Abstract
Viruses exploit the host cell machinery for their own profit. To evade innate immune sensing and promote viral replication, HIV type 1 (HIV-1) subverts DNA repair regulatory proteins and induces G2/M arrest. The preintegration complex of HIV-1 is known to traffic along microtubules and accumulate near the microtubule-organizing center. The centrosome is the major microtubule-organizing center in most eukaryotic cells, but precisely how HIV-1 impinges on centrosome biology remains poorly understood. We report here that the HIV-1 accessory protein viral protein R (Vpr) localized to the centrosome through binding to DCAF1, forming a complex with the ubiquitin ligase EDD-DYRK2-DDB1DCAF1 and Cep78, a resident centrosomal protein previously shown to inhibit EDD-DYRK2-DDB1DCAF1. Vpr did not affect ubiquitination of Cep78. Rather, it enhanced ubiquitination of an EDD-DYRK2-DDB1DCAF1 substrate, CP110, leading to its degradation, an effect that could be overcome by Cep78 expression. The down-regulation of CP110 and elongation of centrioles provoked by Vpr were independent of G2/M arrest. Infection of T lymphocytes with HIV-1, but not with HIV-1 lacking Vpr, promoted CP110 degradation and centriole elongation. Elongated centrioles recruited more γ-tubulin to the centrosome, resulting in increased microtubule nucleation. Our results suggest that Vpr is targeted to the centrosome where it hijacks a ubiquitin ligase, disrupting organelle homeostasis, which may contribute to HIV-1 pathogenesis.
Keywords: centrosome, centriole, ubiquitin ligase, human immunodeficiency virus (HIV), viral protein, microtubule, Cep78, CP110
Introduction
Viruses are pathogens that infect all life forms and reproduce inside living cells. To do so, they must be able to counteract and evade immune defenses, as well as utilize cellular machinery from the host for their own replication. HIV type 1 (HIV-1), which belongs to the lentivirus subgroup of retroviruses, is the causal agent of AIDS (1, 2). The HIV-1 genome encodes five proteins essential for viral replication and four accessory proteins, namely viral infectivity factor (Vif), viral protein U (Vpu), negative regulatory factor (Nef), and viral protein R (Vpr)2 (3, 4). These accessory proteins are not absolutely essential for viral replication in vitro but nevertheless play critical roles in viral infection, survival, and propagation in vivo (5–12). Vpr is among the least characterized in terms of function and mechanism of action. As a predominantly nuclear protein, Vpr has multiple effects on host cells by interacting with a cohort of cellular proteins (13–24). Among these, viral protein R–binding protein (VprBP/DCAF1) is the first protein identified as binding Vpr (15, 25). Current evidence suggests that DCAF1 functions as a protein kinase (26), a transcriptional repressor (27), and a substrate recognition subunit of two distinct multi-subunit ubiquitin ligases, EDD-DYRK2-DDB1DCAF1 and CRL4DCAF1 (28). EDD-DYRK2-DDB1DCAF1 is composed of the DYRK2, EDD, DDB1, and DCAF1 subunits (29), whereas CRL4DCAF1 consists of Roc1, Cullin4, DDB1, and DCAF1 (30–32). Upon binding to a ubiquitin ligase, Vpr directs the ubiquitination of novel substrates and accelerates the ubiquitination of native substrates, leading to their premature degradation (16, 18, 20, 33–35).
In contrast to CRL4DCAF1, which is present in the nucleus, EDD-DYRK2-DDB1DCAF1 exists in two distinct subcellular compartments, the nucleus and the centrosome; the latter comprises a pair of centrioles surrounded by pericentriolar material from which microtubules emanate and elongate (36, 37). In the nucleus, EDD-DYRK2-DDB1DCAF1 functions to suppress telomerase activity by targeting telomerase reverse transcriptase (TERT) for ubiquitination and degradation (36). The down-regulation of TERT is further enhanced by Vpr binding to EDD-DYRK2-DDB1DCAF1 (19). On the other hand, EDD-DYRK2-DDB1DCAF1, at the centrosome, is known to ubiquitinate and induce the degradation of CP110, a protein that controls centriole length (37–41). The ability of EDD-DYRK2-DDB1DCAF1 to ubiquitinate CP110 is subjected to regulation by Cep78, a resident centrosomal protein that directly associates with and inhibits EDD-DYRK2-DDB1DCAF1 in a cell cycle–dependent manner (37). It is currently unknown whether Vpr has the capacity to hijack EDD-DYRK2-DDB1DCAF1 at the centrosome.
The centrosome is the major microtubule-organizing centers in most eukaryotic cells and acts as a central hub for coordinating a multitude of cellular events. Various molecules and cargos are known to transit through this organelle (42). The viral core of HIV-1 disassembles upon entry into the host cells, and the resulting preintegration complex traffics along microtubules and accumulates near the microtubule-organizing center (43–46). Another study reports that HIV-1 subviral particles accumulate at the centrosome under resting T-cells through an unknown mechanism, and infection resumes upon stimulation (47). Interestingly, Vpr has been observed to disrupt certain protein interactions at the centrosome (48) and induce centrosome amplification and multipolar spindle formation (49, 50), suggesting that this viral protein is capable of exerting an effect on the centrosome either directly or indirectly. Despite these observations, the extent to which Vpr modulates different aspects of centrosome biology and the underlying mechanisms have not been studied in detail.
Results
Vpr binds to Cep78 and EDD-DYRK2-DDB1DCAF1 and localizes to the centrosome
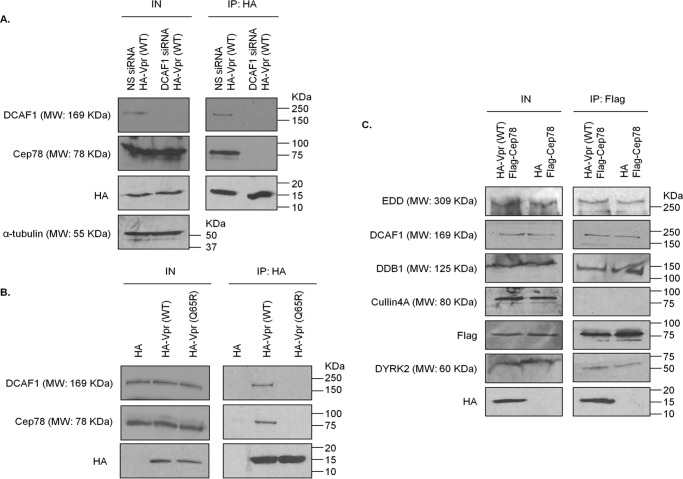
We recently demonstrated that Cep78 forms a complex with EDD-DYRK2-DDB1DCAF1 through DCAF1 (37). Given that Vpr is known to associate with DCAF1 (15, 25), we first asked whether Vpr and Cep78 interact. Endogenous Cep78 and DCAF1 co-immunoprecipitated with HA-Vpr in HEK293 cells (Fig. 1, A and B). When DCAF1 was depleted with siRNA, very little Cep78 was detected in Vpr immunoprecipitates (Fig. 1A). Moreover, endogenous DCAF1 and Cep78 bound to WT Vpr, but neither protein interacted with a Vpr mutant refractory to DCAF1 binding (Vpr(Q65R)) (Fig. 1B). Thus, Vpr likely associates with Cep78 through DCAF1, results that are consistent with the findings that the Vpr- and Cep78-binding sites of DCAF1 are nonoverlapping. Vpr binds to the WD40 domain of DCAF1 (15, 51), whereas Cep78 binds to the acidic domain of DCAF1 (37).
Figure 1.
Vpr interacts with Cep78 and EDD-DYRK2-DDB1DCAF1 through DCAF1. A, HEK293 cells were transfected with nonspecific (NS) or DCAF1 siRNA followed by plasmid expressing HA-Vpr(WT). Lysates were immunoprecipitated (IP) with an anti-HA antibody and Western-blotted with the indicated antibodies. IN, input. α-Tubulin was used as loading control. B, HEK293 cells were transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr mutant refractory to DCAF1 binding (Q65R). Lysates were immunoprecipitated with an anti-HA antibody and Western-blotted with the indicated antibodies. C, HEK293 cells were co-transfected with plasmids expressing FLAG-Cep78 and HA or HA-Vpr(WT). Lysates were immunoprecipitated with an anti-FLAG antibody and Western-blotted with the indicated antibodies.
