Abstract
When the SUMO-1 protein is localized on the genome, it is found on proteins bound to the promoters of the most highly active genes and on proteins bound to the DNA encoding exons. Inhibition of the SUMO-1 modification leads to reductions in initiation of mRNA synthesis and splicing. In this review, we discuss what is known about the SUMOylation of factors involved in transcription initiation, pre-mRNA processing and polyadenylation. We suggest a mechanism by which SUMO modification of factors at the promoters of high activity genes trigger the formation of an RNAPII complex that coordinates and integrates the stimulatory signals for each process to catalyze an extremely high level of gene expression.
Keywords: SUMO, mRNA splicing, transcription, RNA polymerase II
Introduction to SUMO
The Small Ubiquitin-like Modifier (SUMO), which was discovered by its association with human RAD51 and RAD52 using a yeast two-hybrid screen 1, 2, has important regulatory function in many cellular processes. In most mammalian cell types SUMO has three isoforms, SUMO-1 to -3, and one isoform in yeast. As is the case with ubiquitin, SUMO proteins are conjugated to substrate proteins via isopeptide bonds linking the carboxy-terminus of the mature SUMO protein and the ε-amino moiety of lysines in the target protein. SUMO proteins must be cleaved to expose the carboxy-terminus of the mature protein, which is then bound by a thio-ester linkage to its E1 factor, transferred to UBC9, which is the one E2 factor for all SUMO proteins, and together with E3 proteins transfer the SUMO protein to acceptor lysines on the substrate protein 3, 4. SUMO-1 is normally conjugated to its substrate as a monomer while SUMO-2 and SUMO-3, which are 45% identical to SUMO-1 and 96% identical to each other, are often conjugated in poly-SUMO chains. The major SUMO conjugated protein in the cell is RanGAP1, a 90 kDa protein when SUMO modified, and which binds to the nuclear pore complex 5. SUMO-conjugation to RanGAP1 is essential to mediate nuclear protein transport 6.
SUMOylation is the conjugation of SUMO protein to target proteins. SUMOylation regulates multiple cellular processes, including DNA double strand break (DSB) repair 7–10. The different isoforms of human SUMO function in specific pathways of DSB repair: SUMO-1 stimulates all DSB pathways, and in some pathways its function is independent of conjugation to a substrate protein by UBC9. The SUMO-2/3 isoforms are specific to the conservative NHEJ pathway of DSB repair 11. Other processes regulated by SUMOylation include mitochondrial fission 12, synapse function 13, and cell cycle progression 14, 15. Transcriptional regulation has many steps regulated by SUMO modification 16–18. The effect of SUMOylation of many transcription factors is to repress transcription via the recruitment of transcription repressors, such as histone deacetylases 19, 20. Examples of transcription factors that have their positive signal repressed due to SUMOylation include C/EBP, ELK-1, SREBP, SP3 and Jun 21–25. Conversely, SUMOylation of transcription factors have been found to stimulate transcription; examples of such transcription factors include Tcf-4, HSF1/2, and CREB 26–29. In this review article, we will focus on a pathway by which SUMO-1 stimulates the gene expression process via regulation of mRNA synthesis and processing.
Localization in the genome of SUMO modified chromatin associated factors
Histone SUMOylation
In yeast, SUMO modifies all four core nucleosomal histones, and this SUMOylation represses transcription genome-wide, with highest concentration of SUMO at the telomeres 30. As part of the DNA repair process, SUMO modifies H2A.Z in yeast 31. In human cells, among the four core histones, only SUMOylation of H4 has been observed, and H4-SUMO is associated with gene repression via recruitment of histone deacetylases 20. A variety of other chromatin associated (non-histone) proteins are marked by SUMO, including transcriptional regulatory factors. Specific examples of factors stimulated by SUMOylation will be discussed below.
