Abstract
Methionine (Met) is an amino acid essential for many important cellular and biosynthetic functions, including the initiation of protein synthesis and S-adenosylmethionine–mediated methylation of proteins, RNA, and DNA. The de novo biosynthetic pathway of Met is well conserved across prokaryotes but absent from vertebrates, making it a plausible antimicrobial target. Using a systematic approach, we examined the essentiality of de novo methionine biosynthesis in Salmonella enterica serovar Typhimurium, a bacterial pathogen causing significant gastrointestinal and systemic diseases in humans and agricultural animals. Our data demonstrate that Met biosynthesis is essential for S. Typhimurium to grow in synthetic medium and within cultured epithelial cells where Met is depleted in the environment. During systemic infection of mice, the virulence of S. Typhimurium was not affected when either de novo Met biosynthesis or high-affinity Met transport was disrupted alone, but combined disruption in both led to severe in vivo growth attenuation, demonstrating a functional redundancy between de novo biosynthesis and acquisition as a mechanism of sourcing Met to support growth and virulence for S. Typhimurium during infection. In addition, our LC-MS analysis revealed global changes in the metabolome of S. Typhimurium mutants lacking Met biosynthesis and also uncovered unexpected interactions between Met and peptidoglycan biosynthesis. Together, this study highlights the complexity of the interactions between a single amino acid, Met, and other bacterial processes leading to virulence in the host and indicates that disrupting the de novo biosynthetic pathway alone is likely to be ineffective as an antimicrobial therapy against S. Typhimurium.
Keywords: methionine, S-adenosylmethionine (SAM), biosynthesis, transporter, Salmonella enterica, peptidoglycan, in vivo infection, virulence
Introduction
Methionine (Met) is a sulfur-containing proteinogenic amino acid that is required for the initiation of protein synthesis (1). S-adenosylmethionine (SAM),7 a downstream derivative of Met, acts as a major methyl donor in the cell and methylates a variety of macromolecules, such as DNA, RNA, protein, and lipids (2). A de novo pathway for Met biosynthesis is present in the vast majority of prokaryotes, albeit with variations in the enzymes that drive the biosynthetic cascade (3, 4). In contrast, the full de novo Met biosynthesis is absent from vertebrates, which must obtain this amino acid through external sources, such as diet and gut flora (5, 6). In recent years, it has been increasingly recognized that central metabolism represents a promising yet underexploited area for the development of antimicrobial drugs (7). The apparent essentiality in microbes and absence in mammals makes the Met biosynthetic pathway an especially attractive target for antimicrobial therapy.
Salmonella enterica is a Gram-negative, facultative intracellular bacterium that causes gastrointestinal and systemic diseases in animals and humans. There are ∼2500 serovars in this species, which include the human-adapted enteric fever pathogens S. enterica var. Typhi and Paratyphi, alongside non-typhoidal Salmonella (NTS) serovars that cause debilitating gastroenteritis, including S. enterica var. Typhimurium (8, 9). For all Salmonella serovars, the capacity to grow in the host is central to bacterial virulence (10). In mammalian hosts, S. enterica grows in the blood and reticuloendothelial system largely within a defined, membrane-bound endocytic compartment called the Salmonella-containing vacuole (SCV) (11, 12). The extent to which S. enterica salvages essential nutrients from the lumen of the SCV and the nutrient composition of this vacuole remains poorly defined, and the micronutrient environment of the SCV has not been fully resolved. Ex vivo, S. enterica is capable of growth in minimal medium containing glucose as a carbon source and key ions, and the metabolic potential of this species has been recognized and mapped (13). It is less clear, however, how this considerable capability is exploited for maximal growth in vivo. The essentiality of the pathways can only be determined by systematic analysis in vitro and in animal models, and fine mapping is required to determine whether essential pathways contain novel targets for antibiotic development.
The de novo biosynthetic pathway for Met has been previously reported in S. enterica (Fig. 1). This pathway is regulated by the transcription factors MetJ and MetR (14). With its co-repressor SAM, MetJ represses the transcription of all met genes except metH (15). In contrast, MetR is an autoregulated transcriptional activator that controls the expression of metA, metF, metE, and metH (16). In addition to de novo biosynthesis, S. enterica is able to acquire Met from extracellular sources through a high-affinity transporter that is encoded by the metD locus, encoding an ATP-binding cassette (ABC) transporter composed of three subunits: the ATPase (MetN), a transmembrane permease (MetI), and a periplasmic Met-binding protein (MetQ). Biochemical analysis has shown that MetNIQ transports both the d- and l-enantiomers of Met (17, 18). In the absence of the high-affinity MetNIQ transporter system, genetic analysis has suggested that S. enterica is able to transport Met at much lower affinity, through a putative and cryptic low-affinity Met transporter system termed “MetP” (19–21). In contrast to MetNIQ, MetP transports l-Met but not d-Met, and the coding gene(s) for MetP remains unidentified to date.
Figure 1.
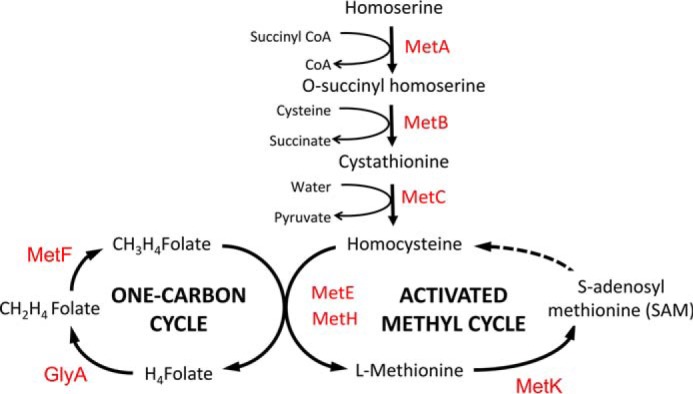
Schematic diagram of the biosynthetic pathway of Met in S. Typhimurium. The first committed step of the pathway is catalyzed by homoserine transsuccinylase (MetA), which succinylates homoserine to form O-succinyl-l-homoserine, which then undergoes a condensation reaction with cysteine to form l-cystathionine, catalyzed by cystathionine γ-synthase (MetB). Cystathionine β-lyase (MetC) catalyzes the conversion of l-cystathionine to l-homocysteine, pyruvate, and ammonia. The final biosynthetic step is catalyzed by distinct Met synthases, MetH and MetE, which are vitamin B12-dependent and -independent synthases, respectively (31, 32, 64). In this final step, l-homocysteine is condensed with 5,10-methyltetrahydrofolate (CH3H4Folate), which donates the methyl group. 5,10-Methyltetrahydrofolate is provided from the one-carbon cycle through a methylenetetrahydrofolate reductase (MetF)-mediated reaction, producing tetrahydrofolate (H4Folate) and Met (65). In S. enterica, Met is recycled through an activated methyl cycle. The primary methyl donor, SAM, is formed by the activation of Met through an ATP-dependent condensation reaction catalyzed by the SAM synthase (MetK)(2, 66, 67). Catalytic enzymes are shown in red; arrows indicate the direction of catalytic reactions.
S. enterica may encounter markedly different nutrient levels during local (e.g. the gut) and systemic (e.g. the spleen and liver) infections, which, in turn, may vary the bacterium's dependence on de novo biosynthesis and nutrient import pathways for growth in different tissue niches. The enzymes required for Met biosynthesis and the Met transporter have been implicated in the virulence of S. enterica and other bacteria (22–28). However, these pathways have not been systematically investigated for their role in the virulence of S. enterica. The aim of this study was to investigate the essentiality of de novo biosynthesis and transport of Met in the growth and virulence of S. Typhimurium in vitro and in vivo.
Results
In vitro validation of the Met biosynthetic pathway in S. Typhimurium and growth of defined Met auxotrophs in mice
Previous transcriptional analyses of S. Typhimurium grown in medium and in tissue culture cells have demonstrated that the genes involved in the de novo Met synthesis pathway (metA, metB, metC, metE, metF, and metH) are expressed under a variety of growth conditions (Fig. S1; deduced from Ref. 29). Whereas the expression level of the de novo pathway is relatively low in rich medium, much stronger expression was observed under growth conditions where the Salmonella pathogenicity island-2 (SPI-2) genes were expressed or when the bacteria were inside macrophages (30).