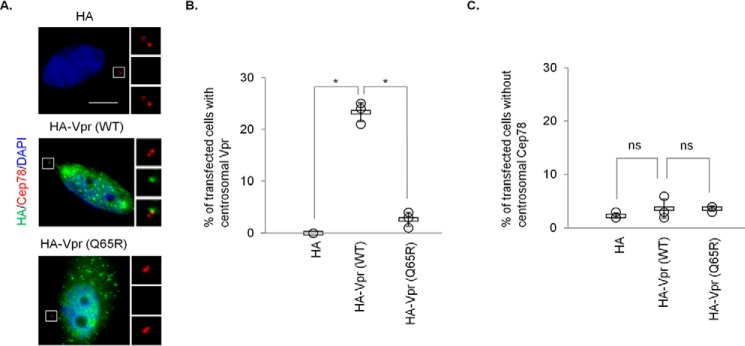
Next, we explored whether Vpr might bind specifically to the Cep78·EDD-DYRK2-DDB1DCAF1 complex, which normally forms at the centrosome. We expressed FLAG-Cep78 and HA-Vpr in HEK293 cells, performed anti-FLAG immunoprecipitations, and demonstrated that Cep78 binds to EDD-DYRK2-DDB1DCAF1 but not to CRL4DCAF1, which is expected (37), and to Vpr (Fig. 1C). WT Vpr co-localized with endogenous Cep78 in ∼20–25% of transfected HeLa cells, indicating that this viral protein is targeted to the centrosome in some contexts (Fig. 2, A and B). In contrast, Vpr(Q65R) did not exhibit centrosomal localization (Fig. 2, A and B). Taken together, these data suggest that Vpr engages in a complex with EDD-DYRK2-DDB1DCAF1 and Cep78 at the centrosome through binding to DCAF1.
Figure 2.
Vpr but not Vpr(Q65R) localizes to the centrosome. A, HeLa cells transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(Q65R) were processed for immunofluorescence and stained with antibodies against HA (green) and Cep78 (red). DNA was stained with DAPI (blue). Scale bar, 2 μm. B and C, the percentage of HA-expressing cells showing centrosomal localization of Vpr (B) or no centrosomal Cep78 staining (C) was determined. For B and C, at least 100 cells were scored for each condition in each experiment, and the mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01; ns, nonsignificant.
Vpr hijacks EDD-DYRK2-DDB1DCAF1 to enhance ubiquitination and degradation of CP110
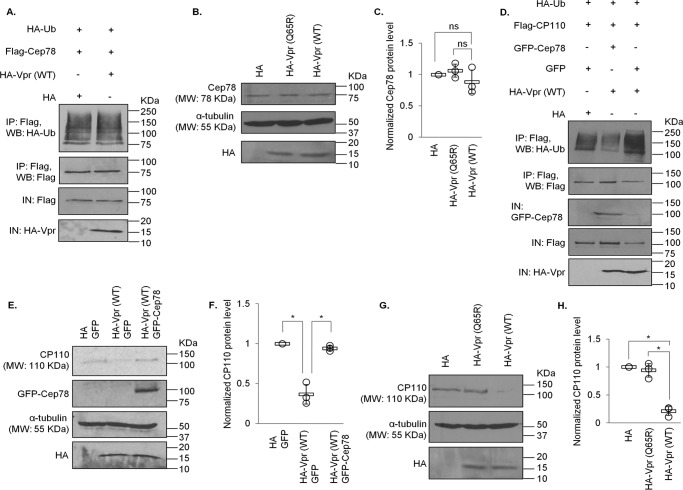
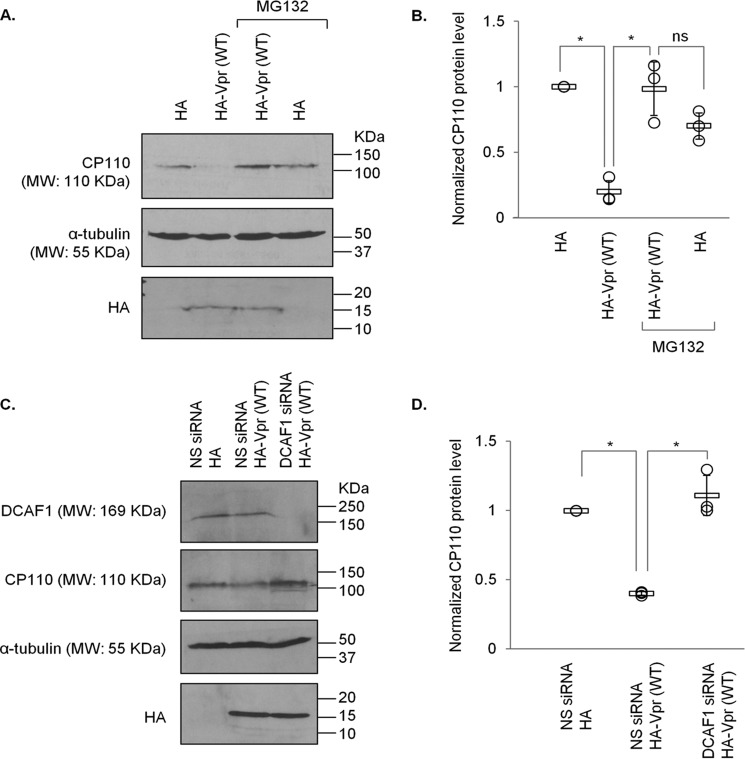
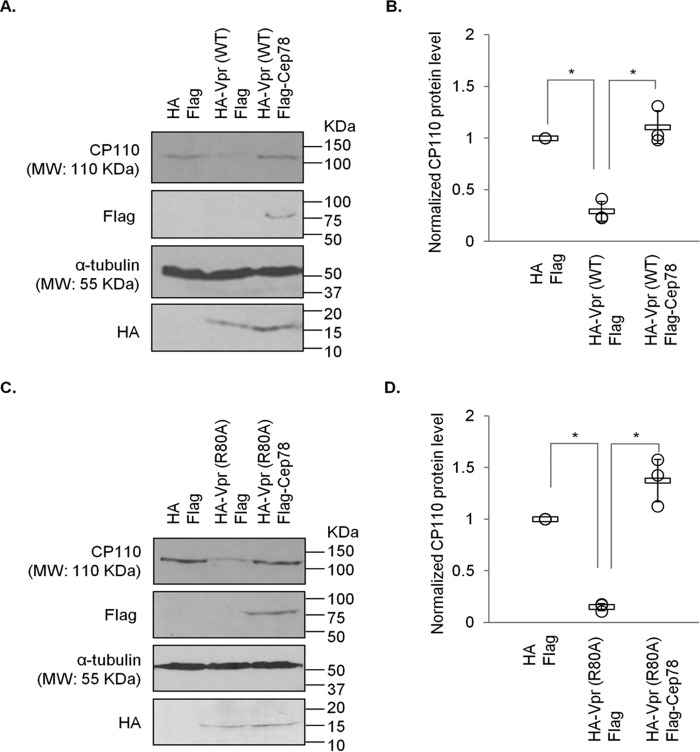
To explore the relevance of Vpr binding to EDD-DYRK2-DDB1DCAF1 and Cep78, we tested whether Vpr might promote ubiquitination of proteins at the centrosome. The ubiquitination levels of Cep78, an inhibitor and nonsubstrate of EDD-DYRK2-DDB1DCAF1 (37), remained the same upon Vpr expression (Fig. 3A). Likewise, centrosomal localization and steady-state levels of Cep78 were not altered by WT Vpr or Vpr(Q65R) (Figs. 2, A and C, and 3, A–C). On the contrary, ubiquitination of CP110, a known centrosomal EDD-DYRK2-DDB1DCAF1 substrate (37), became greatly enhanced by Vpr (Fig. 3D). This was accompanied by a decrease in CP110 protein levels (Fig. 3, D–H) and a loss of centrosomal CP110 staining by immunofluorescence (Fig. 4, A and B). Notably, diminished levels of CP110 were specifically induced by WT Vpr but not Vpr(Q65R) mutant (Figs. 3, G and H, and 4, A and B) and could be rescued by the addition of a proteasome inhibitor, MG132 (Fig. 5, A and B), or depletion of DCAF1 (Fig. 5, C and D). Furthermore, co-expression of Cep78 drastically reduced ubiquitination of CP110 (Fig. 3D) and restored endogenous CP110 to WT levels (Figs. 3, E and F, and 8, A and B). These data indicate that Vpr subverts centrosomal EDD-DYRK2-DDB1DCAF1 to accelerate ubiquitination and proteasomal degradation of a native substrate, CP110; and these effects can be counteracted by overexpression of Cep78.
