues, and to date, it has been technically challenging to determine the abundance and distribution of iron and other elements within a particular tissue.
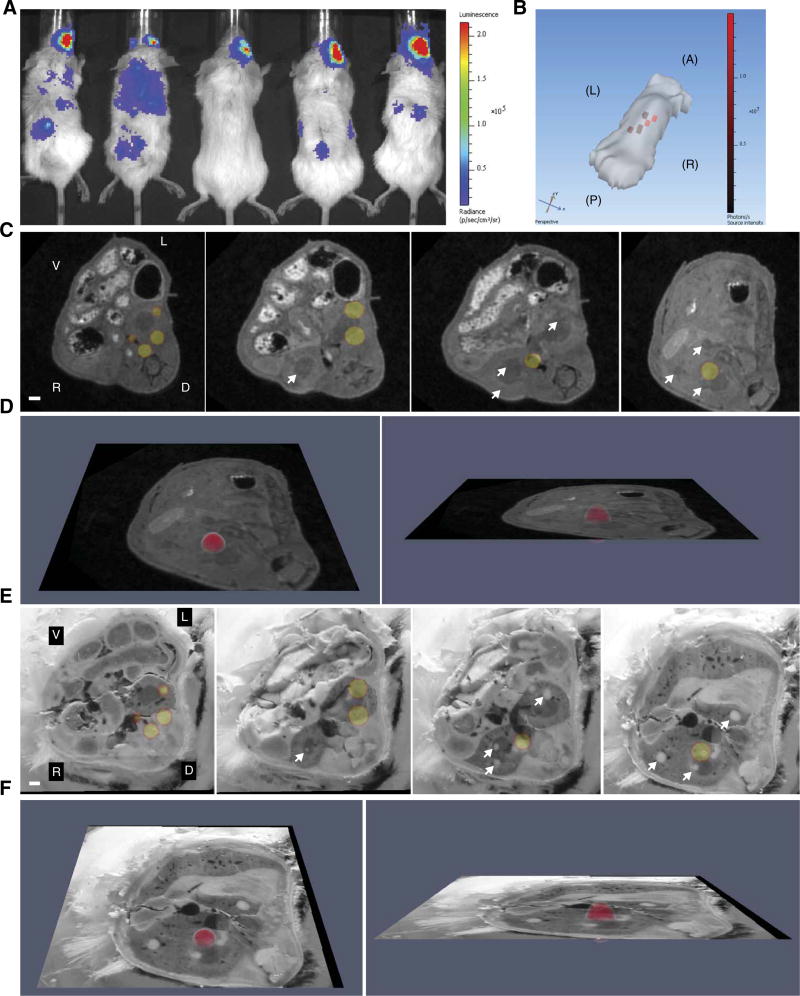
We hypothesized that S. aureus experiences iron starvation in host tissues, but the magnitude of iron starvation is heterogeneous and depends upon the bacterial niche within an organ. To test this hypothesis, mice were intravenously infected with a strain of S. aureus containing an iron-responsive luminescent construct (11). The infection was allowed to proceed for 96 hours, after which BLI was performed on the IVIS200 platform. Two-dimensional (2D) BLI revealed distinct puncta of light production, consistent with disseminated infection and bacterial iron starvation (Fig. 1A). One mouse was selected for 3D BLI analysis to more accurately determine the localization and depth of luminescent signal. 3D BLI effectively clarified the depth, anatomic location, and extent of infectious foci harboring iron-starved bacteria (Fig. 1B). Collectively, these data reveal that S. aureus experiences iron limitation during disseminated infection and define a BLI platform to evaluate other bacterial nutrient stress responses in vivo.
Fig. 1. Nutrient stress responses in murine tissue abscesses revealed by 3D BLI.
(A) BLI of mice (n = 5) infected 4 days before with S. aureus Newman pisdI.Xen1, in which the luxABCDE operon is under control of the iron starvation–responsive isdI promoter. Scale indicates luminescence in photons s−1 cm−2 sr−1. Luminescent signal in the right eye reflects the site of inoculation. (B) A surface topography of the leftmost mouse in (A) was generated using the IVIS200 3D platform. Note that the tail and limbs fail to generate a topographic image in this analysis. Red pixels correspond to the depth and location of bioluminescent signal. Scale indicates luminescence value in photons s−1. A, anterior; P, posterior; L, left; R, right. (C) Transverse MRI images of the torso (including the kidneys) of the leftmost mouse in (A), co-registered to the 3D BLI data. Four representative images are depicted. Arrows depict abscesses without luminescence. V, ventral; D, dorsal; L, left; R, right. White bar represents 1-mm scale. (D) The rightmost image in (C) tilted in the Z plane to demonstrate luminescence in 3D. (E) The mouse was sectioned transversely, and digital images of each section were obtained. This blockface image volume was co-registered to the 3D BLI data. Four representative images are depicted, corresponding to the four images in (C). Arrows depict abscesses without luminescent signal. White bar represents 1-mm scale. (F) The rightmost image in (E) tilted in the Z plane to demonstrate luminescence in 3D. See also movies S1 to S3.
Co-registration of BLI and magnetic resonance imaging defines heterogeneity in bacterial metal starvation within abscesses
BLI revealed distinct foci of light production consistent with disseminated infection by bacteria experiencing metal starvation. However, these experiments did not reveal if all bacteria within distinct tissue lesions experience equivalent metal stress, or if instead host-dependent metal starvation is heterogeneous. Given that bacteria express metal acquisition systems to combat host sequestration, it was hypothesized that not all bacteria in abscesses are metal-starved, and therefore, some abscesses would not exhibit bioluminescence. Testing this hypothesis required further definition of the tissue architecture using magnetic resonance imaging (MRI). Immediately after BLI, MRI was performed on the portion of the mouse encompassing the kidneys, a site of maximal pathology in the S. aureus disseminated infection model (movie S1) (12). Upon completion of MRI, the BLI data set was co-registered to MRI using an iterative closest point algorithm (13). The coordinated imaging data set revealed the presence of multiple abscesses, only a subset of which had bioluminescent signal (Fig. 1, C and D, and movie S2). These data demonstrate heterogeneity in bacterial metal starvation not only among different organs but also within distinct inflammatory lesions in the same tissue. Collectively, the integrated MRI and BLI data suggest that S. aureus is differentially metal-starved when growing in distinct abscesses within the same tissue.
Iterative blockface imaging and histochemical staining reveal the bacterial niche within abscesses
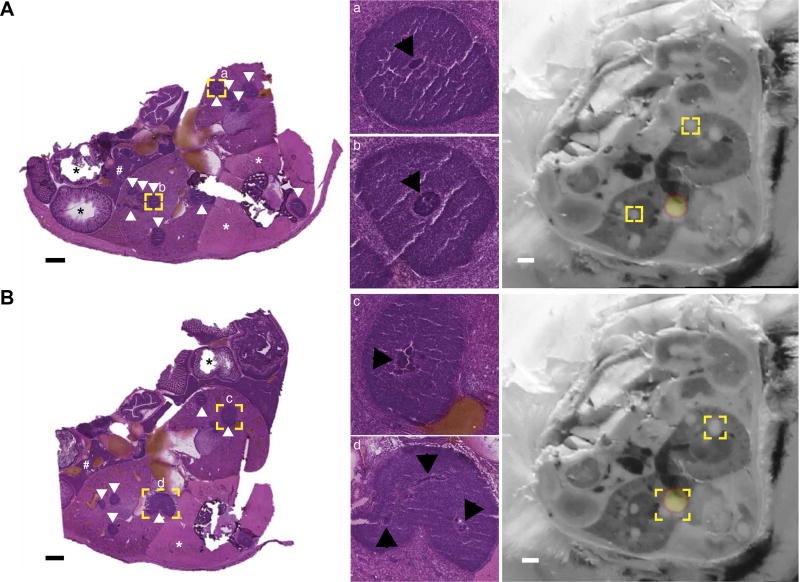
S. aureus exists as a microcolony in mature abscesses (1). Therefore, an alternative hypothesis for the observed lack of light production in abscesses may reflect the absence of suitable numbers of bacteria to reach the detection threshold for BLI. To test this possibility, direct visualization of bacteria within abscesses was necessary. Because this bacterial structure is not visible on MRI, additional analyses were performed to pinpoint the bacterial niche. At the completion of MRI, the infected mouse was euthanized and flash-frozen in a manner that minimized movement from the initial imaging position. The middle third of the mouse was then transversely sectioned at 30-µm increments to include both kidneys in their entirety. Adjacent sections were serially imaged with a high-resolution digital camera (a process henceforth referred to as blockface imaging) and then subjected alternatively to hematoxylin and eosin (H&E) staining or downstream imaging analyses. Blockface imaging of adjacent sections was archived as a volume and then manually co-registered to the BLI data set. Similar to MRI, co-registration of blockface images and BLI data revealed that only a subset of abscesses exhibited luminescence (Fig. 1, E and F, and movie S3). To determine whether this heterogeneity was due to alterations in metal-dependent gene expression or the absence of bacteria, adjacent tissue sections were stained with H&E and imaged under high resolution. Staphylococcal microcolonies could be visualized in many abscesses throughout the infected kidneys, including those without bioluminescent signal (Fig. 2). These results suggest that S. aureus experiences varying metal starvation in abscesses within the same organ despite a similar histopathologic appearance of the lesions. Co-registration of BLI, MRI, and blockface imaging data sets therefore provides a powerful system for analysis of the heterogeneous bacterial response to stress within the host.