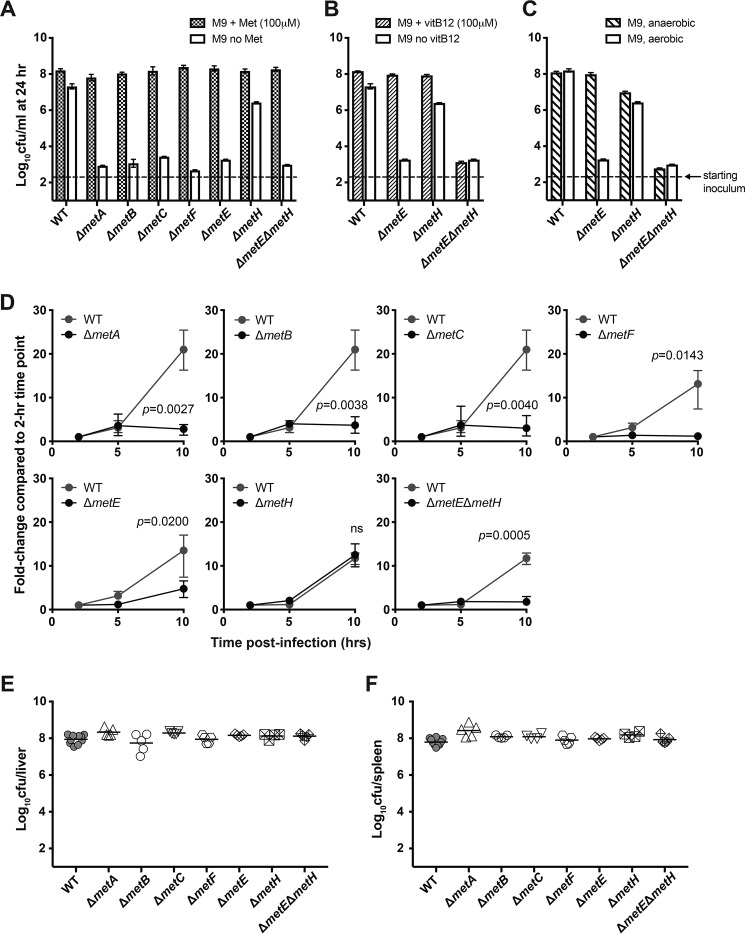
Defined mutants in the Met biosynthetic pathway (ΔmetA, ΔmetB, ΔmetC, ΔmetE, ΔmetF, ΔmetH, and ΔmetEΔmetH) were generated in S. Typhimurium SL1344, sequenced, and tested for their ability to grow in M9 minimal medium, with or without added l-Met, and in nutrient-rich Luria–Bertani (LB) medium. All mutants grew in LB medium at a rate comparable with WT SL1344 (data not shown). In M9 minimal medium supplemented with Met, Met mutants grew as efficiently as SL1344 over a 24-h period (Fig. 2A), suggesting that the absence of the de novo biosynthetic pathway was compensated by the transport of exogenously available Met. As expected, the ΔmetA, ΔmetB, ΔmetC, ΔmetE, and ΔmetF mutants exhibited Met auxotrophy in M9 medium, whereas ΔmetH showed substantial growth over the 24-h period (Fig. 2A), confirming that MetH is not essential for de novo biosynthesis of Met under aerobic conditions (31, 32). The growth of the ΔmetH mutant in M9 medium was apparently supported by a functional Met synthase (i.e. MetE), as ΔmetEΔmetH failed to grow in M9 medium in the absence of exogenous Met (Fig. 2A). It has been suggested that the functionality of MetH in E. coli is vitamin B12–dependent (33). To test whether there is an equivalent dependence in S. Typhimurium, ΔmetE, ΔmetH, and ΔmetEΔmetH mutants were grown in M9 medium with or without vitamin B12. All three mutants grew in the presence of vitamin B12 (Fig. 2B). The ΔmetE mutant, which depended on MetH to synthesize Met, grew only when vitamin B12 was added (Fig. 2B), confirming that MetH activity in S. Typhimurium is vitamin B12–dependent. As S. Typhimurium synthesizes vitamin B12 only under anaerobic conditions (31), growth of the ΔmetE mutant in M9 medium should have been restored when oxygen was depleted. Indeed, ΔmetE, ΔmetH, and ΔmetEΔmetH both grew in M9 medium under anaerobic conditions and phenocopied their growth in M9 medium with vitamin B12 under aerobic conditions (Fig. 2C). These observations are consistent with an understanding that S. Typhimurium synthesizes vitamin B12 under anaerobic conditions and support a level of functional redundancy between MetE and MetH during de novo biosynthesis of Met in S. Typhimurium.
Figure 2.
The de novo biosynthetic mutants demonstrate Met-dependent growth in M9 minimal medium and in HeLa cells but remain fully virulent in mice. S. Typhimurium WT SL1344 and mutant strains were grown shaking at 37 °C in M9 minimal medium with or without Met (A), vitamin B12 (B), or oxygen (C), and the number of viable bacteria (cfu) was determined after 24 h. Bars, mean cfu; error bars, data range. Dotted lines represent the bacterial concentration at the time of inoculation. Data are pooled from two independent experiments. D, the de novo Met biosynthetic mutants become defective for intracellular replication in HeLa cells when Met is absent in the DMEM. HeLa cells were grown to a monolayer and infected with S. Typhimurium WT or mutant strains at an MOI of 5–10, in Met-free DMEM. The intracellular bacterial load at 2 h post-infection is expressed as “1” and used as the reference point to calculate -fold change of intracellular bacterial number at subsequent time points. Data are pooled from three independent experiments. Bars, mean cfu; error bars, data range. Unpaired t tests were used to compare the intracellular load of WT and mutant strains at 10 h post-infection, and p values shown were corrected for multiple comparisons using the Bonferroni–Dunn method; p values greater than 0.05 were deemed not significant (ns). For assessing virulence in vivo, age- and sex-matched C57BL/6 mice were intravenously infected with 200 cfu of the indicated strains of S. Typhimurium, and the bacterial load in the liver (E) and spleen (F) was determined at day 5 post-infection. Symbols represent data from individual animals, and horizontal lines represent the geometric mean of each group. Data are pooled from two independent experiments. One-way ANOVA with Bonferroni post-tests was used for statistical analyses comparing each pair of data groups, and none of the comparisons yielded a p value < 0.05.
To assess the requirement for de novo Met synthesis for intracellular bacterial growth, HeLa cells were infected with S. Typhimurium WT and mutants, and the intracellular bacterial load was assessed after 2, 5, and 10 h. When HeLa cells were cultivated in DMEM growth medium lacking Met, the intracellular growth of ΔmetA, ΔmetB, ΔmetC, and ΔmetF was significantly attenuated compared with the WT control (Fig. 2D). The ΔmetE mutant only showed obvious growth after 5 h. In contrast, the ΔmetH mutant showed intracellular growth comparable with SL1344, suggesting that a functional MetE is sufficient for supporting intracellular replication in the SCV (Fig. 2D). Importantly, when HeLa cells were cultivated in DMEM containing physiological levels of l-Met, none of the mutants showed any significant defect in intracellular growth (Fig. S2), demonstrating that exogenous Met can rescue Met auxotrophy, presumably reflecting direct or indirect transport of Met from the culture medium to the lumen of the bacteria-occupied SCV.
Previous reports have proposed that Met biosynthesis might be required for virulence of S. Typhimurium in mice or chickens (24, 34). To test this requirement in mice, the virulence of individual de novo biosynthetic mutants was studied in C57BL/6 mice, in which WT SL1344 is fully virulent and invariably results in a systemic, lethal infection (35, 36). Groups of mice were infected with 200 cfu of the different Met auxotrophic mutants intravenously, and, at day 5 post-infection, mice were culled, and the bacterial load in the liver (Fig. 2E) and spleen (Fig. 2F) was determined by viable count. The number of bacteria recovered from mice infected with the mutants was comparable with those from mice infected with SL1344. Hence, none of the mutants showed virulence defects in mice following intravenous infection. Another group of mice were infected orally with ΔmetB, ΔmetE, ΔmetH, or ΔmetEΔmetH. The bacterial load in the spleen and liver of mice infected with the mutant strains was comparable with those from mice infected with the WT S. Typhimurium (Fig. S3). These data showed that de novo Met biosynthesis is not essential for the growth of S. Typhimurium in C57BL/6 mice, regardless of the route of infection.
Met auxotrophs deficient in the high-affinity transporter (MetNIQ) are attenuated in mice
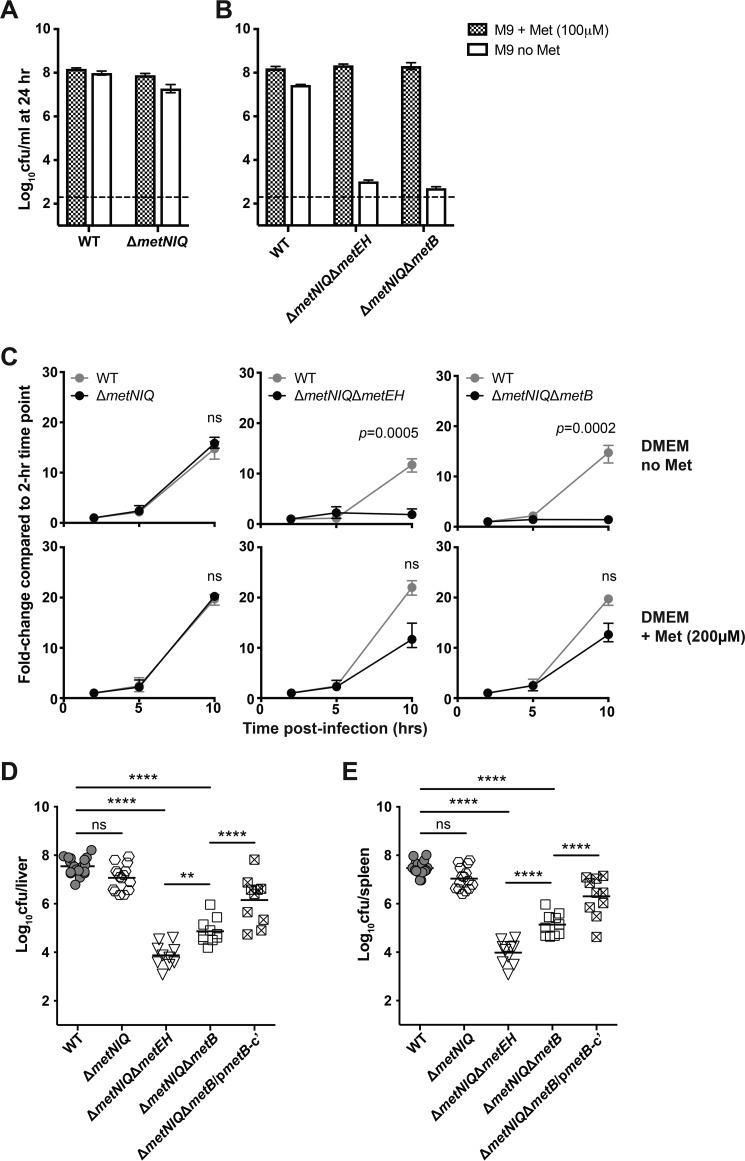
Our observation that S. Typhimurium mutants were able to grow intracellularly and in vivo without a functional de novo Met biosynthetic pathway suggested that Met acquisition via the transporters may play an important role in bacterial virulence. To test this hypothesis, a high-affinity transporter mutant of S. Typhimurium, ΔmetNIQ, was constructed. As expected, the ΔmetNIQ mutant grew in M9 minimal medium in the absence of Met supplementation (Fig. 3A), suggesting that the high-affinity Met transporter is not essential for growth in M9 medium provided that de novo synthesis is intact.
