Abstract
G protein–coupled receptors (GPCRs) can be differentially activated by ligands to generate multiple and distinct downstream signaling profiles, a phenomenon termed biased agonism. The glucagon-like peptide-1 receptor (GLP-1R) is a class B GPCR and a key drug target for managing metabolic disorders; however, its peptide agonists display biased signaling that affects their relative efficacies. In this study, we combined mutagenesis experiments and mapping of surface mutations onto recently described GLP-1R structures, which revealed two major domains in the GLP-1/GLP-1R/Gs protein active structure that are differentially important for both receptor quiescence and ligand-specific initiation and propagation of biased agonism. Changes to the conformation of transmembrane helix (TM) 5 and TM 6 and reordering of extracellular loop 2 were essential for the propagation of signaling linked to cAMP formation and intracellular calcium mobilization, whereas ordering and packing of residues in TMs 1 and 7 were critical for extracellular signal–regulated kinase 1/2 (pERK) activity. On the basis of these findings, we propose a model of distinct peptide–receptor interactions that selectively control how these different signaling pathways are engaged. This work provides important structural insight into class B GPCR activation and biased agonism.
Keywords: G protein-coupled receptor (GPCR), glucagon, receptor structure-function, cell signaling, site-directed mutagenesis, biased agonism, GLP-1 receptor, glucagon-like peptide-1, class B peptide hormone, ERK kinase, arrestin, transmembrane domain
Introduction
GPCRs7 are ubiquitous integrators of extracellular signals for the control of cellular responses. As such, they are key drug targets, and ∼40% of approved therapeutics act via this receptor class (1). Nonetheless, many potential drugs fail in late-stage clinical trials due to lack of predicted efficacy, indicating gaps in our understanding of drug action and/or the specific contributions of signaling events to the control of diseases.
It has become increasingly evident that GPCRs are highly dynamic proteins that can adopt numerous ligand-specific conformational ensembles with distinct impact on signaling and regulatory profiles, even with ligands acting via a common binding pocket, a phenomenon termed biased agonism (2, 3). This not only provides an unprecedented opportunity to sculpt biological responses for therapeutic benefit, but it also creates increased challenges for drug discovery and developmental programs to identify the spectra of ligand behavior and to elucidate structure–activity relationships linking observed behavior to physiology and disease processes.
Class B peptide hormone GPCRs bind important physiological peptides of about 30–40 amino acids, including calcitonin, amylin, adrenomedullin, calcitonin gene-related peptide, secretin, parathyroid hormones, vasoactive intestinal peptide, gastric inhibitory peptide, glucagon, and the glucagon-like peptides (4, 5). As such, these receptors are crucial targets for treatment of chronic diseases, notably osteoporosis, migraine, obesity, and type 2 diabetes. Class B GPCRs are pleiotropically coupled, and biased agonism is commonly observed when signaling is studied across multiple pathways, creating novel therapeutic opportunities. However, optimally exploiting this property requires detailed mechanistic understanding of the drivers of bias (4).
The glucagon-like peptide-1 (GLP-1) receptor (GLP-1R) is a class B GPCR that is both critical to the natural incretin response of the body and a major target for treatment of metabolic disorders. It is among the best studied for biased agonism, and such bias is readily observed for both naturally occurring and synthetic peptide mimetics (6–11). This biased agonism alters responses in pancreatic β-cells (11) and disease models in vivo (10). The prevailing view for class B GPCR peptide binding and receptor activation is a two-domain theory with initial binding of the peptide C terminus to the receptor extracellular domain (ECD) that positions the peptide N terminus relative to the receptor core to facilitate receptor activation (12). The prevalence of GLP-1R biased agonism and the therapeutic implications of this behavior make understanding of the triggers for, and propagation of, bias important for rational drug design and development. We recently revealed that the extracellular loops (ECLs) of the GLP-1R play a crucial role in the biased agonism of exendin-4, oxyntomodulin, and GLP-1(7–36)-NH2 (GLP-1): the first clinically approved GLP-1 mimetic, a biased endogenous GLP-1R peptide, and the most common circulating form of GLP-1, respectively (11, 13). Nonetheless, interpretation of the data was limited by lack of experimentally determined structure for the GLP-1R core and, indeed, any full-length and active class B GPCR structures. Recently, several near-atomic resolution structures of the GLP-1R have been published that include structures of a stabilized inhibitor-bound human receptor transmembrane (TM) domain (14), a stabilized full-length human receptor bound to a modified 11-mer peptide agonist (15), and GLP-1Rs in complex with heterotrimeric Gs protein and either GLP-1 (16) or a newly identified biased agonist, exendin-P5 (17). In the meantime, the structure of a full-length glucagon receptor (GCGR) in complex with a weak partial agonist (18) and the full-length active calcitonin receptor in complex with peptide agonist and heterotrimeric Gs were also solved (19). Collectively, this work has provided novel insights into gross conformational changes linked to the dynamics of class B GPCR activation, including marked kinking of transmembrane helix (TM) 6, outward movement of helices 6 and 7, and the interlinking ECL3, required for full activation, as well as manifest reorganization of other ECLs (16, 17, 19).
The ECD is structurally linked to the receptor core through extension of TM1, and there is accumulating evidence that the far N terminus of the ECD may have a dynamic role in class B GPCR function, both with respect to maintenance of an inactive state, as has been suggested for the glucagon receptor (GCGR) (20), and in ligand-dependent signaling (21, 22). Notably, the TM1 stalk domain is unstructured in complexes of class B receptors complexed to agonist and G protein, but it maintains an extended α-helix in GCGR bound to a partial agonist but without G protein (18). In this study, we performed alanine-scanning analysis of the GLP-1R surface of the far N-terminal 7 amino acids (residues 24–30, immediately after the receptor signal peptide) and the 21 amino acids that link TM1 and the ECD (residues 128–148), and we assessed cell-surface expression, peptide affinity, and peptide efficacy for activation of pathways linked to cAMP accumulation, intracellular calcium ([Ca2+]i) mobilization and phosphorylation of extracellular signal-regulated kinase 1/2 (pERK). Each of these pathways is of physiological relevance for GLP-1R signaling (4, 23), and both these regions are dynamically involved in GLP-1R peptide affinity and signal transduction. The overlay of the new data and that from our prior mutagenic analysis of the ECLs (11) onto the recently solved GLP-1R structures revealed that changes to the conformation of TMs 5 and 6 and reordering of ECL2 were essential for propagation of cAMP formation and [Ca2+]i mobilization signaling, whereas ordering and packing of residues in TMs 1 and 7 were critical for pERK that is principally driven by the Gi–Gβγ–arrestin interaction in the WT receptor.
Results
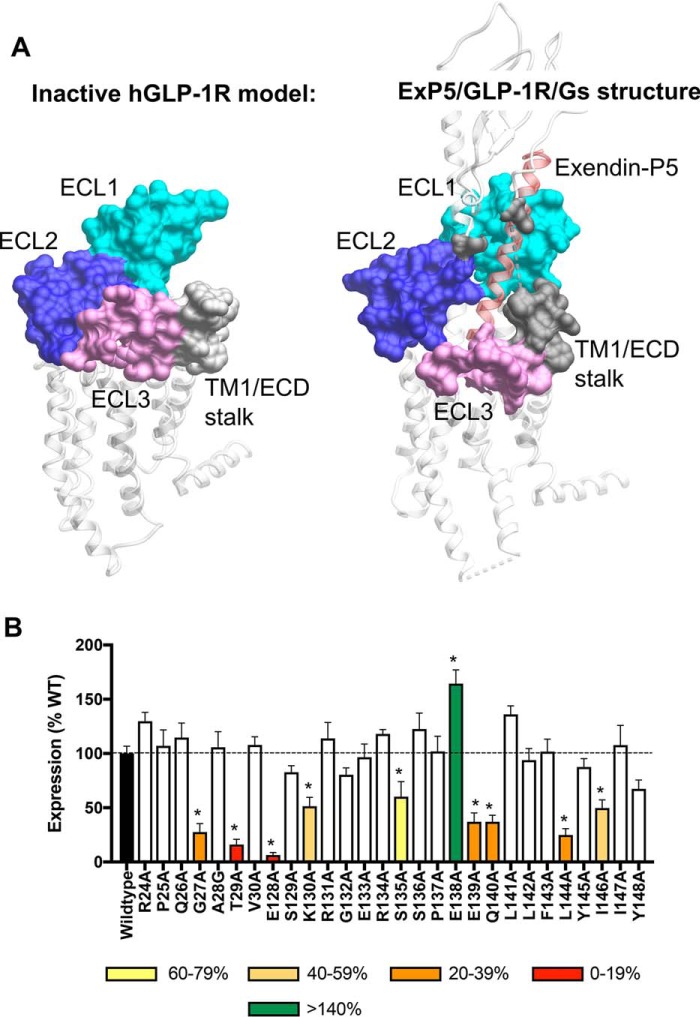
The far N-terminal ECD (Arg-24–Val-30) and TM1/ECD linker (Glu-128–Tyr-148) residues of GLP-1R were mutated to alanine by site-directed mutagenesis and stably expressed in CHO–Flp-In cells by isogenic recombination, with the exception of Ala-28 that was mutated to glycine. The location of these amino acids within the GLP-1R extracellular surface is illustrated on inactive (TM1/ECD linker only) and active hGLP-1R models in Fig. 1A.
Figure 1.
Location of the TM/N-terminal ECD interface in active and inactive models of the human GLP-1R and expression of GLP-1R constructs in stable cell lines. A, surface residues of the GLP-1R. Left-hand panel, inactive model of the hGLP-1R TM domain (residues 134–422) based on the inactive glucagon receptor (PDB code 4L6R). Right-hand panel, active full-length exendin-P5 (ExP5)–hGLP-1R–Gs complex (PDB code 6B3J). The far N-terminal ECD (residues 24–30) and the TM1/ECD stalk (residues 128–148) are illustrated in gray. ECL residues mutated in previous analyses (11) are colored by ECL. ECL1 (residues 201–223; cyan), ECL2 (residues 285–307; dark blue), ECL3 (residues 372–387; light purple) are shown. The position of exendin-P5 is shown as red ribbon representation. B, cell-surface expression was determined by ELISA to the N-terminal c-Myc epitope tag on the hGLP-1R and mutant hGLP-1R isogenically expressed in CHO–Flp-In cells. Data were normalized to the WT receptor. All values are mean ± S.E. of four to six independent experiments, conducted in duplicate. One-way ANOVA and Dunnett's post-test were performed to determine statistical differences (*, p < 0.05).