Figure 3.
Vpr enhances ubiquitination and degradation of CP110 but not Cep78. A, HEK293 cells were co-transfected with plasmids expressing HA-Ub, FLAG-Cep78, and HA or HA-Vpr(WT). Lysates were immunoprecipitated (IP) with an anti-FLAG antibody in 1% SDS and Western-blotted (WB) with the indicated antibodies. IN, input. B, HEK293 cells were transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(Q65R). Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. C, normalized Cep78 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. ns, nonsignificant. D, HEK293 cells were co-transfected with plasmids expressing HA-Ub, FLAG-CP110, GFP or GFP-Cep78, and HA or HA-Vpr(WT). Lysates were immunoprecipitated with an anti-FLAG antibody in 1% SDS and Western-blotted with the indicated antibodies. E, HEK293 cells were co-transfected with plasmids expressing HA and GFP, HA-Vpr(WT) and GFP, or HA-Vpr(WT) and GFP-Cep78. Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. F, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01. G, HEK293 cells were transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(Q65R). Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. H, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
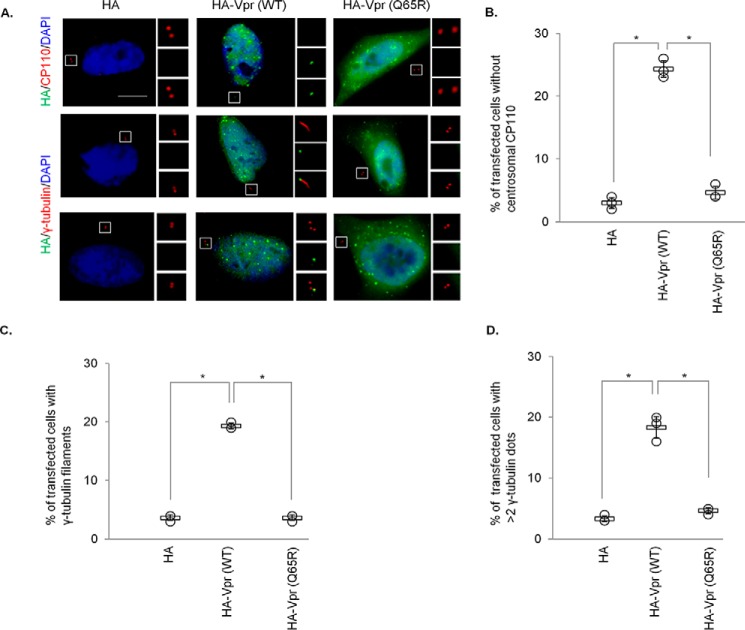
Figure 4.
Vpr induces CP110 loss, centriole elongation, and centrosome amplification. A, HeLa cells transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(Q65R) were processed for immunofluorescence and stained with antibodies against HA (green) and CP110 or γ-tubulin (red). DNA was stained with DAPI (blue). Scale bar, 2 μm. B, the percentage of HA-expressing cells with no centrosomal CP110 staining was determined. C and D, the percentage of HA-expressing cells with elongated centrioles (γ-tubulin filaments) (C) or centrosome amplification (>2 γ-tubulin dots) (D) was determined. For B–D, at least 100 cells were scored for each condition in each experiment, and the mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
Figure 5.
Vpr-induced proteasomal degradation of CP110 occurs in a DCAF1-dependent manner. A, HEK293 cells transfected with plasmid expressing HA or HA-Vpr(WT) were treated with or without 10 μm MG132 for 6 h. Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. B, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01; ns, nonsignificant. C, HEK293 cells were transfected with nonspecific (NS) or DCAF1 siRNA followed by plasmid expressing HA or HA-Vpr(WT). Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. D, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
Figure 8.
Degradation of CP110 induced by Vpr or Vpr(R80A) can be overcome by Cep78 expression. A, HEK293 cells were transfected with plasmids expressing HA and FLAG, HA-Vpr(WT) and FLAG, or HA-Vpr(WT) and FLAG-Cep78. Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. B, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01. C, HEK293 cells were transfected with plasmids expressing HA and FLAG, HA-Vpr(R80A) and FLAG, or HA-Vpr(R80A) and FLAG-Cep78. Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. D, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
Vpr induces centriole elongation through CP110 degradation
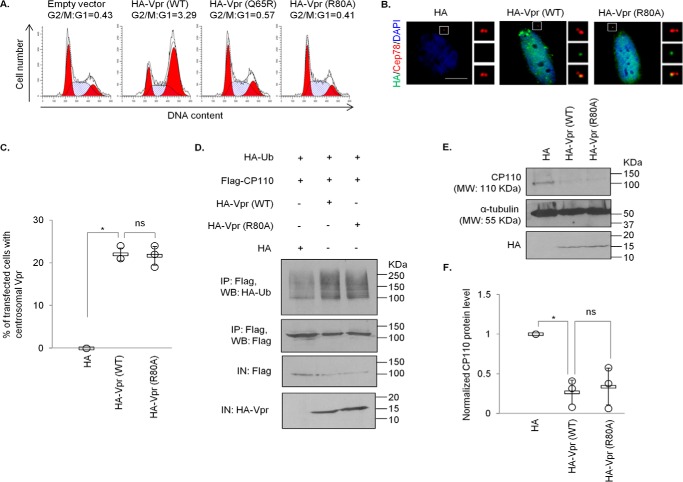
Previously, it has been shown that depletion of CP110 induces the formation of overly long or elongated centrioles, represented by γ-tubulin filaments, in nonciliated or poorly ciliated cells including HeLa (38–41). This phenotype can also be recapitulated by CP110 loss resulting from ablation of Cep78 or overexpression of EDD-DYRK2-DDB1DCAF1 (37). To further substantiate our observations that Vpr enhances degradation of CP110, we found that WT Vpr provokes centriole elongation, whereas Vpr(Q65R) mutant cannot (Fig. 4, A and C). Of note, WT Vpr also induced centrosome amplification (>2 γ-tubulin foci, Fig. 4, A and D), consistent with a previous report (50), but this phenotype is unlikely to be a consequence of CP110 loss because excessive CP110, rather than loss of CP110, drives centrosome amplification (52).