Location of SUMO marks in genome
SUMO proteins had been found associated with both telomeres 32, 33 and with centromeres 34, 35. Genome-wide analysis, which excludes the repetitive sequence elements that are present in centromeres and telomeres, revealed that the preponderance of SUMO-1 bound sites localize to genes with high transcriptional activity 36. The SUMO-1 mark is found at promoters and on exons of active genes. Strikingly, the RNA polymerase II (RNAPII) genes with the highest level of transcription have the highest amount of SUMOylation of chromatin proteins at the promoter; these include genes encoding ribosomal protein subunits and translation regulatory factors 36. These SUMO-1 marks are correlated with other stimulatory marks, such as H3K4me3, and depletion of either SUMO-1 or the E2 factor UBC9 results in decreases in mRNA abundance for these genes. The magnitude of the decrease in SUMO-1 depleted cells in ribosomal protein gene mRNA abundance is in most cases less than 2-fold, indicating that these genes are still transcribed but at lower levels 36.
Preformed transcriptosome versus independent assembly of processing modules
The RNAPII complex is the central player in the process that initiates mRNA synthesis, elongates, caps, splices, and polyadenylates the mRNAs, and SUMOylation has a role in each step. The concept of the “transcriptosome” has emerged whereby the RNAPII is just a component of a larger complex that includes RNA processing factors that will provide synergy to facilitate mRNA production 37. RNAPII had been observed in large complexes, called the RNAPII holoenzyme, that include transcriptional co-activators 38, but the transcriptosome concept goes further likening the RNAPII complex to a coordinately regulated cellular machine. Consistent with this idea, preassembled RNA transcription and processing complexes are assembled in the nuclear Cajal body (CJ) and the transcriptosome translocates to the gene 39. The idea is that the cellular machines are all present to coordinately synthesize and process mRNA.
More recently, the concept has emerged that rather than a preformed transcriptosome, transcription is mediated by the stepwise recruitment of protein complexes that execute different steps on the growing RNA transcript 40. Individual steps of the total process have been linked. The largest subunit of RNAPII has a domain called the Carboxy-Terminal Domain (CTD), which consists of a repeating unit of seven amino acids 41, 42. With initiation of RNA synthesis, the RNAPII CTD is phosphorylated on the serine-5 position of the heptad repeat, and this phosphorylated CTD binds and recruits the splicing factors to process the nascent RNA 43. With regard to the transcriptosome, the question is whether the phosphorylation of the CTD associated with transcription initiation catalyzes the independent assembly of the cellular machines that regulate RNA processing in an ordered fashion as the RNAPII elongates or whether the factors all pre-assemble into a transcriptosome. One possibility that will be developed in the following text is that SUMOylation of specific proteins bound to the promoter can drive the efficient assembly of RNA processing factors along with RNAPII and coactivators, and coordinate the steps to generate a mature mRNA.
SUMO and Initiation
Genome-wide analysis of SUMO-1 bound to chromatin associated factors found that SUMO-1 in human cells, or SUMO in yeast, is associated with the promoters of the most active RNAPII genes in the cell and mapping to about -150 relative to the transcription start site 36, 44, 45. Two different SUMOylated proteins have been found to bind to this site: RAP1 in yeast 44 and SAFB in human 46. SUMOylation of these factors have been shown to stimulate the recruitment of transcription factor TFIID by RAP1 and RNAPII by SAFB.
In the case of SUMO-1, the SAFB1 and SAFB2 proteins were identified by mass spectrometry as SUMO-1 modified chromatin associated factors. SAFB was found to localize to promoters coincident with SUMO-1. Depletion of SAFB or of SUMO-1 had similar effects decreasing RNAPII initiation events on the promoters, decreasing mRNA abundance of SUMO-1 marked genes, and decreased splicing 46. In this study, along with SAFB, the most abundantly SUMO-1 modified proteins associated with chromatin were splicing factors. SAFB is known to interact with RNAPII and splicing factors, suggesting the possibility that SUMO-1 and SAFB collaborate to regulate high expression genes at the level of initiation of mRNA synthesis and processing 46.