Cell-surface expression levels for WT and mutant GLP-1Rs in the CHO–Flp-In stable cell lines were measured through ELISA of anti–c-Myc antibody binding to the N-terminal c-Myc epitope. All receptors were expressed at the cell surface; however, the expression of G27A, T29A, E128A, K130A, S135A, E139A, Q140A, L144A, and I146A mutants was significantly lower and that of E138A was markedly higher than WT GLP-1R. All the other mutant receptors were not significantly different from the WT (Fig. 1B and Table 1).
Table 1.
Effects of human GLP-1R TM1/N-terminal mutants on peptide ligand binding and cell-surface expression
Binding data were analyzed using a three-parameter logistic equation and normalized to the maximal binding of the radiolabeled antagonist 125I-exendin-4(9–39) and the nonspecific binding in the presence of 1 μm exendin-4(9–39). pKi is the negative logarithm of peptide affinity. All the values for binding are mean ± S.E. of three independent experiments, conducted in triplicate. Cell-surface expression was accessed through ELISA detecting the N-terminal c-Myc epitope label on the receptor. Mutant data are compared with the wildtype human GLP-1R expression and shown as percentage. The data for cell-surface expression are mean ± S.E. of four to six independent experiments, conducted in duplicate. One-way ANOVA and Dunnett's post-test were used to determine statistical differences (*, p < 0.05). ND, not determined.
| Whole-cell competition radioligand binding (pKi) |
Cell-surface expression (% wildtype) | ||||
|---|---|---|---|---|---|
| GLP-1(7–36)-NH2 | Exendin-4 | Oxyntomodulin | Exendin(9–39) | ||
| Wildtype | 8.12 ± 0.06 | 9.31 ± 0.06 | 7.52 ± 0.08 | 7.85 ± 0.05 | 100 ± 7 |
| R24A | 8.10 ± 0.08 | 9.54 ± 0.13 | 7.69 ± 0.08 | 7.74 ± 0.06 | 130 ± 8 |
| P25A | 8.17 ± 0.09 | 9.56 ± 0.12 | 7.80 ± 0.08 | 7.71 ± 0.06 | 107 ± 15 |
| Q26A | 7.96 ± 0.09 | 9.23 ± 0.11 | 7.56 ± 0.10 | 7.72 ± 0.07 | 115 ± 13 |
| G27A | ND | ND | ND | ND | 28 ± 8* |
| A28G | 7.67 ± 0.08* | 8.89 ± 0.11* | 7.07 ± 0.08* | 7.61 ± 0.06 | 106 ± 15 |
| T29A | ND | ND | ND | ND | 16 ± 5* |
| V30A | 8.16 ± 0.09 | 9.31 ± 0.11 | 7.72 ± 0.08 | 7.77 ± 0.08 | 108 ± 8 |
| E128A | ND | ND | ND | ND | 6 ± 2* |
| S129A | 8.10 ± 0.11 | 9.94 ± 0.10 | 7.71 ± 0.15 | 7.79 ± 0.10 | 83 ± 6 |
| K130A | 8.12 ± 0.08 | 9.25 ± 0.07 | 7.52 ± 0.12 | 7.64 ± 0.09 | 51 ± 8* |
| R131A | 8.28 ± 0.12 | 9.37 ± 0.09 | 7.72 ± 0.14 | 7.78 ± 0.09 | 114 ± 15 |
| G132A | ND | ND | ND | ND | 80 ± 6 |
| E133A | 8.16 ± 0.07 | 9.45 ± 0.10 | 7.70 ± 0.09 | 7.84 ± 0.10 | 97 ± 12 |
| R134A | 8.12 ± 0.09 | 9.14 ± 0.08 | 7.60 ± 0.12 | 7.65 ± 0.11 | 118 ± 4 |
| S135A | 8.35 ± 0.09 | 9.40 ± 0.10 | 7.79 ± 0.09 | 7.82 ± 0.09 | 60 ± 14* |
| S136A | 7.84 ± 0.11 | 9.29 ± 0.07 | 7.39 ± 0.13 | 7.59 ± 0.15 | 123 ± 15 |
| P137A | 7.18 ± 0.13* | 8.54 ± 0.09* | 6.94 ± 0.12* | 6.98 ± 0.18* | 102 ± 14 |
| E138A | 8.22 ± 0.07 | 9.19 ± 0.10 | 7.21 ± 0.07 | 7.87 ± 0.07 | 164 ± 13* |
| E139A | ND | ND | ND | ND | 37 ± 8* |
| Q140A | 7.84 ± 0.15 | 8.65 ± 0.13* | 7.68 ± 0.21 | 8.15 ± 0.16 | 37 ± 6* |
| L141A | 7.50 ± 0.06 | 8.53 ± 0.06* | 6.17 ± 0.28* | 7.17 ± 0.06* | 136 ± 8 |
| L142A | 7.89 ± 0.07 | 9.23 ± 0.07 | 6.89 ± 0.10* | 7.84 ± 0.07 | 94 ± 11 |
| F143A | 8.19 ± 0.06 | 9.33 ± 0.09 | 7.35 ± 0.09 | 7.65 ± 0.06 | 102 ± 11 |
| L144A | 7.94 ± 0.15 | 9.41 ± 0.17 | 7.15 ± 0.19 | 8.03 ± 0.15 | 25 ± 6* |
| Y145A | 8.25 ± 0.09 | 9.51 ± 0.09 | 7.38 ± 0.10 | 7.92 ± 0.07 | 88 ± 8 |
| I146A | 8.07 ± 0.09 | 9.32 ± 0.12 | 7.57 ± 0.14 | 7.91 ± 0.09 | 50 ± 8* |
| I147A | 7.78 ± 0.05 | 9.00 ± 0.08 | 7.26 ± 0.08 | 7.67 ± 0.08 | 108 ± 18 |
| Y148A | 6.79 ± 0.15* | 8.06 ± 0.06* | 6.26 ± 0.25* | 8.09 ± 0.08 | 67 ± 8 |
Peptide agonist affinity
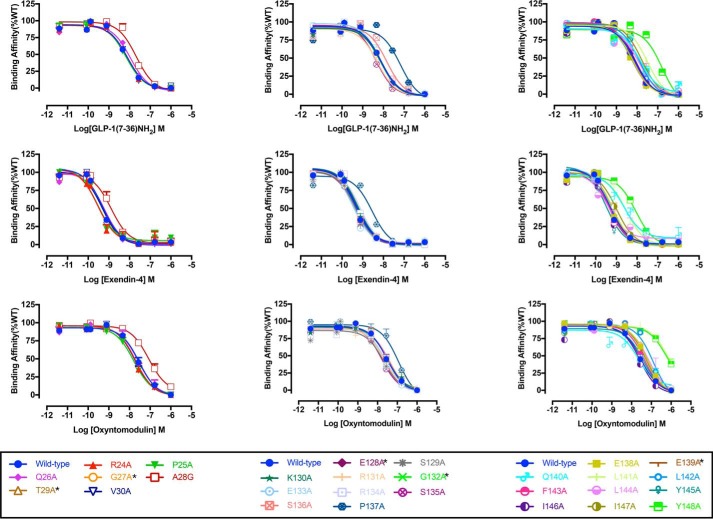
Heterologous whole-cell competition binding with 125I–exendin-4(9–39) was performed to determine peptide agonist affinity for the WT and mutant GLP-1Rs. As reported previously (6), at the WT receptor exendin-4 had the highest affinity (pKi ≈9.31), followed by GLP-1 (pKi ≈8.12), whereas oxyntomodulin had the lowest affinity (pKi ≈7.52) (Table 1 and Fig. 2). No specific binding window could be established for the G132A mutant, despite good cell–surface expression by ELISA, or for the low-expressing mutants G27A, T29A, E128A, and E139A; for all others, competitive binding isotherms were established for each of the peptides. Of the far N-terminal ECD residues that exhibited specific binding, only the A28G mutation modified affinity (<5-fold) for GLP-1 and exendin-4, with a similar trend for oxyntomodulin (Figs. 2 and 3A and Table 1). Of the TM1/stalk residues, P137A, L141A, and Y148A mutations reduced affinity for each of the agonist peptides, whereas the P137A and L141A mutations also reduced affinity of the antagonist exendin-4(9–39). Ligand-specific reductions in affinity were seen for Q140A (exendin-4) and L142A (oxyntomodulin). All other mutations were without significant effect on peptide affinity (Figs. 2 and 3A and Table 1).
Figure 2.
Competitive inhibition of 125I-exendin(9–39) binding by peptide agonists for Ala mutants of the hGLP-1R N-terminal ECD and TM1 and linker region. Binding affinity data are expressed as a percentage of measured bound versus bound in the absence of peptide, each corrected for nonspecific binding (measured in the presence of 1 μm unlabeled exendin(9–39)). Inhibition curves of WT and mutant receptors were stimulated by GLP-1(7–36)NH2 (upper panels), exendin-4 (middle panels), or oxyntomodulin (lower panels) in CHO–Flp-In cells stably expressed WT or mutant receptors. Data were fitted with a three-parameter logistic equation. All values are means ± S.E. of four to six independent experiments, conducted in duplicate.