Figure 3.
S. Typhimurium mutants deficient in the de novo biosynthesis and high-affinity transport of Met are highly attenuated in mice. A and B, S. Typhimurium strains were grown shaking at 37 °C in M9 minimal medium with or without Met, and the number of viable bacteria (cfu) was determined after 24 h. Bars, mean cfu; error bars, data range. Dotted lines, bacterial concentration at the time of inoculation. Data are pooled from two independent experiments. C, HeLa cells were grown to a monolayer and infected with S. Typhimurium WT or mutant strains at an MOI of 5–10, in DMEM with or without Met. The intracellular bacterial load at 2 h post-infection is expressed as “1” and used as the reference point to calculate -fold change of intracellular bacterial number at subsequent time points. Bars, mean cfu; error bars, data range. Data are pooled from three independent experiments. Unpaired t tests were used to compare the intracellular load of WT and mutant strains at 10 h post-infection, and p values shown were corrected for multiple comparisons using the Bonferroni–Dunn method; p values > 0.05 were deemed not significant (ns). For assessing virulence in vivo, age- and sex-matched C57BL/6 mice were intravenously infected with 200 cfu of the indicated strains of S. Typhimurium, and the bacterial load in the liver (D) and spleen (E) was determined at day 5 post-infection. ΔmetNIQΔmetB/pmetB-c′ denotes the complemented strain ΔmetNIQΔmetB pACYC184metB. Symbols represent data from individual animals, and horizontal lines represent the geometric mean of each group. Data are pooled from three independent experiments. One-way ANOVA with Bonferroni post-tests was used for statistical analyses. ****, p < 0.0001; ns, p > 0.05.
To determine whether dual mutations in de novo biosynthesis and the high-affinity transporter of Met affected the growth of S. Typhimurium in vitro and in vivo, ΔmetNIQΔmetEH and ΔmetNIQΔmetB mutants were generated. As expected, the ΔmetNIQΔmetEH and ΔmetNIQΔmetB mutants were unable to grow in M9 minimal medium without added Met (Fig. 3B); this Met auxotrophy confirms that a combined deficiency in both biosynthesis and high-affinity transport is inhibitive for growth. Interestingly, the addition of l-Met into M9 medium restored the growth of all of these mutant strains, consistent with the presence and activity of the putative and functional low-affinity transporter MetP (19, 20). Hence, these data support the theory that MetP activity enables sufficient Met uptake to facilitate efficient growth in vitro.
To determine whether the observed in vitro growth attenuation was reflected by reduced growth of S. Typhimurium inside mammalian cells, ΔmetNIQ, ΔmetNIQΔmetB, and ΔmetNIQΔmetEH mutants were used to infect HeLa cells in Met-free DMEM. Whereas ΔmetNIQ mutant was capable of intracellular growth to a level similar to SL1344 over a 10-h period, ΔmetNIQΔmetB and ΔmetNIQΔmetEH mutants were unable to grow (Fig. 3C, top row). The presence of l-Met in DMEM fully restored the growth of the mutant strains in HeLa cells (Fig. 3C, bottom row), suggesting that the activity of the cryptic transporter MetP was sufficient to support intracellular growth in HeLa cells.
To determine whether dual mutations in de novo biosynthesis and the high-affinity transporter of Met reduced S. Typhimurium virulence in vivo, C57BL/6 mice were intravenously infected with ΔmetNIQ, ΔmetNIQΔmetB, and ΔmetNIQΔmetEH mutant strains or WT. At day 5 post-infection, mice infected with ΔmetNIQ had a similarly high bacterial load in the liver (Fig. 3D) and spleen (Fig. 3E), comparable with that of SL1344-infected mice, indicating that loss of the high-affinity Met transporter alone does not reduce S. Typhimurium virulence in vivo. In contrast, infection with ΔmetNIQΔmetB and ΔmetNIQΔmetEH mutants resulted in a significantly reduced bacterial load in the liver (Fig. 3D) and spleen (Fig. 3E), and this attenuation was reversed when the ΔmetNIQΔmetB mutant was complemented by metB (Fig. 3, D and E). Similar results were obtained when the ΔmetNIQΔmetB and ΔmetNIQΔmetEH mutants were used to infect mice via the oral route (Fig. S4), confirming that ΔmetNIQΔmetB and ΔmetNIQΔmetEH are attenuated in mice regardless of the route of infection. Taken together, these results demonstrate that restriction of Met availability has a strong impact on the virulence of S. Typhimurium in vivo, and the presence of the putative cryptic transporter (MetP) alone is insufficient to sustain maximal bacterial growth in the permissive murine host.
Metabolite profiling in S. Typhimurium Met biosynthetic and high-affinity transporter mutants
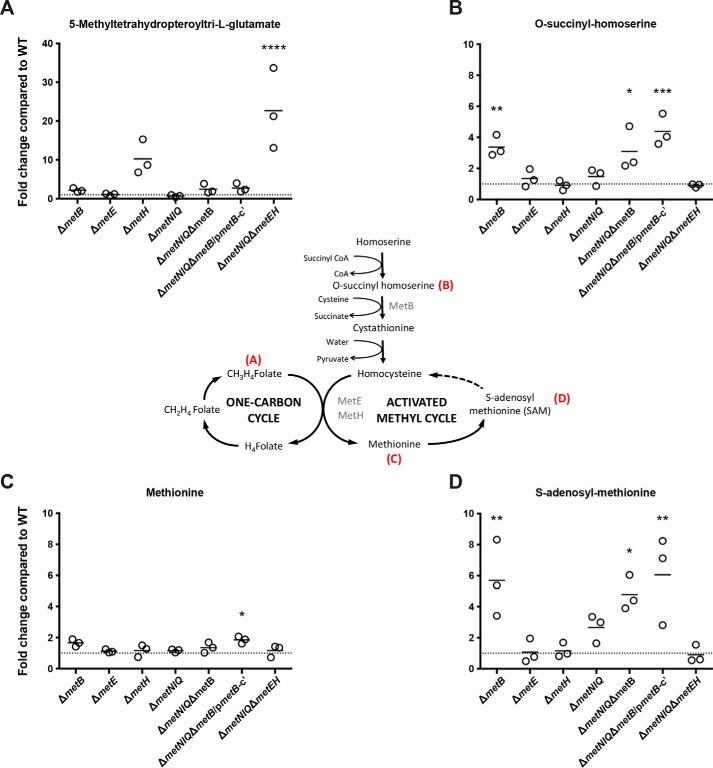
To further define the impact of Met pathway gene disruptions on bacterial metabolism, S. Typhimurium WT and mutants lacking either or both of the de novo biosynthesis pathway and the high-affinity Met transporter were grown in LB medium, and polar metabolites were extracted and analyzed by LC-MS. The intracellular accumulation of 5-methyl-tetrahydropteroyltri-l-glutamate, a member of the polyglutamate forms of 5-methyltetrahydrofolate (37), was significantly increased in ΔmetH (∼10-fold) and further increased in ΔmetNIQΔmetEH (∼25-fold), but not in ΔmetE, compared with SL1344 (Fig. 4). This result suggests that both MetE and MetH are functionally active, as S. Typhimurium grew in LB broth, but MetH is either more abundant or more efficient in the formation of 5-methyltetrahydrofolate. On the other hand, the ΔmetB mutant exhibited a significantly elevated pool of O-succinylhomoserine compared with the WT, consistent with a defect in the MetB-catalyzed conversion of O-succinylhomoserine to cystathionine (Fig. 4).
Figure 4.
