Abstract
Conformations adopted by long stretches of single-stranded DNA (ssDNA) are of central interest in understanding the architecture of replication forks, R loops, and other structures generated during DNA metabolism in vivo. This is particularly so if the ssDNA consists of short nucleotide repeats. Such studies have been hampered by the lack of defined substrates greater than ∼150 nt and the absence of high-resolution biophysical approaches. Here we describe the generation of very long ssDNA consisting of the mammalian telomeric repeat (5′-TTAGGG-3′)n, as well as the interrogation of its structure by EM and single-molecule magnetic tweezers (smMT). This repeat is of particular interest because it contains a run of three contiguous guanine residues capable of forming G quartets as ssDNA. Fluorescent-dye exclusion assays confirmed that this G-strand ssDNA forms ubiquitous G-quadruplex folds. EM revealed thick bead-like filaments that condensed the DNA ∼12-fold. The bead-like structures were 5 and 8 nm in diameter and linked by thin filaments. The G-strand ssDNA displayed initial stability to smMT force extension that ultimately released in steps that were multiples ∼28 nm at forces between 6 and 12 pN, well below the >20 pN required to unravel G-quadruplexes. Most smMT steps were consistent with the disruption of the beads seen by EM. Binding by RAD51 distinctively altered the force extension properties of the G-strand ssDNA, suggesting a stochastic G-quadruplex-dependent condensation model that is discussed.
Keywords: single-molecule biophysics, telomere, G-quadruplex, DNA, electron microscopy (EM)
Introduction
DNA replication, transcription, recombination and repair all involve the generation of single-stranded DNA (ssDNA)4 segments (1–3). In some cases, such as resection of DNA ends during repair or creation of an uncoupled replication fork, these ssDNA segments potentially could span several hundred nucleotides or more in length (4). Mixed sequence ssDNA segments are assumed to behave as random coils with some (weak) secondary structure caused by local base pairing (5, 6). However, if the ssDNA is long and consists of arrays of nucleotide repeats that could fold into macroscopic structures this could profoundly affect subsequent events of DNA metabolism. One well-known example is the trinucleotide repeats linked to the triplet repeat diseases in which it is thought that as the replication fork passes these repeats, the ssDNA can fold into long hairpin structures and generate repeat expansions. The telomeric hexanucleotide repeat (5′-TTAGGG-3′) found in all mammals and many eukaryotes (7) provides another example of a repeating sequence with the potential of folding into large macroscopic structures when present in a single-stranded state. The telomeric repeat is a member of a subset of repetitive DNAs found widely across nature containing runs of three or four contiguous guanine bases that as ssDNA have the ability to form G-quadruplexes (8). Other examples include the replication origin of Kaposi's sarcoma-associated Herpesvirus (KSHV), which contains 26 and 22 short repeats each with three or four guanines each (9), as well as the C9orf72 gene locus, in which expansion of 5′-GGGGCC-3′ repeats from 2 to 24 in the genome to sometimes several thousand copies is the cause of the most common form of familial amyotropic lateral sclerosis and frontotemporal dementia (10). G-quadruplexes form when runs of two or more guanine residues in a DNA stack on each other in a square planar structure stabilized by Hoogsteen base pairing (8, 11, 12). They are further stabilized by certain cations, in particular K+ (13, 14). The identification of telomeric G-quadruplexes in vitro was first reported in 1989 (15, 16). The existence of G-quadruplexes in vivo was initially recognized by immunohistochemistry in the telomeric DNA of the ciliate Stylonychia lemnae (17, 18) and later throughout the human genome (19). Only a portion of the human cellular G-quadruplex immunohistochemistry signal was detected at telomeres consistent with the presence of multiple and likely different repeat sequences throughout the genome capable of forming G quadruplexes (19).
There has been growing interest in the potential role of G quadruplex formation at the ssDNA termini of mammalian and many other eukaryotic telomeres in which the G-rich strand extends beyond the dsDNA–ssDNA junction as a 3′-single-stranded overhang. In humans, this ssDNA extension is ∼150–300 nt in length (7), and studies have pointed to the formation of G quadruplexes in the overhang and their role in inhibiting telomerase, as well as regulating binding and action by the shelterins (20).
A 24-nt ssDNA consisting of (3′-TTAGGG-5′)4 has been crystalized as a model for the telomeric overhang. In the resulting structure, the DNA is arranged into a flat disk 0.6 nm in height and 4 nm on a side with the DNA adopting a parallel G-quadruplex conformation and with TTA triplets looping out in a propeller fashion (21). However, NMR studies and other solution approaches have argued that the 24-nt units exist in hybrid or anti-parallel conformations, suggesting that the parallel arrangement may be promoted by crystallization (22). Potential condensation of G-quadruplex folds into larger structures have been proposed but not experimentally demonstrated (23).
Telomeric repeats also exist in extremely long nucleic acid chains in the form of G-rich transcripts of telomeres termed TERRA (5′-UUAGGG-3′)n. In human cells, TERRA molecules of nearly 9000 nt in length have been observed (24, 25). We previously demonstrated that TERRA is arranged into chains of 24-nt particles joined by 3-nt linkers and provided evidence that these particles are stabilized by G-quadruplex folds (26). Thus, a 24-nt particle stabilized by G-quadruplex formation may be shared by both the telomeric G-rich ssDNA and RNA forms. Moreover, some TERRA RNA remains present in telomeric R-loops following transcription in many organisms (27). Telomeric R-loops have been shown to promote homologous recombination (HR) and are prevalent in a significant number of cell lines and tumors exhibiting the alternative lengthening of telomeres (ALT) phenotype (27).
It is important that we understand the kinds of macroscopic folding that ssDNA segments anywhere from 200 to 2000 nt or more in length can adopt at sites of transcription, recombination, or uncoupled replication forks. To do so requires generating very long ssDNA substrates so that any effect of DNA ends on the internal folding is minimized. In this study, we generated very long ssDNA (up to 20,000 nt) composed of the G-strand telomeric repeat (3′-TTAGGG-5′), the C-strand complement repeat (3′-AATCCC-5′), and a mutant G-strand repeat (3′-TTAGTG-5′). We then interrogated their structure using EM and single molecule magnetic tweezers (smMT) force extension analysis, two methods capable of providing structural information about very large nucleic acid molecules. We demonstrate that the G-strand telomeric ssDNA spontaneously condenses into a thick bead-like filament as imaged by EM and resolved by stepwise elongation following force extension analysis by smMT. Moreover, we show that switchable ATP-dependent RAD51 ssDNA binding could be used to probe the structure of these higher-order G-strand configurations.
Results
Preparation of long G-strand telomeric single-stranded DNA
Tandem copies of ssDNA containing the human G-strand (5′-TTAGGG-3′)n, repeat, the complementary C-strand (5′-CCCTAA-3′)n, repeat, and a mutant repeat disrupting the three-G run (5′-TTAGTG-3′)n were generated using a φ29 DNA polymerase rolling circle replication scheme from a mini-circle template as previously described (Fig. S1a) (28). The size distribution of the ssDNAs was determined by alkaline gel electrophoresis and displayed a broad distribution from 100 to >20,000 nt (Fig. S1b).
G-strand single-stranded DNA contains G-quadruplex structures
Numerous studies have indicated that telomeric G-strand ssDNA folds into G-quadruplex structures (15, 16, 29). CD analysis is frequently used to demonstrate the presence of G-quadruplex structures within DNA (30). However, CD studies require high molar concentrations of a single DNA species, and the long ssDNAs prepared by the rolling-circle method contain a mixture of many different lengths even following alkaline agarose gel isolation and purification, resulting in very low molar concentrations of any individual species. The spectra obtained and further discussion of the limitations are presented in Fig. S2 and Note S1.
