Abstract
Spermatogenesis is precisely controlled by complex gene expression programs and involves epigenetic reprogramming, including histone modification and DNA methylation. SET domain–containing 2 (SETD2) is the predominant histone methyltransferase catalyzing the trimethylation of histone H3 lysine 36 (H3K36me3) and plays key roles in embryonic stem cell differentiation and somatic cell development. However, its role in male germ cell development remains elusive. Here, we demonstrate an essential role of Setd2 for spermiogenesis, the final stage of spermatogenesis. Using RNA-seq, we found that, in postnatal mouse testes, Setd2 mRNA levels dramatically increase in 14-day-old mice. Using a germ cell–specific Setd2 knockout mouse model, we also found that targeted Setd2 knockout in germ cells causes aberrant spermiogenesis with acrosomal malformation before step 8 of the round-spermatid stage, resulting in complete infertility. Furthermore, we noted that the Setd2 deficiency results in complete loss of H3K36me3 and significantly decreases expression of thousands of genes, including those encoding acrosin-binding protein 1 (Acrbp1) and protamines, required for spermatogenesis. Our findings thus reveal a previously unappreciated role of the SETD2-dependent H3K36me3 modification in spermiogenesis and provide clues to the molecular mechanisms in epigenetic disorders underlying male infertility.
Keywords: spermatogenesis, histone modification, histone methylation, testis, reproduction, acrosome biogenesis, histone-to-protamine transition, Setd2, spermiogenesis
Introduction
Mammalian spermatogenesis is a highly specialized differentiation process that can be typically divided into a mitotic phase, a meiotic phase, and a phase of spermiogenesis (1, 2). During the mitotic phase, spermatogonial stem cells either undergo self-renewal to maintain the stem cell pool or enter the differentiation pathway to generate differentiating spermatogonia, which are committed to produce primary spermatocytes (3). In meiosis, spermatocytes then undergo one round of replication followed by meiotic homologous recombination and two rounds of chromosome segregation to give rise to haploid round spermatids (4). During spermiogenesis, round spermatids undergo dramatic morphological and molecular changes, including high chromatin condensation, acrosome formation, flagellum development, and cytoplasm elimination, to transform into elongated mature spermatozoa (5–9). According to morphological characteristics, mouse spermiogenesis is subdivided into 16 steps. Steps 1–8 include round spermatids, whereas steps 9–16 include elongating spermatids (5–9). These processes are characterized by the dynamic changes to gene expression patterns and the dramatic changes of chromatin structure, which have been shown to involve epigenetic regulatory mechanisms (e.g. histone modification, DNA methylation, RNA methylation, noncoding RNAs, etc.) (10–15).
Emerging evidence has emphasized a crucial role of histone posttranslational modifications in mammalian spermatogenesis. Knockout mice carrying mutations in genes encoding histone modification enzymes display male sterility with a broad range of defects of spermatogenesis, revealing a crucial role of histone modifications in mammalian spermatogenesis (11, 16–28). The trimethylation of histone H3 lysine 36 (H3K36me3)4 is a well-characterized feature of transcriptionally active genes and usually enriched in the gene body regions. H3K36me3 is also implicated in pre-mRNA splicing, DNA mismatch repair, and recruitment of Dnmt3b. SET domain–containing 2 (SETD2; also known as HYPB and KMT3A) is the major methyltransferase responsible for H3K36me3 in mammals (29–34). Setd2 ablation in mice leads to global loss of H3K36me3 and embryonic vascular remodeling defects, indicating a critical role of SETD2-mediated H3K36me3 during development (32, 34–36). Although the functions of Setd2 and its associated H3K36me3 have been studied in diverse biological processes, the requirement for Setd2 and H3K36me3 during spermatogenesis remains elusive.
In this report, we show the spatiotemporal distribution of SETD2 and its associated H3K36me3 in mouse testes. We demonstrate that loss of H3K36me3 by germ cell–specific disruption of Setd2 leads to impaired spermiogenesis due to significant down-regulation of its direct target genes, which include many critical master regulators for spermiogenesis. This study thus reveals a crucial role of Setd2 and its associated H3K36me3 in spermiogenesis and uncovers Acrbp1 and protamine genes as direct targets of SETD2-dependent H3K36me3 during this process.
Results
Spatial and temporal patterns of Setd2 and H3K36me3 distribution during spermatogenesis
To explore the role of Setd2 and the associated H3K36me3 during spermatogenesis, we first examined the expression pattern of Sedt2 in mouse testes. In postnatal mouse testes, the Setd2 mRNA level dramatically increased at 14 days old, a stage that corresponds to the appearance of pachytene spermatocytes in the seminiferous epithelium (Fig. S1A), and remained at a high level afterward, suggesting that Setd2 is abundantly expressed in pachytene spermatocytes and spermatids. We then performed immunofluorescence staining to further investigate the subcellular localization of the SETD2 protein in the testis. Consistent with the mRNA expression, we found that SETD2 protein is mainly detected in the nucleus of pachytene spermatocytes and round spermatids, which were confirmed by the counterstaining of peanut agglutinin (PNA), a marker for the acrosome, and γH2AX, a marker for the XY body, respectively (Fig. 1A and Fig. S1B) (37). Moreover, SETD2 is also present in Sertoli cells as confirmed by counterstaining of GATA4, a marker for Sertoli cells (and round spermatids) (38) (Fig. S1C). Collectively, Setd2 is predominantly expressed in pachytene spermatocytes and postmeiotic germ cells in the adult mouse testes.
Figure 1.
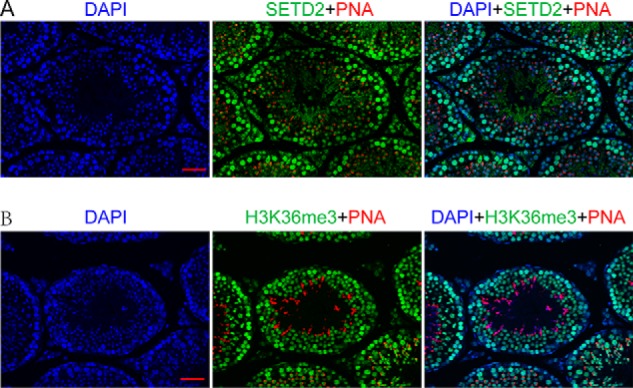
SETD2 expression and H3K36me3 distribution during spermatogenesis. A, immunofluorescence staining for SETD2 (green), rhodamine-labeled PNA (a marker for the acrosome; red), and DAPI (blue) in sections of adult mouse testes. SETD2 immunoreactivity was mostly seen in the nucleus of pachytene spermatocytes and round spermatids. B, immunofluorescent detection for H3K36me3 (green), rhodamine-labeled PNA (red), and DAPI (blue) in sections of adult mouse testes. H3K36me3 staining was detected in the nucleus of all stage of spermatocytes and round as well as elongating spermatids (scale bar, 40 μm).