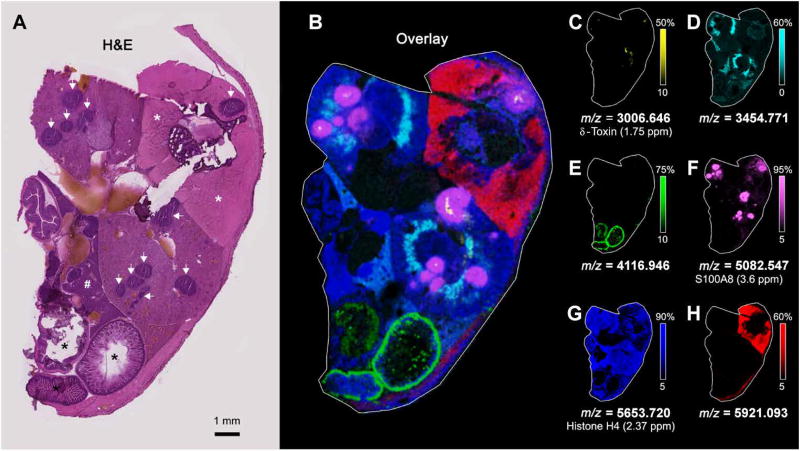
Fig. 2. Visualization of the bacterial nidus demonstrates equivalent bacteria across tissue abscesses.
Serial transverse sections of a mouse intravenously infected with S. aureus Newman pisdI.Xen1 were subjected to downstream imaging, reserved for future analyses, or stained with H&E. (A and B) Two representative H&E sections at 1× (left) and 10× (a + b), 6× (c), or 5× (d) zoom (middle). Black bars represent 1-mm scale. White arrowheads delineate tissue abscesses. Black asterisks indicate intestinal tissue. White asterisks denote paravertebral skeletal muscle. The symbol “#” denotes hepatic parenchyma. Yellow boxes indicate areas chosen for magnification, which are annotated with lowercase letters (a to d). Black arrowheads delineate the bacterial nidus within each abscess. The corresponding blockface images are shown at far right, with bioluminescent signal overlaid. The bioluminescent signal is depicted as a yellow sphere outlined in orange. Yellow boxes on the blockface images correspond to the same abscesses on H&E sections.
LA-ICP-MS characterizes inflammation-associated shifts in nutrient distribution
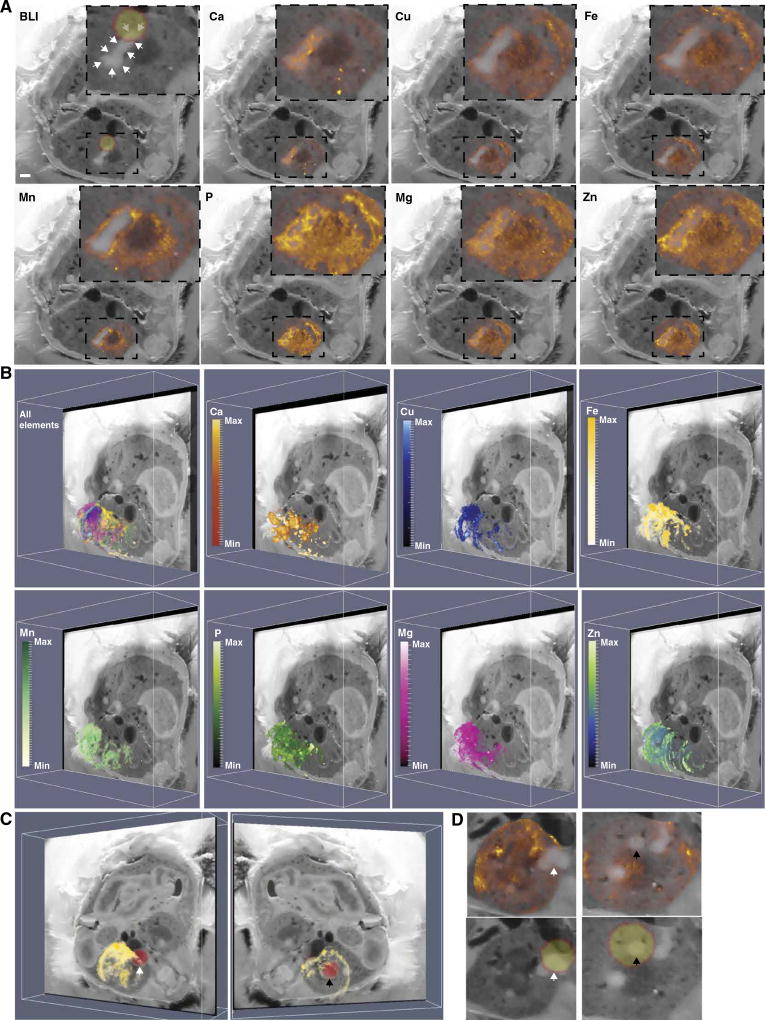
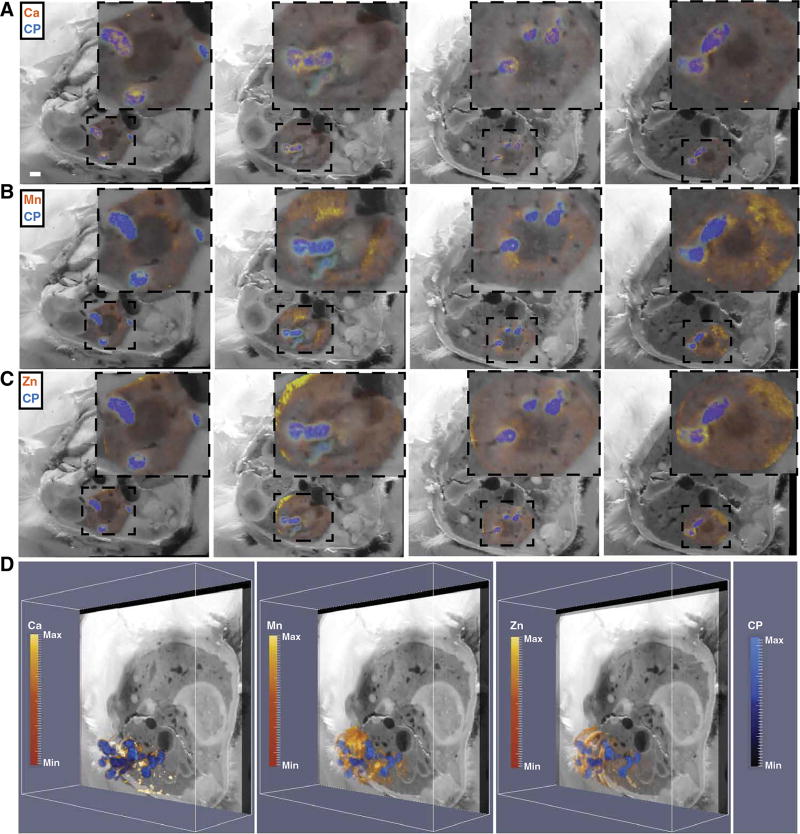
BLI, MRI, and blockface imaging revealed heterogeneity in metal starvation responses during staphylococcal infection. This observation suggests that iron, and perhaps other elements, is heterogeneously distributed throughout a given host tissue and both within and among abscesses. To more globally define shifts in element availability during staphylococcal infection, LA-ICP-MS was performed. LA-ICP-MS enables determination of the abundance and distribution of elements in a tissue section. Iterative LA-ICP-MS was performed on adjacent tissue sections to create a 3D volume of element abundance and distribution throughout the entire kidney. Distributions of calcium, copper, magnesium, manganese, phosphorous, zinc, and iron were determined, and the resultant element maps were co-registered to MRI and blockface volumes. These results demonstrated that abscesses are rich in calcium and relatively devoid of manganese, iron, and zinc (Fig. 3, A and B, and movie S4). Integration of LA-ICP-MS with MRI and blockface imaging also revealed that copper was largely excluded from abscesses. Conversely, magnesium and phosphorous showed a heterogeneous yet similar distribution within abscessed tissue (Fig. 3, A and B). Co-registration of the bioluminescent signals with LA-ICP-MS confirmed that bacterial iron starvation signals corresponded to areas relatively devoid of iron (Fig. 3, C and D, and movie S5). Collectively, the application of LA-ICP-MS to a murine model of S. aureus infection delineates shifts in element abundance and distribution across a whole organ in response to inflammation. These data corroborate the heterogeneous bacterial metal starvation responses depicted by BLI and demonstrate variability in element distribution not only within an entire organ but also within individual tissue lesions in response to infection.
Fig. 3. Characterization of inflammation-associated shifts in element distribution by LA-ICP-MS.
LA-ICP-MS performed on serial transverse sections of the right kidney from a mouse infected with S. aureus Newman pisdI.Xen1. (A) A representative blockface image illustrating element distribution and abundance surrounding an abscess. LA-ICP-MS data integrated with the blockface volume and co-registered images for BLI, calcium (Ca), copper (Cu), iron (Fe), manganese (Mn), phosphorous (P), magnesium (Mg), and zinc (Zn). The intensity of the color signal indicates relative abundance of the element in arbitrary units. White arrows outline the entire abscess under study. The bioluminescent signal is depicted as a yellow sphere outlined in orange. Insets: Enlarged images showing only the abscess and immediate surrounding tissue. White bar represents 1-mm scale. (B) The blockface volume oriented obliquely for each element throughout the kidney. Heat maps depict minimum and maximum threshold values in arbitrary units. Elements are grouped according to similar patterns. An integrated image displaying all elements co-registered to the blockface volume is shown in the top left panel. (C) The elemental imaging volume for iron (Fe) encompassing the infected right murine kidney is co-registered to the blockface and BLI images. Two different images representing rotation around the Y axis demonstrating iron distribution (in yellow) relative to the BLI signal (in red) for bacterial iron starvation are shown. White and black arrowheads depict corresponding abscesses in (D). (D) 2D planar views of the annotated abscesses in (C) with panels for BLI signal (yellow sphere outlined in orange) and Fe elemental imaging (orange heat map). White and black arrowheads depict corresponding abscesses in (C). See also movies S4 and S5.