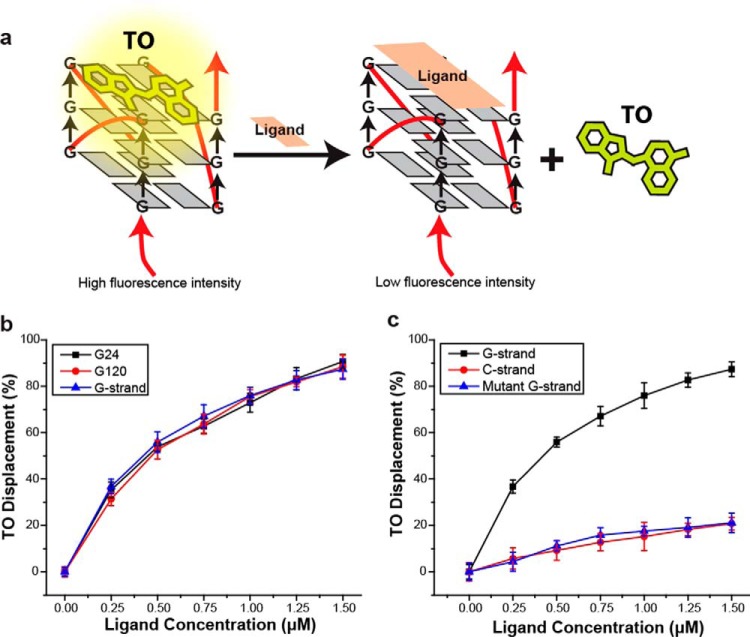
As an alternative, we utilized structure-specific G-quadruplex fluorescence intercalator displacement (G4-FID) analysis to probe the underlying structure of the long ssDNA generated by rolling circle replication (Fig. 1a) (31). The G4-FID assay was developed from similar assays of dsDNA. When thiazole orange (TO) binds DNA, it produces a strong fluorescence signal with high quantum yield while remaining nonfluorescent in the absence of DNA. Displacement from DNA may be monitored as a decrease in fluorescence. This assay is now used widely to determine selectivity of a candidate compound through its ability to displace TO from quadruplex DNA (31–33). We used the 3,6,9-trisubstituted acridine ligand BRACO-19, which was designed by molecular modeling as a G-quadruplex binding-specific candidate molecule. Previous work has shown that BRACO-19 inhibits telomerase activity, which results in telomere shortening and end-to-end chromosomal fusions in cancer cells as a consequence of quadruplex stabilization at the ssDNA overhang (34, 35). Indeed a crystal structure of the human telomeric G-quadruplex and BRACO-19 has been solved (36) and resembles the native unimolecular and bimolecular human telomeric quadruplexes structure (21) bound by TMPyP4, a well-known porphyrin based G-quadruplex binder (37). BRACO-19 was used earlier by another group as candidate G-quadruplex binder for G4-FID assays (33). We introduced increasing BRACO-19 to long G-strand ssDNA, which resulted in reduced TO fluorescence and increased the calculated TO displacement (Fig. 1b, blue triangle; EC50 = 0.44 μm). Moreover, the TO displacement of equivalent concentrations of the smaller (5′-TTAGGG-3′)4 (G24; Fig. 1b, black square; EC50 = 0.43 μm) and (5′-TTAGGG-3′)20 (G120; Fig. 1b, red circle; EC50 = 0.45 μm) G-quadruplex forming oligonucleotides by BRACO-19 appeared nearly identical to the long G-strand DNA. Importantly, we found that BRACO-19 displaced TO from the G-strand ssDNA (Fig. 1c, black square; EC50 = 0.42 μm) but not the complementary C-strand (Fig. 1c, red circle; EC50 = 7.3 μm) or the mutant G-strand ssDNA that disrupts the G-quadruplex repeat (Fig. 1c, blue triangle; EC50 = 7.2 μm). These observations are consistent with the conclusion that the long G-strand ssDNA forms G-quadruplex structures similar to well-defined G-quadruplex forming oligonucleotides that are known to bind the G-quadruplex specific ligand BRACO-19. In contrast, long C-strand and mutant G-strand ssDNA, which lack the G-quadruplex structure, do not bind BRACO-19.
Figure 1.
Fluorescence intercalator displacement analysis of G-quadruplex DNA. a, schematic representation of G4-FID analysis. The displacement of TO by the G-quadruplex binding-specific BRACO-19 ligand results in reduced fluorescence intensity. b, G4-FID analysis performed with 24 nt (G24), 120 nt (G120), and rolling circle-generated (G-strand) G-strand DNA. c, G4-FID analysis of rolling circle-generated G-strand, C-strand, and mutant G-strand DNA.
G-strand single-stranded DNA forms bead-like structures
The G-strand, C-strand, and mutant G-strand ssDNAs were prepared for EM by direct adsorption to thin carbon supports followed by tungsten metal shadow casting. The C-strand DNA appeared thin and lacked any discernable organization, typical of phage ssDNAs prepared by these methods (Fig. 2a). The mutant G-strand DNA (Fig. 2b) was also disorganized, although these molecules appeared to display more compaction that the C-strand DNA. We determined the width of tungsten metal shadow casting for the C-strand (1.8 ± 0.6 nm, n = 268) and found that it appeared approximately half the width of M13 dsDNA (2.9 ± 0.5 nm, S.E., n = 125) (Note S2). The results suggest that in these preparations both the C-strand ssDNA and M13 RF dsDNA were coated with 0.9 nm of tungsten metal to account for their known ∼1- and 2-nm width, respectively.
Figure 2.
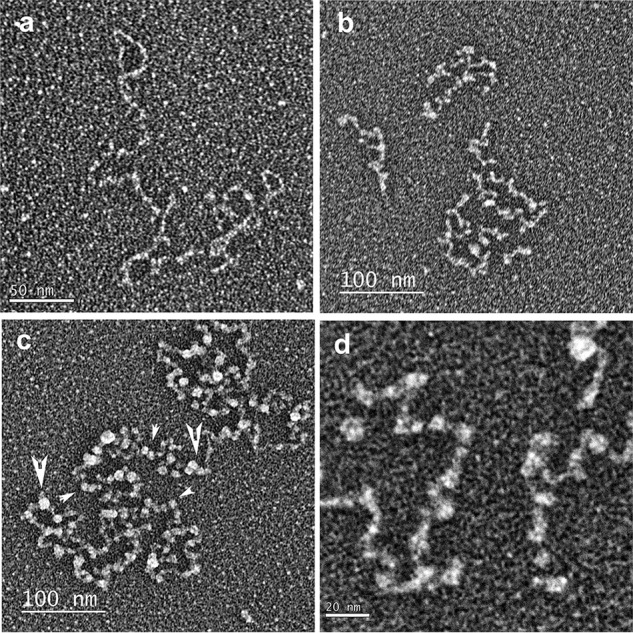
Electron microscopic visualization of long telomeric ssDNAs. Long telomeric ssDNA was synthesized by rolling circle replication and prepared for EM by direct adsorption to thin carbon supports, dehydration, and rotary metal shadow casting with tungsten. a, C-strand ssDNA. b, mutant (TTAGTG)n G-strand ssDNA. c, G-strand DNA. d, higher magnification of G-strand DNA. Small arrowheads indicate small bead-like structures; large arrowheads indicate large bead-like structures.
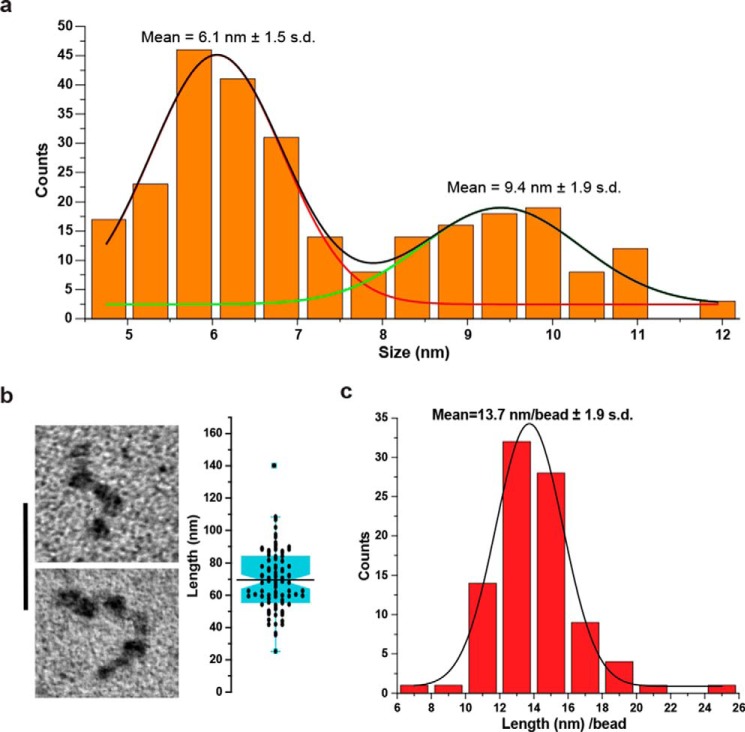
In the EM images, the G-strand ssDNA was remarkably different and consisted of thick condensed fibers with a bead-like appearance that were often linked by thin filaments similar in thickness to the C-strand DNA (Fig. 2, c and d, and Fig. S3). The diameter of these bead-like particles from multiple preparations revealed two size classes (Fig. 3a). The larger bead-like particles (9.4 ± 1.9 nm, S.D.; Fig. 2, c and d, large arrowheads) when corrected for tungsten metal deposition had an estimated mean diameter of 8.5 nm, whereas the smaller bead-like particles (6.1 ± 1.5 nm, S.D.; Fig. 2, c and d, small arrowheads) when corrected for tungsten metal deposition had an estimated mean diameter of 5.2 nm. In numerous cases, the larger particles appeared conjoined in a way that appeared to suggest an association of two smaller bead-like particles might generate the larger ones (see also Fig. S3 and Note S2). In all of the fields of the G-strand ssDNA, some molecules were less organized, suggesting either that other conformations may be possible or that the process of forming the beaded structures may be incomplete.
Figure 3.
Size and length distribution of G-strand bead-like structures. a, the diameter of G-strand bead-like structures was determined by EM (n = 271; see Fig. 2). The binned histogram was fit to two Gaussian curves, and the means and S.D. were determined (see “Experimental procedures”). b, representative EM examples of G-strand DNA saturated with large bead-like particles (left panel). The bar is equivalent to 50 nm. The box plot shows length measures determined by EM for G-strand DNA saturated with large bead-like particles (right panel; n = 91). c, binned histogram of individual G-strand DNA saturated with large bead-like particles length measures divided by the number of bead-like particles observed on each individual molecule (n = 91).
The G-strand ssDNA was subjected to alkaline-agarose gel electrophoresis, and a region corresponding to 1–1.5 knt was isolated, neutralized, and purified. This size-selected G-strand DNA (average of 1.25 knt) was prepared for EM in the presence of K+ cation, and the length and number of beads were determined for molecules that appeared fully saturated with mostly larger bead-like particles and no significant linker DNA (Fig. 3b, left panel). Compared with the calculated length of 875 nm for a random sequence 1.25 knt ssDNA (0.7 nm/nt for ssDNA) (38), the mean length of the saturated G-strand ssDNA (69.5 ± 18.9 nm, S.D.; n = 91; Fig. 3b, right panel) appeared compacted 12.6-fold. By dividing the length of each individual molecule by its distinct number of beads, we determined the mean length per bead from the binned values (13.7 ± 1.9 nm/bead, S.D.; n = 91; Fig. 3c); equivalent to 5 beads/1250 nt. Together, these observations suggest that there are ∼250 nt of G-strand ssDNA per large bead particle.