As SETD2 is considered the main histone H3K36me3 methyltransferase in mammals, we next analyzed the distribution of H3K36me3 in mouse testis. Interestingly, we found that H3K36me3 distribution did not show a pattern identical to that of Setd2 in the testes. As shown in Fig. 1B, H3K36me3 exhibited strong staining in the nucleus of all stages of spermatocytes and the nucleus of round spermatids, whereas weak H3K36me3 signal was detected in spermatogonia and elongated spermatids. Taken together, these observations indicated that SETD2 might be responsible for cellular H3K36me3 in late-stage spermatocytes (pachytene/diplotene spermatocytes) and spermatids. However, whether it is predominantly responsible for H3K36me3 in the early-stage spermatocytes, such as preleptotene spermatocytes, remains to be determined in the future.
Setd2 is essential for murine spermiogenesis
To determine the role of Setd2 during mammalian spermatogenesis, we used a conditional knockout approach that generated a Setd2 floxed line (Setd2fl/fl) in which exons 6 and 7 of the Setd2 allele are flanked by loxP sites (Fig. S2A). To generate germline-specific deletion, mice bearing loxP-flanked alleles of Setd2 were crossed with Stra8-GFPCre knockin mice (Setd2fl/del, Stra8-GFPCre) (Fig. S2A). The Stra8-GFPCre knockin mice express CRE from A1 spermatogonia onward in which both alleles of Setd2 could be excised at A1 spermatogonia (Setd2fl/del, Stra8-GFPCre) (10). Thus, we designate the two different genotypes of mice as control (Setd2fl/del) and Setd2 cKO (Setd2fl/del, Stra8-GFPCre). Immunostaining verified that SETD2 protein was absent in the mutant germ cells (Fig. S2B). Notably, in Setd2 cKO mice, H3K36me3 signals were also completely absent in pachytene spermatocytes and round spermatids, whereas signal remained in preleptotene spermatocytes. These results further confirmed SETD2 as the major H3K36me3 methyltransferase in late-stage spermatocytes and round spermatids but not in early-stage spermatocytes (Fig. S2C). We also found that the levels of H3K9me2, H3K9me3, H3K27me3, H3K79me3, and H3K27Ac were not affected in the mutants, indicating that Setd2 loss did not cause global changes of other histone modifications (Fig. S2D).
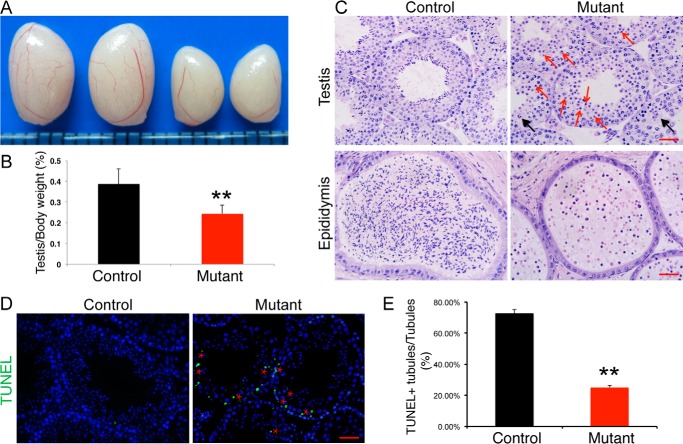
Although the Setd2 cKO males copulate normally, they were completely sterile. Testes from adult Setd2 cKO mice were significantly smaller than those of the littermate controls (Fig. 2, A and B; age, 8 weeks; control, 0.3867 ± 0.0746 g; Setd2 cKO mice, 0.2425 ± 0.0416 g; p < 0.001, n = 8). Histological examinations of adult germ cell mutant testes show that the seminiferous tubules are devoid of mature spermatozoa and contain exclusively round-headed spermatids. Moreover, round spermatids in Setd2 cKO mice did not undergo cellular elongation, indicating a developmental arrest before step 8 spermatids (Fig. 2C). Consistent with this, no sperm was found in the epididymis of germ cell mutants (Fig. 2C). These results suggest that Setd2 plays an essential role in spermatogenesis, and its loss leads to developmental arrests at round-spermatid stage.
Figure 2.
Impaired spermiogenesis in Setd2 cKO mice. A, gross morphology of representative testes from an 8-week-old control and age-matched Setd2 cKO mice. B, comparison of testis weight from 8-week-old controls and Setd2 cKO mice. Data are presented as mean ± S.D. (n = 8 for each genotype). C, H&E staining of testis and epididymis from control and Setd2 cKO adult mice. Red arrows indicate apoptotic spermatocytes and spermatids; black arrows represent multinucleated giant cells (formed by arrested spermatids). D, TUNEL staining of representative testes from adult control and age-matched Setd2 cKO mice. Apoptotic cells were labeled by TUNEL staining (green). The red stars indicate the spermatocyte and spermatid layer. E, comparison of TUNEL-positive seminiferous tubules from adult controls and Setd2 cKO mice. Data are presented as mean ± S.D. (n = 3 for each genotype). Error bars represent S.D. **, p < 0.01, t test (scale bars, 40 μm).
Close examination of the mutant seminiferous tubules showed a high frequency of degenerated germ cells in the spermatocyte and spermatid layers. TUNEL staining further identified a dramatic increase of apoptotic spermatocytes and spermatids in the mutant testes (Fig. 2, D and E). To distinguish whether the defects in spermatocytes resulted from an intrinsic requirement of Sedt2 function or secondary effects of impaired spermiogenesis, we examined the first occurrence of the apoptotic germ cells in Setd2 cKO mouse testes. At 2 weeks old, when the pachytene spermatocytes start to occur in the testis, we found that there was no significant difference between Setd2 cKO and control mouse testes (Fig. S3, A and C). In 4-week-old mice, when the first population of spermatids reaches the elongated spermatid in the control testes, elevated apoptosis in germ cells was observed in Setd2 cKO mouse testes (Fig. S3, B and C). Similar apoptotic spermatocyte defects are also observed in Crem, Trf2, Miwi, Tdrd5, and Mrg15 mutants, although these mutants show impaired spermatid differentiation (39–45). Taken together, these findings suggest that, in Setd2 cKO mouse testes, spermatocyte defects resulted from the spermiogenic arrest.
To verify the requirement for Setd2 in spermiogenesis but not in the early stages of spermatogenesis, we conditionally deleted Setd2 allele in germ cells as early as embryonic day 15 using the Vasa-Cre transgenic line (Setd2fl/del, Vasa-Cre). Inactivation of the Setd2 in gonocytes resulted in a similar phenotype, and the loss of H3K36me3 was only observed in pachytene spermatocytes and spermatids as shown in Setd2fl/del, Stra8-GFPCre mice (Fig. S4, A–C). Thus, these results further demonstrated an essential role for Setd2 and its associated H3K36me3 in spermiogenesis.