Metabolite perturbations following genetic disruption to Met biosynthesis, one-carbon cycle, and activated methyl cycle. Total metabolite pools in each strain were detected and quantitated using liquid chromatography-quadrupole TOF. Key intermediates of the Met biosynthetic pathway, activated methyl cycle, and one-carbon cycle were identified and quantified for the indicated S. Typhimurium strains and compared with the WT level; -fold changes are shown for 5-methyltetrahydropteroyltri-l-glutamate, a member of the polyglutamate forms of 5-methyltetrahydrofolate (A), O-succinylhomoserine (B), Met (C), and SAM (D). ΔmetNIQΔmetB/pmetB-c′ denotes the complemented strain ΔmetNIQΔmetB pACYC184metB. Individual data points represent biological replicates, and the mean for each group is shown. One-way ANOVAs with Bonferroni post-tests were used to compare the metabolite level in each mutant with WT; p values < 0.05 are shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
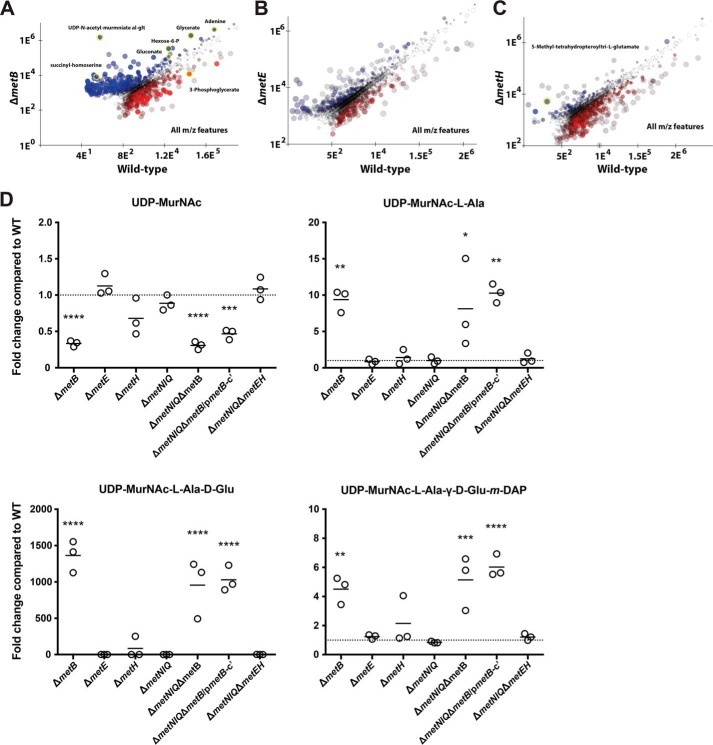
An untargeted mass/charge (m/z) feature analysis of the mutant lines led to the identification of a number of unexpected metabolite changes. Pairwise comparisons were performed to identify differences that were statistically significant (p < 0.05) and to filter interesting m/z features. Deletion of metB led to significant increases in the intracellular levels of SAM (5.5-fold increase compared with WT), and a number of other metabolites, including glycerate, adenine, hexose 6-phosphate, 3-phosphoglycerate, and gluconate, were also increased compared with the WT (Fig. 5A and Table 1). By comparison, ΔmetE did not show any significantly different m/z features (Fig. 5B), whereas only the m/z feature corresponding to 5-methyl-tetrahydropteroyltri-l-glutamate, a representative form of 5-methyltetrahydrofolate, was significantly different in the ΔmetH mutant (Fig. 5C).
Figure 5.
Metabolite perturbations following genetic disruption to metB, metE, and metH. Scatter plots depict WT compared with ΔmetB (A), ΔmetE (B), and ΔmetH (C). Each dot represents a single extracted m/z feature with each axis depicting the arbitrary ion count. m/z features that are no different between conditions plot along the diagonal. The color intensity indicates the confidence of the difference (greater intensity equates to lower variance across biological replicates). Yellow highlight, a subset of confirmed metabolites with a statistically significant difference in a pairwise comparison (p < 0.05 with Benjamini correction). D, the levels of four metabolites associated with peptidoglycan synthesis were significantly altered in ΔmetB and ΔmetNIQΔmetB mutants. UDP-MurNAc, UDP-N-acetylmuraminate; UDP-MurNAc-l-Ala, UDP-N-acetylmuramoyl-l-alanine; UDP-MurNAc-l-Ala-d-Glu, UDP-N-acetylmuramoyl-l-alanyl-d-glutamate; UDP-MurNAc-l-Ala-γ-d-Glu-m-DAP, UDP-N-acetylmuramoyl-l-alanyl-d-glutamyl-meso-diaminopimelate. ΔmetNIQΔmetB/pmetB-c′, the complemented strain ΔmetNIQΔmetB pACYC184metB. Individual data points represent biological replicates, and the mean for each group is shown. One-way ANOVAs with Bonferroni post-tests were used to compare the metabolite level in each mutant with WT; p values <0.05 are shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Table 1.
List of metabolites with significant changes in ΔmetB mutant compared with WT
This table presents results from a one-way ANOVA performed with Tukey's honestly significant difference tests (p < 0.01). FDR, false discovery rate.
| XCMS ID | Metabolite name | m/z | -Fold change (ΔmetB to WT) | p value | FDR |
|---|---|---|---|---|---|
| M900T1200 | 4-Hydroxyphenylacetyl-CoA | 900.15 | 3247.21 | 4.17E − 07 | 4.01E − 05 |
| M878T1201 | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate | 878.17 | 1646.67 | 7.63E − 08 | 1.45E − 05 |
| M880T1200 | 3-Hydroxyhexanoyl CoA | 880.18 | 220.30 | 6.08E − 07 | 5.15E − 05 |
| M749T1087 | UDP-N-acetylmuramoyl-l-alanine | 749.13 | 12.24 | 7.22E − 06 | 2.49E − 04 |
| M163T480 | Rhamnose | 163.06 | 10.30 | 3.99E − 08 | 1.33E − 05 |
| M275T1210 | 6-Phosphogluconate | 275.02 | 8.93 | 1.01E − 04 | 1.74E − 03 |
| M105T825 | d-Glycerate | 105.02 | 8.15 | 8.18E − 06 | 2.67E − 04 |
| M259T1165 | Hexose-phosphate | 259.02 | 7.36 | 7.32E − 04 | 8.43E − 03 |
| M266T579 | Adenosine | 266.09 | 6.13 | 1.44E − 04 | 2.33E − 03 |
| M195T999 | Gluconate | 195.05 | 6.13 | 7.04E − 07 | 5.68E − 05 |
| M1050T1225 | UDP-N-acetylmuramoyl-l-alanyl-d-γ-glutamyl-meso-2,6- diaminopimelate | 1050.26 | 5.63 | 1.75E − 06 | 9.97E − 05 |
| M189T1246 | Diaminoheptandioate | 189.09 | 4.73 | 6.05E − 07 | 5.15E − 05 |
| M620T1224 | UDP-N-acetyl-2-amino-2-deoxy-d-glucuronate | 620.05 | 4.67 | 9.09E − 09 | 7.57E − 06 |
| M134T607 | Adenine | 134.05 | 4.44 | 6.48E − 06 | 2.31E − 04 |
| M686T707 | Dephospho-CoA | 686.14 | 4.22 | 1.36E − 05 | 3.75E − 04 |
| M218T1006 | O-Succinyl-l-homoserine | 218.07 | 3.75 | 5.85E − 05 | 1.14E − 03 |
| M145T567 | 2-Dehydropantoate | 145.05 | 3.72 | 2.60E − 05 | 6.15E − 04 |
| M130T567 | Hydoxyproline | 130.05 | 3.19 | 2.46E − 04 | 3.57E − 03 |
| M273T1080 | Succinyl-arginine | 273.12 | 2.55 | 1.28E − 04 | 2.11E − 03 |
| M678T1139 | UDP-N-acetylmuraminate | 678.09 | 2.45 | 1.41E − 07 | 2.10E − 05 |
| M227T987 | 4-Phosphopantoate | 227.03 | 2.11 | 3.55E − 04 | 4.74E − 03 |
| M116T860 | Valine | 116.07 | 2.03 | 1.21E − 04 | 2.02E − 03 |
Interestingly, several m/z features corresponding to peptidoglycan biosynthetic intermediates were significantly different for the ΔmetB mutant, including UDP-N-acetylmuraminate-alanine-glutamate (Fig. 5D). This, in turn, suggested an association between Met biosynthesis and peptidoglycan biosynthesis. Disruption of metB led to a decrease in intracellular concentrations of UDP-N-acetylmuraminate (∼0.4-fold) and a large increase of UDP-N-acetylmuramoyl-l-alanine (12-fold), UDP-N-acetylmuramoyl-l-alanyl-d-glutamate (1400-fold), and UDP-N-acetylmuramoyl-l-alanyl-d-γ-glutamyl-meso-2,6-diaminopimelate (4-fold) (Fig. 5D). A decrease in UDP-N-acetylmuraminate and an increase in UDP-N-acetylmuramoyl-l-alanine (9-fold), UDP-N-acetylmuramoyl-l-alanyl-d-glutamate (1000-fold), and UDP-N-acetylmuramoyl-l-alanyl-d-γ-glutamyl-meso-2,6-diaminopimelate (5-fold) was also observed in the double mutant ΔmetNIQΔmetB (Fig. 5D).
Ex vivo, the ΔmetB mutant had a reduced growth rate compared with the WT in bile salts. To study the impact of the altered levels of peptidoglycan-related metabolites, the ΔmetB mutant and WT were tested in LB broth in the presence of EDTA, SDS, lysozyme, and fasted state–simulated intestinal fluid, but no significant differences in growth were found (Table S1). A small increase (∼2 mm on 15 mm) in β-lactam antibiotic sensitivity, using a validated disc sensitivity method, was observed for the ΔmetB mutant (data not shown). In addition, when the ΔmetB and WT were tested in MacConkey broth supplemented with 0.6% bile salt, a small but consistent delay in the growth of ΔmetB was observed (Fig. S5).
Discussion
There is increased interest in identifying metabolic pathways in bacterial pathogens, which are essential and distinct from those in their mammalian and human hosts, as potential antibiotic targets (38–42). Previous studies suggested that perhaps 400 S. enterica enzymes are dispensable and that essential pathways are often protected against random mutation by redundancy, reflecting the selective pressure placed on metabolism as a key virulence trait (13). Met biosynthesis and transport is an important part of the interconnected and interdependent amino acid metabolism (43). In this study, the essentiality of Met biosynthesis and transport in the mammalian virulence of S. Typhimurium was investigated.