In the EM experiments described above, the DNA was adsorbed to the thin carbon foils in a buffer containing 2.5 mm spermidine (“Experimental procedures”) followed by dehydration through water-ethanol solutions and air-drying. Although this approach has been found to faithfully reveal structures in DNA in past studies and provides the highest density of material on the substrate, we also mounted the DNA using a new method being developed in this laboratory. Here the DNA is adsorbed to the supports in a low salt buffer containing no divalent metals or spermidine followed by very rapid freezing, freeze-drying, and finally rotary metal shadow casting. Although the images were not as crisp because of residual salt remaining after freeze-drying, the basic beaded appearance of the ssDNA was the same as when prepared by the procedure used in the experiments above (Fig. S4).
The G-strand single-stranded DNA is resistant to extension force
To further examine the properties of the G-strand ssDNA, we developed a smMT system (39–41) in which one end of the G-strand ssDNA was attached to an engineered flow cell surface via a biotin–NeutrAvidin linkage, whereas the other end of the DNA was attached to a superparamagnetic (SPM) bead via a digoxygenin-antidigoxygenin linkage (42). Controlled positioning of permanent neodymium magnets allowed the application of a well-defined force that results in DNA extension, which was monitored by the location of the SPM bead relative to the flow-cell surface (39, 41, 43).
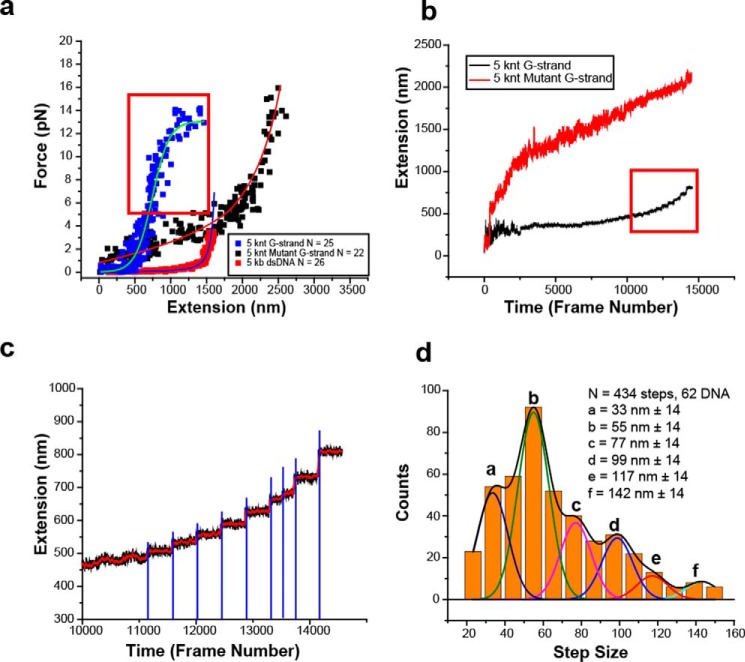
Alkaline-agarose gel electrophoresis was used to size-select ∼5-knt length C-strand, G-strand, and mutant G-strand ssDNA in which the ligated rolling-circle contained biotin and the 5′-thymine residue of the primer oligonucleotide contained a digoxigenin (Fig. S1). The C-strand and mutant G-strand ssDNAs displayed a force extension curve consistent with a freely jointed chain model that is characteristic for ssDNA (Fig. 4a, black squares with red curve, and Fig. S5) (44). In contrast, a 5-kb dsDNA displayed a force extension that is consistent with a worm-like chain model as previously described (Fig. 4a, red squares with blue curve, and Fig. S5) (45). Remarkably, the G-strand ssDNA extension stalled at ∼600 nm that resulted in a force-extension curve that adopted a sigmoidal character (Fig. 4a, blue squares with turquois curve, and Fig. S5). These results are inconsistent with a mechanical extension that is caused by the natural ordering of a disordered polymer molecule under force (39) but rather implies a compacted ordered DNA structure that is relatively resistant to forces below 12 pN (Fig. 4a and Fig. S5). Interestingly, at forces above 5 pN (Fig. 4, a and b, red box, and Fig. S5), we observed stepwise extension of the G-strand ssDNA rather than the continuous smooth extension exhibited by the mutant G-strand (Fig. 4, b and c, and Fig. S5).
Figure 4.
Single molecule magnetic tweezer force extension analysis of G-strand ssDNA. a, force extension analysis of rolling circle and size-selected 5-knt human G-strand ssDNA (n = 25, blue squares), size-selected 5-knt mutant G-strand ssDNA (n = 22, black squares), and 5-kb dsDNA (n = 28, red square). The red box indicates forces where discrete steps are observed during force extension. b, representative extension versus time (frame number at a frame rate of 14.2 ms/frame) of 5 knt G-strand ssDNA (black) and mutant G-strand ssDNA (red). The red box in b indicates forces/frames shown in the red box outlined in a. c, representative magnification of discrete steps observed in red boxes of a and b. Red triangles indicate the time of magnet steps (2-ms movement). Blue lines indicate large change in the second derivative of movement where extension steps are determined. d, binned 5-knt G-strand ssDNA extension steps (n = 434; 62 individual DNA molecules). Multiple Gaussian peaks are shown in different colors with the smoothed curve shown in black. Each Gaussian peak is labeled with a letter (peaks a–f) and the mean step size with standard deviation.
A large positive change in the second derivative of the extension versus time plot (Fig. 4c) was used to identify 434 steps from 62 individual DNA molecules (Fig. 4d). When binned to the resolution of the smMT instrument (10 nm), we observed a distribution of steps that appeared to contain multiple embedded peaks (Fig. 4d). Employing coefficient of determination (46) and root-mean-squared goodness-of-fit (46) analysis appeared to indicate six embedded Gaussian peaks that when smoothed describe the binned histogram data as corresponding to six step-release sizes (Fig. 4d and Fig. S6).
Interestingly, we observed a weak positive trend between the step size and the introduced force, with the largest steps occurring at higher forces (Fig. S7). It is important to note that even at very slow load rates, the release of folded G-quadruplex structure only begins to occur at forces above 20 pN and peaks at 35–55 pN, depending on the force spectroscopy method (47, 48). Together, these results seem to imply that the application of magnetic force results in the stepwise release of higher-order structure(s) within the G-strand ssDNA, while maintaining the intrinsic array of G-quadruplex folds.
Modeling
Several models have been proposed for the structure of the 3′-ssDNA overhang at the terminus of mammalian telomeres, which are largely based on parallel or hybrid G-quadruplex structures and result in somewhat different outcomes (23). For example, the crystal structure of a 22-nt d[AG3(T2AG3)3] ssDNA (21) revealed a flattened disk 0.61 nm in height and 4.2 nm in diameter with the four unpaired TTA triplet nucleotides extending out to the edges of the disk similar to propellers. Other crystal structures have been derived from similar short DNAs bound with different ligands, where with the ligand dominates the outcome (23).
Multimeric G-quadruplex structures are largely based on beads-on-a-string models (29), where multiple G-quadruplex units formed from (3′-TTAGGG-5′)4–20 are arranged into individual thermodynamically unique beads-on-a-string that do not interact with one other, can move freely, and are only restricted by their TTA linker. These models are founded on studies of limited length DNAs (120 nt or less) and do not address the possibility of a variation in the length of intervening repeat ssDNA or interactions between G-quadruplex units via stacking interactions of the G-tetrad cores (23). Perhaps more importantly, there are several types of G-quadruplex folds, as well as suggestions that these different folds may exist together under identical conditions (49). Nevertheless, once folded, all G-quadruplex structure appear exceedingly stable (50).
To understand the distribution of step sizes in the force extension analysis, we performed G-quadruplex folding simulations with variable lengths of G-strand ssDNA (Fig. S8). In these simulations, a G-quadruplex may fold randomly along the length of an ssDNA between four contiguous telomeric repeats. The type of G-quadruplex fold is irrelevant for these simulations, although we assume that once formed these folds are stable, and no subsequent remodeling may occur. These random folding events result in an array of G-quadruplex structures along the G-strand ssDNA that are separated by 0, 1, 2, or 3 repeats with distinct probabilities (Fig. S8).