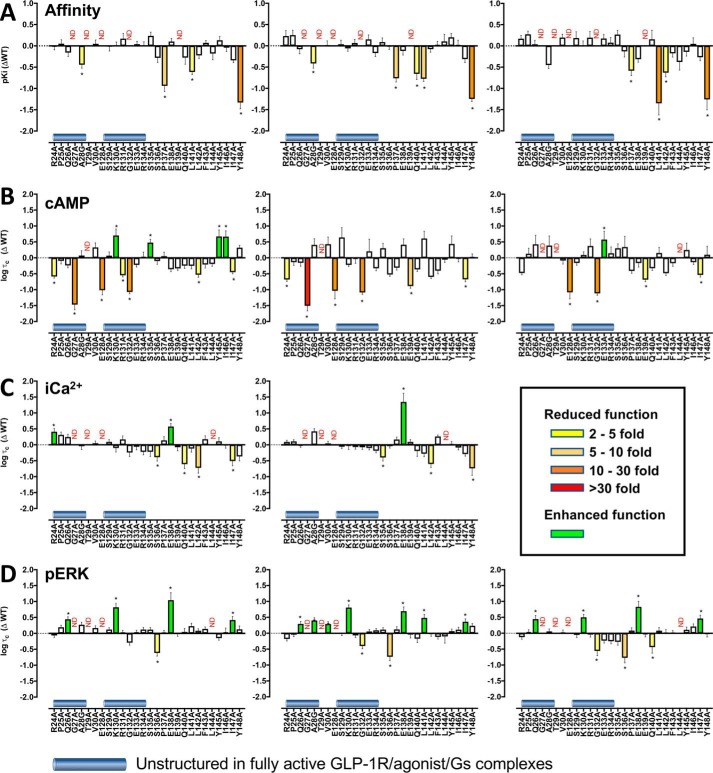
Figure 3.
Changes in affinity (A) and efficacy (B–D) of the agonists GLP-1, exendin-4, and oxyntomodulin at mutant GLP-1Rs. A, pKi values for the agonist peptides were derived from competition of 125I–exendin-4(9–39) binding. Data were plotted as differences in pIC50 of the alanine mutants compared with the wildtype (WT) hGLP-1R for GLP-1 (left panel), exendin-4 (middle panel), and oxyntomodulin (right panel). All pIC50 values were mean ± S.E. of three independent experiments performed in triplicate. B–D, Δlogτc values are the difference in the coupling efficacy (logτc) for cAMP accumulation (B), [Ca2+]i mobilization (C), and ERK phosphorylation (pERK) (D) of the alanine mutant GLP-1Rs compared with the WT receptor for GLP-1 (left panels), exendin-4 (middle panels), and oxyntomodulin (right panels). All functional values are mean ± S.E. of four to six independent experiments, conducted in duplicate. One-way ANOVA and Dunnett's post-test were performed to determined statistical differences (*, p < 0.05), and the bars are colored according to the fold-change between WT and mutant receptors (yellow, 2–5-fold decrease; light orange, 5–10-fold decrease; dark orange, 10–30-fold decrease; red, >30-fold decrease; green, increased affinity (A) or efficacy (B–D)). N.D., not defined.
Agonist efficacy
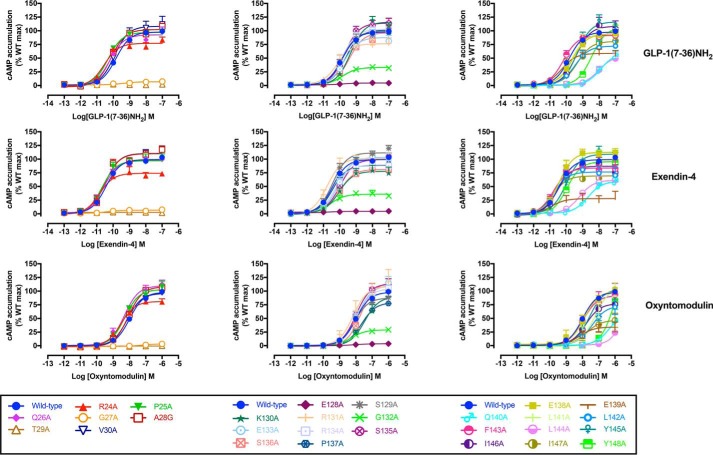
Concentration-response curves for each of the peptides were established to determine agonist potency and maximal responses for canonical signaling endpoints, cAMP accumulation, [Ca2+]i mobilization, and pERK1/2 (Figs. 3, B–D, and 4–6 and Tables 2–4). These data were subject to operational modeling (26) to determine affinity-independent measures of efficacy (τ for each pathway). The operational efficacy term “τ” is a measure of the number of receptors that need to be occupied to give a specified response. The τ values were normalized to receptor expression to derive a receptor expression-independent and quantitative measure of pathway activation “τc” for individual peptides at each of the mutants (27, 28).
Figure 4.
cAMP concentration-response curves for Ala mutants of the hGLP-1R N-terminal ECD and TM1 and linker region. Concentration-response curves for cAMP accumulation of WT and mutant receptors were stimulated by GLP-1(7–36)NH2 (upper panels), exendin-4 (middle panels), or oxyntomodulin (lower panels) in CHO–Flp-In cells stably expressing WT or mutant receptors. Data were normalized to the response elicited by the WT and analyzed with an operational model of agonism. All values are means ± S.E. of four to six independent experiments, conducted in duplicate.
Figure 5.
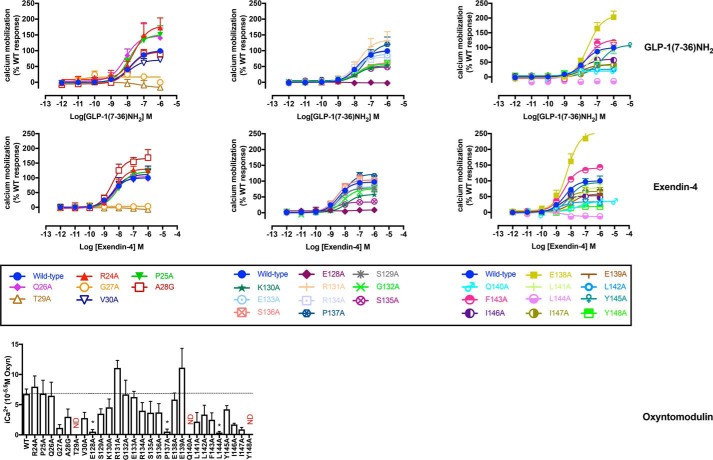
[Ca2+]i mobilization concentration-response curves for Ala mutants of the hGLP-1R N-terminal ECD and TM1 and linker region. Concentration-response curves of [Ca2+]i mobilization of WT and mutant receptors were stimulated by GLP-1(7–36)NH2 (upper panels), exendin-4 (middle panels), or oxyntomodulin (lower panels) in CHO–Flp-In cells stably expressing WT or mutant receptors. Data were normalized to the response elicited by the WT and analyzed with an operational model of agonism. All values are means ± S.E. of four to six independent experiments, conducted in duplicate.
Figure 6.
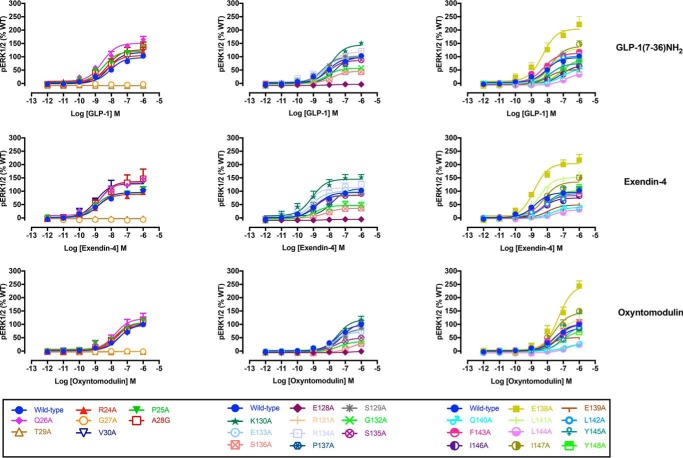
pERK1/2 concentration-response curves for Ala mutants of the hGLP-1R N-terminal ECD and TM1 and linker region. Concentration-response curves of pERK of WT and mutant receptors were stimulated by GLP-1(7–36)-NH2 (upper panels), exendin-4 (middle panels), or oxyntomodulin (lower panels) in CHO–Flp-In cells stably expressing WT or mutant receptors. Data were normalized to the response elicited by the WT and analyzed with an operational model of agonism. All values are means ± S.E. of four to six independent experiments, conducted in duplicate.
Table 2.
Effects of human GLP-1R TM1/N-terminal mutants on agonist-mediated cAMP accumulation
cAMP accumulation data were analyzed using a three-parameter logistic equation to determine pEC50 and Emax values. pEC50 is the negative logarithm of the molar concentration of agonist that induced half the maximal response. Emax for mutants is expressed as a percentage of wildtype. Logτc is the operational efficacy value (determined via the Black and Leff operational model (24)), corrected for cell-surface expression of GLP-1R. All values for cAMP accumulation are mean ± S.E. of four to six independent experiments, conducted in duplicate. One-way ANOVA and Dunnett's post-test were used to determine statistical differences (*, p < 0.05). ND, not determined.