To further validate these conclusions and determine whether element distribution and heterogeneity in the response to metal starvation change as abscesses develop, we conducted imaging analyses on 16 additional infected mice. To demonstrate the adaptability of this imaging platform to other reporter systems (fluorescence versus luminescence) as well as other metal starvation responses, we created two additional bacterial reporter constructs in which iron or manganese limitation drives green fluorescent protein (GFP) or red fluorescent protein (RFP) production, respectively. To determine how the development of infectious lesions over time alters bacterial responses and element distribution, we obtained kidneys from mice 4, 7, or 10 days after infection with each construct. Infected kidneys were immediately flash-frozen, cryosectioned, and then subjected to fluorescent imaging followed by LA-ICP-MS or H&E staining. The combination of fluorescent imaging and LA-ICP-MS again revealed heterogeneity of bacterial iron starvation responses relative to element distribution, both at 4 days after infection and at later time points (fig. S1A). Moreover, the bacterial responses to manganese limitation also varied from lesion to lesion in a manner largely dependent on element distribution in the infected organs (fig. S1B). Collectively, these data demonstrate that bacterial metal starvation responses are heterogeneous within infected organs at multiple time points after infection, and solidify the feasibility of the imaging platform to detect interlesional variation in bacterial stress responses and host element redistribution.
MS imaging delineates bacterial and host proteins at the infectious interface
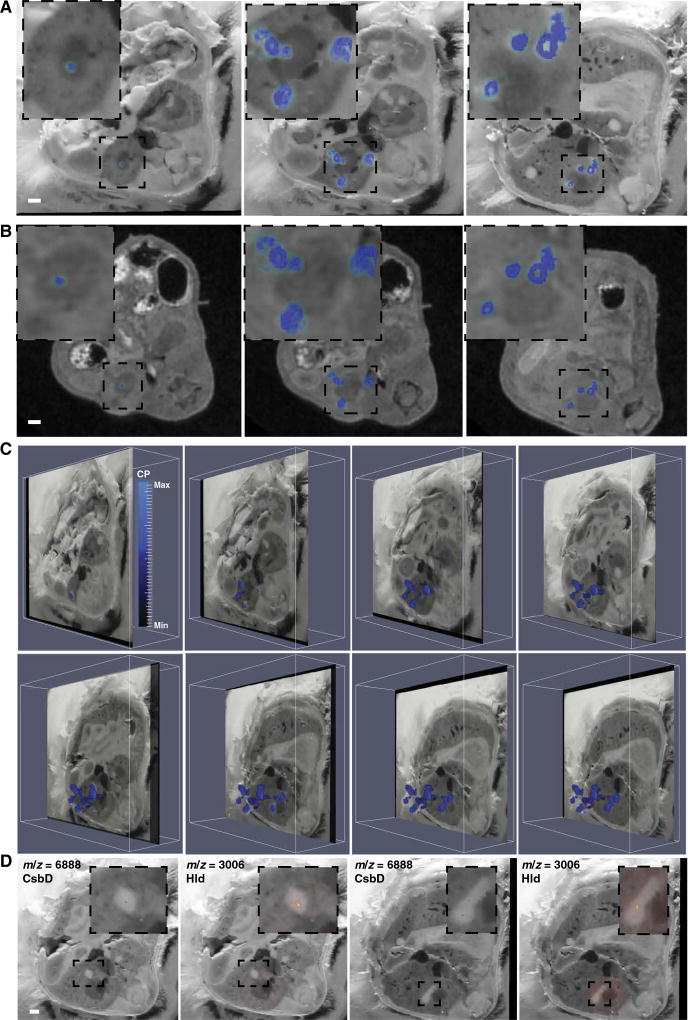
MRI and blockface imaging created 3D volumes to which additional imaging modalities could be co-registered, providing a detailed anatomic backdrop for discovery-based analyses. Co-registration of BLI with each anatomic volume revealed heterogeneity in bacterial starvation responses, despite the homogeneous appearance of tissue abscesses on MRI, blockface imaging, and histopathology. To identify host and bacterial proteins involved in these stress responses, we performed MALDI IMS on serial sections of the infected mouse. The objectives of this approach were to define the host proteins that respond to staphylococcal infection, to determine whether abscesses in the same tissue demonstrate molecular heterogeneity, and to identify staphylococcal proteins that mark the bacterial niche in infected tissues. MALDI IMS detected numerous analytes localized to tissue abscesses, including one protein at m/z (mass/charge ratio) 10,164 that was identified as the S100A8 subunit of calprotectin by subsequent proteomic analysis (Fig. 4, A to C, and movies S6 to S8). Calprotectin is a heterodimeric protein that is abundant in neutrophils and contributes to nutritional immunity by binding the essential nutrient metals zinc and manganese (2, 14–18). Two additional analytes (m/z 3006 and m/z 6888) were of particular interest given that they localized to the center of abscesses in a pattern that seemed to colocalize with the staphylococcal microcolony (Fig. 4D). High-resolution MALDI IMS analysis of an entire transverse section of the infected mouse identified these analytes in multiple abscesses in different organs and provided further anatomic detail through the delineation of tissue-specific signals (Fig. 5).
Fig. 4. MS imaging delineates bacterial and host proteins at the infectious interface.
MALDI IMS performed on serial transverse sections of the region of tissue encompassing the right kidney in a mouse infected with S. aureus 4 days before. A spectral peak at m/z = 10,164, subsequently identified as the S100A8 subunit of the heterodimeric protein calprotectin, colocalized with abscesses when MALDI IMS data were co-registered to blockface imaging (A) or MRI (B). Three representative sections are shown. Insets: Enlarged image showing only the right kidney. White bars represent 1-mm scale. (C) Obliquely oriented serial transverse blockface images with overlaid MALDI IMS data to depict the 3D orientation of S100A8 (m/z = 10,164). Heat map depicts minimum and maximum threshold values in arbitrary units. (D) Blockface images overlaid with MALDI IMS for m/z = 3006 (subsequently identified as δ-hemolysin) and m/z = 6888 (subsequently identified as CsbD-like protein), which are bacterial proteins colocalizing to the nidus within an abscess. Two representative blockface sections are shown. White bar represents 1-mm scale. Insets: Enlarged images showing only the abscess and immediately surrounding tissue. See also movies S6 to S8.
Fig. 5. Molecular histology of tissue microanatomy by MALDI IMS.
(A) H&E mid-thoracic transverse section of a mouse infected with S. aureus 4 days before. White arrows delineate tissue abscesses. Black asterisks indicate intestinal tissue. White asterisks denote paravertebral skeletal muscle. The symbol “#” denotes hepatic parenchyma. (B) MALDI IMS performed on an entire transverse mouse section, corresponding to the H&E section in (A), using a Fourier transform ion cyclotron resonance (FTICR) MS. (C) A spectral peak at m/z 3006.646, localized to the bacterial nidi within several abscesses, was subsequently identified as δ-hemolysin. Additional spectral peaks are overlaid to highlight different regions of the transverse mouse section, including (D) m/z 3454.771 in the outer medulla of the kidneys, (E) m/z 4116.946 in the gastrointestinal tract, (F) a spectral peak at m/z 5082.547 that localizes to the abscess regions and was subsequently identified as the doubly charged form of the S100A8 subunit of the heterodimeric protein calprotectin, (G) m/z 5653.720, subsequently identified as histone H4, in the kidneys and liver, and (H) m/z 5921.093 in muscle. All proteins are in the 2+ charge state.
Given the exciting possibility that these signals could represent bacterial proteins and therefore provide a mechanism for label-free identification of S. aureus in intact tissues, we performed proteomics on tissues from additional infected mice. To generate larger abscesses and facilitate protein identification, infection of additional mice was allowed to proceed for 9 days, after which time the kidneys were removed and processed for proteomics. This approach identified m/z 6888 as S. aureus NWMN_0783, a CsbD-like superfamily protein, and m/z 3006 as S. aureus NWMN_2624 encoding the virulence factor δ-hemolysin. To further confirm the identity of NWMN_0783, mice were infected with wild-type (WT) S. aureus or a strain inactivated for NWMN_0873. At 4 days after infection, the kidneys were removed and subjected to MALDI IMS. A signal at m/z 6888 was only detected in mice infected with WT, not the strain inactivated for NWMN_0873. The role of NWMN_0783 in staphylococcal physiology is unknown, but CsbD-like proteins have been proposed to have roles in stress responses and metal starvation (19, 20). To further confirm the identity of m/z 3006 as NWMN_0873 encoding δ-hemolysin, mice were infected with WT S. aureus or a strain inactivated for δ-hemolysin. However, mice infected with the S. aureus strain inactivated for δ-hemolysin produced abscesses that were too small to be analyzed by MALDI IMS. We therefore grew WT S. aureus or a strain inactivated for δ-hemolysin overnight in vitro and collected bacterial pellets for protein extraction and MS/MS analysis (21). MS/MS analysis of the S. aureus strain inactivated for δ-hemolysin revealed a loss of signal at m/z 3006, further confirming the identity of this peptide. δ-Hemolysin is a broadly conserved virulence factor in S. aureus, capable of lysing host cells and contributing to mortality during septicemia (21–23). Collectively, these data reveal the power of MALDI IMS, when part of a multimodality imaging platform, to identify host and bacterial proteins involved in the pathogenesis of invasive infection.
To investigate how host and bacterial protein distribution and abundance change over time in response to infection, we performed MALDI imaging on kidneys from 16 additional mice infected for 4, 7, or 10 days (fig. S2). These experiments demonstrated the presence of S. aureus δ-hemolysin and the CsbD-like protein in the centers of abscesses from multiple mice at multiple time points, further solidifying the conclusion that MALDI imaging of bacterial proteins could potentially facilitate culture- and label-free microbiologic diagnoses in intact tissues. Furthermore, these experiments demonstrated analytes that change in distribution over time during the course of infection and abscess development (fig. S2). Of particular interest was an analyte at m/z 5047, which had a homogeneous distribution in infected kidneys at days 4 and 7 after infection, but then coalesced around the periphery of abscesses by day 10. Collectively, these data demonstrate the power of multimodal imaging to investigate lesional dynamics during invasive S. aureus infection.