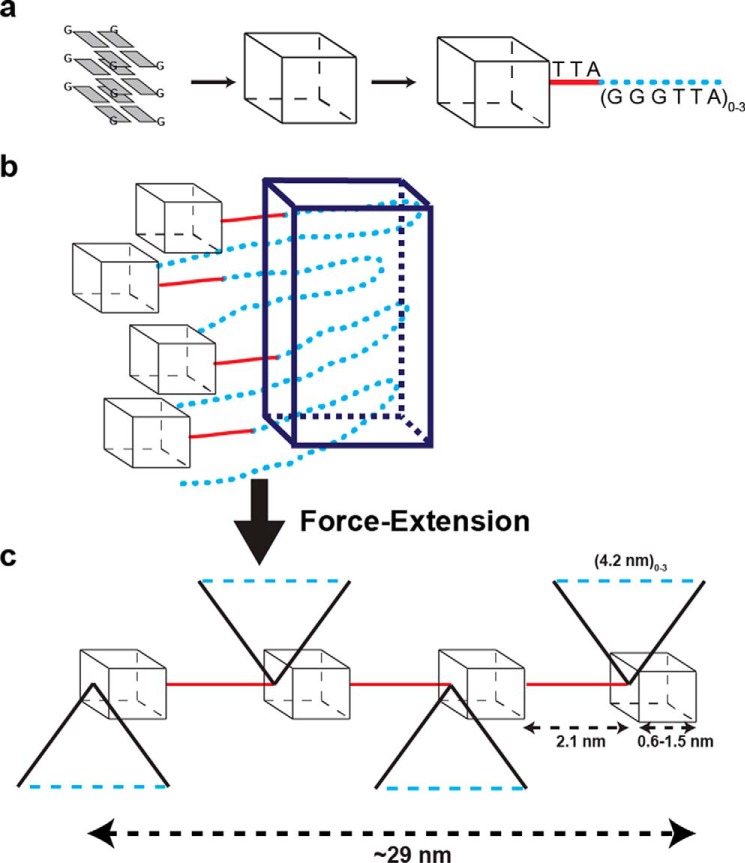
We determined that a G-quadruplex embedded in a 1- or 6-knt G-strand ssDNA is on average separated from the adjacent G-quadruplex by one complete telomeric repeat (Fig. S8). When subjected to smMT at forces below those that disrupt the G-quartet structure, a single G-quadruplex may be pulled from two different corners that adds different lengths depend on the type of fold (51). This corner-to-corner extension would account for 0.6–1.5 nm that with a free TTA tail (2.1 nm) and on average one telomeric repeat (4.2 nm) totals of 6.9–7.8 nm. The projected force extension peaks appear to be tetramer multiples of the ∼7.3 nm unit (calculated: 29, 58, 87, 117, and 146 nm; observed 33, 55, 77, 117, and 142 nm; Fig. 4d). The extra 99-nm (or 77-nm) extension peaks might reflect intermediate extensions and/or overfitting of multiple Gaussian curves.
A plausible model for the higher-order structures exploits the possibility that random condensation along G-strand DNA results in G-quadruplexes that are separated by, on average, one additional G-strand repeat (Fig. 5a). These intervening repeats contain guanine nucleotides that could form Hoogsteen base pairing with nearby intervening G-repeats that are not folded into a G-quadruplex (11, 14). It is our hypothesis that four intervening repeats condensed into a G-residue pseudo-fold, which is the basis for stable higher-order packing (Fig. 5b, dark blue box). In this scenario, four intervening repeats could come from any combination of 1–3 repeats between two embedded G-quadruplexes linked with the additional necessary nearby intervening repeats, such that a total of four repeats are condensed into a G-residue pseudo-fold. The extension of these G-residue pseudo-fold higher-order structures under the force conditions utilized in our studies would leave the bona fide G-quadruplexes intact while stretching the terminal TTA linkers, as well as the intervening G-strand repeats on average ∼29 nm (Fig. 5c). Such a G-residue pseudo-fold formed between intervening repeats could account for the increased force required to extend the G-strand ssDNA that is significantly more than random-coil ssDNA but less than a true G-quadruplex fold (Fig. 4d). Importantly, this model predicts that the G-residue pseudo-folds formed between multiple intervening repeats is more exposed than the G-strand ssDNA folded into a compact G-quadruplex.
Figure 5.
Hypothetical model of higher-order G-strand structures. a, cartoon illustration of G-quartet as a black box, with extended connector non-G-residue nucleotides between G-quadruplex folds (red) and nonfolded G-rich repeats between G-quadruplex folds (blue dots). G-rich repeats may fold into several types of G-quadruplex structures (parallel, anti-parallel, hybrid, etc.) with non-G-residue nucleotide loops connecting the corners of the hypothetical box. In an array of G-quadruplex folds, regardless of the type of folded structure, the non-G-residue nucleotides of the last G-rich repeat may extend to the adjacent G-quadruplex. For the human telomeric repeat (TTAGGG)n, the non-G-residue nucleotides TTA (red) comprise this extended connection, with on average at least one nonfolded G-rich repeat (TTAGGG; blue dots). b, the results suggest that multiples of four nonfolded intervening G-rich repeats (blue dots) may condense into G-residue pseudo-folds (blue box). The condensation into G-residue pseudo-folds is expected to be completely independent of the type of G-quartet fold(s) within the G-quadruplex array because they involve only the intervening G-rich repeats. c, under magnetic force G-residue pseudo-folds may extend in multiples of ∼29 nm.
RAD51 systematically alters higher order G-quadruplex associations
To further probe the structural characteristics, we examined the force extension properties of G-strand ssDNA in the presence of the central HR component RAD51. Unlike strong ssDNA-binding proteins that singularly disrupt G-quadruplex folds formed in ssDNA (52), the RAD51 protein displays a switchable weak ssDNA binding in the absence of ATP (or in the presence of ADP) that is transformed into a robust nucleoprotein filament in the presence of ATP, which firmly coats a ssDNA (53). This cycle of RAD51 ATP binding and hydrolysis has been connected to nucleoprotein filament reordering and turnover during the HR homology search (54) and may play a role in ALT telomere maintenance (55). Importantly, the formation of an ATP-bound RAD51 nucleoprotein filament appears capable of generating sufficient binding forces to provoke nucleosome disassembly in model chromatin substrates (56).
We first determined that multiple cycles of magnetic extension and release of a single G-strand ssDNA resulted in largely identical force extension curves (Fig. 6a and Fig. S9a). This observation strongly suggests that the G-strand ssDNA spontaneously refolds into comparable stable higher order structures following release from magnetic extension. When RAD51 is included in the force extension analysis without ATP (−ATP), the curves gradually flatten following multiple extension and release cycles, demonstrating significantly more extension at low forces (Fig. 6b and Fig. S9b). Combined with the G-quadruplex modeling (see “Modeling” above and Fig. S8), we interpret these results to imply that RAD51 increasingly binds to intervening ssDNA repeat(s) embedded between G-quadruplex folds, eventually inhibiting spontaneous condensation into the G-residue pseudo-folds that constitute the higher order structures. We note that the weak and reversible ssDNA-binding activity of RAD51 (−ATP) does not appear to fully inhibit higher order structure refolding and results in a DNA–protein complex that does not exhibit ssDNA or dsDNA force extension properties (Fig. 6b and Fig. S9b). In contrast, RAD51 in the presence of ATP (+ATP) appears to rapidly form a stable nucleoprotein filament that extends longer than a similar length dsDNA at low forces and displays no intermediate force extension curves regardless of the number of extensions (Fig. 6c and Fig. S9c). The longer extension of the RAD51 nucleoprotein filament is consistent with historical studies demonstrating that a helical ATP-bound RAD51 nucleoprotein filament extends an ssDNA 1.5 times the length of a corresponding dsDNA (57, 58).
Figure 6.
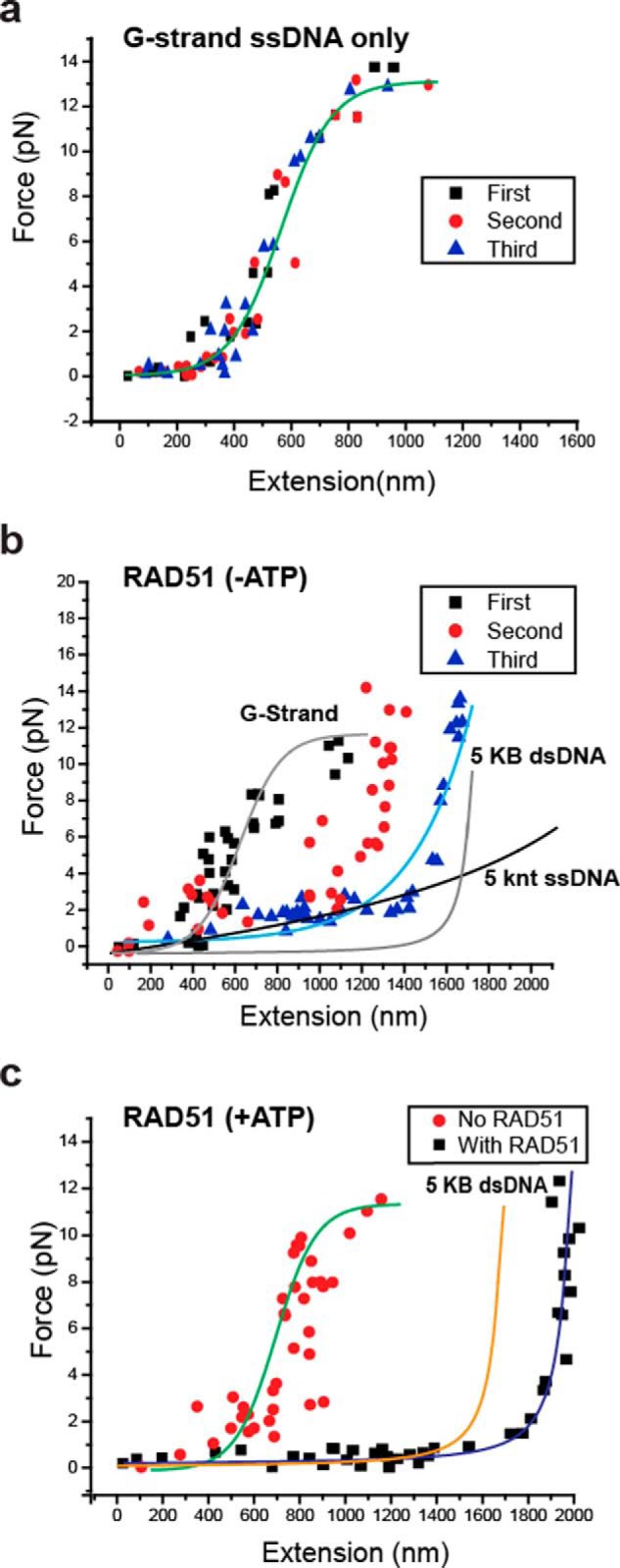
The binding of RAD51 to G-Strand ssDNA. a, representative force-extension of a single G-strand ssDNA in which the applied force is released following extension and then reapplied for a total of three successive cycles. b, representative force-extension of a single G-strand ssDNA in which the applied force is released following extension and then reapplied for a total of three successive analysis in the presence of human RAD51 and in the absence of ATP (−ATP). c, representative force-extension of a single G-strand ssDNA in which the applied force is released following extension and then reapplied for a total of three successive analysis and in the presence of ATP (+ATP).