Vpr-induced CP110 degradation and centriole elongation are independent of G2/M arrest
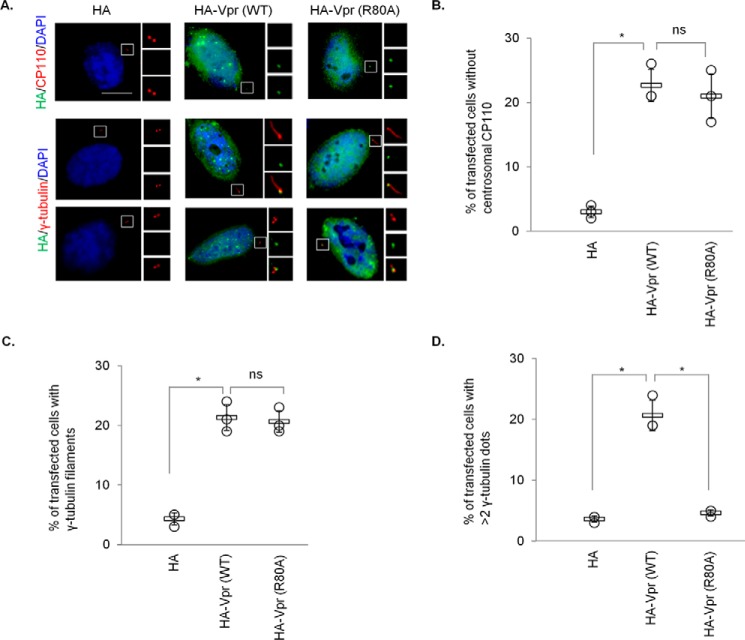
A recent study shows that Vpr associates with the SLX4 complex to induce chromosomal instability, triggering DNA damage response (DDR) and cell cycle arrest at the G2/M phase (20), although this is in debate (53). Coincidentally, CP110 has been documented to undergo ubiquitination by SCFcyclin F and EDD-DYRK2-DDB1DCAF1 (37, 52), and subsequently proteasomal degradation, in G2/M. Thus, we sought to address whether down-regulation of CP110 induced by Vpr is due to prolonged G2/M arrest. For this purpose, we utilized a well-characterized Vpr mutant, Vpr(R80A), which, in contrast to Vpr(Q65R), can bind to DCAF1 but is unable to provoke G2/M arrest (Ref. 54, 55 and Fig. 6A). Similar to WT Vpr, Vpr(R80A) was detected at the centrosome in ∼20–25% of the transfected cells (Fig. 6, B and C). Next, we investigated the consequences of expressing Vpr(R80A) on CP110 and its effect on centriole length. WT Vpr and Vpr(R80A) were equally able to enhance CP110 ubiquitination (Fig. 6D), causing a diminution of CP110 levels (Fig. 6, E and F) and immunostaining at the centrosome (Fig. 7, A and B). Furthermore, WT Vpr and Vpr(R80A) induced γ-tubulin filament formation to similar extent (Fig. 7A, C). The down-regulation of CP110 provoked by WT Vpr or Vpr(R80A) was rescued by ectopic expression of Cep78 (Fig. 8). Remarkably, unlike WT Vpr, Vpr(R80A) did not induce centrosome amplification (Fig. 7, A and D). These data suggest that CP110 down-regulation and centriole elongation could be attributed to the subversion of EDD-DYRK2-DDB1DCAF1 rather than to G2/M arrest. On the contrary, the other phenotype caused by Vpr, namely centrosome amplification, depends on cell cycle arrest at the G2/M phase.
Figure 6.
Vpr centrosomal localization and Vpr-induced ubiquitination and degradation of CP110 are independent of G2/M arrest. A, HEK293T cells were co-transfected with plasmids expressing GFP and HA (Empty vector), HA-Vpr(WT), HA-Vpr(Q65R), or HA-Vpr(R80A). Cell cycle profiles were determined by flow cytometry gating on the GFP+ population. The G2/M:G1 ratio is presented for each condition. B, HeLa cells transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(R80A) were processed for immunofluorescence and stained with antibodies against HA (green) and Cep78 (red). DNA was stained with DAPI (blue). Scale bar, 2 μm. C, the percentage of HA-expressing cells showing centrosomal localization of Vpr was determined. At least 100 cells were scored for each condition in each experiment, and the mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01; ns, nonsignificant. D, HEK293 cells were co-transfected with plasmids expressing HA-Ub, FLAG-CP110, and HA, HA-Vpr(WT), or HA-Vpr(R80A). Lysates were immunoprecipitated (IP) with an anti-FLAG antibody in 1% SDS and Western-blotted (WB) with the indicated antibodies. IN, input. E, HEK293 cells were transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(R80A). Lysates were Western-blotted with the indicated antibodies. α-Tubulin was used as loading control. F, normalized CP110 protein level. The mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
Figure 7.
Vpr-induced centrosome amplification, but not CP110 loss or centriole elongation, is dependent of G2/M arrest. A, HeLa cells transfected with plasmid expressing HA, HA-Vpr(WT), or HA-Vpr(R80A) were processed for immunofluorescence and stained with antibodies against HA (green) and CP110 or γ-tubulin (red). DNA was stained with DAPI (blue). Scale bar, 2 μm. B, the percentage of HA-expressing cells with no centrosomal CP110 staining was determined. C and D, the percentage of HA-expressing cells with elongated centrioles (γ-tubulin filaments) (C) or centrosome amplification (>2 γ-tubulin dots) (D) was determined. For B–D, at least 100 cells were scored for each condition in each experiment, and the mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01; ns, nonsignificant.
Vpr induces CP110 degradation and centriole elongation in infected T-cells
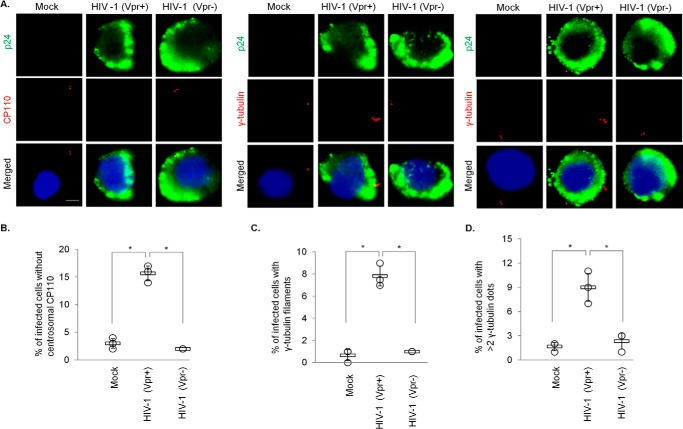
We have thus far shown that Vpr induces the loss of CP110 in two model cell lines, HEK293 and HeLa. However, it remains unknown whether this accessory protein could trigger the same response in CD4+ T lymphocytes that HIV-1 normally infects. To interrogate the relationship between Vpr and CP110 in a more physiologically relevant cell line, we infected CD4+ MT4 T-cells, which are highly susceptible to and permissive for infection with HIV-1. We found that a significant percentage of cells infected with WT HIV-1 (HIV-1 Vpr+) exhibit CP110 loss and centriole elongation (Fig. 9, A–C) in addition to centrosome amplification (Fig. 9A, D). In contrast, very few mock-infected cells or cells infected with HIV-1 lacking Vpr (HIV-1 Vpr−) possess these phenotypes (Fig. 9). These results indicate that HIV-1 can also induce CP110 degradation and centriole elongation in T-cells in a Vpr-dependent manner.
Figure 9.
HIV-1 Vpr induces CP110 loss, centriole elongation, and centrosome amplification in infected T-cells. A, MT4 cells, mock-infected or infected with either WT HIV-1 (Vpr+) or HIV-1 missing Vpr (Vpr−), were processed for immunofluorescence and stained with antibodies against p24 (green) and CP110 or γ-tubulin (red). DNA was stained with DAPI (blue). Scale bar, 2 μm. B, the percentage of p24-positive cells with no centrosomal CP110 staining was determined. C and D, the percentage of p24-positive cells with elongated centrioles (γ-tubulin filaments) (C) or centrosome amplification (>2 γ-tubulin dots) (D) was determined. For B–D, at least 100 cells were scored for each condition in each experiment, and the mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
Elongated centrioles enhance microtubule nucleation
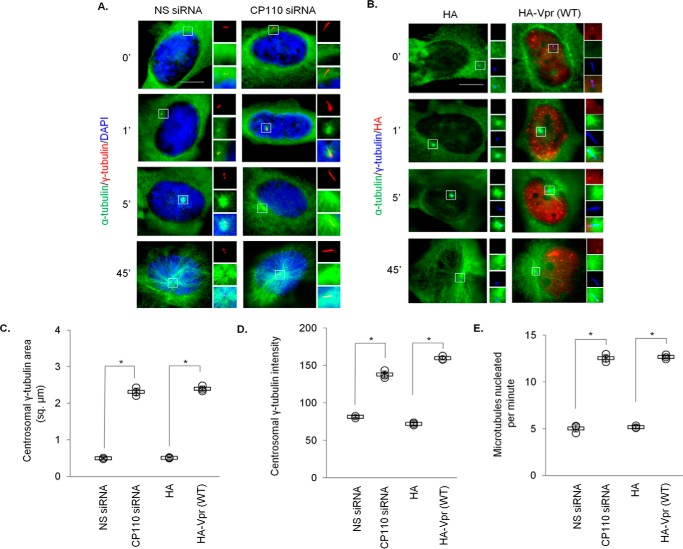
To shed light on the net effects of down-regulating CP110 for Vpr, we studied how elongated centrioles might influence centrosome function. When a centriole becomes abnormally long, the surrounding pericentriolar material becomes distorted into the shape of a filament (38–41). Given that γ-tubulin present in the pericentriolar material plays a crucial role in microtubule nucleation, we addressed the question of whether elongated centrioles might alter nucleation. We quantified the staining area occupied by γ-tubulin and found that it is substantially bigger in CP110-depleted or Vpr-expressing cells than in control cells (Fig. 10, A–C). γ-Tubulin staining intensity was likewise higher upon depletion of CP110 or expression of Vpr (Fig. 10, A, B, and D). Next, we performed microtubule regrowth assays following microtubule depolymerization with nocodazole. Shortly after the removal of the nocodazole, control cells nucleated an aster of microtubules emanating from the centrosome (Fig. 10, A and B, 1′). With time, the aster enlarged, signifying an increase in the length and number of microtubules (Fig. 10, A and B, 5′). Strikingly, cells depleted of CP110 or expressing Vpr formed a bigger aster and nucleated more microtubules at comparable time points (Fig. 10, A and B, 1′ and 5′, and E). In contrast, no gross microtubule-anchoring defects were observed (Fig. 10, A and B, 45′). Together, these data strongly suggest that elongated centrioles have the capacity to recruit more γ-tubulin, resulting in enhanced microtubule nucleation.