Integration of MALDI IMS and LA-ICP-MS enables a molecular map of the struggle for metal between host and pathogen
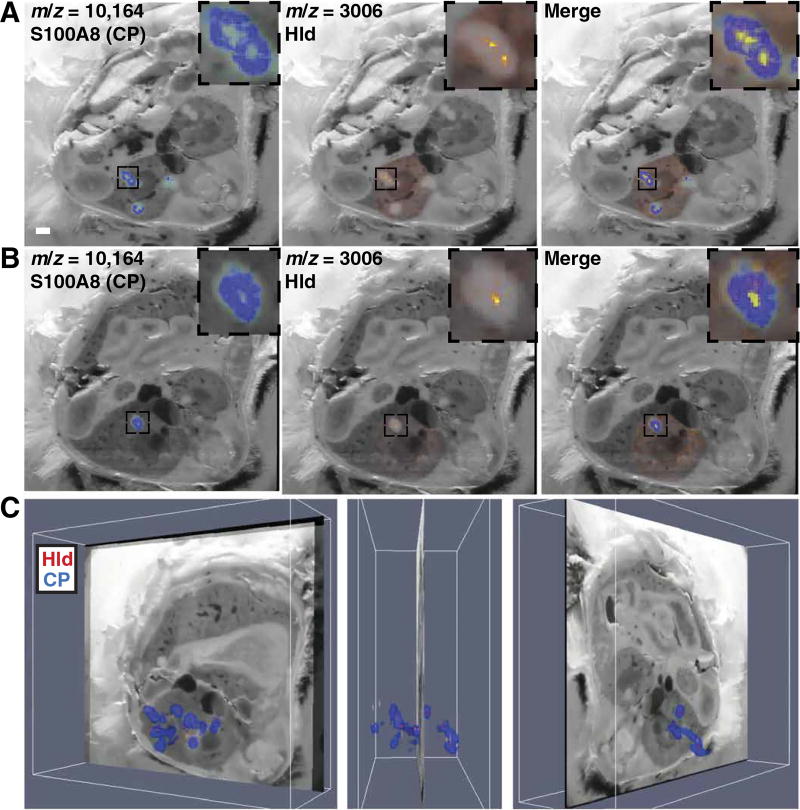
Co-registration of MALDI IMS and LA-ICP-MS further characterized the host-pathogen interface and revealed protein signals that are spatially associated with metal distribution. Calprotectin, a known calcium-binding protein, generally colocalized with calcium throughout the infected organ (Fig. 6A and movie S9) (16). Conversely, the distribution of calprotectin was distinct from that of manganese and zinc, suggesting the possibility that metal-bound calprotectin is preferentially removed from abscesses or, alternatively, that other metal-sequestering proteins participate in element trafficking in abscesses (Fig. 6, B to D) (14). In areas with a strong bacterial-specific protein signal (m/z 3006), there was a paucity of calprotectin signal (Fig. 7, A to C, and movie S10). The segregation of bacterial and calprotectin protein signals suggests mechanisms of calprotectin destruction or exclusion from the staphylococcal microcolony in vivo, providing a partial explanation for how bacteria persist in the presence of an antibacterial protein (14, 15, 24–26). Together, these data reveal the power of integrated MALDI IMS and LA-ICPMS for delineation of element and macromolecular abundance in intact tissues, and provide a molecular map of the competition for nutrients between host and pathogen.
Fig. 6. Integrating MALDI IMS and LA-ICP-MS reveals spatial orientation of the metal-binding host protein calprotectin in relation to nutrient metals in infected mouse tissues.
(A to C) Four representative blockface images (left to right) with integration of MALDI IMS signal for the S100A8 subunit of calprotectin (CP) (m/z = 10,164, blue) and LA-ICP-MS signal (orange-yellow) for calcium (Ca; A), manganese (Mn; B), and zinc (Zn; C). Insets: Enlarged images of the kidney and immediate surrounding tissue. (D) The MALDI IMS imaging volume for calprotectin encompassing the infected right kidney was co-registered to the LA-ICP-MS imaging volume for Ca, Mn, or Zn, displayed obliquely to delineate calprotectin and element distribution throughout the kidney. Heat maps depict minimum and maximum values in arbitrary units. See also movie S9.
Fig. 7. Analysis of the bacterial nidus within tissue abscesses by MALDI IMS demonstrates a paucity of calprotectin signal.
(A and B) Two representative blockface images co-registered to MALDI IMS data for S100A8 (left), δ-hemolysin (middle), or both signals (right). White bar represents 1-mm scale. (C) Co-registration of MALDI IMS data for both S100A8 (blue) and δ-hemolysin (red) with blockface imaging across the entire infected kidney from one mouse infected with S. aureus 4 days before. Three representative images are shown. See also movie S10.
DISCUSSION
Here, we describe an imaging platform that leverages MS for the delineation of macromolecular and element distribution in intact tissues. Through the integration of MALDI IMS, LA-ICP-MS, BLI, MRI, and blockface imaging, this platform achieves whole-organ mapping of protein and metal abundance at high resolution, reveals intrinsic heterogeneity of inflammatory responses, and enables analyses of bacterial transcriptional responses. Together, this workflow provides a technological framework for an integrated, 3D imaging platform capable of characterizing disease-associated alterations in macromolecular abundance and distribution in intact tissues. To demonstrate broad utility in characterizing heterogeneous tissue pathology, we applied the imaging platform to a model of disseminated staphylococcal infection. Multimodality imaging successfully depicted the distribution and abundance of antibacterial host proteins in S. aureus–infected tissues while also enabling the detection of a bacterial virulence factor that marks the intra-abscess niche. A major discovery stemming from this platform is that abscesses within the same organ have heterogeneous molecular architectures despite having a homogeneous appearance by traditional imaging. Consistent with this observation, bacterial pathogens also exhibit heterogeneous stress responses both within and between tissue lesions.
Integration of LA-ICP-MS into the imaging workflow enabled mapping of element abundance throughout an entire organ and illustrated infection-associated shifts in nutrient metal. Consistent with previous studies, tissue abscesses were rich in calcium yet relatively devoid of manganese, iron, and zinc (14). Here, we also discovered that the antimicrobial element copper is effectively excluded from abscesses. Moreover, the resolution achieved by the integrated imaging platform allowed for identification of distinct “microdomains” of calcium, phosphorous, and manganese within an abscess. The data therefore define heterogeneity in elemental nutrients not only among tissue types but also within an individual inflammatory lesion. Although calprotectin colocalized with calcium, it did not strictly colocalize with Mn and Zn, which may suggest that metal-loaded calprotectin is preferentially removed from the abscess or, alternatively, that other metal-sequestering proteins participate in Mn, Zn, Fe, and Cu exclusion from the abscess.
BLI revealed that not all bacteria within abscesses experience equivalent metal limitation, corroborating the patterns of metal distribution observed by LA-ICP-MS. These findings are particularly relevant to the field of microbial pathogenesis, in which nutrient sufficiency and bacterial stress responses are often assessed using transcript or protein abundance from whole-organ homogenates, or using fluorescent reporters that lack the resolution to clarify intralesional heterogeneity. By performing LA-ICP-MS on an entire organ, we provide a new platform for tracking disease-associated shifts in element abundance. The addition of MALDI IMS to elemental imaging enables further characterization of bacterial and host stress responses to shifting element abundance. Moreover, traditional luminescent and fluorescent-based probes can then be integrated into the imaging platform to provide a balance of targeted and discovery-based applications.
We used multimodality imaging to characterize organ-level element and macromolecular alterations in response to disseminated bacterial infection. This imaging platform is readily adaptable to any disease process in which protein or element distribution is perturbed. The results show that this imaging platform can be used not only with BLI but also with fluorescent reporters, both of which can be adapted to the study of several different bacterial nutrient stress responses. This imaging approach could be particularly useful for the analysis of diseases characterized by other discrete lesions, such as solid tumors, in which the heterogeneity within the lesion itself and molecular responses at the interface of healthy and diseased tissue can be assessed. However, the imaging workflow is inherently limited by the requirement for specialized resources such as small-animal imaging equipment, MALDI IMS and LA-ICP-MS instruments, and computational resources for co-registration of imaging modalities. Furthermore, before translation into clinical practice, the imaging platform should be tested in additional animal models with different microbial pathogens and then analyzed in the context of human biopsy specimens. Although this multimodal imaging technology could ultimately enable culture-free diagnosis of microbial pathogens in infected tissues, such an approach would, by definition, require invasive procedures to procure biopsy specimens. Nevertheless, MS-driven, multimodality imaging enables successful delineation of host and bacterial proteins in intact tissues while also detailing element abundance across an entire organ. Collectively, this technology is a promising new research tool for the label-free identification of tissue pathology characterized by altered protein or element abundance.
MATERIALS AND METHODS
Study design
The objective of this study was to delineate bacterial-host interactions in murine tissue sections through the application of a multimodal imaging platform. We hypothesized that individual inflammatory lesions within infected tissues would demonstrate heterogeneity in bacterial and host protein abundance and localization, as well as element distribution. We also hypothesized that multimodal MS imaging could facilitate the culture-free diagnosis of invasive infection in intact tissues through the label-free in situ detection of bacterial proteins. To test this, we performed 3D bioluminescent, magnetic resonance, digital blockface, and MS imaging of a single female BALB/c mouse aged 7 weeks. To further address infection dynamics and demonstrate adaptability of the imaging platform to different bacterial reporters, we also performed imaging analysis on 16 additional mice at various times after infection. Given the nature of the study, sample size calculations and statistical analyses were not relevant in the animal studies. All experiments involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Vanderbilt University. All experiments were performed according to National Institutes of Health (NIH) guidelines, the Animal Welfare Act, and U.S. Federal law. At the completion of imaging procedures, all imaging data were co-registered to either the MRI or digital blockface volume to provide a detailed anatomic backdrop for the analysis of host and bacterial analyte distribution.