Loss of Setd2 results in malformation of acrosome
To elucidate the nature of the defects in spermiogenesis of Setd2 cKO mice, we performed immunostaining using FITC-conjugated PNA, capable of binding the outer acrosomal membrane, and anti-Golgi 58K antibody, a marker for the proacrosome in spermatids (46). Compared with a single acrosomal vesicle in the Golgi phase at steps 2 and 3 of spermiogenesis in controls (Fig. 3A), we found several acromsomal vesicle-like structures in Golgi-phase spermatids of Setd2 cKO mice (Fig. 3B), indicating that disruption of Setd2 could lead to acrosomal malformation as early as in the Golgi phase during spermiogenesis. In the cap phase at steps 4–7 of spermiogenesis, the acrosome expanded to form a caplike structure covering the nucleus of the round spermatids (Fig. 3A). However, in Setd2 cKO mouse spermatids, the caplike acrosome failed to form, resulting in a discontinuous acrosomal structure in the anterior region of the nucleus (Fig. 3B).
Figure 3.
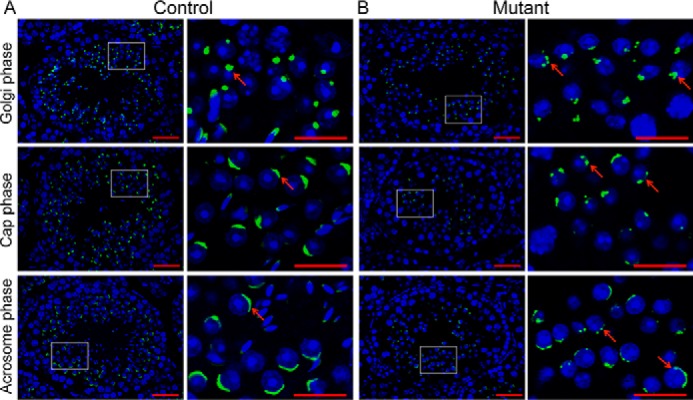
Abnormal acrosome biogenesis in Setd2 cKO mouse spermatids. Shown is histochemical staining of FITC-conjugated PNA (green) and DAPI (blue) in sections of adult control (A) and Setd2 cKO (B) mouse testes. The Golgi, cap, and acrosome phases of spermiogenesis are shown in order. A, PNA staining of representative testes from adult controls. Acrosomes are labeled by PNA (green). The arrow indicates a representative acrosome. In the Golgi phase, the acrosome displays a single granule close to the nucleus. In the cap phase, the acrosome grows to form the cap structure covering the nucleus. In the acrosome phase, the acrosome forms the moon-shaped structure covering the nucleus. B, PNA staining of representative testes from adult Setd2 cKO mice. There are multiple PNA-positive structures (as indicated by the arrows) in Setd2 cKO mouse spermatids throughout spermiogenesis. Right panels show higher-magnification views of the framed areas in the left panels (scale bars, 40 μm).
To characterize the defective acrosome biogenesis in spermiogenesis of Setd2 cKO mice in further detail, we performed a transmission EM (TEM) study (Fig. 4). Consistent with immunofluorescence analyses, the first acrosomal defects were detected in the Golgi-phase spermatids in which several acrosomal structures appeared in Setd2 cKO mouse round spermatids, whereas a single uniformed acrosomal structure was present in control spermatids (Fig. 4, A and B). Compared with control spermatids, acrosomal cap structures were discontinuous and abnormally fragmented in Setd2 cKO mouse spermatids at the cap phase (Fig. 4, C and D). It is of note that the fragmented acrosome persisted in all subsequent stages of spermiogenesis in Setd2 cKO mutants. Taken together, these observations reveal that Setd2 plays crucial roles in acrosome biogenesis and is required for spermiogenesis.
Figure 4.
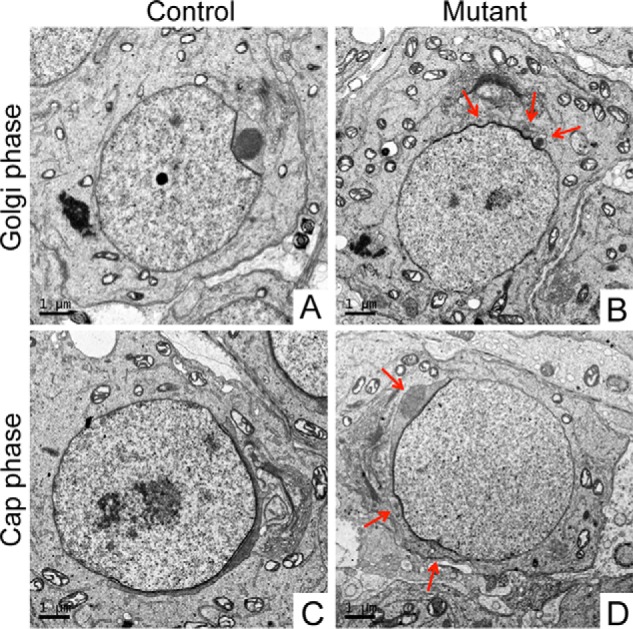
Abnormal acrosomal morphology of Setd2 cKO mouse spermatids. Shown are TEM analyses in ultrathin sections of spermatids from adult control (A and C) and Setd2 cKO (B and D) mice. A, in the Golgi phase of control spermatids, a single acrosomal granule is observed attached to the nuclear envelope. B, in the Golgi phase of Setd2 cKO mouse spermatids, as indicated by the arrows, multiple acrosomal vesicular structures are seen. C, in the cap phase, the acrosome flattens and forms a cap covering one pole of the nucleus. D, in the cap phase of Setd2 cKO mouse spermatids, as indicated by arrows, multiple acrosomal vesicular structures are observed (scale bars, 1 μm).
Setd2 inactivation led to complete loss of H3K36me3 in pachytene spermatocytes and round spermatids
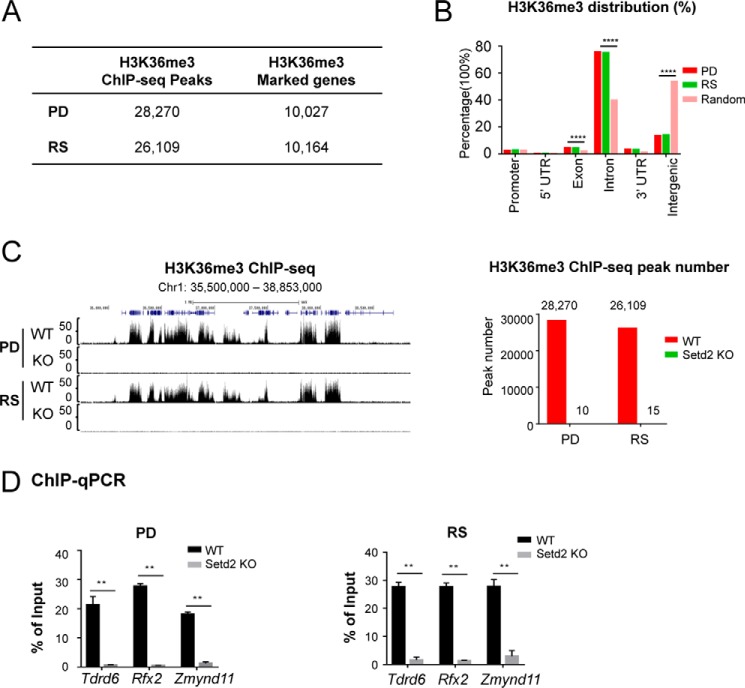
To better gain mechanistic insights into the spermiogenesis defects caused by Setd2 loss, we set out to determine the genome-wide distribution of H3K36me3 by chromatin immunoprecipitation (ChIP)-seq in the pachytene/diplotene spermatocytes (PDs) and round spermatids (RSs) from the control and mutant mice. Our H3K36me3 ChIP-seq analyses identified 28,270 and 26,109 peaks, which cover 10,027 and 10,164 genes in PD and RS cells, respectively (Fig. 5A). In both PD and RS cells, we found that H3K36me3 is mainly enriched in the gene bodies, especially the exonic, intronic, and 3′-UTR regions, in agreement with the canonical understanding of this histone mark (Fig. 5B). Consistent with our hypothesis of SETD2 being the major H3K36me3 methyltransferase in the late stages of spermatogenesis, we found that the mutant cells showed a drastic loss of H3K36me3 (Fig. 5C, normalized by spike-in approach; see “Experimental procedures”). The ChIP-seq results were also confirmed by ChIP-quantitative PCR (ChIP-qPCR) of a few select targets (Fig. 5D).