Figure 10.
Depletion of CP110 or expression of Vpr enhances microtubule nucleation. A, HeLa cells transfected with nonspecific (NS) or CP110 siRNA were subjected to a microtubule regrowth assay. Cells were processed for immunofluorescence at the indicated time points after release and stained with antibodies against α-tubulin (green) and γ-tubulin (red). DNA was stained with DAPI (blue). B, HeLa cells transfected with plasmid expressing HA or HA-Vpr(WT) were subjected to a microtubule regrowth assay. Cells were processed for immunofluorescence at the indicated time points after release and stained with antibodies against HA (red), α-tubulin (green), and γ-tubulin (blue). C, the staining area of γ-tubulin at the centrosome was quantitated. D, the staining intensity of γ-tubulin at the centrosome was quantitated. E, the number of cytoplasmic microtubules emanated from the centrosome was determined at the 1-min time point. For C–E, at least 20 cells were scored for each condition in each experiment, and the mean (thick open line) and standard error (bar) of three independent experiments (○) are shown in the graph. *, p < 0.01.
Discussion
In this work, we sought to obtain molecular insights into how HIV-1 Vpr exploits host machinery at the centrosome. Although an intimate connection exists among HIV-1, Vpr, and centrosomes (42, 47–50, 56), the extent to which Vpr orchestrates its effects on this organelle remains poorly understood. Our data show that Vpr associates with a resident centrosomal protein, Cep78, through DCAF1 and that it localizes to the centrosome by engaging in a complex with the ubiquitin ligase EDD-DYRK2-DDB1DCAF1 and Cep78. Because Vpr and Vpr(R80A) localize to the centrosome with similar efficiency and EDD-DYRK2-DDB1DCAF1 components are known to be present at this organelle throughout the cell cycle (37), it seems likely that centrosomal localization of Vpr is independent of G2/M arrest. Vpr is able to hijack EDD-DYRK2-DDB1DCAF1, accelerating the ubiquitination and degradation of a native centrosomal substrate, CP110. Down-regulation of CP110 triggers the formation of abnormally long centrioles, which recruit excess γ-tubulin, and as a consequence, the nucleation of cytoplasmic microtubules becomes greatly enhanced. In addition, Vpr provokes other centrosome anomalies such as amplification (Ref. 50 and this study), indicating that proteins involved in the regulation of organelle copy number might also be affected. It would therefore be interesting to identify novel Vpr-interacting partners and/or EDD-DYRK2-DDB1DCAF1 substrates and to test whether any of these might be responsible for the centrosome amplification phenotype.
Although Vpr triggers centriole elongation and centrosome amplification, it is clear that these phenotypes occur through two distinct mechanisms. We show that centriole elongation as a result of Vpr-mediated CP110 loss is independent of G2/M arrest, whereas centrosome amplification necessitates G2/M arrest. How then does Vpr-induced G2/M arrest result in centrosome amplification? It is reported that Vpr targets DNA repair factors such as HLTF and UNG (33, 35) for degradation and inappropriately activates the SLX4 complex in the nucleus (20), conditions that could contribute to replication stress and the induction of DDR (57). The DDR protein ataxia telangiectasia-mutated (ATM) and Rad3-related protein (ATR), once activated, initiates downstream signaling cascades that involve activation of checkpoint kinase 1 (CHK1) and inhibition of cell division cycle 25C (CDC25C) and cyclin B/cyclin-dependent kinase 1 (CDK1), ultimately leading to G2/M arrest (58). Curiously, other studies have shown that DNA damage alone is sufficient to induce centrosome amplification (59), and several DDR proteins, such as ATM, ATR, and CHK1, are found in the nucleus and at the centrosome (60). Although the precise functions of DDR proteins at the centrosome await future investigation, it is plausible that a DDR signal originating from the nucleus impinges on the centrosome through the DDR proteins, causing amplification to occur.
What are the benefits that HIV-1 might receive by hijacking EDD-DYRK2-DDB1DCAF1 at the centrosome? The regulation of microtubule dynamics and microtubule-associated proteins such as end-binding proteins and motor proteins is an important facet of the HIV-1 replication cycle. For example, HIV-1 promotes the formation of stable microtubules, an event crucial for early infection and translocation of the viral core in the cytoplasm en route to the nucleus (45). Intact microtubules are needed to facilitate HIV-1 uncoating, and disruption of microtubules by nocodazole impairs this process (46). In macrophages, HIV-1 Vpr perturbs the localization of end-binding protein 1 (EB1) to impair the maturation of phagosomes, leading to defects in innate immunity (61). Moreover, HIV-1 Tat can promote or hinder microtubule stability in a context-dependent fashion (62–64). Thus, it is clear that HIV-1 employs different strategies to remodel the host microtubule network during infection. Further studies will be needed to decipher how CP110 loss, elongated centrioles, and enhanced microtubule nucleation provoked by Vpr might affect various stages of HIV-1 infection.
One interesting finding from our studies is that Cep78 counteracts the effects of Vpr on CP110, raising the possibility that it might have antiviral properties. It would therefore be interesting to test whether this protein might safeguard the centrosome to inhibit viral infection.
Experimental procedures
Cell culture and plasmids
HeLa, HEK293, and HEK293T cells were grown in DMEM (Wisent Inc., 319-005-CL) supplemented with 5% fetal bovine serum (Wisent Inc, 080150) at 37 °C in a humidified 5% CO2 atmosphere. MT4 T-cells were grown in RPMI 1640 (Wisent Inc., 350-000-CL) supplemented with 10% fetal bovine serum at 37 °C in a humidified 5% CO2 atmosphere. The following proteins were expressed from plasmids in mammalian cells: HA-Ub (65), pCBF-FLAG-Cep78 (37), pEGFP-C1-Cep78 (37), pCBF-FLAG-CP110 (66), pQBI25, SVCMV-HA-Vpr, SVCMV-HA-Vpr(Q65R), and SVCMV-HA-Vpr(R80A) (55).
Viral production and infection
Infectious GFP-marked HIV-1 NL4.3 or NL4.3ΔVpr viruses were generated by calcium phosphate transfection of HEK293T cells. Virus-containing supernatants were recovered 2 days post-transfection, clarified, pelleted by ultracentrifugation, and titrated by analyzing the percentage of GFP-positive MT4 T-cells using flow cytometry. MT4 T-cells were infected with the different GFP-expressing NL4.3 viruses at a multiplicity of infection of 0.75. Three days post-infection, cells were plated on coated coverslips and processed for immunofluorescence.
Antibodies
Antibodies used in this study included anti-CP110 (Bethyl Laboratories, A301-344A), anti-Cep78 (Bethyl Laboratories, A301-799A and IRCM6 (37)), anti-DCAF1 (Proteintech, 11612-1-AP), anti-EDD (Bethyl Laboratories, A300-573A), anti-DDB1 (Bethyl Laboratories, A300-462A), anti-cullin 4A (Bethyl Laboratories, A300-739A), anti-GFP (Roche, 11814460001), anti-HA (Santa Cruz Biotechnology, sc-7392, and Novus Biologicals, NB600-362), anti-FLAG (Sigma-Aldrich, F7425 and F3165), anti-α-tubulin (Sigma-Aldrich, T5168), anti-γ-tubulin (Sigma-Aldrich, T3559 and T6557), and anti-DYRK2 (Abcam, ab37912). The anti-p24 monoclonal antibodies were produced from hybridomas 31-90-25 (HB9725) obtained from the American Type Culture Collection.