To confirm the hypothesis that RAD51 forms a stable filament that fully extends the G-strand ssDNA, we examined the RAD51 nucleoprotein filament in the presence of ATP by EM following negative staining. We observed fields of extended RAD51 nucleoprotein filaments with a helical pitch of 10.7 nm (n = 59 measurements) in agreement with the previous studies (Fig. 7). Similar filaments were not present when G-strand ssDNA was incubated with RAD51 in the absence of ATP (not shown). We conclude that RAD51 forms a stable nucleoprotein filament in the presence of ATP that fully extends the G-strand ssDNA, unraveling both the embedded G-quadruplex folds and the higher-order G-residue pseudo-fold structures.
Figure 7.
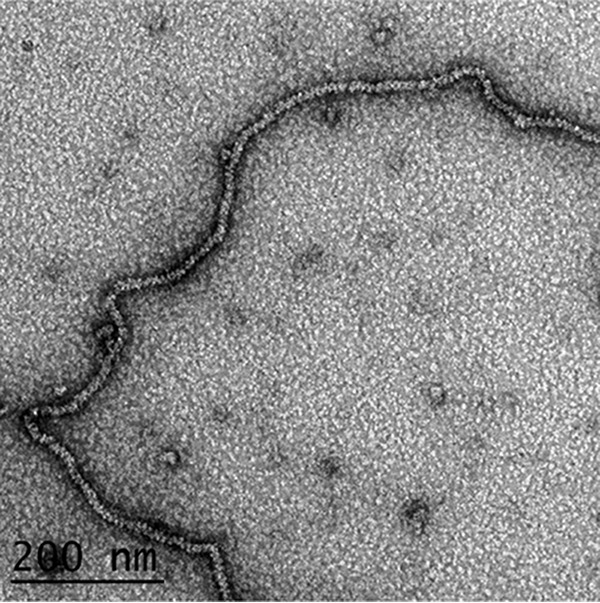
Visualization of RAD51 extension of G-strand DNA. Long G-strand ssDNA was incubated with RAD51 protein in the presence of ATP and then prepared for EM by negative staining (“Experimental procedures”). The ssDNA appears highly extended in agreement with the smMT analysis, and the helical pitch in the filaments (107 A) is typical of RAD51–ssDNA complexes formed with ATP.
Discussion
Extending the structural studies of short (∼20–150 nt) G-strand repeat ssDNAs to the analysis of much longer G-strand repeat ssDNAs has been hampered by the lack of well-defined substrates. To overcome this technical issue, we further developed a rolling circle replication system as previously described (59, 60) and adapted it for generating long ssDNA containing distinct repeating nucleotide sequences. This method allowed us to produce G-strand, C-strand, and mutated G-strand ssDNAs containing different forms of the human telomeric repeat sequence ranging in size from 100 to >20,000 nt (Fig. S1).
G4-FID analysis confirmed the presence of G-quadruplex structures within the long human G-strand ssDNA (5′-TTAGGG-3′)n, but not in the C-strand ssDNA (5′-AATCCC-3′)n or a mutant G-strand ssDNA (5′-TTAGTG-3′)n that interrupts the contiguous G-triplet within the telomere repeat. Using EM we showed that very long human telomeric G-strand ssDNA spontaneously condenses into chains of large discrete bead-like particles with 5- and 8-nm diameters. These bead-like particles compact the ssDNA nearly 12-fold in length. Using size-selected DNA, we determined that the larger beads contain ∼240 nt and inferred that the smaller beads contained half this amount (∼120 nt).
Unlike EM, which requires a final dehydration step, smMT studies may be performed in solution at physiological ionic strength and with predominantly intracellular K+ cation. Force extension analysis indicated several discrete G-strand ssDNA elongation step sizes. Because it requires extension forces >20 pN to unravel a folded 24-nt G-quadruplex (47, 48), we interpreted these results to indicate that the extension steps represented fundamental units of higher-order interaction structures. The weak positive correlation between step size and extension force (Fig. S7) suggests that the higher-order structures are intrinsically more stable the larger they become. In EM fields of the compacted G-strand ssDNA, we noted some particles appeared less organized than others. This might suggest that in addition to the principal structures, other arrangements may be possible that might contribute to the force-extension “noise” that extends outside the baseline 10-nm resolution of the smMT instrument. Taken together, these observations represent the first physical evidence for intramolecular higher-order structures formed by G-strand ssDNA containing embedded G-quadruplexes. These findings appear broadly applicable for the long repetitive blocks of G-rich sequences present in many genomes including the human genome.
We calculated that the major tetramer and octamer higher order structures observed as initial step sizes by smMT would on average contain ∼120 nt (four G-quadruplexes plus four extra repeat loops) and ∼240 nt (eight G-quadruplexes and eight extra repeat loops), respectively. The amount of the octamer higher order structure DNA content is remarkably consistent with the calculated amount of ssDNA associated with the large EM beads (247 nt) and by extension the DNA content of the smaller beads that appear approximately half the size. These two initial smMT extension steps account for 63% of the total events, which appears consistent with the two major bead sizes observed by EM analysis.
We developed a model based on the stochastic formation of G-quadruplexes, followed by condensation of intervening G-repeats (Fig. 5). An intermolecular side-by-side pairing of a G-repeat ssDNA, termed G-wires, has been described that utilized small oligonucleotides (5′-GGGGTTGGGG-3′) (61). In theory, G-wires might provide a plausible structure for the association(s) of intervening telomeric ssDNA repeats. However, we note that the intermolecular G-quadruplex folds predicted by G-wires should display comparable stability to force extension as bona fide intramolecular G-quadruplexes. A less robust interaction between the intervening repeat G-residue pseudo-folds appears more consistent with our studies, which showed that both magnetic force and the weak ssDNA binding activity of RAD51 (−ATP) were capable of disrupting the higher-order structures. For example, we interpret the observation that RAD51 (−ATP) increasingly inhibits condensation following multiple release of magnetic force as consistent with increased shielding of intervening repeat ssDNA by weakly bound RAD51, supporting the intervening repeat G-residue pseudo-fold condensation model (Fig. 5).
We performed extensive nuclease digestion analysis of the long G-strand ssDNA exploiting multiple nucleases with the goal of isolating higher order G-strand structures (not shown). However, under the best conditions, we observed a 24-nt band consistent with a protected G-quadruplex along with a smear of higher molecular weight ssDNA. We concluded that the higher-order G-strand structures are only partially resistant to nucleases, which also appears consistent with exposed features of intervening repeat G-residue interactions as proposed in our model (Fig. 5).
We calculated the probability (P) that a higher-order G-strand structure might condense within various lengths of human telomere 3′-ssDNA by assuming that a minimum of four individual ssDNA G-strand repeats separated by at least three G-quadruplex folds are required to form a stable particle (Fig. S8e and “Experimental procedures”) (62). This analysis suggested that significant fractions of 100 nt (p = 0.13) and 200 nt (p = 0.32) human telomeric G-strand ssDNA may stochastic fold into G-quadruplexes leaving at least four intervening G-repeats, which could condense into a G-residue pseudo-fold. This probability dramatically increased with increasing ssDNA length (Fig. S8e). Moreover, these estimates likely represent a minimum because we do not include the possibility that adjacent ssDNA repeats might nucleate into a G-residue pseudo-fold or consider any potential phasing by the telomere ssDNA–dsDNA junction. Taken as a whole, these studies suggest that in the absence of other factors, higher-order G-strand structures may form naturally on a 3′-ssDNA (63, 64).
We consider the possibility that a higher order telomeric G-strand structure could serve as a molecular switch that might inhibit or facilitate the interactions of telomeric protein factors such as the shelterin protein complex or telomerase. Compaction into these large structures could aid in shielding the telomere from double-stranded break sensing and repair factors prior to binding by telomeric protein components (65), or they might act as a roadblock for telomerase-mediated telomere elongation (13). Cech and co-workers (66) have examined a 144-nt (3′-TTAGGG-5′)n ssDNA bound by hPOT1 and found highly compact DNA-protein particles, suggesting that POT1 is likely to disrupt any higher order G-strand condensation. Indeed POT1 and its partner TPP1 appear to overcome G-quadruplex roadblocks during telomere metabolic processes.