Figure 5.
Setd2 inactivation led to global loss of H3K36me3 in pachytene spermatocytes and round spermatids. A, table summarizing the numbers of H3K36me3 ChIP-seq peaks and H3K36me3-covered genes in PD and RS cells. B, genomic distribution of H3K36me3 ChIP-seq peaks in PD and RS cells compared with genome background. ****, p < 0.0001, one-sided binomial test. C, global loss of H3K36me3 in both PD and RS cells from Setd2 cKO mice. Left, UCSC Genome Browser track showing H3K36me3 ChIP-seq signals at a select genomic region in control and Setd2 cKO mice. Right, numbers of H3K36me3 ChIP-seq peaks in PD and RS cells from control and Setd2 cKO. D, global loss of H3K36me3 in Setd2 cKO mice was confirmed by ChIP-qPCR. q-PCR data are represented as mean ± S.D. from two biological replicates. **, p < 0.01, t test. Error bars represent S.D.
Setd2 loss led to misregulation of critical regulators in spermiogenesis
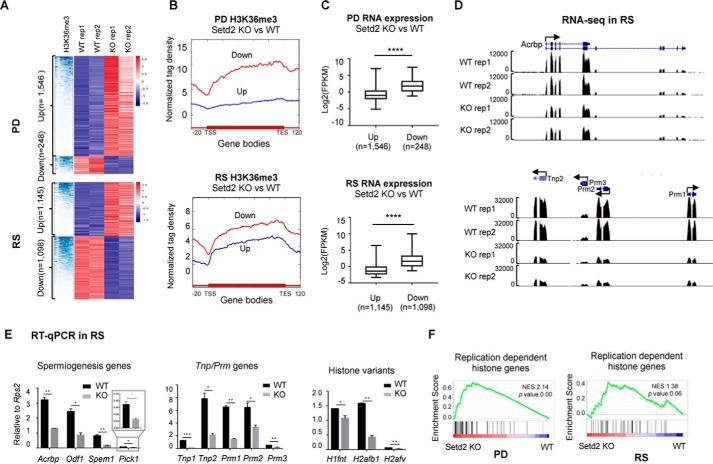
To further investigate the expression changes caused by Setd2 loss, we profiled the transcriptomes of the PD and RS from control and Setd2 cKO mouse testes, respectively. We found that Setd2 loss led to a total of 2,48 and 1,546 transcripts and of 1,088 and 1,145 transcripts significantly (false discovery rate <0.05, >2-fold difference) down- and up-regulated in PD and RS cells, respectively (Fig. 6A and Fig. S5A). Further analyses found that the down-regulated genes are generally marked with higher levels of H3K36me3 in contrast to the up-regulated genes, suggesting that the down-regulated expression is likely the direct effect of Setd2 loss (Fig. 6B). Consistent with the higher H3K36me3 level, the down-regulated genes were also expressed at higher levels than the up-regulated genes in the wide-type cells (Fig. 6C). Gene ontology (GO) analysis of the top-ranked differential genes indicated enrichment in genes associated with system development and motility for up-regulated genes and cell–cell signaling and cytoskeleton for down-regulated genes in RS, respectively (Fig. S5B), suggesting that the developmental defects caused by Setd2 loss might be due to the compromised functions of these pathways in RS.
Figure 6.
Compromised expression of key spermiogenesis regulators and genes involved in histone-to-protamine replacement in PD and RS upon Setd2 loss. A, hierarchical clustering analyses of H3K36me3 ChIP-seq and RNA-seq of the differentially expressed genes in PD and RS cells from control and Setd2 cKO. B, signal plot showing the H3K36me3 distribution over the up- and down-regulated genes in PD (p = 1.54e−92, analysis of variance test) and RS (p = 1.23e−34, analysis of variance test) cells upon Setd2 loss. C, box plot analyses of the expression levels of the up- and down-regulated genes in PD and RS cells upon Setd2 loss. ****, p < 0.0001 by unpaired t test. D and E, expression analyses of the indicate genes in PD and RS cells upon Setd2 loss. D, snapshots showing RNA-seq results. E, RT-qPCR confirmation. Data are represented as mean ± S.D. from two biological replicates. *, p < 0.05; **, p < 0.01, ***, p < 0.001 by t test. Error bars represent S.D. F, gene set enrichment analyses of histone gene expression in PD and RS cells from Setd2 cKO versus control. Normalized enrichment score (NES) and nominal p value are provided according to gene set enrichment analyses. TSS, transcription start site; TES, transcription end site.
It is of note that the expressions of many critical regulators for spermiogenesis, such as Acrbp1, Pick1, Odf1, and Spem1, were also significantly impaired upon Setd2 loss (Fig. 6D and Fig. S5C), which was also confirmed by RT-qPCR (Fig. 6E) (47–50). Among these genes, both Acrbp1 and Pick1 are known to be required for acrosome development, and knockout of either Acrbp1 or Pick1 led to fragmentation of the acrosome, which is similar to that seen in Setd2 mutants. Moreover, loss of Spem1 resulted in sperm deformation. Thus, the malformation of the acrosome caused by Setd2 loss could be due to the misregulation of these genes, which are likely SETD2 direct targets.