Reagents
Acetic acid, trifluoroacetic acid (TFA), 2,5-dihydroxyacetphenone (DHA), and 2-2-tribromoethanol (Avertin) were purchased from Sigma-Aldrich Chemical Co. High-performance liquid chromatography (HPLC)–grade acetonitrile (ACN), ethanol, and chloroform were purchased from Thermo Fisher Scientific.
Bacterial strains and growth conditions
The S. aureus clinical isolate Newman served as the WT genetic background for all experiments unless otherwise noted. Newman pisdI.Xen1 was previously described (11). A Newman csbD transposon mutant was created by φ85-mediated transduction of the csbD::erm allele from strain NE946, which was obtained from the Network on Antimicrobial Resistance in S. aureus transposon library at BEI Resources. A S. aureus strain inactivated for δ-hemolysin was provided by M. Otto (21). Bacteria were grown in tryptic soy broth (TSB) at 37°C with shaking at 180 rpm. Chloramphenicol was added to the media at a final concentration of 10 µg/ml where indicated. To prepare an inoculum for the disseminated infection model, overnight cultures were back-diluted 1:100 and grown for an additional 3 hours, after which time the bacteria were harvested by centrifugation and washed in phosphate-buffered saline (PBS). Washed bacteria were resuspended to a final concentration of about 5 × 108 colony-forming units (CFU) per milliliter and kept on ice until used for retro-orbital injection.
Construction of S. aureus fluorescent reporter strains
The sequence for codon-optimized gfp was amplified from pGFP-F (27) using primers 5′-ACGATCCGGGGAATTCCATATGTTATTTGTAGAGCTCATCCATGCCATG-3′ and 5′-CTTGTTTGGATCCTCGAGGGATCCATGCCCGGGAGCAAAGGA-3′ and subsequently assembled using New England Biolabs (NEB) Gibson assembly into pOS1 that was digested with Nde I and Bam HI. PisdA was amplified from S. aureus Newman genomic DNA using primers 5′-GCCGAAGAATTCCAAAACATAATCCTCCTTTTTATG-3′ and 5′-GCCAAGCATATGGTTGTTTTCCTCCTAAGGATACAA-3′ and was ligated into pOS1 gfp after digestion with Eco RI and Nde I to produce pOS1 PisdAgfp. This plasmid was subsequently confirmed by sequencing. The reporter’s fluorescent response to iron starvation was confirmed in vitro by measuring fluorescent intensity after growth in TSB supplemented with chloramphenicol (10 µg/ml) and the iron chelator 2,2-dipyridyl (1 mM) or an equal volume of ethanol (vehicle).
The sequence for codon-optimized rfp was amplified from pRFP-F (27) using primers 5′-GTATAAACGTCATATGATGGATAATACAGAAGATGTTATTAAAG-3′ and 5′-AAACACTACCCCCTTGTTTGGATCCTTATAAAAACAAATGATGACGAC-3′. PmntA was amplified from S. aureus Newman genomic DNA using primers 5′-AAAGACGATCCGGGGAATTCACTTTCACCTCACATACATTGTCTA-3′ and 5′-CATCTTCTGTATTATCCATCATATGACGTTTATACCTCCTAATTAAAAGTTTAGGCTAAC-3′. The fragments were assembled into pOS1 plasmid that was digested with Eco RI and Bam HI using NEB HiFi Assembly to form pOS1 PmntArfp. The resulting plasmid was confirmed by sequencing. The reporter’s fluorescent response to low manganese conditions was confirmed in vitro by measuring fluorescent intensity after growth in TSB supplemented with chloramphenicol (10 µg/ml) and 60% calprotectin buffer [100 mM NaCl, 3 mM CaCl2, 5 mM β-mercaptoethanol, and 20 mM tris (pH 7.5)] containing calprotectin (250 µg/ml), calprotectin + 10.4 µM MnCl2 (250 µg/ml), or an equal volume of calprotectin buffer (vehicle) in 96-well black-walled, clear-bottom plates.
Murine model of S. aureus–disseminated infection
Disseminated infection was induced in 6- to 8-week-old BALB/cJ (imaging) or C57BL6/J (proteomic identification) female mice by intravenous injection of S. aureus. Briefly, mice were anesthetized with 2-2-tribromoethanol (Avertin) and then subjected to retro-orbital injection of 5 × 107 CFU of S. aureus in 100 µl of PBS. Upon recovery from anesthesia, the mice were returned to the animal housing facility and provided food and water ad libitum. At 4, 7, or 10 days after infection, the mice were subjected to imaging procedures. One infected mouse was used for the full 3D workflow, whereas 16 additional mice were infected with either of two fluorescent reporters (eight mice each) to examine lesional dynamics and establish the feasibility of using additional reporters in the workflow. For proteomic identification of proteins imaged by MALDI IMS, a separate group of mice was infected and analyzed after 9 days.
Bioluminescent imaging
BLI was performed using an IVIS 200 optical imaging system. Mice were anesthetized with 2% isoflurane, shaved, secured to a rigid restraint device, and placed in the imaging chamber of the IVIS 200 system. Spectral imaging was obtained by imaging bacterial luciferase emission through four 20-nm band-pass filters at wavelengths from 560 to 620 nm. The surface topography of the mouse was generated using a structured light image. Tomographic source reconstruction was performed using the Living Image Software 3D Analysis software package. The emission spectrum of bacterial luciferase and tissue optical properties of muscle were used for reconstruction. An emission lower threshold of 2000 photons s−1 was used. Final reconstructed source voxel size was 2 mm.
Fluorescent imaging
Fluorescent imaging was performed using an IVIS Spectrum system for direct imaging of flash-frozen, sectioned (20 µm) kidneys mounted on glass, indium tin oxide–coated float glass slides (Delta Technologies) or poly-l-lysine–coated vinyl slides to facilitate downstream imaging analyses (H&E staining, MALDI IMS, and LA-ICP-MS, respectively) on serial tissue sections. Tissue sections from mice infected with the PisdAgfp construct were imaged with an excitation/emission filter pair of 465/520 nm. Tissue sections from mice infected with the PmntArfp construct were imaged with an excitation/emission filter pair of 570/620 nm. Photographic images were also obtained with a subject height of 0.12, binning of 2, and F/stop of 8. Frozen slides were allowed to equilibrate to about 20°C before imaging and then subsequently frozen again before MALDI IMS or LA-ICP-MS imaging. Fluorescent images were automatically thresholded using Living Image Software and displayed on “yellow hot” (manganese reporter) or “green” (iron reporter) colorimetric heat maps.
Magnetic resonance imaging
After BLI, mice were maintained under anesthesia in the rigid restraint device and carefully transported to the MRI area. Mice were subsequently secured in the prone position in a 38–mm–inner diameter radiofrequency (RF) coil and placed in a Varian 9.4T horizontal bore imaging system for data collection. Respiration rate and internal body temperature were continuously monitored. A constant body temperature of 37°C was maintained using heated airflow. Scout images were collected in all three imaging planes for localization of the kidneys, using a gradient echo sequence with repetition time (TR) of 75 ms, echo time of 5 ms, slice thickness of 2 mm, flip angle of 35°, and an average of four acquisitions. Additional parameters include field of view (FOV) of 32 mm × 32 mm and data matrix of 128 × 128.
After localization of the kidneys, 3D multiple gradient echo imaging data were acquired with FOV of 25.6 mm × 25.6 mm × 25.6 mm, TR of 40 ms, echo spacing of 1.7 ms, echo train length of 7, flip angle of 25°, data matrix of 128 × 128 × 128, with four acquisitions per phase encode step, for a total acquisition time of about 43 min. The acquired data were zero padded and reconstructed using MATLAB 2014a at 256 × 256 × 256 matrix using an inverse Fourier transform, resulting in a nominal isotropic resolution of 100 µm. The mice were euthanized with a 5% isoflurane overdose after imaging and then submerged for about 30 s in liquid nitrogen. Care was taken to avoid movement of mice within the rigid restraint device. The samples were then stored in a −80°C freezer until removed for sectioning, blockface imaging, and MALDI IMS/LA-ICP-MS.
Preparation of samples for IMS
The frozen mouse body was manually truncated by removing both the head and tail regions, leaving a section of the torso encompassing both kidneys. The posterior side was affixed to a cryostat chuck with optimal cutting temperature polymer. Transverse 30-µm-thick sections were acquired throughout the length of the kidney using a Leica CM 1900 cryostat. Blockface digital images were acquired after every section using a digital camera (Canon EOS 60D SLR digital camera). Sectioning was performed by collecting serial tissue sections in batches of five onto the following types of slides (in order, labeled A to E): (A) gold-coated stainless steel plate for MALDI IMS, (B) glass slide for histological analysis, (C) nitric acid–washed poly-l-lysine–coated vinyl slides (Electron Microscopy Sciences) for LA-ICP-MS imaging, (D) gold-coated stainless steel plate for MALDI IMS backup, and (E) nitric acid–washed poly-l-lysine–coated vinyl for LA-ICP-MS imaging backup. This set of five serial tissue sections (A to E) constitutes a single “voxel” in 3D image reconstruction. Because the tissue was sectioned at 30-µm thickness, a single voxel transversed 150 µm, resulting in a z-dimension spatial resolution of 150 µm. A total of 240 sections were ultimately collected, or 48 voxels. For the MALDI analyses, two sections were collected onto a single gold plate (voxel 1, section A and voxel 2, section A were collected together).