We showed that de novo Met biosynthesis is not essential when the bacteria are able to obtain Met from their environment. The growth rate of the bacteria with or without de novo Met biosynthesis appeared similar in Met-rich media (e.g. LB broth), suggesting that transport systems can compensate for the loss of de novo biosynthesis. Met biosynthetic mutants require the presence of Met in the culture medium in order to grow inside of HeLa cells (Fig. 2D and Fig. S2), suggesting that extracellular Met becomes available to bacteria in the SCV within a few hours of infection. It is likely that Met is relatively abundant in the SCV during in vivo infection, as all of the Met biosynthetic mutants were as virulent as WT S. Typhimurium in mice (Fig. 2, E and F), presumably because the Met acquisition systems have access to a sufficient source of Met for supporting bacterial growth in the infected animal.
It is only when de novo biosynthesis and high-affinity transport of Met are both disrupted that we observed severe growth attenuation in HeLa cells (Fig. 3C) and, importantly, in infected mice (Fig. 3, D and E). This result strongly suggests functional redundancy between transport and biosynthesis as a source of Met for S. Typhimurium inside of the host. Whether Met is acquired (e.g. ΔmetB) or synthesized (e.g. ΔmetNIQ and ΔmetNIQΔmetB complemented with metB), the growth in vivo appears to be comparable, suggesting that either de novo biosynthesis or the high-affinity transport system of Met alone can provide Met in excess to what is required for S. Typhimurium to grow at maximal capacity in the host. Hence, at least in the case of S. Typhimurium, it is probably quite difficult to inhibit growth by targeting the Met biosynthetic pathway. The observation that ΔmetNIQΔmetB and ΔmetNIQΔmetEH were still able to grow in mice is suggestive of a Met acquisition system independent of MetNIQ, which is consistent with the presence of a low-affinity transporter MetP (19, 20). Hence, our study also provides additional support for the presence of a putative, low-affinity transporter, MetP, which has yet to be identified.
Our description of this apparent redundancy between de novo synthesis and high-affinity transport of Met conflicted with previous studies that showed Met auxotrophs are defective for intracellular survival in macrophages and epithelial cells (22, 23) and have reduced virulence in mice (24, 28) and in 1-day-old chickens (34). This may be at least in part due to differences in the study design. In our study, C57BL/6 mice were infected with a single strain, whereas previous studies typically used competition assays (i.e. infecting the same host with WT and mutant strains), which may put more pressure on the mutant to grow in the competitive environment. It is not uncommon to have divergent results from single infections and competitive infections (44, 45). Another study has shown that the S. Typhimurium lacking the high-affinity transporter MetNIQ is attenuated in C3H/HeN mice (25). C3H/HeN mice carry the resistant allele of Nramp1, which controls Fe3+ availability in the phagosome and is a major determinant of murine susceptibility to S. Typhimurium; consequently, C3H/HeN mice are much more resistant to S. Typhimurium infection than the Nramp1-deficient C57BL/6 mice (46, 47). It is possible that the differences in attenuation observed in earlier studies, but not ours, relate to differences in genetic susceptibility of the mice. Finally, earlier animal studies did not complement the bacterial Met mutation nor sequence the mutant strains, and it is possible that secondary mutations, not mutations in Met biosynthesis per se, were responsible for the observed attenuation in mice.
To generate novel insights about the Met biosynthesis pathway in S. Typhimurium, the changes in bacterial concentrations of key metabolites were examined by LC-MS (Figs. 4 and 5). Mass spectrometry revealed substrate accumulation in Met biosynthetic mutants ΔmetB, ΔmetE, ΔmetH, and the high-affinity transporter mutant ΔmetNIQ and mutants deficient in both (ΔmetNIQΔmetB and ΔmetNIQΔmetEH). This analysis revalidated the pathway model for Met biosynthesis shown in Fig. 1, which has been proposed since the 1980s but to the best of our knowledge, has not been systematically studied in S. enterica until now.
The observed accumulation of substrates provides interesting insights into the kinetics of enzyme activities. The deletion of both Met synthases (i.e. ΔmetNIQΔmetEH) leads to a profound difference in 5-methyltetrahydrofolate concentration, a metabolite in the one-carbon cycle (Fig. 4). In the ΔmetH mutant, there is a 10-fold higher intracellular concentration of the substrate compared with ΔmetE, indicating that MetE is much less efficient than MetH, supporting previous findings (48). Presumably, perturbation of the one-carbon cycle led to an accumulation of homocysteine, which is toxic for bacterial cells (49, 50). This is probably why we consistently observed that ΔmetNIQΔmetEH grew slower in vivo compared with ΔmetNIQΔmetB (Fig. 3 and Fig. S4).
The disruption of the metB gene led to several unexpected observations. First, the intracellular pool of SAM was increased in ΔmetB compared with WT (Fig. 4), indicating a defect in SAM catabolism following perturbation of the activated methyl cycle in this mutant. This observation argues that in the ΔmetB mutant, homocysteine, which is converted into SAM, cannot be derived through de novo biosynthesis in ΔmetB and hence is not fed into the activated methyl cycle. Further comparisons of the LC-MS/MS data between ΔmetB and SL1344 revealed differences in many metabolites from disparate pathways; how disruption to a single enzyme in Met biosynthesis caused perturbations in other pathways was unclear, but the data demonstrate the complexity of modeling intracellular metabolite fluxes in bacteria. Interestingly, this analysis revealed that the concentrations of several metabolites linked with peptidoglycan synthesis were grossly increased in ΔmetB and ΔmetNIQΔmetB (Fig. 5D). Enterobacteriaceae peptidoglycan usually consists of alternating molecules of GlcNAc and N-acetylmuramic acid that are linked by a tetrapeptide of l-alanine, d-glutamate, diaminopimelic acid (DAP), and d-alanine. Because Met biosynthesis and m-DAP biosynthesis are linked through aspartate metabolism (43) and l-cystathionine can substitute for DAP (51), the perturbation of the intracellular metabolite pools related to Met biosynthesis might also impact peptidoglycan synthesis. Whereas LC-MS revealed significant changes in the levels of peptidoglycan intermediates, these changes were not reflected by increased sensitivity to agents (e.g. β-lactam antibiotics) that act through peptidoglycan synthesis or directly on peptidoglycan with the exception of growth in bile. Bile is known to remodel S. enterica peptidoglycan (52). However, it is recognized that Enterobacteriaceae with altered cell wall physiology are very robust. For example, E. coli with decreased m-DAP, which reduced the peptidoglycan density by 50%, did not show any detectable alteration in morphology or growth characteristics (53).
The data obtained from the LC-MS analysis with the complemented mutant ΔmetNIQΔmetB (Figs. 4 and 5) suggested that complementation by pACYC184metB did not fully restore the metabolite levels seen in the ΔmetNIQ mutant. However, this is not unusual because, as related, genetic complementation using a plasmid will differ from the native level due to plasmid copy number and regulation, accounting for the partially complemented phenomenon that is observed.
This study supports earlier models of S. enterica metabolism that have suggested significant redundancy in key pathways linked with growth (13). The research reported here was a systematic analysis of Met metabolism in response to earlier investigations, which suggested that Met auxotrophy was attenuating for bacterial growth in animals and that de novo Met metabolism might therefore provide new antibiotic targets. The regulation of Met synthesis, a complex regulon comprising MetR and MetJ (16, 54), was not investigated because the aim of the study was to determine the essentiality of de novo Met synthesis in in vivo growth. We found that mutants unable to synthesize Met efficiently obtained the amino acid from their intracellular niche via their high-affinity Met transporter, suggesting that the SCV contains sufficient Met to sustain normal bacterial growth even in the absence of de novo synthesis. Severe reductions in virulence in vivo were only observed when both de novo Met biosynthesis and high-affinity Met transport were lost. The impact of mutations in the Met biosynthesis and transporter genes on other pathways was revealed, reaffirming the complexity of the bacterial metabolome and the interactions between metabolites that have yet to be mapped. Considerably more basic science on bacterial metabolism will be needed to identify novel, functionally nonredundant targets.
Experimental procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. All mutant strains were constructed on the S. Typhimurium SL1344 genetic background, and SL1344 was used as the WT strain in all experiments. The SL1344 strain has been described previously (36); its virulence is well defined and is resistant to streptomycin. Appropriate antibiotics, including streptomycin (50 μg/ml), chloramphenicol (30 μg/ml), kanamycin (50 μg/ml), and ampicillin (100 μg/ml), were added to growth medium as required. To obtain mid-exponential growth phase, S. Typhimurium and E. coli strains were grown in 10 ml of LB broth (BD Difco) overnight, and 100 μl of the overnight culture was subcultured into 10 ml of fresh LB broth and grown with shaking (180 rpm) at 37 °C for 3–4 h until the optical density reading at 600 nm reached 0.6–0.8. Growth phenotypes were characterized in LB broth or M9 minimal medium (2 mm MgSO4, 0.1 mm CaCl2, 0.4% glucose, 8.5 mm NaCl, 42 mm Na2HPO4, 22 mm KH2PO4, 18.6 mm NH4Cl, and 100 μm histidine). Met or vitamin B12 was supplemented at 100 μm. For assessing growth under anaerobic conditions, cultures were grown shaking in air-tight jars with AnaeroGen (ThermoFisher) for depletion of oxygen.
Table 2.