Impaired activation of transition nuclear protein and protamine genes in Setd2 cKO mutants
Among the deregulated genes identified in Setd2 cKO mice, we also noticed aberrant expressions of many replication-dependent histone genes (Fig. 6F). It is well-known that, during late stages of sperm differentiation, the majority of the core histones are subsequently replaced by transition nuclear proteins (TNPs) and protamines (PRMs), leading to the formation of highly condensed chromatin in elongated spermatids (51). Either Tnp- or Prm-deficient mice are infertile due to disruption of chromatin condensation in elongating spermatids (52, 53). Such processes are also accompanied with a coordinated and gradual transcriptional silencing of the canonical replication-dependent histone genes and activation of the germline-specific histone and Tnp genes. Consistent with the spermatid developmental defects of the Setd2 cKO mice, we identified significant down-regulation of Tnps, Prm1/2/3, and several testis-specific histone variant genes, such as H1fnt, H2afb1, and H2afv, accompanied with up-regulation of many replication-dependent histone genes (Fig. 6F) in both isolated pachytene spermatocyte and purified round spermatids. These observations were further validated by RT-qPCR (Fig. 6E). These findings indicate that the transition from histone-to-protamine replacement could also be compromised upon Setd2 loss, thereby contributing to the defective spermiogenesis.
Discussion
SETD2 is the major methyltransferase of histone H3K36me3, a mark associated with active transcription. Here, we show that SETD2 is the primary H3K36me3 methyltransferase in PD and RS and is essential for spermiogenesis. Loss of Setd2 led to complete loss of H3K36me3 in PD and RS cells and stalled development at round-spermatid stage, thereby resulting in sterility. Mechanistically, we found that the activation of several critical master regulators for spermiogenesis is impaired in Setd2 cKO germ cells accompanied with compromised expression of motility-related genes. Interestingly, we also found that the expressions of many histone genes and Tnp as well as protamine genes were significantly affected, indicating an impaired histone-to-protamine transition during spermiogenesis. Taken together, we have revealed an essential role of Setd2 during spermiogenesis but not early stages of spermatogenesis.
SETD2-mediated H3K36me3 is a transcriptionally coupled histone methylation mark, enriched over gene bodies undergoing active transcription (31). Although the enzymology and molecular function of SETD2 have been well-documented, the biological consequences of Sedt2 inactivation in animals and at the organism level have not been well-explored. To understand its biological significance during spermatogenesis, we generated a Setd2 cKO model in this study. Consistent with previously reports, we found that H3K36me3 patterns in PD and RS are also enriched in gene bodies in PD and RS cells. Setd2 loss leads to expression changes of hundreds to thousands of genes in PD and RS cells (Fig. 6A; false discovery rate <0.05, >2-fold difference). However, because H3K36me3 is a mark associated with active transcription, we were surprised to find fewer down-regulated genes than up-regulated genes in both PD and RS cells from Setd2 cKO mice. Further analyses found that the down-regulated genes are generally marked by higher levels of H3K36me3 in the WT germ cells (Fig. 6, B and C) and are expressed at higher levels. These observations not only suggest that the down-regulated expression is likely the direct effect of Setd2 loss but also indicate that Setd2 and H3K36me3 are required for transcriptional activities of more highly expressed genes, which may play important roles in spermiogenesis. In contrast, the up-regulated genes are generally expressed at low levels, and the elevated expression may be due to the compromised distribution of transcriptional machineries upon global loss of H3K36me3. Indeed, we observed significant impaired expression of critical spermiogenesis regulators, including Acrbp1, Pick1, Odf1, Spem1, Tnp, and Prm genes, and misexpression of histone genes, which normally undergo transcriptional silencing at these stages. For example, loss of either Acrbp1 or Pick1 resulted in acrosomal fragmentation similar to that observed in Setd2 mutants. This suggests that malformation of the acrosome in Setd2 mutants results from decreased expression of Acrbp1 and Pick1. Moreover, down-regulation of Tnp and Prm genes could disrupt histone-to-protamine transition, thereby causing the spermatid chromatin condensation defects. However, we cannot directly observe the spermatid chromatin condensation phenotype because arrest of spermiogenesis in Setd2 mutants occurred long before spermatid chromatin condensation. Thus, dysregulation of multiple pathways might together contribute to the spermatid developmental defects in Setd2 cKO mice.
Notably, at global levels, we found that SETD2 and H3K36me3 patterns displayed dissimilarities during spermatogenesis. The highest level of SETD2 was observed in pachytene spermatocytes and postmeiotic germ cells in the adult mouse testes, whereas strong H3K36me3 levels are maintained throughout all stages of spermatocytes and round spermatids with spermatogonia and elongated spermatids showing weaker H3K36me3. Importantly, we found that the H3K36me3 level was largely unchanged in preleptotene spermatocytes of the Setd2 cKO mice when compared with control littermates. Such findings suggested that H3K36me3 might be subjected to the regulation of other methyltransferases in the early stages of spermatogenesis. In support of this, Prdm9 was recently reported to mediate noncanonical H3K36me3 at hot spots of recombination sites during meiosis (54). As the epigenome undergoes rapid programming during spermatogenesis, whether there are other unknown methyltransferases responsible for H3K36me3, especially in the early stages of spermatogenesis, will need further investigation.
Experimental procedures
Mice
The Setd2 conditional knockout mouse line was produced by Shanghai Biomodel Organism Co. as shown in Fig. S2A. The germline-specific deletion of Setd2 was generated by crossing with Stra8-GFPCre mice or Vasa-Cre mice as described previously (10). All mice were maintained on the C57BL/6J background. All animal experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee at Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
Histology, immunofluorescence, and TUNEL staining
For testis histology, Bouin's fixed sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E). For immunofluorescence analysis, sections were retrieved in 10 mm sodium citrate buffer (pH 6.0) by boiling in a microwave and further washed in PBS with 0.1% Triton X-100. The sections were blocked with blocking buffer (10% donkey serum and 0.1% Triton X-100 in PBS) and then incubated with the primary antibodies overnight at 4 °C followed by Alexa Fluor 488– and Alexa Fluor 594–conjugated donkey secondary antibodies (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. The fluorescent sections were mounted with Prolong Gold Antifade medium containing DAPI (Molecular Probes) and then analyzed by fluorescence microscopy (Olympus) or confocal microscopy (Olympus). Apoptotic cells were analyzed using the In Situ Cell Death Detection kit, Fluorescein (Roche Applied Science) according to the manufacturer's instructions. The following primary antibodies were used in this study: rabbit anti-SETD2 (LS-B12260 and LS-C332416, LSBio), rabbit anti-H3K36me3 (4909S, Cell Signaling Technology), mouse anti-γH2AX (05-636, Millipore), goat anti-GATA4 (SC-1237, Santa Cruz Biotechnology), and FITC-conjugated PNA (L7381, Sigma).