Sections obtained for MALDI analysis (A and D) were thaw-mounted on gold-coated stainless steel MALDI target plates for 30 s. All plates for MALDI IMS analyses were washed to remove interfering lipids and salts as previously described (28, 29). Briefly, sections were sequentially washed in 70% ethanol for 30 s, 100% ethanol for 30 s, Carnoy’s fluid (60% ethanol, 30% chloroform, and 10% acetic acid) for 2 min, 100% ethanol for 30 s, water for 30 s, and 100% ethanol for 30 s. The plates were allowed to dry before being placed in petri dishes, flooded with nitrogen gas, sealed, and placed in a −80°C freezer until analysis.
The slides for histological analysis (B) were stained with H&E. Optical images of stained sections were obtained at ×20 magnification using a Leica SCN400 Brightfield Slide scanner. Sections obtained for LA-ICP-MS analysis (C and E) were thaw-mounted onto nitric acid–washed poly-l-lysine–coated vinyl slides as previously described (29–31). Slides were placed in a slide box and stored at −80°C until analysis.
LA-ICP-MS imaging
Trace element imaging was performed as previously described (30, 31). Briefly, samples were ablated using the LSX-213 laser ablation system and analyzed using a coupled Element 2 high-resolution sector field ICP-MS. Vinyl slides were placed into a sealed chamber and were ablated in multiline mode (line by line) at a rate of 30 µm/s using a focused Nd:YAG laser beam (213 nm). The laser had a spot size of 100 µm with a laser shot frequency of 5 Hz. Only the region encompassing the target kidney (right) was ablated. Helium gas was used to assist in laser ablation of the sample and to transport ablated sample particles online from ablation chambers into the ICP-MS. Particles in helium were mixed with argon gas at a flow rate of ~1 liter/min before online introduction to the ICP-MS. Flow rates of both helium and argon gas were tuned and optimized for each sample to obtain the same signal response with SRM 612 standard from the National Institute of Standards and Technology (NIST). The mass spectrometer was operated in medium-resolution mode.
Matrix-assisted laser desorption/ionization imaging mass spectrometry
Plates for MALDI IMS were removed from the −80°C freezer and allowed to warm up to room temperature for 30 min before opening the sealed petri dish to minimize water condensation on the samples. After warming, digital images of each plate were acquired on a desktop scanner at 2400 dots per inch. DHA MALDI matrix was prepared at a concentration of 15 mg/ml in 90% ACN with 0.2% TFA. Matrix was applied to the plate robotically using TM-Sprayer. The sprayer was operated at a flow rate of 0.2 ml/min with a 10-psi nitrogen flow gas and a nozzle temperature of 85°C. Samples were sprayed at a velocity of 1100 mm/min and a track spacing of 2 mm. Six total spray passes were applied to sections, rotating and offsetting every other pass by 1 mm to ensure even coverage of matrix. Just before MS analysis, the plates were rehydrated by first heating the plate to 37°C in a laboratory oven for 2 min, followed by 3 min at 37°C in a petri dish with 1 ml of 50 mM acetic acid (28, 29).
Sections for 3D analysis were analyzed on a MALDI TOF MS (SimulTOF 200 Combo) in positive ion linear mode. This system is equipped with a 349-nm, diode-pumped, frequency-tripled Nd:YLF laser. Ion images were acquired in typewriter mode (acquisition in one lateral direction only) using continuous raster sampling at a 50-µm calculated horizontal spatial resolution and a 50-µm vertical step (28, 32, 33). Only the region encompassing the right kidney was analyzed. Sections were analyzed using 2-kHz laser repetition rate, stage speed of 2 mm/s, and 50 hardware averages for a 40-Hz digitizer acquisition rate. The effective pixel rate is slightly slower because of both the time required to return the stage to the beginning of the next raster line and the built-in delay to flush acquired spectra from the acquisition card to the hard disk at the end of each line scan. Over the 48 plains of analysis, 930,398 pixels were acquired in 22.2 hours of instrument time (for an effective pixel rate of ~11.6 Hz).
After data acquisition, all spectra were preprocessed for baseline subtraction, intensity normalization, and m/z alignment. All 48 individual 2D IMS data sets were concurrently loaded into MATLAB to establish a single 3D IMS data set. Each 2D data set consists of spectra that contain around 30,000 m/z bins, each describing a range from m/z 600 to 49,000. Given that each 2D IMS experiment reports its own m/z axis, and subsequent preprocessing algorithms need to approach the spectra that were collected over all cutting depths as a single (3D) IMS data set with a single m/z axis, the spectra of all 2D data sets were resampled to a single consensus m/z axis. The msresample() function from the Bioinformatics Toolbox of MATLAB was used to resample uniformly. To reduce the size and dimensionality of the overall data set, this step was also used to down-sample the number of bins per spectrum to 7944. The resampling step ensures that the spectra collected across different 2D experiments can be directly compared along the chemical (m/z) domain. To ensure that ion intensity values can be compared along the (inter-experiment) z dimension as well as along the (intra-experiment) x and y dimensions, all spectra across the different cutting planes were preprocessed as one all-encompassing data set. Although this induces an increased computational and memory resource cost over preprocessing 2D IMS data serially on a plane-by-plane basis, it makes 3D ion volumes more robust and less susceptible to cutting plane-to-plane signal variation. The preprocessing steps entail normalization, baseline correction, m/z alignment, and peak picking. The spectra were normalized using common ion current as a reference, disregarding differential peaks (34). Baseline correction was accomplished using a spline approximation of the baseline at the 0.1 quantile of ion intensities per spectrum, using a window size of 500 and a step size of 250. Furthermore, the spectra were aligned along the m/z axis to reduce peak drift, allowing a maximum m/z shift of 60. These steps were implemented using the msbackadj() function and the msalign() function provided by the Bioinformatics Toolbox of MATLAB. The subtraction of the baseline estimate can sometimes result in small negative intensity values in nonpeak areas of the mass spectral profile when the estimate overshoots the true baseline. Such artifacts were removed by setting negative ion intensities to zero after baseline correction. The m/z alignment was driven by five reference peaks that were present across the majority of cutting depths: m/z 5653; 10,164; 11,306; 14,050; and 14,956 (35). The preprocessing phase was finalized by a feature extraction step that reduces the high-dimensional full profile spectra to distinct peaks, retrieving the corresponding peak intensities across all cutting depths. This peak-picking step uses the mspeaks() function provided by MATLAB and uses nearest-neighbor interpolation to extract peak heights from each spectrum (36).
High mass resolution and accuracy IMS data were collected on a 15T MALDI FTICR MS (Bruker Daltonics) from the kidney region of a single transverse tissue section (section 34D). The instrument is equipped with an Apollo II dual MALDI/ESI ion source and a Smartbeam II 2 kHz Nd:YAG (355 nm) laser. Data were collected from m/z 900 to 18,000. Tuning of the funnel RF amplitude [190 Vpp (peak-to-peak voltage)], accumulation hexapole (1.4 MHz, 1200 Vpp), transfer optics (1 MHz, 310 Vpp), time of flight delay (2.0 ms), and ICR cell (sweep excitation power, 40%) were required for high m/z analysis. Imaging data were collected using the small laser setting (~50 µm) with a pixel spacing of 75 µm in both x and y dimensions and 500 laser shots averaged per pixel. External calibration was performed before analysis using CsI clusters and internally after analysis using a series of previously identified proteins. FlexImaging 4.1 (Bruker Daltonics) was used to visualize ion images (28).
Frozen kidney sections from mice infected with the iron- and manganese-responsive promoters were analyzed by MALDI IMS after fluorescent imaging using a rapifleX MALDI Tissuetyper (Bruker Daltonics). The rapifleX is equipped with a Smartbeam 3D Nd:YAG (355 nm) laser capable of a 10-kHz repetition rate. Images were collected in beam scan mode with a pixel size of 30 µm and the laser in single spot mode (diameter ≈ 5 µm.) Data were collected from m/z 2000 to 23,000 with 600 laser shots averaged per pixel.
Imaging acquisitions of biological replicate sections were imported in SCiLS imaging software (Bruker Daltonics). Data sets were background-subtracted using the convolution algorithm and normalized together for comparison using the total ion current. Peaks for each imaging acquisition were conditionally located in SCiLS using a spectral stride of 16 with a limit of 100 maximum peaks per spectrum. Using the Find Discriminative m/z Values function, signals from samples at days 4 and 10 after infection were compared using a receiver operating characteristic (ROC) curve. Ion intensity maps were chosen from values corresponding to an area under the ROC curve greater than 0.75 on a scale of 0 to 1.
Protein identification
Select protein ions of interest from the MALDI images were identified using an indirect strategy (37, 38). Briefly, common spatial localizations between an ion image acquired on the high mass resolution FTICR instrument and ion images acquired on the low mass resolution TOF instrument were used to link high mass accuracy measurements made on the FTICR to the protein ions observed in the TOF data. These accurate mass measurements (<5 parts per million) provided values to subsequently target for top-down liquid chromatography (LC)–MS/MS proteomics.
For top-down proteomics, on-tissue protein microextractions were first performed on abscesses using a tissue section containing clear infectious foci in a manner similar to that previously described (37). Briefly, protein was extracted from abscesses using 0.5 µl of a solution containing 30% ACN and 0.1% TFA, and microextracts from four sections were combined. A second extraction was performed sequentially on the same four sections using 0.5 µl of a 50% ACN, 5% formic acid solution. The microextraction (2 or 2.3 µl) was diluted at least 10-fold in 0.1% formic acid and subsequently bomb-loaded onto a reversed-phase capillary trap column using a helium-pressurized cell as previously described (28, 37). Eluted proteins were mass analyzed using an electron transfer dissociation (ETD)–enabled LTQ Orbitrap Velos mass spectrometer equipped with a nanoelectrospray ionization source and operated using a data-dependent method. A full-scan spectrum of m/z 400 to 2000 [resolving power (RP), 60,000 at m/z 200] was acquired as the initial scan event per duty cycle. After the full scan, four data-dependent scan events were selected for fragmentation using ETD in the Velos ion trap using the four most abundant ions in each MS scan. Dynamic exclusion was enabled, allowing a repeat count of 1 within 20 s. ETD tandem mass spectra were acquired sequentially using the LTQ Velos ion trap followed by the Orbitrap (RP, 15,000 at m/z 200) for mass analysis. An isolation width of 3 Da and an ETD reaction time of 90 to 100 ms were used for MS/MS spectra. The MSn automatic gain control (AGC) target value in the ion trap was set to 2 × 104, the MSn AGC target for Orbitrap scan events was 8 × 105, and the ETD reagent ion (fluoranthene) AGC target was set to 1 × 105. Parent ions of interest were selected from the full-scan LC-MS data and matched ions detected in FTICR IMS data sets. Fragmentation spectra from these ions were manually de novo sequenced and searched using BLAST databases. Targets of interest isolated from data-dependent methods were targeted for fragmentation by customizing the instrumental method with targeted scan events to ensure ETD MS/MS of selected ions.