Strains and plasmids used for this study
Str, streptomycin; Ap, ampicillin; Chl, chloramphenicol; Km, kanamycin; Ara, arabinose; Flp, flippase; FRT, fippase recombinase target.
| Strain or plasmid | Relevant phenotypes and genotypes | Source/Reference |
|---|---|---|
| Strains | ||
| S. enterica Typhimurium SL1344 | Wild-type strain rpsL hisG46; StrR | Ref. 36 |
| S. enterica Typhimurium BRD666 | Restriction negative modification positive (r−m+) SL1344; StrR | Ref. 68 |
| E. coli DH5α | Cloning strain | Ref. 69 |
|
| ||
| Plasmids | ||
| pGEM-T Easy | High-copy number cloning vector for PCR products; ApR | Promega |
| pACBSR | Medium-copy number, mutagenesis plasmid; p15A ori; Ara-inducible I-SceI and λ Red recombinase; ChlR | Ref. 55 |
| pACYC184 | Medium-copy number cloning vector, p15A ori; TetR, ChlR | Ref. 70 |
| pACYC184metB | S. Typhimurium SL1344 metB cloned into pACYC184; ChlR | This study |
| pCP20 | FLP recombinase, temperature-sensitive replicon; ApR, ChlR | Ref. 71 |
| pKD4 | FRT-flanked KmR cassette; ApR, KmR | Ref. 72 |
Construction of strains and plasmids
Defined deletions of sequence encoding the met biosynthesis genes metA, metB, metC, metE, metF, and metH and Met high-affinity transporter metNIQ were generated in S. Typhimurium SL1344. The biosynthetic mutant and high-affinity transporter mutant were combined together to generate the double mutant ΔmetNIQΔmetB and the triple mutant ΔmetNIQΔmetEH. The double mutant ΔmetNIQΔmetB was complemented by introducing the de novo biosynthetic metB gene into ΔmetNIQΔmetB strain in trans. This strain is named ΔmetNIQΔmetB pACYC184metB. Gene deletions and concomitant insertions of an antibiotic resistance cassette were constructed using a Lambda Red-mediated “gene gorging” method (55). All constructs were verified by PCR and moved to an SL1344 or relevant background via P22 phage transduction (36, 56). Primers used to construct mutants are listed in Table 3. Genetic complementation of mutant ΔmetNIQΔmetB was achieved by cloning metB into pACYC184 via the BamHI and SalI sites and then introducing pACYC184metB into the ΔmetNIQΔmetB mutant. All constructs were verified by restriction analysis and DNA sequencing. Key mutants were also sequenced using Illumina whole-genome sequencing and analyzed using the pipeline at the Wellcome Sanger Institute (Hinxton, UK).
Table 3.
Oligonucleotide primers used in this study for construction of mutants and complementation
Endonuclease restriction sites are underlined; kanamycin resistance cassette-specific sequences are in boldace type. F, forward (5′) primer; R, reverse (3′) primer.
| Mutants | Sequence (5′–3′) |
|---|---|
| ΔmetA-ISceI-F | TAGGGATAACAGGGTAATCGCCAGTGTTAACGCATGTTC |
| ΔmetA-ISceI-R | TAGGGATAACAGGGTAATCGGAATACCACGAATCTGCC |
| ΔmetA-Kan-F | CTAAGGAGGATATTCATATGCGCAGCCACGGTAATTTACTG |
| ΔmetA-Kan-R | GAAGCAGCTCCAGCCTACACAACCTGATAACCTCACGACATACG |
| ΔmetB-ISceI-F | TAGGGATAACAGGGTAATCGCAGATCGGCATCATCC |
| ΔmetB-ISceI-R | TAGGGATAACAGGGTAATCTTCATCAACCTGCGGCTG |
| ΔmetB-Kan-F | CTAAGGAGGATATTCATATGCGGTATTGAAGATGGCGAAG |
| ΔmetB-Kan-R | GAAGCAGCTCCAGCCTACACACAGCCGTATTGTTCGTCATCG |
| ΔmetC-ISceI-F | TAGGGATAACAGGGTAATCCTTCGTTATCTTCGCTGCC |
| ΔmetC-ISceI-R | TAGGGATAACAGGGTAATCAGCAGAGTGCGGACAAACG |
| ΔmetC-Kan-F | CTAAGGAGGATATTCATATGGCTGGTTCGGGTGCATATTG |
| ΔmetC-Kan-R | GAAGCAGCTCCAGCCTACACACACTATTCACTGAGCCAAGCG |
| ΔmetE-ISceI-F | TAGGGATAACAGGGTAATCTACCTGCGGCCAGCTTG |
| ΔmetE-ISceI-R | TAGGGATAACAGGGTAATCAATGCGGTCGCCACTCTG |
| ΔmetE-Kan-F | CTAAGGAGGATATTCATATGGGCGTTAGCGAACATGGTC |
| ΔmetE-Kan-R | GAAGCAGCTCCAGCCTACACATCAACTCGCGACGCAGG |
| ΔmetF-ISceI-F | TAGGGATAACAGGGTAATGCAGCCTGATGGAGCATGG |
| ΔmetF-SceI-R | TAGGGATAACAGGGTAATGCCACGACCATCAATAGAACG |
| ΔmetF-Kan-F | CTAAGGAGGATATTCATATGGCCGTGAAGGAGTGAAGGA |
| ΔmetF-Kan-R | GAAGCAGCTCCAGCCTACACACTTCCGCCAGGCTCTGATTC |
| ΔmetH-ISceI-F | TAGGGATAACAGGGTAATCGGTGAGTCGTGGAATTAGGC |
| ΔmetH-ISceI-R | TAGGGATAACAGGGTAATCGTCAGGGCGACAAGATCC |
| ΔmetH-Kan-F | CTAAGGAGGATATTCATATGGAGGATGTTGAGCGGTGGC |
| ΔmetH-Kan-R | GAAGCAGCTCCAGCCTACACAGCCGTCCAGCACCAGAATAC |
| ΔmetNIQ-ISceI-F | TAGGGATAACAGGGTAATCACAGCTGTGCAGCAGG |
| ΔmetNIQ-ISce-R | TAGGGATAACAGGGTAATACTGCCCTGCGGATGG |
| ΔmetNIQ-Kan-F | CTAAGGAGGATATTCATATGTCCCCTGCTGGAACACTT |
| ΔmetNIQ-Kan-R | GAAGCAGCTCCAGCCTACACAGTCTGATGAAGTGTACGAAGCC |
| metB-BamHI-F | TGGATCCGTCGCAGATGTGCGCTAATG |
| metB-SalI-R | TGTCGACCATAATGCCTGCGACACGC |
Infection of epithelial cells
Infection of HeLa cells (sourced from American Type Culture Collection (ATCC)) was conducted using established gentamicin protection assay protocols (57, 58). Briefly, HeLa cells were grown in DMEM supplemented with 10% fetal calf serum and 2 mm l-GlutaMAX (Life Technologies), in a humidified 37 °C, 5% CO2 incubator. One day before infection, HeLa cells were seeded in 24-well plates at 2 × 105 cells/well. S. Typhimurium strains were grown to mid-exponential phase before and were frozen in LB with 10% glycerol at −80 °C. Bacteria were thawed immediately before infection, washed in antibiotic-free tissue culture medium, and diluted in DMEM with or without l-Met and then added to HeLa cell monolayers at a multiplicity of infection (MOI) of 5–10. The cfu in the inoculum was estimated by plating on LB agar plates. Infected HeLa cells were centrifuged at 600 × g for 5 min immediately after the addition of bacteria and then incubated for 1 h at 37 °C. After 1 h, the tissue culture medium was replaced with DMEM with or without Met and containing 100 μg/ml gentamicin to kill extracellular bacteria. The concentration of gentamicin was reduced to 10 μg/ml at 2 h post-infection and maintained for the remainder of the experiment. To enumerate intracellular bacteria, cells were washed twice with PBS and lysed with 1% Triton X-100 (Sigma) for 15 min to release the intracellular bacteria. The bacteria were enumerated by plating appropriate dilutions on LB agar plates.
Ethics statement
All animal research conducted in this study was approved by the Animal Ethics Committee (Biochemistry and Molecular Biology, Dental Science, Medicine, Microbiology, and Immunology) at the University of Melbourne, under project number 1413141. All experiments were conducted in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 8th edition, 2013.
Mouse infections
Age- and sex-matched C57BL/6 mice were used between 6 and 8 weeks of age to assess the virulence of mutant strains, using either the intravenous or oral route of infection. For intravenous infections, 200 cfu of each bacterial strain was prepared in 200 μl of PBS and injected into the lateral tail vein. For oral infections, mice were orally gavaged with 100 μl of 10% sodium bicarbonate immediately before oral gavage of 200 μl containing ∼5 × 107 cfu of S. Typhimurium strains. To prepare the inoculum, all strains of S. Typhimurium were grown shaking at 180 rpm in M9 minimal medium supplemented with 100 μm Met, at 37 °C for 24 h, and stored in 10% glycerol at −80 °C until use. Immediately before infection, the stored aliquots were thawed and diluted in PBS to the required concentration. At designated time points post-infection, the spleen and liver were removed aseptically, homogenated using the Stomacher 80 Biomaster paddle blender (Seward), and serial dilutions were plated on LB agar plates with streptomycin to determine the bacterial load.
Preparation of stock solutions and standards for LC-MS
Stock solutions of the related underivatized metabolites were prepared at 1.0 mg/ml in an appropriate solvent. Before using, the solutions were combined and diluted with water to give an appropriate standard metabolite mix solution. 13C15N-Aspartate (Cambridge Bioscience) was used as an internal standard at 1 μm in Milli-Q water. All stock solutions were stored at −20 °C.
Sample harvest (metabolic arrest) for LC-MS
Each S. Typhimurium strain was grown at 37 °C to mid-exponential phase in 10 ml of LB, with shaking at 180 rpm. The culture was diluted into 30 ml of PBS and chilled in an ice/water slurry for 5 min and subsequently centrifuged (931 × g, 1 °C, 10 min). The pellet was then washed in 1 ml of PBS and centrifuged (17,295 × g). This washing step was repeated before removing the PBS and storing the cell pellet at −80 °C until metabolite extraction was performed.