The self-condensation of long segments of ssDNA containing runs of Gs could have significant global consequences for replication, recombination, and repair. As noted above, if the G-rich strand at the C9orf72 locus (C2G4)n assumed a higher order conformation, this could not only induce replication fork stalling but in itself might lead to further expansion by mechanisms related to replication restart, recombination or repair. Indeed, the expanded repeat has been shown to cause replication fork stalling in vitro (67). The KSHV replication origin contains ∼260 bp of G-rich repeats on one strand. Were this G-rich strand to condense into a higher order structure, this would likely generate a severe bend or knot at the KSHV replication origin with the C-rich strand present in a more open unstructured state. The net result would likely be a complex scaffold with very different protein binding properties than the linear dsDNA. It will be of interest to generate other long ssDNAs, including ones containing the G-rich C9orf72 repeat and the KSHV origin repeats and examine their architecture using the biophysical methods described here.
Experimental procedures
Rolling circle replication
Telomeric ssDNA circles were synthesized using oligonucleotides with either WT G-strand, mutant G-strand, or C-strand sequences (Table S1) containing 20 telomeric repeats (IDT, Coralville, IA) as described earlier (28). In brief, oligonucleotides were ligated into circles using CircLigaseTM, and any remaining linear oligonucleotides were digested with ExoI and ExoIII (New England Biolabs Inc., Ipswich, MA) according to the manufacturer's recommendations (Epicenter Biotechnologies, Madison, WI). Circular products were confirmed by 7% denaturing PAGE.
For size selection of long G-strand DNA we used alkaline agarose gel electrophoresis. In brief, 0.8% alkaline agarose gels were prepared in alkaline buffer (10× buffer contains 10 n NaOH and 0.5 m EDTA, pH 8.0). The sample DNA was digested with S1 nuclease and mixed with alkaline gel loading buffer. After electrophoresis, the gel was soaked in neutralizing solution (containing 1 m Tris-HCl and 1.5 m NaCl), stained with SYBR® Green I nucleic acid gel stain (Invitrogen) followed by isolation of target DNA length using a QIAquick gel extraction kit (Qiagen).
G-quadruplex fluorescent intercalator displacement (G4-FID)
A constant temperature (20 °C) SPEX Fluorolog-3 (Horiba) spectrofluorometer with thermostatted cell holders (3 ml) was used to perform G4-FID studies in 10 mm lithium cacodylate buffer (pH 7.3) and 100 mm KCl. Briefly, 0.25 μm prefolded DNA in lithium cacodylate KCl buffer was mixed with TO (0.50 μm). Each ligand addition step (from 0.5 to 10 equivalents) was followed by a 3-min equilibration period, after which the fluorescence spectrum was recorded. The percentage of displacement was calculated from the fluorescence area (FA, 510–750 nm, λex = 501 nm), using the following equation: TO displacement (%) = 100 − [(FA/FA0) × 100], where FA0 is the fluorescence of TO bound to DNA without added ligand. The TO displacement (%) was plotted as a function of the concentration of added ligand.
Electron microscopy
DNA was adsorbed onto grid supports covered with a thin glow discharge-treated carbon film for 30 s to 1 min in the presence of the buffer present with the DNA or a buffer containing 2.5 mm spermidine, 10 mm Tris-HCl, 50 mm KCl, 75 mm NaCl, and 1 mm MgCl (pH 7.5). The samples were washed in water followed by a series of ethanol dehydration steps, air-drying, and rotary shadow casting with tungsten at 1 × 106 torr (68). The samples were visualized using a Tecnai 12 TEM (FEI Inc., Hillsboro, OR) at 40 kV, and the images were collected with a Gatan Orius charge-coupled device camera (Gatan Inc., Pleasanton, CA) with digital micrograph–supporting software (Gatan Inc., Pleasanton, CA). Dimensions of telomeric DNA filaments were measured from digital micrographs using digital micrograph (Gatan Inc., Pleasanton, CA) and ImageJ (National Institutes of Health, Bethesda, MD). Negative staining of the G-rich ssDNA bound by human RAD51 (purified as previously described (56)) was carried out by incubating 50 ng of G-rich ssDNA with 1500 ng of RAD 51 in a buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 0.5 mm DTT, and 2 mm MgCl2 for 30 min at 37 °C in the presence or absence of 2.5 mm ATP. Drops of the sample were adsorbed to glow charged thin carbon supports for 3 min followed by washed with 2% uranyl acetate, air drying, and imaging in a Tecnai 12 as above at 80 kV. Images for publication were arranged and contrast-optimized using Adobe Photoshop CS5 (Adobe Systems, San Jose, CA).
Magnetic tweezers preparation
Flow cells were engineered with glass cover slides affixed with double-sided tape to an aluminum foundation that maximized SPM bead imaging. Telomeric C-strand, G-strand or mutant G-strand ssDNA (30 pm final) was mixed with bead formation buffer (50 mm Tris-Cl, pH 7.6, 100 mm KCl). The combined sample was boiled for 10 min followed by snap chilling on ice for 1–2 h. Prior to attachment, the glass slides were treated with (3-aminopropyl) triethoxysilane followed by a 1:100 mixture of Biotin-PEG SVA to mPEG-SVA (Invitrogen). NeutrAvidin (500 μm; Invitrogen) was injected in the flow cell at a rate of 8 μl min−1, followed by the ssDNA. Tosyl activated M-280 SPM Dynabeads (ThermoFisher Scientific) were coated with anti-digoxigenin antibodies (Roche). Stock beads were removed and resuspended in a 0.1 m borate, pH 9.5 (buffer A). The beads were then resuspended in a mixture of buffer A and anti-digoxigenin at a 20 μg:1 mg, antibody:beads ratio. The beads were incubated for 12–17 h at 37 °C with slow tilt rotation. 3 m ammonium sulfate in buffer A (buffer B) was mixed into the beads. The beads were then resuspended in 10 mm Na/K-phosphate (pH 7.4), 140 mm NaCl/KCl (PBS) with 0.5% (w/v) acetylated BSA (Sigma) (buffer C). After incubation at 37 °C, 1 h, the coated beads were resuspended in PBS (pH 7.4) with 0.1% (w/v) acetylated BSA (buffer D). Buffer D was removed and then added to reach the desired bead concentration (2 × 109 beads ml−1).
Anti-digoxigenin–coated beads were mixed with running buffer (50 mm Tris-Cl, pH 7.6, 100 mm KCl, 200 μg μl−1 acetylated BSA, 0.0025% Tween 20; Amresco) and injected into the flow cell containing the ssDNA that was bound to the surface via a biotin–NeutrAvidin linkage; at 8 μl min−1 while agitating the system. The bound ssDNA was washed extensively with running buffer to remove free SPM beads prior to analysis. Force extension measurements used four 1-cm3 rare earth magnets (Neodynium, Magcraft). The SPM beads were imaged using a 530-nm LED lamp (Thorlabs) and a 100× Olympus oil immersion objective, and the images were collected on a 1024 × 1024 pixel charge-coupled device camera (Grasshopper Express 1.0 MP Mono FireWire 1394b) at a frame rate of 70 ms.
The human RAD51 protein was purified and stored as previously described (52). Force extension analysis was performed in running buffer additionally containing 2 mm MgCl2, 100 μm DTT, and human RAD51 (500 nm) with or without 1 mm ATP (Roche).
Magnetic tweezers data analysis
SPM bead analysis was performed with the 3D bead tracking software Video Spot Tracker (Computer Integrated Systems for Microscopy and Manipulation, University of North Carolina-Chapel Hill). Displacement events were determined using the software Edge Detector (Computer Integrated Systems for Microscopy and Manipulation, University of North Carolina-Chapel Hill) and MATLAB and Statistics Toolbox Release 2014b (MathWorks, Inc., Natick, MA). Edge Detector was edited to avoid scoring negative displacement because these can bias the value of large positive displacement. The data were smoothed with the Savitzky–Golay filter3 and an average window size of 20 points using Origin software (OriginLabs, Northhampton, MA). Distributions for displacement events were binned to the resolution size of step size (∼10 nm) and [size of bins (Max − Min/number of bins − 0.5)], where Max is the maximum value of events, Min is the minimum value of events, Bin Start is the Min; Bin End is the (bin size × number of bins). The events were fit to multiple Gaussian distributions to determine several different observed displacement changes. The coefficient of determination for different numbers of peaks was used to select the appropriate number of Gaussians to fit. All fittings were done with Origin software. Force and extension values were determined for various magnet positions above the flow cell surface. Force calculations were determined by measuring the bead's fluctuations transverse to the direction of stretching and equating the fluctuation to the equipartition theorem (43). Double-stranded and mutant G-strand DNA curves were fit to the freely jointed chain model for DNA nonlinear elasticity (56).
Simulation of multiple quadruplex formation
Define an n-dimensional array ā, composed of ai, i = 1, …, n where ai = 0 for all i.
Define Algorithm A.
Create n random numbers in an n-dimensional array ū, composed of ui, i = 1, …, n, where ui = ∼iid U([0,1]). Sort the array S(ū) to create ordered uniformly random numbers ui, i = 1, …, n, where ui is the ith sorted value of ū. Then let I(ui) be the original index of the ith sorted value of ū.
| (1) |
Algorithm A results in ā having a series or set of blocks B, where b ϵ B = {[I(ui), I(ui) + 1, I(ui) + 2, I(ui) + 3]}, for all i = 1, …, n satisfying Σ3j=0a(ui+j) = 0 after potentially reassigning values for ai value through (i − 1) previous iterations. Note that this implies a(I(ui)+j) = 1 for j = 0, 1, 2, and 3 for each element b ϵ B after algorithm A. That is, each element b of B is a set or block of four consecutive indices denoting consecutive positions in the array ā each having a value of 1 with gaps between blocks having size 0, 1, 2, or 3, which have consecutive values of 0.