| Agonist-mediated cAMP accumulation |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| GLP-1(7–36)-NH2 |
Exendin-4 |
Oxyntomodulin |
|||||||
| pEC50 | E max | Logτc | pEC50 | E max | Logτc | pEC50 | E max | Logτc | |
| Wildtype | 9.84 ± 0.04 | 97.77 ± 1.13 | 0.77 ± 0.07 | 10.43 ± 0.03 | 99.32 ± 0.90 | 0.81 ± 0.09 | 7.99 ± 0.03 | 98.16 ± 1.13 | 0.73 ± 0.06 |
| R24A | 10.55 ± 0.14* | 77.09 ± 2.92* | 0.18 ± 0.05* | 10.83 ± 0.16 | 74.16 ± 3.10* | 0.13 ± 0.06* | 8.52 ± 0.11 | 81.17 ± 2.99 | 0.25 ± 0.06 |
| P25A | 10.33 ± 0.08 | 97.02 ± 2.21 | 0.67 ± 0.10 | 10.67 ± 0.18 | 97.12 ± 4.65 | 0.67 ± 0.10 | 8.29 ± 0.11 | 102.90 ± 4.26 | 0.86 ± 0.18 |
| Q26A | 10.20 ± 0.09 | 92.56 ± 2.61 | 0.53 ± 0.07 | 10.56 ± 0.08 | 97.69 ± 2.24 | 0.66 ± 0.11 | 8.31 ± 0.10 | 109.70 ± 3.98 | 1.15 ± 0.28 |
| G27A | 9.36 ± 0.49 | 6.91 ± 0.99* | −0.70 ± 0.16* | 10.39 ± 0.57 | 6.62 ± 0.94* | −0.70 ± 0.16* | ND | ND | ND |
| A28G | 10.14 ± 0.10 | 102.20 ± 3.14 | 0.84 ± 0.15 | 10.51 ± 0.12 | 110.00 ± 3.61 | 1.22 ± 0.19 | 8.10 ± 0.07 | 108.20 ± 3.07 | 1.13 ± 0.30 |
| T29A | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| V30A | 9.93 ± 0.16 | 108.20 ± 5.48 | 1.09 ± 0.14 | 10.50 ± 0.10 | 110.50 ± 3.01 | 1.24 ± 0.22 | 8.14 ± 0.14 | 95.92 ± 5.19 | 0.64 ± 0.11 |
| E128A | 9.87 ± 0.39 | 4.59 ± 0.42* | −0.25 ± 0.14* | 10.62 ± 0.72 | 4.63 ± 0.48* | −0.22 ± 0.25 | 8.08 ± 0.53 | 4.15 ± 0.83* | −0.35 ± 0.20* |
| S129A | 9.85 ± 0.10 | 98.92 ± 3.30 | 0.83 ± 0.12 | 10.44 ± 0.09 | 111.50 ± 2.93 | 1.45 ± 0.31* | 8.10 ± 0.11 | 87.57 ± 3.84 | 0.57 ± 0.07 |
| K130A | 9.35 ± 0.09 | 116.20 ± 3.83* | 1.47 ± 0.20* | 10.05 ± 0.18 | 77.74 ± 4.44* | 0.59 ± 0.05 | 7.46 ± 0.17 | 89.78 ± 7.01 | 0.82 ± 0.10 |
| R131A | 10.15 ± 0.21 | 76.34 ± 4.84* | 0.22 ± 0.05* | 10.78 ± 0.15 | 101.40 ± 4.12 | 0.78 ± 0.14 | 8.16 ± 0.26 | 108.70 ± 10.93 | 1.10 ± 0.22 |
| G132A | 9.83 ± 0.16 | 33.04 ± 1.67* | −0.31 ± 0.06* | 10.58 ± 0.25 | 35.90 ± 2.36* | −0.28 ± 0.06* | 8.29 ± 0.12 | 29.04 ± 1.30* | −0.39 ± 0.06* |
| E133A | 9.74 ± 0.17 | 87.76 ± 4.92 | 0.55 ± 0.07 | 10.09 ± 0.16 | 103.40 ± 5.28 | 1.01 ± 0.38 | 7.90 ± 0.11 | 110.50 ± 5.20 | 1.30 ± 0.26* |
| R134A | 9.86 ± 0.12 | 102.80 ± 4.06 | 0.80 ± 0.15 | 10.28 ± 0.17 | 89.67 ± 4.44 | 0.48 ± 0.08 | 7.97 ± 0.10 | 104.30 ± 4.41 | 0.87 ± 0.20 |
| S135A | 9.72 ± 0.15 | 114.00 ± 5.30* | 1.24 ± 0.11* | 9.99 ± 0.09 | 99.42 ± 2.79 | 1.11 ± 0.15 | 7.99 ± 0.11 | 113.40 ± 5.13 | 1.03 ± 0.14 |
| S136A | 9.42 ± 0.23 | 96.37 ± 7.34 | 0.67 ± 0.10 | 10.00 ± 0.10 | 81.16 ± 2.54* | 0.29 ± 0.05 | 7.70 ± 0.10 | 116.20 ± 5.12 | 1.07 ± 0.33 |
| P137A | 9.64 ± 0.12 | 101.30 ± 3.90 | 0.82 ± 0.13 | 10.45 ± 0.29 | 88.74 ± 7.37 | 0.50 ± 0.07 | 7.65 ± 0.20 | 79.11 ± 6.64 | 0.32 ± 0.07 |
| E138A | 9.63 ± 0.15 | 94.25 ± 4.49 | 0.41 ± 0.08 | 10.46 ± 0.14 | 112.30 ± 4.24* | 1.22 ± 0.18 | 7.86 ± 0.16 | 100.40 ± 6.56 | 0.57 ± 0.13 |
| E139A | 10.29 ± 0.19 | 58.81 ± 3.12* | 0.43 ± 0.06 | 10.90 ± 0.51 | 28.03 ± 3.75* | −0.08 ± 0.07* | 8.16 ± 0.31 | 34.23 ± 3.94* | 0.04 ± 0.06* |
| Q140A | 7.91 ± 0.15* | 59.59 ± 5.48* | 0.52 ± 0.08 | 8.68 ± 0.14* | 59.75 ± 3.60* | 0.45 ± 0.07 | 6.32 ± 0.18* | 63.82 ± 9.80 | 0.40 ± 0.07 |
| L141A | 9.68 ± 0.18 | 95.20 ± 5.60 | 0.51 ± 0.09 | 10.43 ± 0.12 | 113.20 ± 3.68* | 1.42 ± 0.23 | 7.22 ± 0.18* | 106.90 ± 10.38 | 0.88 ± 0.18 |
| L142A | 9.48 ± 0.19 | 72.12 ± 4.84* | 0.24 ± 0.08* | 10.79 ± 0.27 | 76.65 ± 5.75* | 0.21 ± 0.05 | 7.41 ± 0.18 | 72.94 ± 5.98 | 0.25 ± 0.08 |
| F143A | 10.18 ± 0.13 | 91.47 ± 3.55 | 0.56 ± 0.11 | 10.78 ± 0.21 | 84.00 ± 4.91* | 0.41 ± 0.10 | 7.96 ± 0.24 | 91.72 ± 9.10 | 0.56 ± 0.10 |
| L144A | 8.03 ± 0.18* | 53.89 ± 5.51* | 0.57 ± 0.07 | 8.90 ± 0.14* | 68.52 ± 3.71* | 0.75 ± 0.09 | ND | ND | ND |
| Y145A | 9.36 ± 0.13 | 116.40 ± 5.51* | 1.44 ± 0.21* | 10.00 ± 0.09 | 109.30 ± 3.28 | 1.25 ± 0.25 | 7.57 ± 0.11 | 103.90 ± 4.88 | 0.98 ± 0.22 |
| I146A | 9.41 ± 0.16 | 108.30 ± 6.35 | 1.43 ± 0.18* | 10.55 ± 0.21 | 87.23 ± 5.09 | 0.78 ± 0.10 | 7.79 ± 0.27 | 77.14 ± 8.62 | 0.59 ± 0.07 |
| I147A | 9.34 ± 0.20 | 80.68 ± 5.54* | 0.32 ± 0.05* | 10.83 ± 0.39 | 69.46 ± 7.47* | 0.14 ± 0.07* | 7.74 ± 0.36 | 46.61 ± 6.97 | 0.19 ± 0.07* |
| Y148A | 8.45 ± 0.12* | 106.20 ± 6.02 | 1.08 ± 0.07 | 9.96 ± 0.29 | 95.57 ± 9.29 | 0.84 ± 0.10 | 5.95 ± 0.50* | 173.10 ± 106.70* | 0.83 ± 0.27 |
Table 3.
Effects of human GLP-1R TM1/N-terminal mutants on agonist-mediated [Ca2+]i mobilization
[Ca2+]i mobilization data were analyzed using a three-parameter logistic equation to determine pEC50 and Emax values. pEC50 is the negative logarithm of the molar concentration of agonist that induced half the maximal response. Emax for mutants is expressed as a percentage of wildtype. Logτc is the operational efficacy value (determined via the Black and Leff operational model (24)), corrected for cell-surface expression of GLP-1R. All values for cAMP accumulation are mean ± S.E. of four to six independent experiments, conducted in duplicate. One-way ANOVA and Dunnett's post-test were used to determine statistical differences (*, p < 0.05). ND, not determined.