Regulation of splicing
The regulation of splicing is functionally tied to the rate of elongation. SUMOylation regulates one of the RNAPII elongation factors, Transcription Elongation Regulator 1 (TCERG1). Modification of the human TCERG1 with any of SUMO-1, -2, or -3 causes a decrease in the elongation stimulatory activity of TCERG1. Mutation of the SUMO acceptor lysines of TCERG1 resulted in higher elongation rates 47. While in the case of TCERG1 SUMOylation resulted in transcription repression, in the following examples SUMO modification stimulates mRNA processing.
The genome-wide analysis of SUMO-1 conjugation to chromatin associated proteins revealed a striking enrichment of the SUMO-1 mark on exons and no enrichment on introns 36. In light of the data that the most abundantly detected SUMO-1 substrates associated with chromatin after SAFB are splicing factors 46, and independent observations that splicing factors associate with the chromatin 48, 49, it is likely that the SUMO-1 conjugated proteins associated with exons are splicing factors. A key point that is unclear at this point is how the exons are recognized at the DNA level. It is suggested that RNAPII elongates at a slower rate in some exons that are alternatively spliced 50. It has also been suggested that a long noncoding RNA binds to exon encoding DNA and recruits chromatin modifiers that affect the splicing of the exon 51. Though SUMOylation is enriched on chromatin of DNA encoding exons, if it is splicing effector proteins that are the source of the SUMO-1 mark on exonic DNA, then it is unlikely that SUMO-1 has a role in initiating the exon recognition but rather that the SUMO modification is a downstream event. In such a case, the SUMO modification could be stimulating protein-protein interactions that accelerate the RNA processing.
Important splicing factors, such as serine-arginine rich (SR) proteins 55, 56, have been identified as SUMO substrates 46, 57. The SR protein SF2/ASF interacts with UBC9 and enhances SUMOylation of specific substrates as well as promoting SUMO conjugation to RNA processing factors 58. The recruitment of regulatory splicing factors depend on chromatin modification factors, transcriptional regulators as well as RNAPII elongation rate 50, 51, 59, 60, and the splicing process occurs simultaneously with the elongating RNAPII 61–63. SUMOylation may be the glue that binds transcription initiation, transcription elongation, and splicing together.
The events at initiation can affect splicing. SUMO-1 modification of SAFB stimulates initiation by RNAPII. In addition, depletion of either SUMO-1 or of SAFB from cells result in a decrease in the rate of splicing detected in nuclear pre-mRNAs 46. Splicing still occurs, indicating that the SUMOylation of SAFB does not have a required role in splicing, but the splicing is about 40% slower. As discussed above, the genes marked with SAFB-SUMO-1 at promoters are the most highly expressed RNAPII genes in the cell, and it may be anticipated that the splicing of these genes is also at an accelerated pace, leading to the concept of a “fast track” in mRNA synthesis and processing.
SUMO modifications and mRNA polyadenylation
Pre-mRNA 3′ processing requires four multi-subunit protein complexes, CPSF, CstF, CFI and CFII, in addition to the poly(A) polymerase (PAP) 64, which associate with RNAPII and other proteins to form a complex for the pre-mRNA 3′ processing 65. CPSF has been shown to interact with TFIID and together with its counterpart CstF to remain associated with RNAPII throughout the coding region 66. When 3′ processing complexes are purified under cleavage and polyA conditions, all the four main 3′ processing complexes plus other proteins are found in both conditions, confirming the association of these complexes with the early transcriptional machinery. This linkage of transcription initiation with polyadenylation factors supports the concept of transcriptosome formation.
SUMO-2/3 modified forms of CPSF, PAP, and an assembly factor called Simplekin have been detected. SUMOylation of PAP occurs via six SUMO acceptor sites. Lysine to arginine mutation of those sites in effect leads to lower concentration of these factors in the nucleus. The SUMO-2/3 modification is necessary for PAP nuclear localization. Blockage of SUMOylation of CPSF and Simplekin affected the assembly and activity of pre-mRNA 3′ complex and was lethal to cell viability 67, 68.