Let gk be the size of the kth “gap” between the block bk and bk+1, gk ϵ {0,1,2,3} for all k = 1, …, m, where m = ∼M is a random number representing the total number of gaps that depends on algorithm A and each iteration of algorithm A. The distribution of M over iterations of algorithm A is potentially obscure and may not have a simple representation. The frequency of each gap length within an iteration of algorithm A, i.e. N(gk = 0) = N0, N(gk = 1) = N1, N(gk = 2) = N2, N(gk = 3) = N3, are also random numbers from different distributions that are dependent on n. By definition, N0 + N1 + N2 + N3 = m.
Calculating the probability of higher-order G-strand structure formation
A solution to the likelihood that a human telomere G-strand repeat sequence (TTAGGGn) supports an environment compatible with higher-order structure formation begins by defining a condition where four consecutive repeats (TTAGGG4) fold into a G-quadruplex, followed by 1–3 repeats (TTAGGG1–3; termed a “gap”). This chain of G-strand repeats can be thought of as a “unit” containing a G-quadruplex fold with adjacent gap, surrounded by G-quadruplex folds containing no gaps.
Structural analysis has suggested that three gaps with an adjacent G-guadruplex (3 units) plus one additional gap of 1–3 G-strand repeats are minimally required to form a higher-order structure. Thus, we can reframe the probability of a nucleotide sequence having conditions compatible with higher-order structure formation as the probability that at least four successive gaps will occur within the G-strand sequence. If we term developing a single gap as a “success,” then Feller (62) described a simple method for determining the probability of at least r consecutive successes (gaps) in n Bernoulli trials (total gaps),
| (2) |
where p is the probability of success of a single gap, q = 1 − p, and x is the real root,
| (3) |
which cannot equal 1/p.
In our case, the success probability (p) depends on total probability of 1–3 repeats occurring in a defined DNA length and can be empirically determined from simulated quadruplex formation. As an example, for 120 nt of 3′-ssDNA: total probability = 0.28 (1 repeat) + 0.23 (2 repeats) + 0.14 (3 repeats) = 0.65. From simulation, we can also calculate n total gaps in m 3′-ssDNA nucleotides from the average gap size (Ave) expanded to the unit size.
| (4) |
where m/6 is the maximum number of G-repeats in m nucleotides of 3′-ssDNA. As an example, for 120 nt of 3′-ssDNA containing a hexameric repeat (TTAGGG): (120 ÷ 6) × 1/[4 (repeats per G-quadruplex) + 1.15 (average repeats between G-quadruplexes)] = 3.88. For simplicity, 3.88 is rounded to the nearest integer 4, effectively making the resulting probability (P) calculation a minimum.
From Equation 1, we can then determine the probability of at least four consecutive gaps in m nucleotides. As an example, for 120 nt, q × p4 = 0.35 × (0.65)4 = 0.06; x = 1.10, leading to a probability P(r = 4, n = ∼4) = 0.16. These results suggest that within a population of 120 nt, 3′-ssDNA 16% will stochastically condense into a higher order G-strand structures. As expected, Equation 1 becomes more accurate as n increase. The probability of higher-order G-strand structure versus 3′-ssDNA length is plotted.
Author contributions
A. K., N. Ö. A., R. F., and J. D. G. conceptualization; A. K. and J. D. G. resources; A. K., N. J., N. Ö. A., R. F., and J. D. G. formal analysis; A. K., R. F., and J. D. G. supervision; A. K., R. F., and J. D. G. funding acquisition; A. K., N. J., N. Ö. A., R. F., and J. D. G. validation; A. K., N. J., N. Ö. A., R. F., and J. D. G. investigation; A. K., N. J., N. Ö. A., R. F., and J. D. G. visualization; A. K., N. J., N. Ö. A., R. F., and J. D. G. methodology; A. K., N. J., N. Ö. A., R. F., and J. D. G. writing-original draft; A. K. and J. D. G. project administration; A. K., N. J., N. Ö. A., R. F., and J. D. G. writing-review and editing; N. J. and N. Ö. A. data curation; N. J. and N. Ö. A. software.
Supplementary Material
Acknowledgments
We thank Prof. Ashutosh Tripathi (University of North Carolina Macromolecular Interactions Facility) for help with G4-FID studies, Gayan Senavirathne for purified human RAD51 protein, Wendell Jones for G-quadruplex folding simulation and probability guidance, and laboratory colleagues for many helpful discussions.
This work was supported by National Institutes of Health Grants GM31819 and ES013773 (to J. D. G.) and GM080176 (to R. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1, Figs. S1–S9, and Notes S1 and S2.
- ssDNA
- single-stranded DNA
- nt
- nucleotide(s)
- smMT
- single-molecule magnetic tweezers
- KSHV
- Kaposi's sarcoma-associated Herpesvirus
- HR
- homologous recombination
- ALT
- alternative lengthening of telomeres
- G4-FID
- G-quadruplex fluorescence intercalator displacement
- TO
- thiazole orange
- SPM
- superparamagnetic
- FA
- fluorescence area
- knt
- kilonucleotides.
References
- 1. Fujioka, K., Aratani, Y., Kusano, K., and Koyama, H. (1993) Targeted recombination with single-stranded DNA vectors in mammalian cells. Nucleic Acids Res. 21, 407–412 10.1093/nar/21.3.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mitra, S., and Kornberg, A. (1966) Enzymatic mechanisms of DNA replication. J. Gen. Physiol. 49, 59–79 10.1085/jgp.49.6.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tomonaga, T., and Levens, D. (1996) Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 93, 5830–5835 10.1073/pnas.93.12.5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma, W., Westmoreland, J. W., and Resnick, M. A. (2013) Homologous recombination rescues ssDNA gaps generated by nucleotide excision repair and reduced translesion DNA synthesis in yeast G2 cells. Proc. Natl. Acad. Sci. U.S.A. 110, E2895–E2904 10.1073/pnas.1301676110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goddard, N. L., Bonnet, G., Krichevsky, O., and Libchaber, A. (2000) Sequence dependent rigidity of single stranded DNA. Phys. Rev. Lett. 85, 2400–2403 10.1103/PhysRevLett.85.2400 [DOI] [PubMed] [Google Scholar]
- 6. Zhang, Y., Zhou, H., and Ou-Yang, Z. C. (2001) Stretching single-stranded DNA: interplay of electrostatic, base-pairing, and base-pair stacking interactions. Biophys. J. 81, 1133–1143 10.1016/S0006-3495(01)75770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blackburn, E. H. (1991) Structure and function of telomeres. Nature 350, 569–573 10.1038/350569a0 [DOI] [PubMed] [Google Scholar]
- 8. Sen, D., and Gilbert, W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366 10.1038/334364a0 [DOI] [PubMed] [Google Scholar]
- 9. Nicholas, J., Zong, J. C., Alcendor, D. J., Ciufo, D. M., Poole, L. J., Sarisky, R. T., Chiou, C. J., Zhang, X., Wan, X., Guo, H. G., Reitz, M. S., and Hayward, G. S. (1998) Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 79–88 [DOI] [PubMed] [Google Scholar]
- 10. Rademakers, R., Neumann, M., and Mackenzie, I. R. (2012) Advances in understanding the molecular basis of frontotemporal dementia. Nat. Rev. Neurol. 8, 423–434 10.1038/nrneurol.2012.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gellert, M., Lipsett, M. N., and Davies, D. R. (1962) Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 48, 2013–2018 10.1073/pnas.48.12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bochman, M. L., Paeschke, K., and Zakian, V. A. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 10.1038/nrg3296,10.1038/ni.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rhodes, D., and Lipps, H. J. (2015) G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637 10.1093/nar/gkv862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sen, D., and Gilbert, W. (1990) A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 344, 410–414 10.1038/344410a0 [DOI] [PubMed] [Google Scholar]
- 15. Sundquist, W. I., and Klug, A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 342, 825–829 10.1038/342825a0 [DOI] [PubMed] [Google Scholar]
- 16. Williamson, J. R., Raghuraman, M. K., and Cech, T. R. (1989) Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59, 871–880 10.1016/0092-8674(89)90610-7 [DOI] [PubMed] [Google Scholar]
- 17. Paeschke, K., Juranek, S., Simonsson, T., Hempel, A., Rhodes, D., and Lipps, H. J. (2008) Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 15, 598–604 10.1038/nsmb.1422 [DOI] [PubMed] [Google Scholar]
- 18. Schaffitzel, C., Berger, I., Postberg, J., Hanes, J., Lipps, H. J., and Plückthun, A. (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 98, 8572–8577 10.1073/pnas.141229498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biffi, G., Tannahill, D., McCafferty, J., and Balasubramanian, S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palm, W., and de Lange, T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- 21. Parkinson, G. N., Lee, M. P., and Neidle, S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 10.1038/nature755 [DOI] [PubMed] [Google Scholar]
- 22. Li, J., Correia, J. J., Wang, L., Trent, J. O., and Chaires, J. B. (2005) Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 33, 4649–4659 10.1093/nar/gki782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petraccone, L. (2013) Higher-order quadruplex structures. Top. Curr. Chem. 330, 23–46 [DOI] [PubMed] [Google Scholar]