Isolation of pachytene/diplotene spermatocytes and round spermatids
Spermatocytes and round spermatids were enriched using Hoechst 33342/propidium iodide–based fluorescence-activated cell sorting (FACS) as described previously with minor modifications (55). In brief, testes from adults were collected in PBS and placed on ice. After removal of the tunica albuginea and interstitial testicular cells, the seminiferous tubules were digested with PBS containing 120 units/ml collagenase type I (Worthington), further digested with 0.25% trypsin (Gibco) and 5 mg/ml DNase I (Gibco), and then terminated by adding fetal bovine serum (FBS). The dissociated testicular cell suspension was collected and resuspended at a concentration of 1 × 106 cells/ml in DMEM (Hyclone) with 0.1 mg/ml DNase I. The testicular cell suspension was then stained with Hoechst 33342 at a final concentration of 6 μg/ml for 30 min and stained with propidium iodide prior to sorting immediately. Sorting of the spermatocytes and round spermatids was performed using flow cytometric analysis (BD Biosciences). The collected spermatocytes contained ∼85 and ∼10% of pachytene and diplotene spermatocytes, respectively, based on nuclear spreading analysis. About 90% of the sorted spermatids were round spermatids as determined by PNA staining.
Western blotting
The sorted pachytene spermatocytes and round spermatids were lysed in 2× SDS loading buffer, and a total of 10 μg of whole cell extracts were loaded to each lane for Western blot analyses. Antibodies used in this study were H3K9me2 (A2359, ABclonal), H3K9me3 (A2360, ABclonal), H3K27me3 (39158, Active Motif), H3K27ac (39133, Active Motif), H3K36me3 (4909S, Cell Signaling Technology), H3K79me3 (ab2621, Abcam), and H3 (7168-1-AP, Proteintech).
Transmission electron microscopy
Fresh testes were fixed in 2.5% (v/v) glutaraldehyde in 0.1 m phosphate buffer, pH 7.4, for 2 h at 4 °C; washed with 0.1 m phosphate buffer; postfixed in 2% OsO4 for 1.5 h; dehydrated in a graded ethanol series before transfer to acetone; and embedded in Poly/Bed 812. Ultrathin sections were taken with a Leica EM UC7 ultramicrotome (Leica, Inc.), doubly stained with uranyl acetate and Reynold's lead citrate, and then imaged on a FEI Tecnai G2 Spirit TEM (FEI Co.) at 120-kV accelerating voltage.
ChIP and ChIP-seq
ChIP assay was carried out as described previously (56). The sorted pachytene spermatocytes and round spermatids were cross-linked with 1% formaldehyde for 10 min and then stopped by adding 125 mm glycine. Chromatin samples were lysed with lysis buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS) and sonicated using a Qsonica R2. A total of 2 μg of H3K36me3 antibody (4909S, Cell Signaling Technology) was incubated with each chromatin sample overnight at 4 °C together with 0.5 μg of spike-in antibody (61686, Active Motif) and 20 ng of spike-in chromatin (53083, Active Motif) for normalization purposes. The protein–DNA complexes were immobilized on 15 μl of protein A/G beads (SA032005, Smart Lifesciences Inc., Changzhou, China) and then washed four times with lysis buffer, twice with low-salt buffer (10 mm Tris-HCl, 250 mm LiCl, 1 mm EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate), and once with 10 mm Tris-HCl, pH 8.0. Decross-linking was carried out in elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS) at 65 °C for 5 h. Proteinase K and RNase A digestion was performed at 55 °C for 1 h. DNA samples were then purified using a PCR extraction kit (28006, Qiagen) and analyzed by real-time PCR or prepared for deep sequencing according to the manufacturer's guidelines (KK8503, Kapa Biosystems).
RNA-seq and qRT-PCR
RNA samples were prepared using an RNeasy minikit (Qiagen). mRNA pools were enriched from total RNA by poly(A) beads. Barcoded RNA-seq libraries were constructed using an NEBNext Ultra Directional RNA Library Prep kit for Illumina according to the manufacturer's instructions (New England Biolabs). Nonstrand-specific pair end 150-bp sequencing was by X-Ten (Illumina, run by WuXi App Tec). qRT-PCR was performed using a SuperScript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen).
ChIP-seq peak calling and distribution analysis
ChIP-seq reads were mapped with Bowtie (version 0.12.7) (parameters, −m 1). Duplicate reads were removed by Samtools. The mapped unique and monoclonal reads were extended to 350 bp based on the average sonicated chromatin DNA length. H3K36me3 peaks were called using MACS1.4 with default settings. For H3K36me3 distribution analysis, the ChIP-seq signals over gene bodies of the up- and down-regulated genes in Setd2 mutant PD and RS cells were analyzed by a homemade script called Signalplot based on the normalized ChIP-seq tag density; all the gene bodies were aligned as the same length.
Heat map analysis
The RNA-seq heat map were generated based on the expression -fold changes in WT and Setd2 mutant cells using Pheatmap, which is an R package. The H3K36me3 heat map over gene bodies was generated using Deeptools2.0.
RNA-seq–related bioinformatics analysis
Low-quality bases and adapter-containing reads were trimmed from raw data by Trim-galore with default parameters. Then the remaining trimmed sequence were mapped against the reference genome (mm9) with Bowtie/TopHat version 2, which allowed mapping across splicing junctions by read segmentation. All programs were performed with default settings unless otherwise specified. Unique mapped reads were subsequently assembled into transcripts guided by reference RefSeq annotation with Cufflinks. The expression level of each gene was quantified with normalized FPKM with FPKM_count in RSeQC software. Differentially expressed genes were identified by asking for a log2(-fold change) ≥1 or ≤−1 with FPKM >0.3 in at least one sample.
GO analysis
GO analysis for differentially expressed genes was performed on the database for annotation, visualization and integrated discovery (DAVID) website.
Author contributions
X. Z., B. R., and R. L. formal analysis; X. Z. and B. R. investigation; L. L. resources; F. L. and M.-H. T. supervision; F. L. and M.-H. T. funding acquisition; F. L. and M.-H. T. writing-original draft.
Supplementary Material
Acknowledgments
We thank the Histology, Flow Cytometry, and Vivarium services at Shanghai Institute of Biochemistry and Cell Biology.
This work was supported by National Natural Science Foundation of China Grant 31671553, Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB19000000, Ministry of Science and Technology of China Grant 2014CB943101, National Key Research and Development Program of China Grant 2016YFC1000600, Science and Technology Commission of Shanghai Municipality Grant 17JC1420102, and the Shanghai Institutes for Biological Sciences foundation (to M.-H. T.); State Key Development Program Grants 2014CB943103 and 2016YFA0101800, National Natural Science Foundation of China Grant 31371303, and Science and Technology Commission of Shanghai Municipality Major Project 2017SHZDZX01 (to F. L.); and National Natural Science Foundation of China Grant 81772938 and Science and Technology Commission of Shanghai Municipality Grants 16140902100 and 18140902700 (to L. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
H3K36me3 ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE108717.
- H3K36me3
- trimethylation of histone H3 lysine 36
- SETD2
- SET domain–containing 2
- seq
- sequencing
- PNA
- peanut agglutinin
- cKO
- conditional knockout
- H3K9me2
- dimethylation of histone H3 lysine 9
- H3K27Ac
- acetylation of histone H3 lysine 27
- TEM
- transmission EM
- PD
- pachytene/diplotene spermatocyte
- RS
- round spermatid
- qPCR
- quantitative PCR
- GO
- gene ontology
- TNP
- transition nuclear protein
- PRM
- protamine
- qRT-PCR
- quantitative RT-PCR
- FPKM
- fragments per kilobase of transcript per million mapped reads.