mRNA stability
SUMOylation regulates one of the key factors controlling mRNA stability in human cells, nucleolin. Cell stress induces SUMO-1 modification of nucleolin, which in turn causes its binding to and stabilization of the GADD45a mRNA. Expression of a mutant nucleolin with its SUMO-1 acceptor lysine mutated, results in a failure for the mutant nucleolin to bind to the mRNA and a failure to stabilize it 69. Thus, it is likely that the SUMOylation of nucleolin has an important regulatory role in specific stress-induced mRNA stabilization. The stability and export from the nucleus of mRNA is also regulated by SUMOylation of the THO complex in yeast; THO regulates the association of mRNAs into mRNPs for export. SUMOylation of the Hpr1 subunit of the THO complex controls THO binding to mRNPs. Hpr1 mutants defective for SUMOylation fail to bind to mRNPs, leading to destabilization of several stress-induced transcripts 70. In these two experimental settings, SUMO modification of protein factors stabilize subsets of mRNAs.
Concept of fast-track mRNA synthesis and processing
It is striking that the genes with the highest level of expression, such as those encoding ribosomal proteins and translation factors, have the highest level of SUMO-1 modification of chromatin associated proteins at the promoter 36. Results are consistent with the concept that transcription initiation, elongation and splicing still occur in the absence of the SUMO modification of these factors, but perhaps at a slower pace. The idea that transcription and splicing occur in the neighborhood of nuclear pore complex and the product is primed for rapid RNA synthesis, rapid pre-mRNA processing, and ensuing rapid nuclear-cytoplasmic transport, form the basis to postulate that SUMOylation functions to speed up multiple steps, from initiation through to pre-mRNA 3′ processing, hence fast-tracking the process of mRNA synthesis and processing.
Concept of group SUMOylation
Similar to what is proposed in this review for transcription, SUMOylation of multiple DNA repair factors accelerates the repair process, and the SUMO modification of these proteins stimulate protein physical interactions as if it were protein glue 7. Mutation of 11 SUMO-acceptor lysine residues in DNA repair factors in yeast do not eliminate DNA repair, but repair is slowed. These observations led to the notion that DNA repair process requires group SUMOylation of repair proteins to speed repair in a synergistic manner 7.
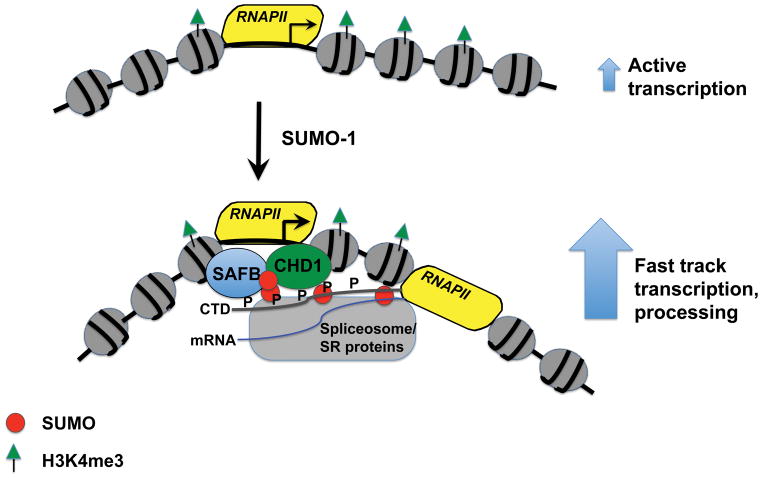
Perhaps the same concept applies to group SUMOylation of transcription, splicing and polyadenylation factors. Group SUMOylation can facilitate the formation of an RNAPII transcription and processing complex. This concept is summarized in Figure 1.
Figure 1.
Model for SUMOylation mediated fast track transcription and processing of mRNA.
In this depiction many factors have been left out to emphasize how “active” transcription may differ from “fast track” expression. We model that on most RNAPII templates, transcription occurs by multiple independent protein-protein and protein-template interactions to yield a pre-mRNA product that is likely processed co-transcriptionally. We model that the key difference with the fast-track synthesis is the SUMO modification of multiple factors, including SAFB, splicing factors and polyadenylation factors to stimulate the physical interaction of these complexes and coordinate the change from an active level of expression to a significantly higher level of expression, driving mRNA levels for these genes to very high abundances.