- 24. Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., and Lingner, J. (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- 25. Schoeftner, S., and Blasco, M. A. (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- 26. Randall, A., and Griffith, J. D. (2009) Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J. Biol. Chem. 284, 13980–13986 10.1074/jbc.M900631200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rippe, K., and Luke, B. (2015) TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 22, 853–858 10.1038/nsmb.3078 [DOI] [PubMed] [Google Scholar]
- 28. Kar, A., Willcox, S., and Griffith, J. D. (2016) Transcription of telomeric DNA leads to high levels of homologous recombination and t-loops. Nucleic Acids Res. 44, 9369–9380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu, H., Gu, X., Nakano, S., Miyoshi, D., and Sugimoto, N. (2012) Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions. J. Am. Chem. Soc. 134, 20060–20069 10.1021/ja305384c [DOI] [PubMed] [Google Scholar]
- 30. Paramasivan, S., Rujan, I., and Bolton, P. H. (2007) Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods 43, 324–331 10.1016/j.ymeth.2007.02.009 [DOI] [PubMed] [Google Scholar]
- 31. Monchaud, D., Allain, C., and Teulade-Fichou, M. P. (2006) Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg Med. Chem. Lett. 16, 4842–4845 10.1016/j.bmcl.2006.06.067 [DOI] [PubMed] [Google Scholar]
- 32. Monchaud, D., and Teulade-Fichou, M. P. (2010) G4-FID: a fluorescent DNA probe displacement assay for rapid evaluation of quadruplex ligands. Methods Mol. Biol. 608, 257–271 10.1007/978-1-59745-363-9_15 [DOI] [PubMed] [Google Scholar]
- 33. Largy, E., Saettel, N., Hamon, F., Dubruille, S., and Teulade-Fichou, M. P. (2012) Screening of a chemical library by HT-G4-FID for discovery of selective G-quadruplex binders. Curr. Pharm. Des. 18, 1992–2001 10.2174/138161212799958350 [DOI] [PubMed] [Google Scholar]
- 34. Burger, A. M., Dai, F., Schultes, C. M., Reszka, A. P., Moore, M. J., Double, J. A., and Neidle, S. (2005) The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 65, 1489–1496 10.1158/0008-5472.CAN-04-2910 [DOI] [PubMed] [Google Scholar]
- 35. Mo, Y., Gan, Y., Song, S., Johnston, J., Xiao, X., Wientjes, M. G., and Au, J. L. (2003) Simultaneous targeting of telomeres and telomerase as a cancer therapeutic approach. Cancer Res. 63, 579–585 [PubMed] [Google Scholar]
- 36. Campbell, N. H., Parkinson, G. N., Reszka, A. P., and Neidle, S. (2008) Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 130, 6722–6724 10.1021/ja8016973 [DOI] [PubMed] [Google Scholar]
- 37. Parkinson, G. N., Ghosh, R., and Neidle, S. (2007) Structural basis for binding of porphyrin to human telomeres. Biochemistry 46, 2390–2397 10.1021/bi062244n [DOI] [PubMed] [Google Scholar]
- 38. Chi, Q. J., Wang, G. X., and Jiang, J. H. (2013) The persistence length and length per base of single-stranded DNA obtained from fluorescence correlation spectroscopy measurements using mean field theory. Physica A 392, 1072–1079 10.1016/j.physa.2012.09.022 [DOI] [Google Scholar]
- 39. Gosse, C., and Croquette, V. (2002) Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 82, 3314–3329 10.1016/S0006-3495(02)75672-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lipfert, J., Hao, X., and Dekker, N. H. (2009) Quantitative modeling and optimization of magnetic tweezers. Biophys. J. 96, 5040–5049 10.1016/j.bpj.2009.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Vlaminck, I., Henighan, T., van Loenhout, M. T., Burnham, D. R., and Dekker, C. (2012) Magnetic forces and DNA mechanics in multiplexed magnetic tweezers. PLoS One 7, e41432 10.1371/journal.pone.0041432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lionnet, T., Allemand, J. F., Revyakin, A., Strick, T. R., Saleh, O. A., Bensimon, D., and Croquette, V. (2012) Magnetic trap construction. Cold Spring Harb. Protoc. 2012, 133–138 [DOI] [PubMed] [Google Scholar]
- 43. Savitzky, A., and Golay, M. J. (1964) Smoothing and differentiation of data by least squares procedures. Anal. Chem. 36, 1627 10.1021/ac60214a047 [DOI] [Google Scholar]
- 44. Reineker, P., Winkler, R. G., and Glatting, G. (1995) Freely jointed chain with variable segment number and length. Colloid Polymer Sci. 273, 32–37 10.1007/BF00655671 [DOI] [Google Scholar]
- 45. Bouchiat, C., Wang, M. D., Allemand, J., Strick, T., Block, S. M., and Croquette, V. (1999) Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 76, 409–413 10.1016/S0006-3495(99)77207-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Freedman, D. A., Pisiani, R., and Purves, R. A. (2007) Statistics, 4th Ed., W. W. Norton, Inc., New York [Google Scholar]
- 47. de Messieres, M., Chang, J. C., Brawn-Cinani, B., and La Porta, A. (2012) Single-molecule study of G-quadruplex disruption using dynamic force spectroscopy. Phys Rev. Lett. 109, 058101 10.1103/PhysRevLett.109.058101 [DOI] [PubMed] [Google Scholar]
- 48. Lynch, S., Baker, H., Byker, S. G., Zhou, D., and Sinniah, K. (2009) Single molecule force spectroscopy on G-quadruplex DNA. Chemistry 15, 8113–8116 10.1002/chem.200901390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ambrus, A., Chen, D., Dai, J., Bialis, T., Jones, R. A., and Yang, D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 10.1093/nar/gkl348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang, Q., Ma, L., Hao, Y.-H., and Tan, Z. (2010) Folding equilibrium constants of telomere G-quadruplexes in free state or associated with proteins determined by isothermal differential hybridization. Anal. Chem. 82, 9469–9475 10.1021/ac102168m [DOI] [PubMed] [Google Scholar]
- 51. Hänsel, R., Löhr, F., Foldynová-Trantírková, S., Bamberg, E., Trantírek, L., and Dötsch, V. (2011) The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 39, 5768–5775 10.1093/nar/gkr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qureshi, M. H., Ray, S., Sewell, A. L., Basu, S., and Balci, H. (2012) Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J. Phys. Chem. B 116, 5588–5594 10.1021/jp300546u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tombline, G., Heinen, C. D., Shim, K.-S., and Fishel, R. (2002) Biochemical characterization of the human RAD51 protein: III. modulation of DNA binding by adenosine nucleotides. J. Biol. Chem. 277, 14434–14442 10.1074/jbc.M109917200 [DOI] [PubMed] [Google Scholar]
- 54. Heyer, W. D., Ehmsen, K. T., and Liu, J. (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cesare, A. J., and Reddel, R. R. (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 10.1038/nrg2763 [DOI] [PubMed] [Google Scholar]
- 56. Senavirathne, G., Mahto, S. K., Hanne, J., O'Brian, D., and Fishel, R. (2017) Dynamic unwrapping of nucleosomes by HsRAD51 that includes sliding and rotational motion of histone octamers. Nucleic Acids Res. 45, 685–698 10.1093/nar/gkw920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Conway, A. B., Lynch, T. W., Zhang, Y., Fortin, G. S., Fung, C. W., Symington, L. S., and Rice, P. A. (2004) Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 11, 791–796 10.1038/nsmb795 [DOI] [PubMed] [Google Scholar]
- 58. Shinohara, A., and Ogawa, T. (1999) Rad51/RecA protein families and the associated proteins in eukaryotes. Mutat. Res. 435, 13–21 10.1016/S0921-8777(99)00033-6 [DOI] [PubMed] [Google Scholar]
- 59. Brockman, C., Kim, S. J., and Schroeder, C. M. (2011) Direct observation of single flexible polymers using single stranded DNA. Soft Matter 7, 8005–8012 10.1039/c1sm05297g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee, K. S., Marciel, A. B., Kozlov, A. G., Schroeder, C. M., Lohman, T. M., and Ha, T. (2014) Ultrafast redistribution of E. coli SSB along long single-stranded DNA via intersegment transfer. J. Mol. Biol. 426, 2413–2421 10.1016/j.jmb.2014.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marsh, T. C., and Henderson, E. (1994) G-wires: self-assembly of a telomeric oligonucleotide, d(GGGGTTGGGG), into large superstructures. Biochemistry 33, 10718–10724 10.1021/bi00201a020 [DOI] [PubMed] [Google Scholar]
- 62. Feller, W. (1968) An Introduction to Probability Theory and Its Applications, Wiley, Hoboken, NJ [Google Scholar]
- 63. Makarov, V. L., Hirose, Y., and Langmore, J. P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88, 657–666 10.1016/S0092-8674(00)81908-X [DOI] [PubMed] [Google Scholar]
- 64. Wright, W. E., Tesmer, V. M., Huffman, K. E., Levene, S. D., and Shay, J. W. (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11, 2801–2809 10.1101/gad.11.21.2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Arnoult, N., and Karlseder, J. (2015) Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 22, 859–866 10.1038/nsmb.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Taylor, D. J., Podell, E. R., Taatjes, D. J., and Cech, T. R. (2011) Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J. Mol. Biol. 410, 10–17 10.1016/j.jmb.2011.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thys, R. G., and Wang, Y. H. (2015) DNA replication dynamics of the GGGGCC repeat of the C9orf72 gene. J. Biol. Chem. 290, 28953–28962 10.1074/jbc.M115.660324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Griffith, J. D., and Christiansen, G. (1978) Electron microscope visualization of chromatin and other DNA–protein complexes. Annu. Rev. Biophys. Bioeng 7, 19–35 10.1146/annurev.bb.07.060178.000315 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.