| Agonist-mediated intracellular calcium mobilization |
||||||
|---|---|---|---|---|---|---|
| GLP-1(7–36)-NH2 |
Exendin-4 |
|||||
| pEC50 | E max | Logτc | pEC50 | E max | Logτc | |
| Wildtype | 7.83 ± 0.04 | 100.00 ± 1.11 | −0.22 ± 0.03 | 8.45 ± 0.04 | 100.00 ± 1.35 | −0.22 ± 0.02 |
| R24A | 7.60 ± 0.27 | 179.60 ± 20.27* | 0.19 ± 0.10* | 8.25 ± 0.12 | 130.40 ± 5.76* | −0.12 ± 0.05 |
| P25A | 7.89 ± 0.17 | 151.30 ± 11.07* | 0.09 ± 0.09 | 8.07 ± 0.14 | 120.50 ± 6.80* | −0.11 ± 0.05 |
| Q26A | 8.14 ± 0.29 | 146.40 ± 16.50* | 0.02 ± 0.09 | 8.31 ± 0.12 | 104.50 ± 4.87 | −0.24 ± 0.05 |
| G27A | ND | 16.77 ± 4.36* | ND | ND | ND | ND |
| A28G | 7.86 ± 0.14 | 96.66 ± 5.98 | −0.27 ± 0.10 | 8.45 ± 0.19 | 165.90 ± 11.23* | 0.21 ± 0.09 |
| T29A | ND | ND | ND | ND | ND | ND |
| V30A | 7.93 ± 0.24 | 70.75 ± 7.07* | 0.17 ± 0.07 | 8.31 ± 0.24 | 111.50 ± 9.64 | −0.17 ± 0.06 |
| E128A | ND | ND | ND | ND | ND | ND |
| S129A | 7.83 ± 0.12 | 101.30 ± 5.03 | −0.13 ± 0.07 | 8.47 ± 0.19 | 80.73 ± 5.52 | −0.27 ± 0.06 |
| K130A | 7.83 ± 0.18 | 52.81 ± 4.09* | −0.32 ± 0.13 | 7.89 ± 0.17 | 59.33 ± 4.30* | −0.25 ± 0.11 |
| R131A | 7.76 ± 0.25 | 133.80 ± 14.52* | −0.05 ± 0.11 | 8.50 ± 0.24 | 98.00 ± 8.13 | −0.28 ± 0.07 |
| G132A | 7.79 ± 0.20 | 58.45 ± 4.75* | −0.47 ± 0.13 | 8.14 ± 0.23 | 76.96 ± 7.21* | −0.29 ± 0.07 |
| E133A | 7.84 ± 0.18 | 89.51 ± 6.77 | −0.28 ± 0.10 | 8.30 ± 0.21 | 84.37 ± 6.62 | −0.31 ± 0.06 |
| R134A | 8.15 ± 0.32 | 79.99 ± 9.90 | −0.44 ± 0.09 | 8.36 ± 0.19 | 84.59 ± 5.60 | −0.40 ± 0.06 |
| S135A | 7.99 ± 0.20 | 48.35 ± 3.85* | −0.44 ± 0.15 | 8.71 ± 0.22 | 34.19 ± 2.48* | −0.61 ± 0.11* |
| S136A | 7.81 ± 0.18 | 61.68 ± 4.72* | −0.61 ± 0.12* | 8.56 ± 0.16 | 106.70 ± 5.75 | −0.26 ± 0.06 |
| P137A | 7.35 ± 0.25 | 124.00 ± 14.18 | 0.08 ± 0.12 | 8.13 ± 0.10 | 126.00 ± 5.21 | −0.04 ± 0.08 |
| E138A | 7.56 ± 0.13 | 209.20 ± 11.92* | 0.35 ± 0.10* | 8.27 ± 0.20 | 253.40 ± 18.09* | 1.13 ± 0.27* |
| E139A | 7.94 ± 0.20 | 42.50 ± 3.51* | −0.30 ± 0.11 | 7.94 ± 0.21* | 58.47 ± 4.87* | −0.12 ± 0.08 |
| Q140A | 8.89 ± 0.30* | 20.06 ± 2.38* | −0.83 ± 0.14* | 7.49 ± 0.42* | 34.38 ± 4.54* | −0.41 ± 0.17 |
| L141A | 7.49 ± 0.25 | 101.00 ± 11.56 | −0.34 ± 0.11 | 7.79 ± 0.18 | 80.46 ± 6.34 | −0.49 ± 0.09 |
| L142A | 8.74 ± 0.32* | 25.97 ± 2.71* | −0.95 ± 0.17* | 8.77 ± 0.23 | 33.56 ± 2.55* | −0.82 ± 0.11* |
| F143A | 7.65 ± 0.12 | 127.20 ± 7.06 | −0.05 ± 0.11 | 8.48 ± 0.08 | 140.70 ± 3.98* | 0.05 ± 0.05 |
| L144A | ND | ND | ND | ND | ND | ND |
| Y145A | 6.76 ± 0.20* | 108.50 ± 10.35 | −0.12 ± 0.12 | 8.10 ± 0.25 | 94.96 ± 9.30 | −0.19 ± 0.09 |
| I146A | 7.97 ± 0.19 | 60.52 ± 4.60* | −0.23 ± 0.10 | 8.80 ± 0.28 | 52.91 ± 5.09* | −0.30 ± 0.07 |
| I147A | 7.60 ± 0.22 | 44.28 ± 4.42* | −0.73 ± 0.15* | 8.50 ± 0.28 | 66.87 ± 6.33* | −0.51 ± 0.07 |
| Y148A | 7.99 ± 0.25 | 40.42 ± 3.79* | −0.59 ± 0.14 | 8.30 ± 0.21 | 18.91 ± 1.50* | −0.95 ± 0.22* |
Table 4.
Effects of human GLP-1R TM1/N-terminal mutants on agonist-mediated pERK1/2
pERK1/2 phosphorylation data were analyzed using a three-parameter logistic equation to determine pEC50 and Emax values. pEC50 is the negative logarithm of the molar concentration of agonist that induced half the maximal response. Emax for mutants is expressed as a percentage of wildtype. Logτc is the operational efficacy value (determined via the Black and Leff operational model (24)), corrected for cell-surface expression of GLP-1R. All values for cAMP accumulation are mean ± S.E. of four to six independent experiments, conducted in duplicate. One-way ANOVA and Dunnett's post-test were used to determine statistical differences (*, p < 0.05). ND, not determined.
| Agonist-mediated ERK1/2 phosphorylation |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| GLP-1(7–36)-NH2 |
Exendin-4 |
Oxyntomodulin |
|||||||
| pEC50 | E max | Logτc | pEC50 | E max | Logτc | pEC50 | E max | Logτc | |
| Wildtype | 8.07 ± 0.05 | 100.00 ± 1.87 | 0.03 ± 0.02 | 8.77 ± 0.05 | 98.37 ± 1.56 | 0.02 ± 0.02 | 7.53 ± 0.04 | 101.40 ± 1.64 | 0.05 ± 0.03 |
| R24A | 8.21 ± 0.19 | 108.50 ± 7.64 | −0.03 ± 0.06 | 8.83 ± 0.18 | 90.87 ± 5.55 | −0.16 ± 0.07 | 7.82 ± 0.11 | 100.40 ± 4.85 | −0.07 ± 0.07 |
| P25A | 8.51 ± 0.13 | 125.30 ± 5.34 | 0.22 ± 0.06 | 8.95 ± 0.18 | 96.53 ± 5.67 | −0.03 ± 0.06 | 7.73 ± 0.14 | 110.80 ± 6.22 | 0.09 ± 0.07 |
| Q26A | 8.57 ± 0.16 | 157.30 ± 7.93* | 0.47 ± 0.08* | 8.68 ± 0.16 | 137.30 ± 7.13* | 0.31 ± 0.08* | 7.76 ± 0.22 | 126.10 ± 12.12 | 0.49 ± 0.12* |
| G27A | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| A28G | 8.25 ± 0.24 | 131.40 ± 11.84* | 0.30 ± 0.08 | 8.71 ± 0.29 | 142.10 ± 13.91* | 0.42 ± 0.09* | 7.66 ± 0.20 | 110.20 ± 9.56 | 0.11 ± 0.08 |
| T29A | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| V30A | 8.16 ± 0.12 | 120.80 ± 5.95 | 0.20 ± 0.07 | 8.96 ± 0.23 | 132.90 ± 10.41* | 0.31 ± 0.07* | 7.45 ± 0.11 | 107.80 ± 5.45 | 0.07 ± 0.08 |
| E128A | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| S129A | 8.42 ± 0.12 | 104.10 ± 4.55 | 0.15 ± 0.06 | 9.00 ± 0.17 | 87.98 ± 5.16 | 0.01 ± 0.06 | 7.45 ± 0.15 | 86.48 ± 5.89 | −0.004 ± 0.08 |
| K130A | 7.93 ± 0.08 | 151.20 ± 5.17* | 0.85 ± 0.11* | 9.08 ± 0.21 | 151.40 ± 9.84* | 0.83 ± 0.09* | 7.51 ± 0.12 | 122.50 ± 6.66 | 0.55 ± 0.09* |
| R131A | 7.93 ± 0.16 | 109.70 ± 7.47 | 0.06 ± 0.07 | 8.88 ± 0.13 | 96.88 ± 4.49 | −0.05 ± 0.06 | 7.50 ± 0.16 | 104.10 ± 7.36 | 0.005 ± 0.08 |
| G132A | 8.19 ± 0.17 | 57.61 ± 4.22* | −0.26 ± 0.08 | 8.93 ± 0.29 | 46.60 ± 5.30* | −0.38 ± 0.09* | 7.45 ± 0.24 | 35.06 ± 4.55* | −0.51 ± 0.15* |
| E133A | 8.37 ± 0.13 | 101.80 ± 5.00 | 0.07 ± 0.06 | 9.08 ± 0.15 | 102.90 ± 5.02 | 0.07 ± 0.06 | 7.76 ± 0.17 | 72.90 ± 5.38 | −0.19 ± 0.08 |
| R134A | 8.09 ± 0.13 | 122.30 ± 6.33 | 0.15 ± 0.07 | 9.36 ± 0.21 | 116.90 ± 7.21 | 0.11 ± 0.06 | 7.63 ± 0.16 | 79.86 ± 5.78 | −0.20 ± 0.08 |
| S135A | 8.06 ± 0.18 | 88.91 ± 6.40 | 0.15 ± 0.07 | 8.90 ± 0.19 | 86.39 ± 5.43 | 0.13 ± 0.06 | 7.57 ± 0.14 | 48.80 ± 3.35* | −0.22 ± 0.10 |
| S136A | 7.81 ± 0.16 | 45.74 ± 3.36* | −0.58 ± 0.10* | 8.26 ± 0.16 | 35.37 ± 2.32* | −0.72 ± 0.13* | 6.73 ± 0.25* | 32.54 ± 6.85* | −0.73 ± 0.16* |
| P137A | 7.64 ± 0.15 | 102.70 ± 6.67 | 0.05 ± 0.07 | 7.80 ± 0.25* | 114.50 ± 11.67 | 0.14 ± 0.08 | 7.17 ± 0.16 | 112.30 ± 9.51 | 0.13 ± 0.09 |
| E138A | 8.29 ± 0.21 | 214.30 ± 16.04* | 1.07 ± 0.24* | 8.82 ± 0.13 | 214.00 ± 9.04* | 0.71 ± 0.13* | 7.30 ± 0.16 | 258.30 ± 20.51* | 0.88 ± 0.18* |
| E139A | 7.95 ± 0.20 | 52.17 ± 4.96* | 0.02 ± 0.08 | 7.98 ± 0.24* | 48.99 ± 5.12* | −0.02 ± 0.11 | 7.90 ± 0.18 | 51.16 ± 4.23* | 0.01 ± 0.08 |
| Q140A | 7.02 ± 0.21* | 57.35 ± 7.44* | 0.08 ± 0.11 | 7.70 ± 0.31* | 38.88 ± 5.68* | −0.15 ± 0.12 | 6.78 ± 0.26* | 27.40 ± 5.10* | −0.39 ± 0.19* |
| L141A | 7.71 ± 0.11 | 137.10 ± 6.84* | 0.26 ± 0.09 | 8.52 ± 0.13 | 158.10 ± 6.82* | 0.50 ± 0.11* | 7.30 ± 0.18 | 126.10 ± 11.22 | 0.13 ± 0.10 |
| L142A | 8.00 ± 0.16 | 106.40 ± 6.71 | 0.12 ± 0.06 | 8.05 ± 0.17* | 94.20 ± 6.46 | 0.00 ± 0.08 | 7.03 ± 0.19 | 97.49 ± 11.29 | 0.06 ± 0.10 |