Extraction of metabolites for LC-MS
Cell pellets were resuspended with 400 μl of 75% ethanol solution containing 1:1000 of internal standard. Cell lysis was ensured by repeated cycles of freeze-thaw, for a total of 10 times, while cooling to −80 °C. Cellular debris was pelleted by centrifugation (17,295 × g, 5 min, 1 °C). The metabolic extract was transferred to a new tube and stored at −80 °C until analysis.
Instrumentation
A SeQuant ZIC-pHILIC column (5 μm, 150 × 4.6 mm; Millipore) coupled to a 1260 series HPLC system (Agilent) was used to separate metabolites. The method used was described previously (59) with slight modifications; a flow rate of 0.3 ml/min with 20 mm ammonium carbonate in water and 100% acetonitrile was used as the mobile phase. A binary gradient was set up as follows: 0.5 min, 80% acetonitrile; 15.5 min, 50% acetonitrile; 17.5 min, 20% acetonitrile; 18.5 min, 5% acetonitrile; 21 min, 5% acetonitrile; 23 min, 80% acetonitrile; held at 80% acetonitrile until 29.5 min. Detection of metabolites was performed on an Agilent Q-TOF mass spectrometer 6545 operating in negative ESI mode. The scan range was 85–1200 m/z between 2 and 28.2 min at 0.8 spectra/s.
Calibration and validation
LC-MS.d files were converted to .mzXML files using MS convert and analyzed using the LCMS R package (60, 61). Following alignment, groups were extracted with a mass window of 10 ppm, and statistical analysis was performed using MetaboAnalyst version 3.0 (62). The data set was uploaded, filtered using the interquartile range, and log-transformed, and a one-way ANOVA was performed with Tukey's honestly significant difference test (p < 0.01). Data were analyzed using MAVEN in parallel to validate the LC-MS results (63). Following alignment, metabolites were assigned using exact mass (<10 ppm) and retention time (compared with a standards library of 150 compounds run the same day). Scatter plots were generated for each pairwise comparison, and statistical significance was determined using a p value < 0.05 with Benjamini correction.
Author contributions
A. U. H. and R. A. S. conceptualization; A. U. H., N. W., S. A. C., H. J. N., J. J. T., M. J. M., and R. A. S. formal analysis; A. U. H., N. W., S. A. C., D. M. H., J. J. W., T. A. S., M. R. D., and J. C. H. investigation; A. U. H., N. W., S. A. C., H. J. N., D. M. H., J. J. W., T. A. S., M. R. D., J. C. H., M. J. M., and R. A. S. methodology; A. U. H. and S. A. C. writing-original draft; N. W., H. J. N., M. J. M., and R. A. S. supervision; N. W. and J. J. T. visualization; N. W., H. J. N., T. L., M. J. M., and R. A. S. writing-review and editing; T. L., M. J. M., and R. A. S. funding acquisition.
Supplementary Material
Acknowledgments
We acknowledge the helpful advice of Dr. Peter Ayling. Sarah Baines assisted with the analysis of the whole-genome sequences. Shruti Gujaran assisted with testing for antibiotic resistance in different bacterial strains.
This work was supported by Australian National Health and Medical Research Council Program (NHMRC) Program Grant 606788 (to T. L. and R. A. S.). This work was also supported by an International Postgraduate Research Scholarship in Australia (to A. U. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S5.
- SAM
- S-adenosylmethionine
- NTS
- nontyphoidal Salmonella
- SCV
- Salmonella-containing vacuole
- LB
- Luria broth
- DMEM
- Dulbecco's modified Eagle's medium
- cfu
- colony-forming unit(s)
- DAP
- diaminopimelic acid
- MOI
- multiplicity of infection
- ANOVA
- analysis of variance.
References
- 1. Kozak, M. (1983) Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 47, 1–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cantoni, G. L. (1975) Biological methylation: selected aspects. Annu. Rev. Biochem. 44, 435–451 10.1146/annurev.bi.44.070175.002251 [DOI] [PubMed] [Google Scholar]
- 3. Kalan, E. B., and Ceithaml, J. (1954) Methionine biosynthesis in Escherichia coli. J. Bacteriol. 68, 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferla, M. P., and Patrick, W. M. (2014) Bacterial methionine biosynthesis. Microbiology 160, 1571–1584 10.1099/mic.0.077826-0 [DOI] [PubMed] [Google Scholar]
- 5. Morowitz, M. J., Carlisle, E. M., and Alverdy, J. C. (2011) Contributions of intestinal bacteria to nutrition and metabolism in the critically ill. Surg. Clin. North Am. 91, 771–785, viii 10.1016/j.suc.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neis, E. P., Dejong, C. H., and Rensen, S. S. (2015) The role of microbial amino acid metabolism in host metabolism. Nutrients 7, 2930–2946 10.3390/nu7042930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murima, P., McKinney, J. D., and Pethe, K. (2014) Targeting bacterial central metabolism for drug development. Chem. Biol. 21, 1423–1432 10.1016/j.chembiol.2014.08.020 [DOI] [PubMed] [Google Scholar]
- 8. Mastroeni, P., and Maskell, D. (eds) (2006) Salmonella Infections: Clinical, Immunological and Molecular Aspects, Cambridge University Press, Cambridge, UK [Google Scholar]
- 9. Gordon, M. A. (2011) Invasive nontyphoidal Salmonella disease. Curr. Opin. Infect. Dis. 24, 484–489 10.1097/QCO.0b013e32834a9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steeb, B., Claudi, B., Burton, N. A., Tienz, P., Schmidt, A., Farhan, H., Mazé, A., and Bumann, D. (2013) Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 9, e1003301 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knodler, L. A., and Steele-Mortimer, O. (2003) Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4, 587–599 10.1034/j.1600-0854.2003.00118.x [DOI] [PubMed] [Google Scholar]
- 12. Steele-Mortimer, O. (2008) The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11, 38–45 10.1016/j.mib.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., and Bumann, D. (2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307 10.1038/nature04616 [DOI] [PubMed] [Google Scholar]
- 14. Somers, W. S., and Phillips, S. E. (1992) Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature 359, 387–393 10.1038/359387a0 [DOI] [PubMed] [Google Scholar]
- 15. Old, I. G., Phillips, S. E., Stockley, P. G., and Saint Girons, I. (1991) Regulation of methionine biosynthesis in the Enterobacteriaceae. Prog. Biophys. Mol. Biol. 56, 145–185 10.1016/0079-6107(91)90012-H [DOI] [PubMed] [Google Scholar]
- 16. Maxon, M. E., Redfield, B., Cai, X. Y., Shoeman, R., Fujita, K., Fisher, W., Stauffer, G., Weissbach, H., and Brot, N. (1989) Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc. Natl. Acad. Sci. U.S.A. 86, 85–89 10.1073/pnas.86.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gál, J., Szvetnik, A., Schnell, R., and Kálmán, M. (2002) The metD D-methionine transporter locus of Escherichia coli is an ABC transporter gene cluster. J. Bacteriol. 184, 4930–4932 10.1128/JB.184.17.4930-4932.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Merlin, C., Gardiner, G., Durand, S., and Masters, M. (2002) The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184, 5513–5517 10.1128/JB.184.19.5513-5517.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ayling, P. D., and Bridgeland, E. S. (1972) Methionine transport in wild-type and transport-defective mutants of Salmonella Typhimurium. J. Gen. Microbiol. 73, 127–141 10.1099/00221287-73-1-127 [DOI] [PubMed] [Google Scholar]
- 20. Ayling, P. D., Mojica-a, T., and Klopotowski, T. (1979) Methionine transport in Salmonella Typhimurium: evidence for at least one low-affinity transport system. J. Gen. Microbiol. 114, 227–246 10.1099/00221287-114-2-227 [DOI] [PubMed] [Google Scholar]
- 21. Shaw, N. A., and Ayling, P. D. (1991) Cloning of high-affinity methionine transport genes from Salmonella Typhimurium. FEMS Microbiol. Lett. 62, 127–131 [DOI] [PubMed] [Google Scholar]
- 22. Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986) Mutants of Salmonella Typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193 10.1073/pnas.83.14.5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leung, K. Y., and Finlay, B. B. (1991) Intracellular replication is essential for the virulence of Salmonella Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 88, 11470–11474 10.1073/pnas.88.24.11470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ejim, L. J., D'Costa, V. M., Elowe, N. H., Loredo-Osti, J. C., Malo, D., and Wright, G. D. (2004) Cystathionine β-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 72, 3310–3314 10.1128/IAI.72.6.3310-3314.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richardson, A. R., Payne, E. C., Younger, N., Karlinsey, J. E., Thomas, V. C., Becker, L. A., Navarre, W. W., Castor, M. E., Libby, S. J., and Fang, F. C. (2011) Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10, 33–43 10.1016/j.chom.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bogard, R. W., Davies, B. W., and Mekalanos, J. J. (2012) MetR-regulated Vibrio cholerae metabolism is required for virulence. MBio 3, e00236–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berney, M., Berney-Meyer, L., Wong, K.-W., Chen, B., Chen, M., Kim, J., Wang, J., Harris, D., Parkhill, J., Chan, J., Wang, F., and Jacobs, W. R., Jr. (2015) Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 112, 10008–10013 10.1073/pnas.1513033112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jelsbak, L., Mortensen, M. I. B., Kilstrup, M., and Olsen, J. E. (2016) The in vitro redundant enzymes PurN and PurT are both essential for systemic infection of mice in Salmonella enterica serovar Typhimurium. Infect. Immun. 84, 2076–2085 10.1128/IAI.00182-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kröger, C., Colgan, A., Srikumar, S., Händler, K., Sivasankaran, S. K., Hammarlöf, D. L., Canals, R., Grissom, J. E., Conway, T., Hokamp, K., and Hinton, J. C. (2013) An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14, 683–695 10.1016/j.chom.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 30. Srikumar, S., Kröger, C., Hébrard, M., Colgan, A., Owen, S. V., Sivasankaran, S. K., Cameron, A. D., Hokamp, K., and Hinton, J. C. (2015) RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 11, e1005262–26 10.1371/journal.ppat.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeter, R. M., Olivera, B. M., and Roth, J. R. (1984) Salmonella Typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J. Bacteriol. 159, 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. González, J. C., Banerjee, R. V., Huang, S., Sumner, J. S., and Matthews, R. G. (1992) Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry 31, 6045–6056 10.1021/bi00141a013 [DOI] [PubMed] [Google Scholar]