A dense network of protein-protein interactions occurs centering on the elongating RNAPII downstream of the start site. A subset of the interactions that occur on the template with elongating RNAPII is shown in Figure 1. Though methylation, acetylation, ubiquitination and SUMOylation all mark the chromatin on an active gene, to keep the model depiction simple, only the histone H3 lysine 4 trimethyl (H3K4me3) and SUMO-1 modifications are shown in the Figure. In the genome-wide study of SUMO-1 bound to chromatin associated proteins, SUMOylation of chromatin bound to promoters is associated with the stimulatory mark H3K4me3 found downstream of the transcription start site 36, 52. The H3K4me3 mark is bound by CHD1, which in turn stimulates RNAPII elongation and recruits the splicing machinery to the chromatin. Depletion of CHD1 decreases splicing and decreases the association of splicing components such as the U2 snRNP with chromatin 48. SAFB, the protein that is conjugated to SUMO-1 at the promoter and stimulates initiation by RNAPII, also binds to CHD1 53. SAFB also binds to the phospho-serine-5 CTD 54, which is also bound by splicing factors 43. The splicing factor SC35 is required for transcription elongation, suggesting a reciprocal functional role between elongation and splicing machineries 49.
We suggest that for most active genes RNAPII transcription synthesizes mRNA, and stimulation of transcription by the actions of CHD1 or by phospho-CTD recruitment of mRNA processing factors may happen, but not in the coordinated complex enabled by SUMOylation. Many of the most active genes in the cell, such as genes encoding ribosomal proteins and translational regulatory factors, are the genes with high abundance mRNAs found to be most affected by the depletion of SUMO-1 or its E2 factor, UBC9 36. Based on available data, we suggest that the SUMOylation of SAFB is a trigger that stimulates the physical association with CHD1 and splicing factors to coordinate a giant complex that synthesizes the mRNA and processes it. Since SUMOylation factors are abundant at the nuclear pore complex, it is possible that the SUMO modifications that trigger the “fast track” transcription localize to the nuclear pore, thus priming these genes for mRNA synthesis, processing and translocation to the cytoplasm. It is possible that following group SUMOylation, re-initiation of highly processive mRNA synthesis is stimulated and drives high levels of transcription consistent with the fast track model. We emphasize that this model is not proven, but rather is based on independent observations of protein-protein interactions that make sense as an integrated mechanism.
References
- 1.Saitoh H, Pu RT, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 2.Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 3.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 4.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 5.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 10.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Parvin JD. Small ubiquitin-like modifier (SUMO) isoforms and conjugation-independent function in DNA double-strand break repair pathways. J Biol Chem. 2014;289:21289–21295. doi: 10.1074/jbc.C114.582122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dadke S, Cotteret S, Yip SC, Jaffer ZM, Haj F, Ivanov A, Rauscher F, 3rd, Shuai K, Ng T, Neel BG, et al. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat Cell Biol. 2007;9:80–85. doi: 10.1038/ncb1522. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo B, Sharrocks AD. Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Molecular and cellular biology. 2009;29:3204–3218. doi: 10.1128/MCB.01128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Molecular cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. The EMBO journal. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics: official journal of the DNA Methylation Society. 2009;4:440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- 20.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano Y, Murata S, Tanaka K, Shimizu M, Sato R. Sterol regulatory element-binding proteins are negatively regulated through SUMO-1 modification independent of the ubiquitin/26 S proteasome pathway. J Biol Chem. 2003;278:16809–16819. doi: 10.1074/jbc.M212448200. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Cantwell CA, Johnson PF, Pfarr CM, Williams SC. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J Biol Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- 23.Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 24.Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell. 2003;12:63–74. doi: 10.1016/s1097-2765(03)00265-x. [DOI] [PubMed] [Google Scholar]
- 26.Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP, Taylor CT. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem. 2001;276:18513–18518. doi: 10.1074/jbc.M008066200. [DOI] [PubMed] [Google Scholar]