| F143A | 8.22 ± 0.16 | 116.70 ± 6.85 | 0.17 ± 0.06 | 8.24 ± 0.28 | 86.81 ± 9.12 | −0.10 ± 0.07 | 7.50 ± 0.22 | 104.80 ± 10.36 | 0.06 ± 0.08 |
| L144A | ND | ND | ND | 7.57 ± 0.36* | 29.28 ± 5.54* | −0.08 ± 0.21 | ND | ND | ND |
| Y145A | 7.78 ± 0.41 | 76.36 ± 13.92 | −0.12 ± 0.06 | 8.13 ± 0.28* | 99.84 ± 11.07 | 0.08 ± 0.06 | 7.32 ± 0.46 | 106.00 ± 24.78 | 0.15 ± 0.09 |
| I146A | 7.14 ± 0.34* | 68.81 ± 13.41* | 0.07 ± 0.13 | 8.30 ± 0.22 | 76.61 ± 6.48 | 0.13 ± 0.08 | 7.38 ± 0.18 | 89.57 ± 8.13 | 0.26 ± 0.08 |
| I147A | 7.77 ± 0.16 | 144.90 ± 9.75* | 0.45 ± 0.11* | 8.21 ± 0.12 | 140.80 ± 6.46* | 0.38 ± 0.09* | 7.58 ± 0.17 | 149.10 ± 11.18* | 0.51 ± 0.11* |
| Y148A | 7.11 ± 0.21* | 93.43 ± 11.28 | 0.16 ± 0.11 | 8.01 ± 0.20* | 105.60 ± 8.68 | 0.25 ± 0.07 | 7.12 ± 0.17 | 78.45 ± 7.84 | 0.03 ± 0.10 |
cAMP accumulation
Within the far N terminus of the ECD, mutants G27A and T29A that had poor expression displayed very weak to no measurable cAMP response. Unlike binding affinity, which was unaltered, the R24A mutant had reduced efficacy for GLP-1 and exendin-4 (Figs. 3B and 4 and Table 2). Within the TM1/ECD linker there was only limited correlation between effects on binding affinity and cAMP efficacy and on cAMP efficacy between peptides. E128A that was poorly expressed responded very weakly to all peptides. The I147A mutant induced modest loss of efficacy, and the G132A mutant exhibited 10–30-fold decreased efficacy for all peptides. The L142A mutant had a significantly attenuated efficacy for GLP-1, with similar fold decreases for exendin-4 and oxyntomodulin, although these latter effects did not achieve significance. The E139A mutant that had undetectable 125I–exendin-4(9–39) binding had reduced efficacy with exendin-4 and to a lesser extent oxyntomodulin but not GLP-1. There were statistically significant increases in efficacy for oxyntomodulin at E133A, whereas K130A, S135A, Y145A, and I146A caused selective increases in GLP-1 efficacy, and R131A induced a weak and selective decrease in GLP-1 efficacy (Figs. 3B and 4 and Table 2).
iCa2+ mobilization
Because of relatively weak [Ca2+]i mobilization by oxyntomodulin, only a single high concentration (∼3 μm) was assessed. For GLP-1 and exendin-4, no measurable response was seen at the poorly expressed G27A, T29A, and E128A mutants, whereas the E138A mutant exhibited increased efficacy for both peptides, even after correction for the higher cell-surface expression (Figs. 3C and 5 and Table 3). There was decreased efficacy for both peptides with the L142A mutant, whereas L144A abolished [Ca2+]i mobilization, despite unaltered cAMP efficacy. There was selective loss of exendin-4 efficacy at the S135A and Y148A mutants and of GLP-1 efficacy at the S136A, Q140A, and I147A mutants, although these effects were relatively small (Figs. 3C and 5 and Table 3). There was also a weak and selective increase in GLP-1 efficacy at the R24A mutant, whereas the opposite effect was seen for GLP-1–dependent cAMP efficacy (Fig. 3, C versus B, and Tables 2 and 3). Of interest, although not quantitative, there was no measurable response for the single high concentration of oxyntomodulin at the P137A and Y148A mutants, despite approximately WT levels of cell-surface receptor expression and limited (for P137A) or no (Y148A) effect of the mutation on oxyntomodulin affinity (Fig. 3C and Table 1).
ERK1/2 phosphorylation
As seen with the other pathways, the poorly expressed mutants G27A, T29A, and E128A had no measurable pERK response to any of the peptides. For the far N-terminal ECD mutations, there was a slightly increased efficacy for all peptides with the Q26A mutant and a selective weak increase in efficacy for exendin-4 at the A28G and V30A mutants (Figs. 3D and 6 and Table 4). For the TM1/ECD linker, there was increased efficacy for all three peptides at the K130A, E138A and I147A mutants and decreased efficacy at the S136A mutant. A weak loss of efficacy for exendin-4 and oxyntomodulin, but not GLP-1, occurred with the G132A mutant, whereas the L144A mutant abolished pERK response to GLP-1 and oxyntomodulin but did not alter efficacy for exendin-4. Of the remaining mutants, only L141A (increased exendin-4 efficacy) and Q140A (weak loss of oxyntomodulin efficacy) had any significant effect (Figs. 3D and 6 and Table 4).
Discussion
Structural insights into GLP-1R biased agonism
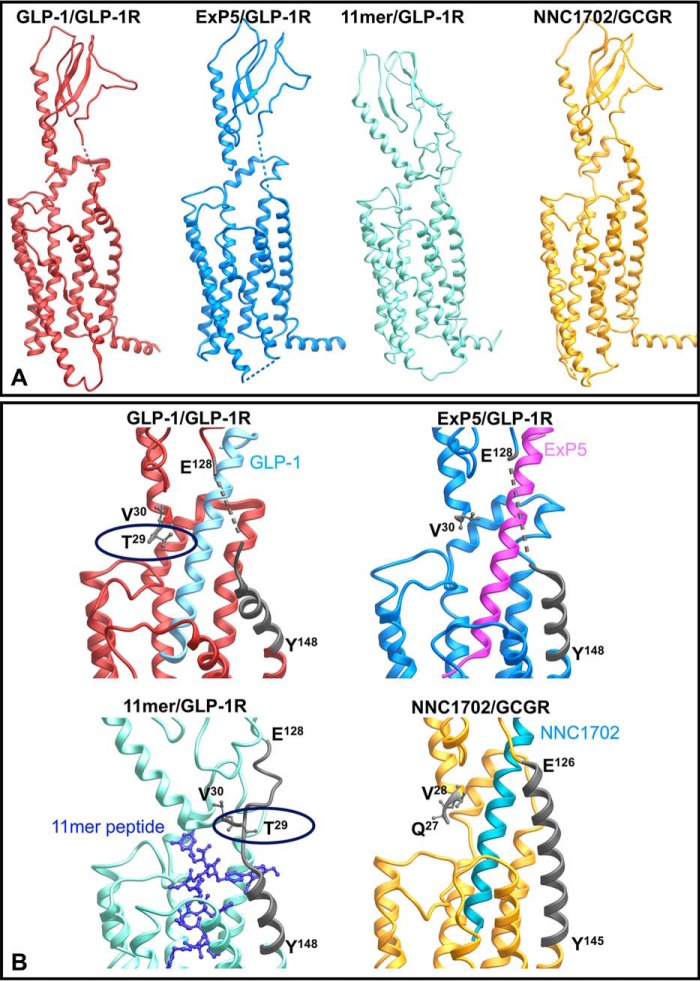
Our results indicate that the far N terminus and the linker region between TM1 and the ECD play discrete roles in receptor stability and expression and in peptide-specific signaling. Recent advances in structural determination for class B GPCRs and particularly the GLP-1R provide an unprecedented opportunity to map key surface residues in three-dimensional space relevant to both inactive and active structures. For the GLP-1R, four new structures have recently been solved, which include a modified human GLP-1R TM domain structure bound to negative allosteric modulators (NAMs) (14), a thermostabilized full-length human GLP-1R bound to a modified 11-mer agonist peptide (15), a fully active rabbit GLP-1R in complex with GLP-1 and the heterotrimeric Gs protein (16), and human GLP-1R in complex with the biased agonist, exendin-P5, and the Gs protein (17). Details of structures, including differences from WT human GLP-1R, are described in Table 5. This work complements the previously published inactive structures of the related GCGR (29, 30). The NAM-bound GLP-1R contains structural alterations, including an introduced cysteine bridge between TM helices 5 and 6, that disrupt key networks of the native inactive receptor. As such, we have used the inactive GCGR (PDB code 4L6R) (29) as a template to model the inactive GLP-1R (Fig. 1A) (28). The active rabbit GLP-1R complex has a global resolution of 4.1 Å, with limited side-chain resolution and ambiguity in potential modeling of ECLs, whereas the exendin-P5–GLP-1R complex has a global resolution of 3.3 Å with good side-chain resolution for most of the receptor; we have used this structure as the principal template for comparative mapping of the effects of mutations between active and inactive states (Fig. 1A). All full-length structures lack density for residues 24–28 of the far N terminus, indicating that this segment is flexible upon ligand binding. However, the position of Val-30 that is resolved in the structures suggests that the far N-terminal residues likely make transient interactions with ECLs 2 and 3 and/or the peptide agonist (Fig. 7). The new data are considered holistically consistent with previously published ECL alanine-scanning mutagenesis studies (11, 13) to yield a comprehensive structure-function analysis of signal propagation networks in the GLP-1R. Nonetheless, as with all such structure–function analyses, our observations are influenced by the recombinant cellular environment, including the relative expression of transducer and regulatory proteins that will differ from endogenous sites of receptor expression.
Table 5.