RNAi and transient expression of recombinant proteins
For RNAi, synthetic siRNA for nonspecific (NS) control, DCAF1, and CP110 were as described previously (29, 67, 68) and purchased from GE Dharmacon. Transfection of siRNA into HEK293 or HeLa cells was performed using siIMPORTER (Millipore, 64-101) per the manufacturer's instructions, and cells were processed for immunoprecipitation, immunoblotting, or immunofluorescence at 72 h post-transfection. For expression of recombinant proteins, expression vector(s) was/were transfected into HEK293 cells using calcium phosphate or HeLa cells using polyethylenimine, and the cells were processed at 72 h post-transfection. For experiments involving both RNAi and recombinant protein expression, HEK293 cells were transfected with siRNA followed by transfection of expression vector 24 h later. Cells were processed 72 h after siRNA transfection. Optimal knockdown and recombinant protein expression were achieved at 72 and 48–72 h, respectively, post-transfection.
Immunoprecipitation, immunoblotting, and immunofluorescence
Immunoprecipitation, immunoblotting, and immunofluorescence were performed as described (67, 68). Cells were lysed in a lysis buffer (50 mm HEPES, pH 7.4, 250 mm NaCl, 5 mm EDTA/pH 8, 0.1% NP-40, 1 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 2 μg of aprotinin, 10 mm NaF, 50 mm β-glycerophosphate, and 10% glycerol) at 4 °C for 30 min. The extracted proteins were recovered in the supernatant after centrifugation at 16,000 × g. For immunoblotting, 100 μg of extract was used as input. For immunoprecipitation, 2 mg of extract was incubated with anti-FLAG-(Sigma-Aldrich, A2220) or anti-HA–agarose (Sigma-Aldrich, A2095) beads at 4 °C for 2 h. The beads were washed three times with a lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotted with primary antibodies and horseradish peroxidase–conjugated secondary antibodies (Rockland Inc, 610-703-002 and 611-7302). For indirect immunofluorescence, cells were fixed with cold methanol and permeabilized with 1% Triton X-100/PBS. Slides were blocked with 3% BSA in 0.1% Triton X-100/PBS prior to incubation with primary antibodies. Secondary antibodies used were Cy3 (Jackson Immunoresearch Laboratories, 711-165-151 and 715-165-152)-, Alexa 647 (Jackson Immunoresearch Laboratories, 711-605-152)-, DyLight 649 (Jackson Immunoresearch Laboratories, 715-495-151)-, or Alexa 488 (Thermo Fisher Scientific, A11008, A11055, and A11001)–conjugated donkey anti-mouse, anti-goat, or anti-rabbit IgG. Cells might also be stained with DAPI (Molecular Probes, D3571), and slides were mounted, observed, and photographed using a Leitz DMRB (Leica) microscope (×100, NA 1.3) equipped with a Retiga EXi cooled camera.
In vivo ubiquitination assay
In vivo ubiquitination assays were performed as described (37, 69). Briefly, HEK293 cells were transfected with various plasmids including HA-Ub. Cells were lysed at 72 h post-transfection, and the desired protein was immunoprecipitated with 1% SDS (Bio Basic Inc., SB0485) to prevent noncovalently linked binding partners from co-immunoprecipitating with the desired protein. After extensive washing, the bound proteins were analyzed by SDS-PAGE and immunoblotting with an anti-HA antibody.
Microtubule regrowth assay
Cells were treated with 10 μm nocodazole (Sigma-Aldrich, M1404) for 1 h at 4 °C. After washing the cells several times with cold medium, they were placed in a prewarmed medium at 37 °C. Cells were fixed at various time points (0, 1, 5, and 45 min) after reaching 37 °C and processed for immunofluorescence.
Cell cycle analysis
HEK293T cells were co-transfected with plasmids expressing GFP (pQBI25) and WT or mutant HA-Vpr. At 48 h post-transfection, cells were fixed, permeabilized, and stained with propidium iodide as described (55). Cell cycle analysis was performed on the GFP+ population by flow cytometry (BD FACSCalibur, Becton Dickinson). The ModFit mathematical model (ModFit LT v4.1.7, Verity Software House) was used to enumerate the proportions of cells in G1 and G2/M phases.
Quantitation of γ-tubulin staining area and intensity
A region of interest (ROI) was drawn around γ-tubulin, which marks the centrosome, and the area of the ROI was calculated using Volocity 6 (PerkinElmer). The area of the ROI was then used to measure the fluorescence intensity of γ-tubulin, again using Volocity 6. Image conditions were identical in all cases, and none of the images were saturated, as confirmed by the pixel intensity range.
Quantitation of cytoplasmic microtubules
The number of microtubules emanated from the centrosome at 0 min after microtubule regrowth was subtracted from that at 1 min after regrowth and presented as microtubules nucleated/min.
Quantitation of Western blotting
Protein bands from Western blotting films were quantitated with ImageJ. Different film exposure lengths were used to prevent saturation. Quantitation was normalized with respect to the loading control.
Data and statistical analysis
Each experiment was conducted three times. The statistical significance of the difference between two means was calculated using a two-tailed Student's t test. Differences were considered significant at p < 0.01.
Author contributions
D. H. and J. A. F. B. data curation; D. H. software; D. H. and J. A. F. B. formal analysis; D. H., J. A. F. B., E. A. C., and W. Y. T. validation; D. H., E. A. C., and W. Y. T. investigation; D. H. visualization; D. H., J. A. F. B., E. A. C., and W. Y. T. methodology; D. H., J. A. F. B., E. A. C., and W. Y. T. writing-review and editing; E. A. C. and W. Y. T. conceptualization; E. A. C. and W. Y. T. supervision; E. A. C. and W. Y. T. funding acquisition; W. Y. T. resources; W. Y. T. writing-original draft; W. Y. T. project administration.
Acknowledgments
We thank all members of the Tsang laboratory for constructive advice, M. Bego and R. Lodge for helpful discussions, and J. Archambault for providing plasmids.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (to W. Y. T.) and a grant from the Canadian Institutes of Health Research (CIHR) (to E. A. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- Vpr
- viral protein R
- TERT
- telomerase reverse transcriptase
- HA
- hemagglutinin
- Ub
- ubiquitin
- DDR
- DNA damage response
- DAPI
- 4′,6-diamidino-2-phenylindole
- ROI
- region of interest.