- 28.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto H, Ihara M, Matsuura Y, Kikuchi A. Sumoylation is involved in beta-catenin-dependent activation of Tcf-4. EMBO J. 2003;22:2047–2059. doi: 10.1093/emboj/cdg204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A. Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Hang LE, Liu X, Cheung I, Yang Y, Zhao X. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol. 2011;18:920–926. doi: 10.1038/nsmb.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B. Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J. 2004;23:3844–3853. doi: 10.1038/sj.emboj.7600394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown PW, Hwang K, Schlegel PN, Morris PL. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum Reprod. 2008;23:2850–2857. doi: 10.1093/humrep/den300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu HW, Zhang J, Heine GF, Arora M, Gulcin Ozer H, Onti-Srinivasan R, Huang K, Parvin JD. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40:10172–10186. doi: 10.1093/nar/gks819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halle JP, Meisterernst M. Gene expression: increasing evidence for a transcriptosome. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- 38.Parvin JD, Young RA. Regulatory targets in the RNA polymerase II holoenzyme. Curr Opin Genet Dev. 1998;8:565–570. doi: 10.1016/s0959-437x(98)80012-9. [DOI] [PubMed] [Google Scholar]
- 39.Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darnell JE., Jr Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. 2013;19:443–460. doi: 10.1261/rna.038596.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 43.Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci U S A. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chymkowitch P, Nguea AP, Aanes H, Koehler CJ, Thiede B, Lorenz S, Meza-Zepeda LA, Klungland A, Enserink JM. Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res. 2015;25:897–906. doi: 10.1101/gr.185793.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes & development. 2010;24:1242–1252. doi: 10.1101/gad.1917910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu HW, Banerjee T, Guan X, Freitas MA, Parvin JD. The chromatin scaffold protein SAFB1 localizes SUMO-1 to the promoters of ribosomal protein genes to facilitate transcription initiation and splicing. Nucleic Acids Res. 2015;43:3605–3613. doi: 10.1093/nar/gkv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Alvarez M, Montes M, Sanchez-Hernandez N, Hernandez-Munain C, Sune C. Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1) J Biol Chem. 2010;285:15220–15233. doi: 10.1074/jbc.M109.063750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22:370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Tai HH, Geisterfer M, Bell JC, Moniwa M, Davie JR, Boucher L, McBurney MW. CHD1 associates with NCoR and histone deacetylase as well as with RNA splicing proteins. Biochem Biophys Res Commun. 2003;308:170–176. doi: 10.1016/s0006-291x(03)01354-8. [DOI] [PubMed] [Google Scholar]
- 54.Nayler O, Stratling W, Bourquin JP, Stagljar I, Lindemann L, Jasper H, Hartmann AM, Fackelmayer FO, Ullrich A, Stamm S. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998;26:3542–3549. doi: 10.1093/nar/26.15.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vassileva MT, Matunis MJ. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol Cell Biol. 2004;24:3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pelisch F, Gerez J, Druker J, Schor IE, Munoz MJ, Risso G, Petrillo E, Westman BJ, Lamond AI, Arzt E, et al. The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc Natl Acad Sci U S A. 2010;107:16119–16124. doi: 10.1073/pnas.1004653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 60.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 62.Iannone C, Valcarcel J. Chromatin’s thread to alternative splicing regulation. Chromosoma. 2013;122:465–474. doi: 10.1007/s00412-013-0425-x. [DOI] [PubMed] [Google Scholar]
- 63.Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 65.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol Cell Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vethantham V, Rao N, Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22:499–511. doi: 10.1101/gad.1628208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang D, Liang Y, Xie Q, Gao G, Wei J, Huang H, Li J, Gao J, Huang C. A novel post-translational modification of nucleolin, SUMOylation at Lys-294, mediates arsenite-induced cell death by regulating gadd45alpha mRNA stability. J Biol Chem. 2015;290:4784–4800. doi: 10.1074/jbc.M114.598219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bretes H, Rouviere JO, Leger T, Oeffinger M, Devaux F, Doye V, Palancade B. Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs. Nucleic Acids Res. 2014;42:5043–5058. doi: 10.1093/nar/gku124. [DOI] [PMC free article] [PubMed] [Google Scholar]