Sequence variations in published structures
| PDB code | Description | Structure fragment | Mutations/differences from hGLP-1R | Missing residues in the structure |
|---|---|---|---|---|
| 5VEW (Song et al. 14) | Crystal structure of human GLP-1R transmembrane domain in complex with negative allosteric modulator PF-06372222 | Residues 128–431. Residues 258–260 at intracellular loop 2 were replaced with T4 lysozyme. Residues 205–214 from ECL1 were replaced by a GSG linker | S193C, I196F, Y205G, ΔThr-207, ΔAla-208, ΔAla-209, ΔGln-210, ΔGln-211, ΔHis-212, ΔGln-213, W214G, S225A, M233X, S271A, I317C, G318I, K346A, C347F, G361C | Glu-128, Ser-129, Lys-130, Arg-131, Gly-132, Glu-133, Arg-134, Ser-135, Met-204, Asp-215, Glu-373, His-374, Ala-375, Arg-376, Gly-377, Thr-378, Leu-379, Glu-423, His-424, Leu-425, His-426, Ile-427, Gln-428, Arg-429, Asp-430, Ser-431 |
| 5NX2 (Jazayeri et al. 15) | Crystal structure of human GLP-1 receptor bound to the 11-mer agonist peptide 5 | Residues 24–432 | T207E, Q211A, D215R, L232F, L260F, G295A, T298A, C329A, P358A, G361A, H363V, V405A | Arg-24, Pro-25, Gln-26, Gly-27, Ala-28, Glu-418, Arg-419, Trp-420, Arg-421, Leu-422, Glu-423, His-424, Leu-425, His-426, Ile-427, Gln-428, Arg-429, Asp-430, Ser-431, Ser-432 |
| 5VAI (Zhang et al. 16) | Cryo-EM structure of active rabbit GLP-1 receptor in complex with GLP-1 and Gs protein | Residues 24–422 | T106A, H112P, Q140R | Arg-24, Pro-25, Gln-26, Gly-27, Ala-28, Ser-129, Arg-130, Arg-131, Gly-132, Glu-133, Ser-134, Leu-422 |
| 6B3J (Liang et al. 17) | Cryo-EM structure of active human GLP-1 receptor in complex with exendin-P5 and Gs protein | Residues 23–466 | None | Ala–23–Thr–29, Ser–129–Ser–136, Asn–338–Thr–342, His–424–Gly–466 |
Figure 7.
Peptide-bound full-length structures of GLP-1R and GCGR. A, full-length structures illustrating the relative position of the N-terminal ECD to the receptor core. B, zoom-in of the resolved far N-terminal residue(s) and TM1/ECD stalk (highlighted in dark gray). The backbones of the peptide agonists are illustrated in ribbon (GLP-1, exendin-P5 (ExP5), and NNC1702) or X-stick (11-mer).
Structural reorganization upon receptor activation
ECD–receptor core interactions
There is accumulating evidence that interactions between the ECD and TM core of class B GPCRs contribute to receptor quiescence and peptide-mediated receptor activation (20, 21, 31). For the related GCGR, interactions of the far N terminus and the residues in ECL3 contribute to maintenance of a quiescent state (20), although ground state interactions at the intracellular face also play a key role (22, 32, 33). As noted above, the far N terminus is dynamic in peptide-bound states (15, 16), making interpretation of mutations in the context of available structures difficult. The dramatic loss of cell-surface expression for G27A and T29A is indicative of the important roles of these amino acids in receptor stability, potentially via loss of stabilizing interaction between Thr-29 and the receptor core that would be consistent with a role of the far N terminus in maintaining receptor quiescence. Nevertheless, the GLP-1R is expressed at the cell surface when the full ECD is truncated (31), suggesting that loss of receptor expression is due to destabilizing interactions of the modified ECD. Glycines provide structural flexibility, and Gly-27 may be required for favorable positioning of Thr-29. In the active GLP-1- and exendin-P5–bound receptors, Thr-29 is proximal to the peptide ligand, although not within hydrogen-bonding distance. In the structure of the modified 11-mer bound receptor, the position of the ECD is not constrained by peptide binding, where Thr-29 interacts with the linker between TM1 and the ECD (Fig. 7B).
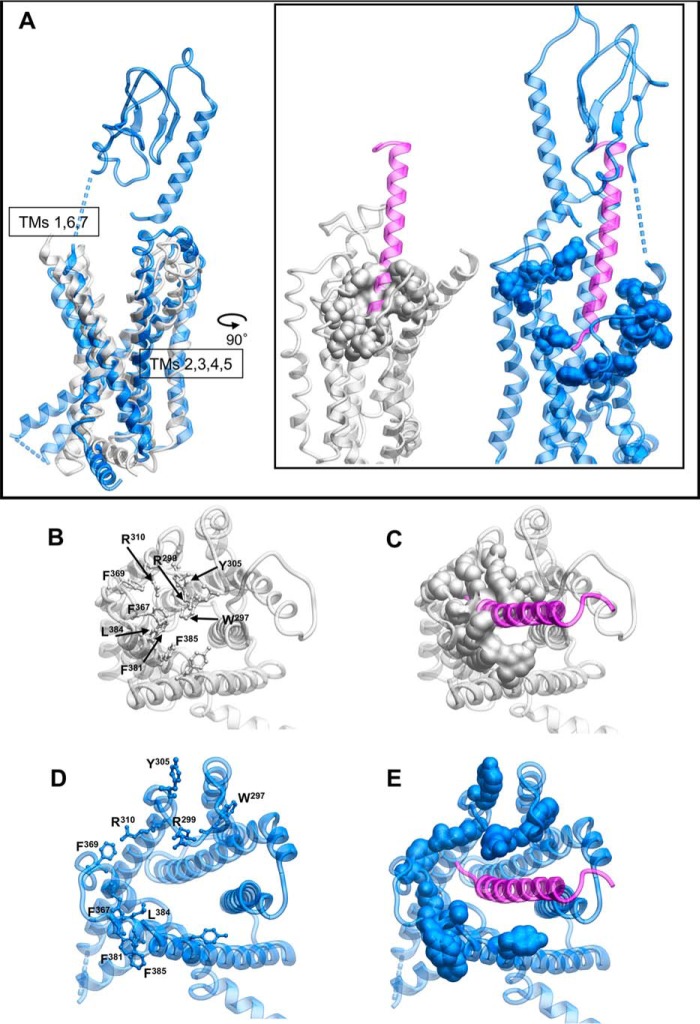
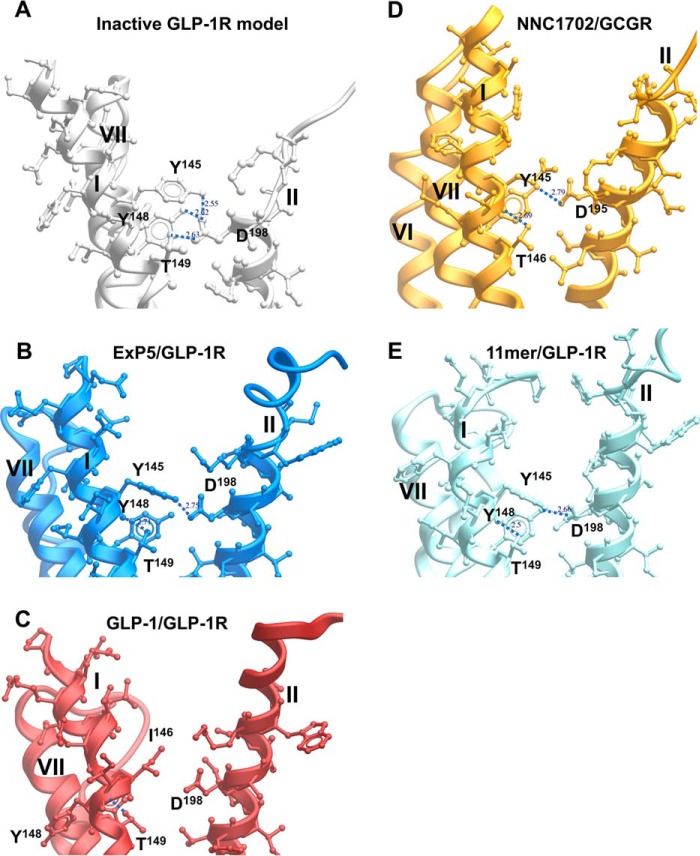
Class B GPCRs, including the GLP-1R, exhibit an extracellular-oriented V-shape cavity within the TM bundle that provides domain separation of the external facing segments of TMs 1, 7, and 6, and TMs 2–5 (Fig. 8A). Nonetheless, in the inactive model, there are key hydrogen-bonded interactions between Asp-198 of TM2 and Tyr-145, Tyr-148, and Thr-149 of TM1 that coordinate Tyr-145 and Tyr-148 away from TM7 (Fig. 9A), and this facilitates tight packing of TMs 1 and 7. Consistent with this, D198A mutation leads to marked loss of receptor expression and/or GLP-1-stimulated cAMP signaling (34–36). In the active, exendin-P5–bound structure, this hydrogen bond network is weakened, with loss of interactions between Asp-198 and both Tyr-148 and Thr-149, facilitating the movement of TM1 toward TM7, whereas the kink in TM1 is stabilized by hydrogen bonding of the side chain of Thr-149 with the backbone oxygen of Tyr-145 (Fig. 9B). A similar pattern of interaction is observed in the 11-mer bound structure (Fig. 9E). Although the position of the kink in TM1 of the GLP-1/GLP-1R structure is likely conserved (17) (although modeled differently in 5VAI), there is further loss of the interaction between Tyr-145 and Asp-198, and this leads to an anti-clockwise rotation of the upper end of TM1, with a parallel rotation of TM7 that is not seen in the exendin-P5 structure (Fig. 9, B and C). This likely contributes to the distinct conformation of ECL3 between the GLP-1– and exendin-P5–bound structures that has been linked to efficacy differences of the two peptides (17).
Figure 8.
Peptide binding to the GLP-1R requires reorganization of aromatic/hydrophobic residues in the receptor core. A, side view of the inactive GLP-1R model (gray) or exendin-P5-bound GLP-1R structure (blue) with the exendin-P5 peptide illustrated in purple. Residues that occupy the core of the receptor in the inactive model are shown as space fill. B–E, top view of the structures where the ECD has been omitted for clarity. B and C, inactive GLP-1R model. D and E, exendin-P5-bound GLP-1R structure.