Further protein purification was needed to isolate certain targets of interest. To maximize the abundance of bacterial proteins in kidney abscesses, C57BL6/J mice were infected with S. aureus. C57BL6/J mice were chosen on the basis of our previous experience in proteomic identification of bacterial and host targets in this mouse strain and to further illustrate the ability of MALDI IMS to identify bacterial proteins in multiple mouse strains (14). At day 9 after infection, the kidneys were excised and homogenized in 1 ml of PBS. To extract proteins, 1 ml of 2,2,2-trifluoroethanol and 2 ml of 50% ACN were added to the homogenate. Kidneys were further homogenized using an ultrasonicator dismembrator (model 150E, Thermo Fisher Scientific). Extracts were centrifuged to remove particulates and were fractionated using a Waters 2690 Separations Module equipped with an offline fraction collector. Proteins were fractionated using a Vydak 208TP 150 mm 5 µm C8 column and an aqueous to organic gradient over 120 min. Fractions were collected every minute. The fractionation procedure was performed four times, and the time-aligned fractions were combined. The fractionation was dried using the Savant SPD131DDA SpeedVac Concentrator (Thermo Fisher Scientific) and reconstituted in 20 µl of 50% ACN with 0.1% TFA. One microliter of each fraction well was spotted onto a Bruker AnchorChip (Bruker Daltonics) with 1 µl of DHA matrix (15 mg/ml) in 40% ACN. A MALDI mass spectrum was obtained for each spot, and fractions of interest were manually selected based on the presence of m/z values of interest. Fractions of interest were subjected to targeted LC-MS/MS analysis as described above. To identify δ-hemolysin, a bottom-up proteomics approach was used. A purified fraction from above was subjected to overnight tryptic digestion. For analysis by LC-MS/MS, peptides were loaded onto a capillary reverse-phase analytical column (360 µm outer diameter × 100 µm inner diameter) using an Eksigent NanoLC Ultra HPLC and autosampler. The analytical column was packed with 20 cm of C18 reverse-phase material (Jupiter, 3-µm beads, 300 Å, Phenomenex) directly into a laser-pulled emitter tip. Peptides were gradient-eluted over a 90-min gradient at a flow rate of 500 nl/min. The mobile phase solvents consisted of water containing 0.1% formic acid (solvent A) and ACN containing 0.1% formic acid (solvent B). The gradient consisted of the following: 0 to 15 min (sample loading via autosampler onto column), 2% B; 15 to 65 min, 2 to 40% B; 65 to 74 min, 40 to 90% B; 74 to 75 min, 90% B; 75 to 76 min, 90 to 2% B; 76 to 90 min (column equilibration), 2% B. Peptides were sequenced using Q Exactive Orbitrap MS. The mass spectrometer collected one full scan followed by MS/MS fragmentation data for the top 12 ions. A mass range of m/z 300 to 2000 was collected per each scan. A targeted scan event was included to obtain MS/MS data for m/z 497.77, representing the [M + 2H] 2+ ion for the tryptic peptides WIIDTVNK. Further confirmation of peptide identity was confirmed using S. aureus strains lacking either CsbD or δ-hemolysin. For CsbD, groups of mice were infected with either WT S. aureus or a strain lacking CsbD (see the “Bacterial strains and growth conditions” section). At 4 days after infection, infected kidneys were harvested and subjected to the same protein identification methods used for identification of δ-hemolysin. To further confirm peptide identity, mice infected with WT S. aureus or a S. aureus strain inactivated for CsbD were analyzed by MALDI IMS, looking for the presence of signal at m/z = 6888. Taking a similar approach for δ-hemolysin, mice infected with a S. aureus strain inactivated for hld were noted to make much smaller kidney abscesses that were not amenable to MALDI IMS. We therefore grew either the WT or hld mutant strains overnight in TSB in tightly capped flasks. Bacterial pellets were then collected for downstream protein extraction and MS/MS analysis. MS/MS profiles were compared to proteins extracted from WT-infected kidneys.
Imaging reconstruction and registration
Care was taken not to move the mouse between optical imaging, MRI, and the freezing process. Thus, a rigid-body image registration was used to transform the MRI data into the blockface images’ coordinate system (13, 24). The resulting transformation was also applied to the optical data, resulting in the optical, MRI, and blockface data in a unified coordinate system.
Imaging the blockface data during tissue sectioning does not create intrinsically registered images. To reconstruct an aligned 3D blockface data set, the imaging data were aligned slice by slice through an image registration that maximizes a normalized mutual information metric using a rigid-body transformation (13, 24). This approach starts with the central blockface image and registers a neighboring slice in the same direction. Registration is continued for the entire volume by registering each successive slice to the proceeding slice until the initial image is reached. Once all blockface slices were aligned, they were combined into one 3D data set, providing the base coordinate system to which all imaging data will be transformed.
To perform the optical to MR registration, a full-body MRI data set of the mouse was required. However, because of limitations of the MRI coil, two separate scans were necessary to obtain full coverage of the specimen. Thus, the upper and lower sections of the mouse were imaged separately with overlapping portions toward the middle of the body. After acquisition, the overlapping data were used to combine the two scans to form one cohesive structural image using image registration.
The transformation between the optical imaging coordinate system and MRI coordinate system was calculated using a surface-based registration. During acquisition, the optical imaging system creates a 3D surface mesh of the mouse. A second mesh was created in the MRI coordinate system by manually segmenting the mouse body using the full-body MRI. Once each surface mesh was created and manually reoriented to similar positions, an iterative closest point algorithm was used to align the optical mesh in the MRI mesh coordinate system (13). This provided the necessary transformation to move the raw optical data into the MRI coordinate system.
Once the 48 2D MALDI data sets were acquired, a single m/z bin was chosen with well-defined anatomical features for each slice. This image was manually registered to the corresponding blockface image. The resulting transformation was then applied to all ion images for this slice. These data were combined to form one 3D MALDI data set for each of the defined m/z bins by inserting data into the appropriate location in an empty matrix of the same dimensions as the blockface data and interpolating the missing data between slices using a weighted interpolation. This resulted in a 3D MALDI image for each of the selected m/z bins within the same coordinate system as the blockface.
Similar to the MALDI IMS to blockface registration, a single element image from LA-ICP-MS was chosen for each slice and manually registered to the corresponding blockface image. The resulting transformation was applied to the remaining element images for the similar slice. Once all data had been registered, 2D data were inserted into the correct location in 3D space, and data between slices were interpolated. The result was a 3D LA-ICP-MS data set for each analyzed element in the same coordinate system as the blockface.
Statistical analysis
The experimental design of this study is focused toward obtaining a multimodal exploratory view into the pathogenesis of invasive infection and the subsequent generation of new research hypotheses. Hence, the study design is concentrated toward ensuring robustness and mitigating outlier influence on the observations and imaging results. We therefore took steps to minimize nonbiological variability through biological, instrumental, and in silico means, using various experimental replicates and data preprocessing methods. Fully independent biological replicate measurements were acquired to verify the veracity of observations made in the 3D molecular imaging data set. These biological replicates consist of 2D IMS experiments at three distinct time points: days 4, 7, and 10 after infection. For both the day 4 and day 7 experimental groups, we acquired three distinct 2D experiments, whereas for the day 10 experimental group, we obtained two distinct 2D IMS experiments. As described in the “Matrix-assisted laser desorption/ionization imaging mass spectrometry” section, the presence of instrumental noise sources and nonbiological signal variation is addressed by several preprocessing methods. Ion intensity noise is addressed through intensity normalization, whereas chemical background noise and drift along the m/z axis are rectified by baseline correction and m/z alignment. Although these methods are commonly used to minimize intra-experiment variation, it is important to note that, in this study, we performed these methods across all cutting planes concurrently as one large IMS data set. This avoids the introduction of algorithm-induced plane-to-plane variation artifacts and allows the preprocessing methods to address both inter-experiment and intra-experiment variation from nonbiological sources.
For the 3D IMS data, observational robustness is also approached through acquisition designs that allow for nonbiological variability between different measurement sets to be detected and minimized. Along the z axis, this is done by sampling the tissue volume at many different cutting planes using distinct 2D IMS experiments (N = 48). Although these experiments cannot be considered technical replicates, there is sufficient correlation between subsequent 2D cutting plane measurements to detect outlier 2D IMS experiments and ensure measurement robustness. Within each 2D IMS experiment, in the x,y plane, nonbiological pixel-to-pixel variation is assessed by sampling the tissue section at many distinct locations (7900 < N < 33000 for the cutting planes in this study). Although pixels cannot be considered technical replicates of each other, using neighboring pixels to detect outlier pixels is warranted because IMS experiments tend to exhibit positive spatial autocorrelation (39, 40).
To validate 3D imaging results, 2D replicate experiments were performed on biological and technical replicates. Data were mined using SCiLS (Bruker Daltonics) to identify discriminative m/z values. This function within the SCiLS software package is a univariate, binary comparison based on individual intensity values and their frequency in an IMS acquisition. In comparing two groupings, a true-positive rate and a true-negative rate are calculated and plotted in a ROC curve. Once plotted, a discriminatory threshold between 0 and 1 can be chosen, with 1 representing a complete discrimination between the two groups (41). Data were filtered for m/z values, where this value exceeded 0.75.