References
- 1. Griswold, M. D. (2016) Spermatogenesis: the commitment to meiosis. Physiol. Rev. 96, 1–17 10.1152/physrev.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clermont, Y. (1972) Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev. 52, 198–236 10.1152/physrev.1972.52.1.198 [DOI] [PubMed] [Google Scholar]
- 3. de Rooij, D. G. (2001) Proliferation and differentiation of spermatogonial stem cells. Reproduction 121, 347–354 10.1530/rep.0.1210347 [DOI] [PubMed] [Google Scholar]
- 4. Zickler, D., and Kleckner, N. (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hermo, L., Pelletier, R. M., Cyr, D. G., and Smith, C. E. (2010) Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 73, 241–278 10.1002/jemt.20783 [DOI] [PubMed] [Google Scholar]
- 6. Hermo, L., Pelletier, R. M., Cyr, D. G., and Smith, C. E. (2010) Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc. Res. Tech. 73, 279–319 10.1002/jemt.20787 [DOI] [PubMed] [Google Scholar]
- 7. Hermo, L., Pelletier, R. M., Cyr, D. G., and Smith, C. E. (2010) Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 5: intercellular junctions and contacts between germs cells and Sertoli cells and their regulatory interactions, testicular cholesterol, and genes/proteins associated with more than one germ cell generation. Microsc. Res. Tech. 73, 409–494 10.1002/jemt.20786 [DOI] [PubMed] [Google Scholar]
- 8. Hermo, L., Pelletier, R. M., Cyr, D. G., and Smith, C. E. (2010) Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 4: intercellular bridges, mitochondria, nuclear envelope, apoptosis, ubiquitination, membrane/voltage-gated channels, methylation/acetylation, and transcription factors. Microsc. Res. Tech. 73, 364–408 10.1002/jemt.20785 [DOI] [PubMed] [Google Scholar]
- 9. Hermo, L., Pelletier, R. M., Cyr, D. G., and Smith, C. E. (2010) Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 3: developmental changes in spermatid flagellum and cytoplasmic droplet and interaction of sperm with the zona pellucida and egg plasma membrane. Microsc. Res. Tech. 73, 320–363 10.1002/jemt.20784 [DOI] [PubMed] [Google Scholar]
- 10. Lin, Z., Hsu, P. J., Xing, X., Fang, J., Lu, Z., Zou, Q., Zhang, K. J., Zhang, X., Zhou, Y., Zhang, T., Zhang, Y., Song, W., Jia, G., Yang, X., He, C., and Tong, M. H. (2017) Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 27, 1216–1230 10.1038/cr.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. An, J., Zhang, X., Qin, J., Wan, Y., Hu, Y., Liu, T., Li, J., Dong, W., Du, E., Pan, C., and Zeng, W. (2014) The histone methyltransferase ESET is required for the survival of spermatogonial stem/progenitor cells in mice. Cell Death Dis. 5, e1196 10.1038/cddis.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Legrand, J. M. D., and Hobbs, R. M. (2018) RNA processing in the male germline: Mechanisms and implications for fertility. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2017.10.006 [DOI] [PubMed] [Google Scholar]
- 13. Stewart, K. R., Veselovska, L., and Kelsey, G. (2016) Establishment and functions of DNA methylation in the germline. Epigenomics 8, 1399–1413 10.2217/epi-2016-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rathke, C., Baarends, W. M., Awe, S., and Renkawitz-Pohl, R. (2014) Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta 1839, 155–168 10.1016/j.bbagrm.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 15. de Mateo, S., and Sassone-Corsi, P. (2014) Regulation of spermatogenesis by small non-coding RNAs: role of the germ granule. Semin. Cell Dev. Biol. 29, 84–92 10.1016/j.semcdb.2014.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dhar, S., Thota, A., and Rao, M. R. (2012) Insights into role of bromodomain, testis-specific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. J. Biol. Chem. 287, 6387–6405 10.1074/jbc.M111.288167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaucher, J., Boussouar, F., Montellier, E., Curtet, S., Buchou, T., Bertrand, S., Hery, P., Jounier, S., Depaux, A., Vitte, A. L., Guardiola, P., Pernet, K., Debernardi, A., Lopez, F., Holota, H., et al. (2012) Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 31, 3809–3820 10.1038/emboj.2012.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayashi, K., Yoshida, K., and Matsui, Y. (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378 10.1038/nature04112 [DOI] [PubMed] [Google Scholar]
- 19. Iwamori, N., Iwamori, T., and Matzuk, M. M. (2013) H3K27 demethylase, JMJD3, regulates fragmentation of spermatogonial cysts. PLoS One 8, e72689 10.1371/journal.pone.0072689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim, S., Namekawa, S. H., Niswander, L. M., Ward, J. O., Lee, J. T., Bardwell, V. J., and Zarkower, D. (2007) A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 3, e62 10.1371/journal.pgen.0030062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambrot, R., Lafleur, C., and Kimmins, S. (2015) The histone demethylase KDM1A is essential for the maintenance and differentiation of spermatogonial stem cells and progenitors. FASEB J. 29, 4402–4416 10.1096/fj.14-267328 [DOI] [PubMed] [Google Scholar]
- 22. Lu, L. Y., Wu, J., Ye, L., Gavrilina, G. B., Saunders, T. L., and Yu, X. (2010) RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell 18, 371–384 10.1016/j.devcel.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mu, W., Starmer, J., Fedoriw, A. M., Yee, D., and Magnuson, T. (2014) Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 28, 2056–2069 10.1101/gad.246124.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okada, Y., Scott, G., Ray, M. K., Mishina, Y., and Zhang, Y. (2007) Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123 10.1038/nature06236 [DOI] [PubMed] [Google Scholar]
- 25. Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schöfer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., Opravil, S., Doyle, M., Sibilia, M., and Jenuwein, T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- 26. Tachibana, M., Nozaki, M., Takeda, N., and Shinkai, Y. (2007) Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 26, 3346–3359 10.1038/sj.emboj.7601767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang, Y., Zhu, T., Li, Q., Liu, C., Han, F., Chen, M., Zhang, L., Cui, X., Qin, Y., Bao, S., and Gao, F. (2015) Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 5, 11031 10.1038/srep11031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan, W., Si, Y., Slaymaker, S., Li, J., Zheng, H., Young, D. L., Aslanian, A., Saunders, L., Verdin, E., and Charo, I. F. (2010) Zmynd15 encodes a histone deacetylase-dependent transcriptional repressor essential for spermiogenesis and male fertility. J. Biol. Chem. 285, 31418–31426 10.1074/jbc.M110.116418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edmunds, J. W., Mahadevan, L. C., and Clayton, A. L. (2008) Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 27, 406–420 10.1038/sj.emboj.7601967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fontebasso, A. M., Schwartzentruber, J., Khuong-Quang, D. A., Liu, X. Y., Sturm, D., Korshunov, A., Jones, D. T., Witt, H., Kool, M., Albrecht, S., Fleming, A., Hadjadj, D., Busche, S., Lepage, P., Montpetit, A., et al. (2013) Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 125, 659–669 10.1007/s00401-013-1095-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner, E. J., and Carpenter, P. B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou, V. W., Goren, A., and Bernstein, B. E. (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet 12, 7–18 10.1038/nrg2905 [DOI] [PubMed] [Google Scholar]