- 33. Banerjee, R. V., Johnston, N. L., Sobeski, J. K., Datta, P., and Matthews, R. G. (1989) Cloning and sequence analysis of the Escherichia coli metH gene encoding cobalamin-dependent methionine synthase and isolation of a tryptic fragment containing the cobalamin-binding domain. J. Biol. Chem. 264, 13888–13895 [PubMed] [Google Scholar]
- 34. Shah, D. H., Shringi, S., Desai, A. R., Heo, E.-J., Park, J.-H., and Chae, J.-S. (2007) Effect of metC mutation on Salmonella Gallinarum virulence and invasiveness in 1-day-old White Leghorn chickens. Vet. Microbiol. 119, 352–357 10.1016/j.vetmic.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 35. Wray, C., and Sojka, W. J. (1978) Experimental Salmonella Typhimurium infection in calves. Res. Vet. Sci. 25, 139–143 [PubMed] [Google Scholar]
- 36. Hoiseth, S. K., and Stocker, B. A. (1981) Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 10.1038/291238a0 [DOI] [PubMed] [Google Scholar]
- 37. Kwon, Y. K., Lu, W., Melamud, E., Khanam, N., Bognar, A., and Rabinowitz, J. D. (2008) A domino effect in antifolate drug action in Escherichia coli. Nat. Chem. Biol. 4, 602–608 10.1038/nchembio.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coates, A. R. M., and Hu, Y. (2007) Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 152, 1147–1154 10.1038/sj.bjp.0707432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harada, E., Iida, K., Shiota, S., Nakayama, H., and Yoshida, S. (2010) Glucose metabolism in Legionella pneumophila: dependence on the Entner-Doudoroff pathway and connection with intracellular bacterial growth. J. Bacteriol. 192, 2892–2899 10.1128/JB.01535-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rossi, M., Amaretti, A., and Raimondi, S. (2011) Folate production by probiotic bacteria. Nutrients 3, 118–134 10.3390/nu3010118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pitchandi, P., Hopper, W., and Rao, R. (2013) Comprehensive database of Chorismate synthase enzyme from shikimate pathway in pathogenic bacteria. BMC Pharmacol. Toxicol. 14, 29 10.1186/2050-6511-14-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bourne, C. (2014) Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics 3, 1–28 10.3390/antibiotics3010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beuzón, C. R., and Holden, D. W. (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3, 1345–1352 10.1016/S1286-4579(01)01496-4 [DOI] [PubMed] [Google Scholar]
- 45. Raman, S. B., Nguyen, M. H., Cheng, S., Badrane, H., Iczkowski, K. A., Wegener, M., Gaffen, S. L., Mitchell, A. P., and Clancy, C. J. (2013) A competitive infection model of hematogenously disseminated candidiasis in mice redefines the role of Candida albicans IRS4 in pathogenesis. Infect. Immun. 81, 1430–1438 10.1128/IAI.00743-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lissner, C. R., Swanson, R. N., and O'Brien, A. D. (1983) Genetic control of the innate resistance of mice to Salmonella Typhimurium: expression of the Ity gene in peritoneal and splenic macrophages isolated in vitro. J. Immunol. 131, 3006–3013 [PubMed] [Google Scholar]
- 47. Nairz, M., Fritsche, G., Crouch, M.-L. V., Barton, H. C., Fang, F. C., and Weiss, G. (2009) Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell Microbiol. 11, 1365–1381 10.1111/j.1462-5822.2009.01337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bertrand, E. M., Moran, D. M., McIlvin, M. R., Hoffman, J. M., Allen, A. E., and Saito, M. A. (2013) Methionine synthase interreplacement in diatom cultures and communities: Implications for the persistence of B12 use by eukaryotic phytoplankton. Limnol. Oceanogr. 58, 1431–1450 10.4319/lo.2013.58.4.1431 [DOI] [Google Scholar]
- 49. Tuite, N. L., Fraser, K. R., and O'Byrne, C. P. (2005) Homocysteine toxicity in Escherichia coli is caused by a perturbation of branched-chain amino acid biosynthesis. J. Bacteriol. 187, 4362–4371 10.1128/JB.187.13.4362-4371.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sikora, M., and Jakubowski, H. (2009) Homocysteine editing and growth inhibition in Escherichia coli. Microbiology 155, 1858–1865 10.1099/mic.0.026609-0 [DOI] [PubMed] [Google Scholar]
- 51. Richaud, C., Mengin-Lecreulx, D., Pochet, S., Johnson, E. J., Cohen, G. N., and Marlière, P. (1993) Directed evolution of biosynthetic pathways: recruitment of cysteine thioethers for constructing the cell wall of Escherichia coli. J. Biol. Chem. 268, 26827–26835 [PubMed] [Google Scholar]
- 52. Hernández, S. B., Cava, F., Pucciarelli, M. G., García-Del Portillo, F., de Pedro, M. A., and Casadesús, J. (2015) Bile-induced peptidoglycan remodelling in Salmonella enterica. Environ. Microbiol. 17, 1081–1089 10.1111/1462-2920.12491 [DOI] [PubMed] [Google Scholar]
- 53. Prats, R., and de Pedro, M. A. (1989) Normal growth and division of Escherichia coli with a reduced amount of murein. J. Bacteriol. 171, 3740–3745 10.1128/jb.171.7.3740-3745.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Augustus, A. M., and Spicer, L. D. (2011) The MetJ regulon in gammaproteobacteria determined by comparative genomics methods. BMC Genomics 12, 558 10.1186/1471-2164-12-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Herring, C. D., Glasner, J. D., and Blattner, F. R. (2003) Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311, 153–163 10.1016/S0378-1119(03)00585-7 [DOI] [PubMed] [Google Scholar]
- 56. Susskind, M. M., and Botstein, D. (1978) Molecular genetics of bacteriophage P22. Microbiol. Rev. 42, 385–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mandell, G. L. (1973) Interaction of intraleukocytic bacteria and antibiotics. J. Clin. Invest. 52, 1673–1679 10.1172/JCI107348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vaudaux, P., and Waldvogel, F. A. (1979) Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob. Agents Chemother. 16, 743–749 10.1128/AAC.16.6.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cobbold, S. A., Chua, H. H., Nijagal, B., Creek, D. J., Ralph, S. A., and McConville, M. J. (2016) Metabolic dysregulation induced in Plasmodium falciparum by dihydroartemisinin and other front-line antimalarial drugs. J. Infect. Dis. 213, 276–286 10.1093/infdis/jiv372 [DOI] [PubMed] [Google Scholar]
- 60. Smith, C. A., Want, E. J., O'Maille, G., Abagyan, R., and Siuzdak, G. (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- 61. Tautenhahn, R., Böttcher, C., and Neumann, S. (2008) Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9, 504 10.1186/1471-2105-9-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xia, J., Sinelnikov, I. V., Han, B., and Wishart, D. S. (2015) MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 10.1093/nar/gkv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clasquin, M. F., Melamud, E., and Rabinowitz, J. D. (2012) LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics, Chapter 14, Unit 14.11 10.1002/0471250953.bi1411s37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. González, J. C., Peariso, K., Penner-Hahn, J. E., and Matthews, R. G. (1996) Cobalamin-independent methionine synthase from Escherichia coli: a zinc metalloenzyme. Biochemistry. 35, 12228–12234 10.1021/bi9615452 [DOI] [PubMed] [Google Scholar]
- 65. Katzen, H. M., and Buchanan, J. M. (1965) Enzymatic synthesis of the methyl group of methionine. VIII. Repression-depression, purification, and properties of 5,10-methylene-tetrahydrofolate reductase from Escherichia coli. J. Biol. Chem. 240, 825–835 [PubMed] [Google Scholar]
- 66. Chiang, P. K., Gordon, R. K., Tal, J., Zeng, G. C., Doctor, B. P., Pardhasaradhi, K., and McCann, P. P. (1996) S-Adenosylmethionine and methylation. FASEB J. 10, 471–480 10.1096/fasebj.10.4.8647346 [DOI] [PubMed] [Google Scholar]
- 67. Struck, A.-W., Thompson, M. L., Wong, L. S., and Micklefield, J. (2012) ChemInform abstract: S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem 13, 2642–2655 10.1002/cbic.201200556 [DOI] [PubMed] [Google Scholar]
- 68. Turner, S. J., Carbone, F. R., and Strugnell, R. A. (1993) Salmonella Typhimurium δ aroA δ aroD mutants expressing a foreign recombinant protein induce specific major histocompatibility complex class I-restricted cytotoxic T lymphocytes in mice. Infect. Immun. 61, 5374–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grant, S. G., Jessee, J., Bloom, F. R., and Hanahan, D. (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649 10.1073/pnas.87.12.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chang, A. C., and Cohen, S. N. (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cherepanov, P. P., and Wackernagel, W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- 72. Datsenko, K. A., and Wanner, B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.