References
- 1. Barré-Sinoussi, F., Chermann, J. C., Rey, F., Nugeyre, M. T., Chamaret, S., Gruest, J., Dauguet, C., Axler-Blin, C., Vézinet-Brun, F., Rouzioux, C., Rozenbaum, W., and Montagnier, L. (1983) Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871 10.1126/science.6189183 [DOI] [PubMed] [Google Scholar]
- 2. Gallo, R. C., Sarin, P. S., Gelmann, E. P., Robert-Guroff, M., Richardson, E., Kalyanaraman, V. S., Mann, D., Sidhu, G. D., Stahl, R. E., Zolla-Pazner, S., Leibowitch, J., and Popovic, M. (1983) Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science 220, 865–867 10.1126/science.6601823 [DOI] [PubMed] [Google Scholar]
- 3. Frankel, A. D., and Young, J. A. (1998) HIV-1: Fifteen proteins and an RNA. Annu. Rev. Biochem. 67, 1–25 10.1146/annurev.biochem.67.1.1 [DOI] [PubMed] [Google Scholar]
- 4. Coffin, J. M., Hughes, S. H., and Varmus, H. E. (1997) Retroviruses: The Interactions of Retroviruses and their Hosts. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 5. Sato, K., Misawa, N., Iwami, S., Satou, Y., Matsuoka, M., Ishizaka, Y., Ito, M., Aihara, K., An, D. S., and Koyanagi, Y. (2013) HIV-1 Vpr accelerates viral replication during acute infection by exploitation of proliferating CD4+ T cells in vivo. PLoS Pathog. 9, e1003812 10.1371/journal.ppat.1003812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krisko, J. F., Martinez-Torres, F., Foster, J. L., and Garcia, J. V. (2013) HIV restriction by APOBEC3 in humanized mice. PLoS Pathog. 9, e1003242 10.1371/journal.ppat.1003242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sato, K., Izumi, T., Misawa, N., Kobayashi, T., Yamashita, Y., Ohmichi, M., Ito, M., Takaori-Kondo, A., and Koyanagi, Y. (2010) Remarkable lethal G-to-A mutations in Vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J. Virol. 84, 9546–9556 10.1128/JVI.00823-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sato, K., Takeuchi, J. S., Misawa, N., Izumi, T., Kobayashi, T., Kimura, Y., Iwami, S., Takaori-Kondo, A., Hu, W. S., Aihara, K., Ito, M., An, D. S., Pathak, V. K., and Koyanagi, Y. (2014) APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 10, e1004453 10.1371/journal.ppat.1004453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dave, V. P., Hajjar, F., Dieng, M. M., Haddad, É., and Cohen, É. A. (2013) Efficient BST2 antagonism by Vpu is critical for early HIV-1 dissemination in humanized mice. Retrovirology 10, 128 10.1186/1742-4690-10-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato, K., Misawa, N., Fukuhara, M., Iwami, S., An, D. S., Ito, M., and Koyanagi, Y. (2012) Vpu augments the initial burst phase of HIV-1 propagation and downregulates BST2 and CD4 in humanized mice. J. Virol. 86, 5000–5013 10.1128/JVI.07062-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watkins, R. L., Zou, W., Denton, P. W., Krisko, J. F., Foster, J. L., and Garcia, J. V. (2013) In vivo analysis of highly conserved Nef activities in HIV-1 replication and pathogenesis. Retrovirology 10, 125 10.1186/1742-4690-10-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zou, W., Denton, P. W., Watkins, R. L., Krisko, J. F., Nochi, T., Foster, J. L., and Garcia, J. V. (2012) Nef functions in BLT mice to enhance HIV-1 replication and deplete CD4+CD8+ thymocytes. Retrovirology 9, 44 10.1186/1742-4690-9-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belzile, J. P., Abrahamyan, L. G., Gérard, F. C., Rougeau, N., and Cohen, E. A. (2010) Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 6, e1001080 10.1371/journal.ppat.1001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gérard, F. C., Yang, R., Romani, B., Poisson, A., Belzile, J. P., Rougeau, N., and Cohen, É. A. (2014) Defining the interactions and role of DCAF1/VPRBP in the DDB1-cullin4A E3 ubiquitin ligase complex engaged by HIV-1 Vpr to induce a G2 cell cycle arrest. PLoS ONE 9, e89195 10.1371/journal.pone.0089195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang, S., Feng, Y., Narayan, O., and Zhao, L. J. (2001) Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene 263, 131–140 10.1016/S0378-1119(00)00583-7 [DOI] [PubMed] [Google Scholar]
- 16. Wen, X., Casey Klockow, L., Nekorchuk, M., Sharifi, H. J., and de Noronha, C. M. (2012) The HIV1 protein Vpr acts to enhance constitutive DCAF1-dependent UNG2 turnover. PLoS ONE 7, e30939 10.1371/journal.pone.0030939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahn, J., Vu, T., Novince, Z., Guerrero-Santoro, J., Rapic-Otrin, V., and Gronenborn, A. M. (2010) HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J. Biol. Chem. 285, 37333–37341 10.1074/jbc.M110.133181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schröfelbauer, B., Yu, Q., Zeitlin, S. G., and Landau, N. R. (2005) Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 79, 10978–10987 10.1128/JVI.79.17.10978-10987.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang, X., Singh, S., Jung, H. Y., Yang, G., Jun, S., Sastry, K. J., and Park, J. I. (2013) HIV-1 Vpr protein inhibits telomerase activity via the EDD-DDB1-VPRBP E3 ligase complex. J. Biol. Chem. 288, 15474–15480 10.1074/jbc.M112.416735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laguette, N., Brégnard, C., Hue, P., Basbous, J., Yatim, A., Larroque, M., Kirchhoff, F., Constantinou, A., Sobhian, B., and Benkirane, M. (2014) Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156, 134–145 10.1016/j.cell.2013.12.011 [DOI] [PubMed] [Google Scholar]
- 21. Sawaya, B. E., Khalili, K., Mercer, W. E., Denisova, L., and Amini, S. (1998) Cooperative actions of HIV-1 Vpr and p53 modulate viral gene transcription. J. Biol. Chem. 273, 20052–20057 10.1074/jbc.273.32.20052 [DOI] [PubMed] [Google Scholar]
- 22. Forget, J., Yao, X. J., Mercier, J., and Cohen, E. A. (1998) Human immunodeficiency virus type 1 vpr protein transactivation function: Mechanism and identification of domains involved. J. Mol. Biol. 284, 915–923 10.1006/jmbi.1998.2206 [DOI] [PubMed] [Google Scholar]
- 23. Schröfelbauer, B., Hakata, Y., and Landau, N. R. (2007) HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. U.S.A. 104, 4130–4135 10.1073/pnas.0610167104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hrecka, K., Gierszewska, M., Srivastava, S., Kozaczkiewicz, L., Swanson, S. K., Florens, L., Washburn, M. P., and Skowronski, J. (2007) Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 11778–11783 10.1073/pnas.0702102104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao, L. J., Mukherjee, S., and Narayan, O. (1994) Biochemical mechanism of HIV-I Vpr function: Specific interaction with a cellular protein. J. Biol. Chem. 269, 15577–15582 [PubMed] [Google Scholar]
- 26. Kim, K., Kim, J. M., Kim, J. S., Choi, J., Lee, Y. S., Neamati, N., Song, J. S., Heo, K., and An, W. (2013) VprBP has intrinsic kinase activity targeting histone H2A and represses gene transcription. Mol. Cell 52, 459–467 10.1016/j.molcel.2013.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim, K., Heo, K., Choi, J., Jackson, S., Kim, H., Xiong, Y., and An, W. (2012) Vpr-binding protein antagonizes p53-mediated transcription via direct interaction with H3 tail. Mol. Cell. Biol. 32, 783–796 10.1128/MCB.06037-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakagawa, T., Mondal, K., and Swanson, P. C. (2013) VprBP (DCAF1): a promiscuous substrate recognition subunit that incorporates into both RING-family CRL4 and HECT-family EDD/UBR5 E3 ubiquitin ligases. BMC Mol. Biol. 14, 22 10.1186/1471-2199-14-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maddika, S., and Chen, J. (2009) Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11, 409–419 10.1038/ncb1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Angers, S., Li, T., Yi, X., MacCoss, M. J., Moon, R. T., and Zheng, N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 31. He, Y. J., McCall, C. M., Hu, J., Zeng, Y., and Xiong, Y. (2006) DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20, 2949–2954 10.1101/gad.1483206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higa, L. A., Wu, M., Ye, T., Kobayashi, R., Sun, H., and Zhang, H. (2006) CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283 10.1038/ncb1490 [DOI] [PubMed] [Google Scholar]