- 33. Zhu, X., He, F., Zeng, H., Ling, S., Chen, A., Wang, Y., Yan, X., Wei, W., Pang, Y., Cheng, H., Hua, C., Zhang, Y., Yang, X., Lu, X., Cao, L., et al. (2014) Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 46, 287–293 10.1038/ng.2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuan, H., Li, N., Fu, D., Ren, J., Hui, J., Peng, J., Liu, Y., Qiu, T., Jiang, M., Pan, Q., Han, Y., Wang, X., Li, Q., and Qin, J. (2017) Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J. Clin. Investig. 127, 3375–3391 10.1172/JCI94292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu, M., Sun, X. J., Zhang, Y. L., Kuang, Y., Hu, C. Q., Wu, W. L., Shen, S. H., Du, T. T., Li, H., He, F., Xiao, H. S., Wang, Z. G., Liu, T. X., Lu, H., Huang, Q. H., et al. (2010) Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 2956–2961 10.1073/pnas.0915033107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang, Y., Xie, S., Zhou, Y., Xie, Y., Liu, P., Sun, M., Xiao, H., Jin, Y., Sun, X., Chen, Z., Huang, Q., and Chen, S. (2014) H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep. 8, 1989–2002 10.1016/j.celrep.2014.08.031 [DOI] [PubMed] [Google Scholar]
- 37. Avilés, M., Castells, M. T., Martínez-Menarguez, J. A., Abascal, I., and Ballesta, J. (1997) Localization of penultimate carbohydrate residues in zona pellucida and acrosomes by means of lectin cytochemistry and enzymatic treatments. Histochem. J. 29, 583–592 10.1023/A:1026432211012 [DOI] [PubMed] [Google Scholar]
- 38. McClusky, L. M., Patrick, S., Barnhoorn, I. E., van Dyk, J. C., de Jager, C., and Bornman, M. S. (2009) Immunohistochemical study of nuclear changes associated with male germ cell death and spermiogenesis. J. Mol. Histol. 40, 287–299 10.1007/s10735-009-9240-3 [DOI] [PubMed] [Google Scholar]
- 39. Blendy, J. A., Kaestner, K. H., Weinbauer, G. F., Nieschlag, E., and Schütz, G. (1996) Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380, 162–165 10.1038/380162a0 [DOI] [PubMed] [Google Scholar]
- 40. Nantel, F., Monaco, L., Foulkes, N. S., Masquilier, D., LeMeur, M., Henriksén, K., Dierich, A., Parvinen, M., and Sassone-Corsi, P. (1996) Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380, 159–162 10.1038/380159a0 [DOI] [PubMed] [Google Scholar]
- 41. Martianov, I., Fimia, G. M., Dierich, A., Parvinen, M., Sassone-Corsi, P., and Davidson, I. (2001) Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7, 509–515 10.1016/S1097-2765(01)00198-8 [DOI] [PubMed] [Google Scholar]
- 42. Zhang, D., Penttila, T. L., Morris, P. L., Teichmann, M., and Roeder, R. G. (2001) Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292, 1153–1155 10.1126/science.1059188 [DOI] [PubMed] [Google Scholar]
- 43. Deng, W., and Lin, H. (2002) miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 10.1016/S1534-5807(02)00165-X [DOI] [PubMed] [Google Scholar]
- 44. Yabuta, Y., Ohta, H., Abe, T., Kurimoto, K., Chuma, S., and Saitou, M. (2011) TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 192, 781–795 10.1083/jcb.201009043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iwamori, N., Tominaga, K., Sato, T., Riehle, K., Iwamori, T., Ohkawa, Y., Coarfa, C., Ono, E., and Matzuk, M. M. (2016) MRG15 is required for pre-mRNA splicing and spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 113, E5408–E5415 10.1073/pnas.1611995113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mortimer, D., Curtis, E. F., and Miller, R. G. (1987) Specific labelling by peanut agglutinin of the outer acrosomal membrane of the human spermatozoon. J. Reprod. Fertil. 81, 127–135 10.1530/jrf.0.0810127 [DOI] [PubMed] [Google Scholar]
- 47. Zheng, H., Stratton, C. J., Morozumi, K., Jin, J., Yanagimachi, R., and Yan, W. (2007) Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc. Natl. Acad. Sci. U.S.A. 104, 6852–6857 10.1073/pnas.0701669104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanemori, Y., Koga, Y., Sudo, M., Kang, W., Kashiwabara, S., Ikawa, M., Hasuwa, H., Nagashima, K., Ishikawa, Y., Ogonuki, N., Ogura, A., and Baba, T. (2016) Biogenesis of sperm acrosome is regulated by pre-mRNA alternative splicing of Acrbp in the mouse. Proc. Natl. Acad. Sci. U.S.A. 113, E3696–E3705 10.1073/pnas.1522333113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao, N., Kam, C., Shen, C., Jin, W., Wang, J., Lee, K. M., Jiang, L., and Xia, J. (2009) PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J. Clin. Investig. 119, 802–812 10.1172/JCI36230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang, K., Meinhardt, A., Zhang, B., Grzmil, P., Adham, I. M., and Hoyer-Fender, S. (2012) The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Mol. Cell. Biol. 32, 216–225 10.1128/MCB.06158-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eddy, E. M. (1998) Regulation of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 9, 451–457 10.1006/scdb.1998.0201 [DOI] [PubMed] [Google Scholar]
- 52. Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y. C., Hecht, N. B., and Eddy, E. M. (2001) Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat. Genet. 28, 82–86 10.1038/88313 [DOI] [PubMed] [Google Scholar]
- 53. Zhao, M., Shirley, C. R., Hayashi, S., Marcon, L., Mohapatra, B., Suganuma, R., Behringer, R. R., Boissonneault, G., Yanagimachi, R., and Meistrich, M. L. (2004) Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis 38, 200–213 10.1002/gene.20019 [DOI] [PubMed] [Google Scholar]
- 54. Powers, N. R., Parvanov, E. D., Baker, C. L., Walker, M., Petkov, P. M., and Paigen, K. (2016) The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet. 12, e1006146 10.1371/journal.pgen.1006146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gaysinskaya, V., Soh, I. Y., van der Heijden, G. W., and Bortvin, A. (2014) Optimized flow cytometry isolation of murine spermatocytes. Cytometry A 85, 556–565 10.1002/cyto.a.22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lan, F., Collins, R. E., De Cegli, R., Alpatov, R., Horton, J. R., Shi, X., Gozani, O., Cheng, X., and Shi, Y. (2007) Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448, 718–722 10.1038/nature06034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.