Supplementary Material
Full MRI image volume.
Co-registration of blockface digital imaging to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164) and LA-ICP-MS imaging of calcium.
Fig. S1. Iron- and manganese-responsive fluorescent bacterial reporters coupled with LA-ICP-MS reveal lesional dynamics during invasive infection.
Fig. S2. MALDI IMS reveals the bacterial niche and lesional dynamics in abscessed tissues during invasive S. aureus infection.
Co-registration of blockface digital imaging to MALDI IMS signals for the S100A8 component of calprotectin (m/z = 10,164) and S. aureus δ-hemolysin (m/z = 3006).
Co-registration of MRI to 3D BLI.
Co-registration of blockface digital imaging to 3D BLI.
Co-registration of blockface digital imaging to LA-ICP-MS imaging of calcium, copper, iron, magnesium, manganese, phosphorous, and zinc.
Co-registration of blockface digital imaging to 3D BLI and LA-ICP-MS imaging of iron.
Co-registration of MRI to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164).
Co-registration of blockface digital imaging to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164).
Co-registration of blockface digital imaging to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164).
Acknowledgments
We thank M. Otto for the gift of the hld mutant strain. We thank members of the Cassat and Skaar laboratories for critical feedback on this manuscript. Funding: J.E.C. is supported by National Institute of Allergy and Infectious Diseases grants R01AI132560 and K08AI113107, as well as a Burroughs Wellcome Fund Career Award for Medical Scientists. This work was also supported by NIH grants R21 AI107233, R01 AI069233, R01 AI101171, and R01 AI073843 to E.P.S. R.M.C., J.M.S., B.M.P., and R.V.d.P. are supported by the NIH/National Institute of General Medical Sciences (2P41 GM103391-07) and Defense Advanced Research Projects Agency (W911NF-14-2-0022). The 15T FTICR MS in the Mass Spectrometry Research Center at Vanderbilt University was acquired through the NIH Shared Instrumentation Grant Program (1S10OD012359). This work was supported by NIH grant 1S10RR026742-01A1 for the LA-ICP-MS, housed in the Vanderbilt Mass Spectrometry Research Center.
Footnotes
Author contributions: Project design: J.E.C. and E.P.S.; methodology: J.E.C., E.P.S., J.L.M., W.J.P., B.M.P., R.M.C., K.J.W., and Z.S.; data processing: R.V.d.P.; completion of experiments: J.E.C., J.L.M., W.J.P., B.M.P., K.J.W., Z.S., Y.Z., J.V., D.C.C., K.L.R., A.M.J., M.L.R., J.M.S., and C.M.G.; writing—original draft: J.E.C., J.L.M., and E.P.S.; writing—review and editing: all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw data have been deposited into Dryad Digital Repository (http://datadryad.org) under doi:10.5061/dryad.q9080.
REFERENCES AND NOTES
- 1.Cheng AG, DeDent AC, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 2011;19:225–232. doi: 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hood MI, Skaar EP. Nutritional immunity: Transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: A role for manganese and zinc. Curr. Opin. Chem. Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skaar EP, Raffatellu M. Metals in infectious diseases and nutritional immunity. Metallomics. 2015;7:926–928. doi: 10.1039/c5mt90021b. [DOI] [PubMed] [Google Scholar]
- 5.Reyzer ML, Caprioli RM. MALDI-MS-based imaging of small molecules and proteins in tissues. Curr. Opin. Chem. Biol. 2007;11:29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sussulini A, Becker JS, Becker JS. Laser ablation ICP-MS: Application in biomedical research. Mass Spectrom. Rev. 2017;36:47–57. doi: 10.1002/mas.21481. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 9.Seeley EH, Caprioli RM. 3D imaging by mass spectrometry: A new frontier. Anal. Chem. 2012;84:2105–2110. doi: 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells WM, III, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Med. Image Anal. 1996;1:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 14.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 15.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zackular JP, Chazin WJ, Skaar EP. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J. Biol. Chem. 2015;290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yui S, Mikami M, Yamazaki M. Purification and characterization of the cytotoxic factor in rat peritoneal exudate cells: Its identification as the calcium binding protein complex, calprotectin. J. Leukoc. Biol. 1995;58:307–316. doi: 10.1002/jlb.58.3.307. [DOI] [PubMed] [Google Scholar]
- 18.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akbar S, Lee SY, Boylan SA, Price CW. Two genes from Bacillus subtilis under the sole control of the general stress transcription factor σB. Microbiology. 1999;145:1069–1078. doi: 10.1099/13500872-145-5-1069. [DOI] [PubMed] [Google Scholar]
- 20.Prágai Z, Harwood CR. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology. 2002;148:1593–1602. doi: 10.1099/00221287-148-5-1593. [DOI] [PubMed] [Google Scholar]
- 21.Wang R, Braughton KR, Kretschmer D, Bach T-HL, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 22.Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013;11:667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung GYC, Yeh AJ, Kretschmer D, Duong AC, Tuffuor K, Fu C-L, Joo H-S, Diep BA, Li M, Nakamura Y, Nunez G, Peschel A, Otto M. Functional characteristics of the Staphylococcus aureus δ-toxin allelic variant G10S. Sci. Rep. 2015;5:18023. doi: 10.1038/srep18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attia AS, Schroeder KA, Seeley EH, Wilson KJ, Hammer ND, Colvin DC, Manier ML, Nicklay JJ, Rose KL, Gore JC, Caprioli RM, Skaar EP. Monitoring the inflammatory response to infection through the integration of MALDI IMS and MRI. Cell Host Microbe. 2012;11:664–673. doi: 10.1016/j.chom.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLOS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLOS Pathog. 2012;8:e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 2013;79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spraggins JM, Rizzo DG, Moore JL, Rose KL, Hammer ND, Skaar EP, Caprioli RM. MALDI FTICR IMS of intact proteins: Using mass accuracy to link protein images with proteomics data. J Am. Soc. Mass Spectrom. 2015;26:974–985. doi: 10.1007/s13361-015-1147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Caprioli RM. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 2011;83:5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakeman CA, Moore JL, Noto MJ, Zhang Y, Singleton MD, Prentice BM, Gilston BA, Doster RS, Gaddy JA, Chazin WJ, Caprioli RM, Skaar EP. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun. 2016;7:11951. doi: 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prentice BM, Chumbley CW, Caprioli RM. High-speed MALDI MS/MS imaging mass spectrometry using continuous raster sampling. J. Mass Spectrom. 2015;50:703–710. doi: 10.1002/jms.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spraggins JM, Caprioli RM. High-speed MALDI-TOF imaging mass spectrometry: Rapid ion image acquisition and considerations for next generation instrumentation. J. Am. Soc. Mass Spectrom. 2011;22:1022–1031. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van de Plas R. Tissue based proteomics and biomarker discovery. Multivariate data mining strategies for mass spectral imaging, thesis. University of Leuven; Leuven, Belgium: 2010. [Google Scholar]
- 35.Monchamp P, Andrade-Cetto L, Zhang JY, Henson R. Signal processing methods for mass spectrometry. In: Alterovitz G, Ramoni MF, editors. Artech House Bioinformatics & Biomedical Imaging Series. pp. xviii–386. (Artech House, 2007), chap. Signal Processing Methods for Mass Spectrometry. [Google Scholar]
- 36.Morris JS, Coombes KR, Koomen J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- 37.Schey KL, Anderson DM, Rose KL. Spatially-directed protein identification from tissue sections by top-down LC-MS/MS with electron transfer dissociation. Anal. Chem. 2013;85:6767–6774. doi: 10.1021/ac400832w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacAleese L, Stauber J, Heeren RMA. Perspectives for imaging mass spectrometry in the proteomics landscape. Proteomics. 2009;9:819–834. doi: 10.1002/pmic.200800363. [DOI] [PubMed] [Google Scholar]
- 39.Van de Plas R, Yang J, Spraggins J, Caprioli RM. Image fusion of mass spectrometry and microscopy: A multimodality paradigm for molecular tissue mapping. Nat. Methods. 2015;12:366–372. doi: 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cassese A, Ellis SR, Ogrinc Potočnik N, Burgermeister E, Ebert M, Walch A, van den Maagdenberg AMJM, McDonnell LA, Heeren RMA, Balluff B. Spatial autocorrelation in mass spectrometry imaging. Anal. Chem. 2016;88:5871–5878. doi: 10.1021/acs.analchem.6b00672. [DOI] [PubMed] [Google Scholar]
- 41.Jones EA, Deininger S-O, Hogendoorn PCW, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. J. Proteomics. 2012;75:4962–4989. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full MRI image volume.
Co-registration of blockface digital imaging to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164) and LA-ICP-MS imaging of calcium.
Fig. S1. Iron- and manganese-responsive fluorescent bacterial reporters coupled with LA-ICP-MS reveal lesional dynamics during invasive infection.
Fig. S2. MALDI IMS reveals the bacterial niche and lesional dynamics in abscessed tissues during invasive S. aureus infection.
Co-registration of blockface digital imaging to MALDI IMS signals for the S100A8 component of calprotectin (m/z = 10,164) and S. aureus δ-hemolysin (m/z = 3006).
Co-registration of MRI to 3D BLI.
Co-registration of blockface digital imaging to 3D BLI.
Co-registration of blockface digital imaging to LA-ICP-MS imaging of calcium, copper, iron, magnesium, manganese, phosphorous, and zinc.
Co-registration of blockface digital imaging to 3D BLI and LA-ICP-MS imaging of iron.
Co-registration of MRI to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164).
Co-registration of blockface digital imaging to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164).
Co-registration of blockface digital imaging to MALDI IMS signal for the S100A8 component of calprotectin (m/z = 10,164).