- 33. Lahouassa, H., Blondot, M. L., Chauveau, L., Chougui, G., Morel, M., Leduc, M., Guillonneau, F., Ramirez, B. C., Schwartz, O., and Margottin-Goguet, F. (2016) HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc. Natl. Acad. Sci. U.S.A. 113, 5311–5316 10.1073/pnas.1600485113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou, X., DeLucia, M., Hao, C., Hrecka, K., Monnie, C., Skowronski, J., and Ahn, J. (2017) HIV-1 Vpr protein directly loads helicase-like transcription factor (HLTF) onto the CRL4-DCAF1 E3 ubiquitin ligase. J. Biol. Chem. 292, 21117–21127 10.1074/jbc.M117.798801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hrecka, K., Hao, C., Shun, M. C., Kaur, S., Swanson, S. K., Florens, L., Washburn, M. P., and Skowronski, J. (2016) HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3921–3930 10.1073/pnas.1605023113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jung, H. Y., Wang, X., Jun, S., and Park, J. I. (2013) Dyrk2-associated EDD-DDB1-VprBP E3 ligase inhibits telomerase by TERT degradation. J. Biol. Chem. 288, 7252–7262 10.1074/jbc.M112.416792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hossain, D., Javadi Esfehani, Y., Das, A., and Tsang, W. Y. (2017) Cep78 controls centrosome homeostasis by inhibiting EDD-DYRK2-DDB1VprBP. EMBO Rep. 18, 632–644 10.15252/embr.201642377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spektor, A., Tsang, W. Y., Khoo, D., and Dynlacht, B. D. (2007) Cep97 and CP110 suppress a cilia assembly program. Cell 130, 678–690 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- 39. Schmidt, T. I., Kleylein-Sohn, J., Westendorf, J., Le Clech, M., Lavoie, S. B., Stierhof, Y. D., and Nigg, E. A. (2009) Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- 40. Kohlmaier, G., Loncarek, J., Meng, X., McEwen, B. F., Mogensen, M. M., Spektor, A., Dynlacht, B. D., Khodjakov, A., and Gönczy, P. (2009) Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19, 1012–1018 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang, C. J., Fu, R. H., Wu, K. S., Hsu, W. B., and Tang, T. K. (2009) CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11, 825–831 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- 42. Afonso, P. V., Zamborlini, A., Saïb, A., and Mahieux, R. (2007) Centrosome and retroviruses: The dangerous liaisons. Retrovirology 4, 27 10.1186/1742-4690-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McDonald, D., Vodicka, M. A., Lucero, G., Svitkina, T. M., Borisy, G. G., Emerman, M., and Hope, T. J. (2002) Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159, 441–452 10.1083/jcb.200203150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fackler, O. T., and Kräusslich, H. G. (2006) Interactions of human retroviruses with the host cell cytoskeleton. Curr. Opin. Microbiol. 9, 409–415 10.1016/j.mib.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 45. Sabo, Y., Walsh, D., Barry, D. S., Tinaztepe, S., de Los Santos, K., Goff, S. P., Gundersen, G. G., and Naghavi, M. H. (2013) HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14, 535–546 10.1016/j.chom.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lukic, Z., Dharan, A., Fricke, T., Diaz-Griffero, F., and Campbell, E. M. (2014) HIV-1 uncoating is facilitated by dynein and kinesin 1. J. Virol. 88, 13613–13625 10.1128/JVI.02219-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zamborlini, A., Lehmann-Che, J., Clave, E., Giron, M. L., Tobaly-Tapiero, J., Roingeard, P., Emiliani, S., Toubert, A., de Thé, H., and Saïb, A. (2007) Centrosomal pre-integration latency of HIV-1 in quiescent cells. Retrovirology 4, 63 10.1186/1742-4690-4-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bolton, D. L., Barnitz, R. A., Sakai, K., and Lenardo, M. J. (2008) 14-3-3 theta binding to cell cycle regulatory factors is enhanced by HIV-1 Vpr. Biol. Direct. 3, 17 10.1186/1745-6150-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chang, F., Re, F., Sebastian, S., Sazer, S., and Luban, J. (2004) HIV-1 Vpr induces defects in mitosis, cytokinesis, nuclear structure, and centrosomes. Mol. Biol. Cell 15, 1793–1801 10.1091/mbc.e03-09-0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Watanabe, N., Yamaguchi, T., Akimoto, Y., Rattner, J. B., Hirano, H., and Nakauchi, H. (2000) Induction of M-phase arrest and apoptosis after HIV-1 Vpr expression through uncoupling of nuclear and centrosomal cycle in HeLa cells. Exp. Cell Res. 258, 261–269 10.1006/excr.2000.4908 [DOI] [PubMed] [Google Scholar]
- 51. Le Rouzic, E., Belaïdouni, N., Estrabaud, E., Morel, M., Rain, J. C., Transy, C., and Margottin-Goguet, F. (2007) HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 10.4161/cc.6.2.3732 [DOI] [PubMed] [Google Scholar]
- 52. D'Angiolella, V., Donato, V., Vijayakumar, S., Saraf, A., Florens, L., Washburn, M. P., Dynlacht, B., and Pagano, M. (2010) SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 466, 138–142 10.1038/nature09140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fregoso, O. I., and Emerman, M. (2016) Activation of the DNA damage response is a conserved function of HIV-1 and HIV-2 Vpr that is independent of SLX4 recruitment. MBio 7, e01433–16 10.1128/mBio.01433-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gaynor, E. M., and Chen, I. S. (2001) Analysis of apoptosis induced by HIV-1 Vpr and examination of the possible role of the hHR23A protein. Exp. Cell Res. 267, 243–257 10.1006/excr.2001.5247 [DOI] [PubMed] [Google Scholar]
- 55. Belzile, J. P., Duisit, G., Rougeau, N., Mercier, J., Finzi, A., and Cohen, E. A. (2007) HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3, e85 10.1371/journal.ppat.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Minemoto, Y., Shimura, M., Ishizaka, Y., Masamune, Y., and Yamashita, K. (1999) Multiple centrosome formation induced by the expression of Vpr gene of human immunodeficiency virus. Biochem. Biophys. Res. Commun. 258, 379–384 10.1006/bbrc.1999.0640 [DOI] [PubMed] [Google Scholar]
- 57. Romani, B., and Cohen, E. A. (2012) Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr. Opin. Virol. 2, 755–763 10.1016/j.coviro.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brégnard, C., Benkirane, M., and Laguette, N. (2014) DNA damage repair machinery and HIV escape from innate immune sensing. Front. Microbiol. 5, 176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bourke, E., Dodson, H., Merdes, A., Cuffe, L., Zachos, G., Walker, M., Gillespie, D., and Morrison, C. G. (2007) DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 8, 603–609 10.1038/sj.embor.7400962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mullee, L. I., and Morrison, C. G. (2016) Centrosomes in the DNA damage response: The hub outside the centre. Chromosome Res. 24, 35–51 10.1007/s10577-015-9503-7 [DOI] [PubMed] [Google Scholar]
- 61. Dumas, A., Lê-Bury, G., Marie-Anaïs, F., Herit, F., Mazzolini, J., Guilbert, T., Bourdoncle, P., Russell, D. G., Benichou, S., Zahraoui, A., and Niedergang, F. (2015) The HIV-1 protein Vpr impairs phagosome maturation by controlling microtubule-dependent trafficking. J. Cell Biol. 211, 359–372 10.1083/jcb.201503124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen, D., Wang, M., Zhou, S., and Zhou, Q. (2002) HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 21, 6801–6810 10.1093/emboj/cdf683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Mareuil, J., Carre, M., Barbier, P., Campbell, G. R., Lancelot, S., Opi, S., Esquieu, D., Watkins, J. D., Prevot, C., Braguer, D., Peyrot, V., and Loret, E. P. (2005) HIV-1 Tat protein enhances microtubule polymerization. Retrovirology 2, 5 10.1186/1742-4690-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aprea, S., Del Valle, L., Mameli, G., Sawaya, B. E., Khalili, K., and Peruzzi, F. (2006) Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J. Neurosci. 26, 4054–4062 10.1523/JNEUROSCI.0603-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Treier, M., Staszewski, L. M., and Bohmann, D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78, 787–798 10.1016/S0092-8674(94)90502-9 [DOI] [PubMed] [Google Scholar]
- 66. Tsang, W. Y., Bossard, C., Khanna, H., Peränen, J., Swaroop, A., Malhotra, V., and Dynlacht, B. D. (2008) CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell 15, 187–197 10.1016/j.devcel.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barbelanne, M., Song, J., Ahmadzai, M., and Tsang, W. Y. (2013) Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum. Mol. Genet. 22, 2482–2494 10.1093/hmg/ddt100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barbelanne, M., Hossain, D., Chan, D. P., Peränen, J., and Tsang, W. Y. (2015) Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum. Mol. Genet. 24, 2185–2200 10.1093/hmg/ddu738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Das, A., Qian, J., and Tsang, W. Y. (2017) USP9X counteracts differential ubiquitination of NPHP5 by MARCH7 and BBS11 to regulate ciliogenesis. PLoS Genet. 13, e1006791 10.1371/journal.pgen.1006791 [DOI] [PMC free article] [PubMed] [Google Scholar]