Figure 9.
Polar residues in the GLP-1R TM1 are coordinated by Asp-198 (TM2) in the inactive model, orienting key side chains away from TM7 and facilitating tight packing of TM1/TM7. View facing the TM1/TM2 boundary is shown. Key TM1 side chains interacting with Asp-198 are depicted in X-stick representation and labeled. A, inactive GLP-1R model. B, exendin-P5 (ExP5)/GLP-1R structure. C, GLP-1/GLP-1R structure. D, NNC1702/GCGR structure. E, 11-mer agonist/GLP-1R structure. TM helices are labeled in roman numerals.
Both Tyr-148 and Thr-149 (37, 38) play important roles in peptide agonist affinity but do not make direct interactions with GLP-1 in the active structure, and the same is true for P137A and L141A in the TM1 stalk. Thus, these residues contribute to the reorganization of TM1 and packing with TM7. These changes are also likely to impact the position of Glu-139. Mutation of this amino acid causes loss of exendin-4(9–39) binding. In the GLP-1–bound active structure, the side chain of Glu-139 is directed toward the GLP-1 peptide, suggesting a direct interaction that may be more prevalent for the truncated exendin peptide (and indeed, this is observed in the exendin-P5/GLP-1R/Gs structure (17)). This would be consistent with the lack of effect of the E139A mutant on GLP-1 signaling, and only limited attenuation of oxyntomodulin and exendin-4–mediated cAMP production (Fig. 3). Of the TM1 stalk residues, only mutations to Thr-149 also translate into a major impact on signaling (37, 38), implying that its role in structural reorganization is also critical for activation transition and effector binding. This loss can be recovered by allosteric modulator binding at the intracellular face of the receptor that is predicted to destabilize ground state interactions at the base of the receptor (14, 37, 38).
In the active and Gs protein-complexed receptor structures, the linker region between the ECD and the core is poorly resolved, suggesting a high degree of flexibility even when the peptide is bound. In the inactive GCGR (PDB 4L6R) (29), the TM1 stalk is present as an extended α-helix, and although this may be partially due to crystal-packing artifacts, an extended α-helix is also present in the structure of the full-length GCGR bound to a partial agonist peptide, NNC1702 (Fig. 7) (18). This suggests that order to disorder transition of the TM1 stalk may be required for full receptor activation. Indeed, this would be required to accommodate the movement of TM1 toward TM7, seen in the active and active-like structures. An important role for the TM1 stalk is supported by our current mutagenesis data (Fig. 3). Polar residues in this region, particularly Ser-136, Lys-130, and Glu-138, had effects on peptide signaling in a pathway- and peptide-specific manner, indicating that formation and disruption of interactions formed by these amino acids contribute to conformational transition for activation. Similar behavior was seen for the G132A mutant, suggesting that backbone flexibility plays a role in these effects. Somewhat surprisingly, E128A had a profound effect on receptor expression, presumably via destabilization of the receptor protein, in a similar manner to that observed for the T29A mutant. Although speculative, it is possible that these amino acids are in close proximity in the inactive receptor and form part of an important network that stabilizes this state. In studies where E128A is further overexpressed by transient expression, there is a <3-fold loss of GLP-1 potency and an ∼5-fold loss of exendin-4 potency for cAMP production (39), consistent with a limited role of this residue for peptide binding.
In the inactive homology model, the deeper binding cavity occupied by agonist peptides is capped by a series of large aromatic residues, particularly Trp-297 and Tyr-305 (Fig. 8, B and C versus D and E), whose position is predicted to be stabilized by H-bonding. In the active structure, these amino acids undergo large-scale movements associated with reorganization of ECL2 that reorients Trp-297 away from the binding pocket, where it forms interactions with other ECL2 residues, accompanied by small outward movements of Arg-299 and Asn-300 that directly contact the peptide in the active structure. Release of ground state interactions in the inactive ECL2 loop structure enables unwinding of the top of TM6 with an ∼180° rotation of Tyr-305 accompanied by an ∼9-Å displacement of the Cα carbon. Previous molecular dynamics simulations on the inactive GLP-1R model suggested that this aromatic cap provided a significant energy barrier to deeper entry of the GLP-1 N terminus, with entry facilitated by Glu-9 of the peptide that forms a salt bridge with Arg-190 of the receptor (11). Intriguingly, mutation of Trp-297 and the adjacent Cys-296 (that is covalently linked to Cys-226 in TM3) markedly attenuated GLP-1 and exendin-4 affinity, but it did not impact oxyntomodulin affinity. Oxyntomodulin contains an uncharged Gln, positionally equivalent to GLP-1 Glu-9, and does not interact with Arg-190 in the receptor core (11). It is therefore possible that oxyntomodulin binds in a shallower orientation. A significant and selective decrease in oxyntomodulin affinity for F381A, L142A, and K202A mutants and lack of an effect for the R380A mutant are consistent with this hypothesis (Fig. 10).
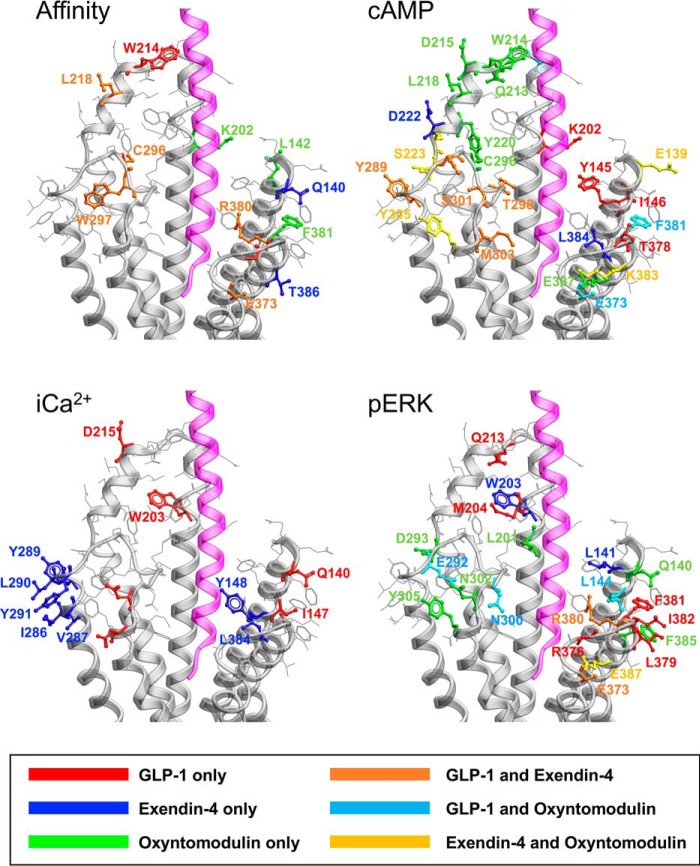
Figure 10.
Peptide-selective effects on agonist affinity, cAMP accumulation, [Ca2+]i mobilization, and pERK. Mutated amino acids with similar effects across peptides are not highlighted. Maps for affinity, cAMP, and pERK include all three peptides. The map for [Ca2+]i includes only GLP-1 and exendin-4. The exendin-P5 peptide is displayed in magenta.
Peptide-mediated signaling and bias
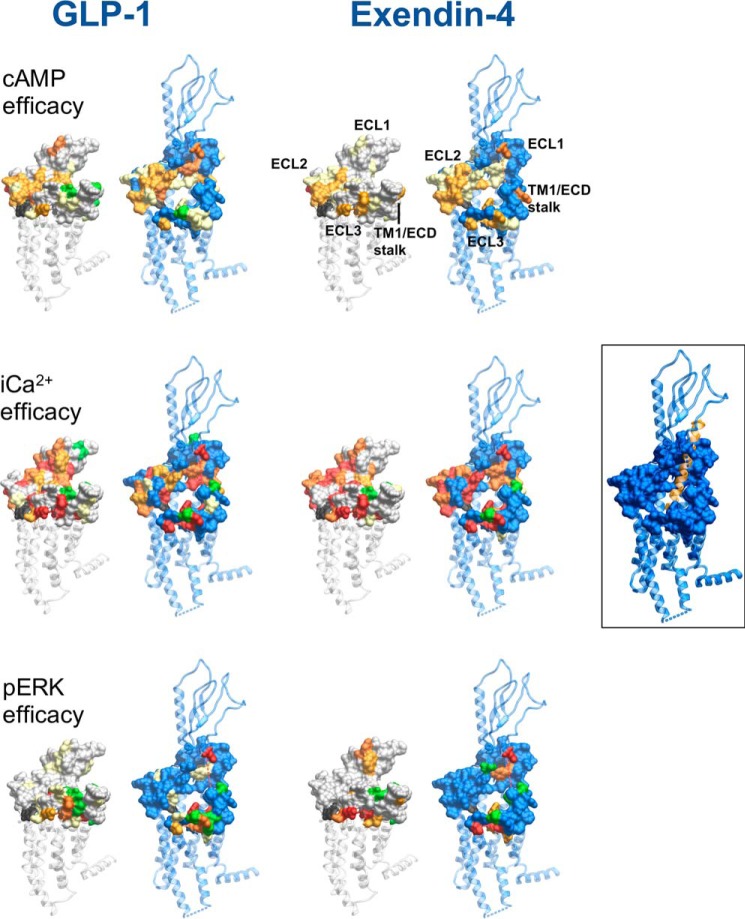
Comparison of the position of mutated residues that affect GLP-1–mediated cAMP formation between fully active and inactive models reveals two major networks involved in GLP-1R function (Figs. 10 and 11). The first includes residues of ECL2 and the membrane-proximal TM regions and the proximal segment of ECL3. K288A impacted ligand binding and was critical for propagation of cAMP signaling (13, 24, 40). It stabilizes the center of the ECL2 network and may coordinate interactions between the ECL2 residues. Both exendin-4 and oxyntomodulin were similarly affected by mutations to ECL2 indicating a general role in propagation of cAMP signaling (Figs. 10 and 11). Despite the lack of effect of W297A on oxyntomodulin binding, it was crucial for signaling of all three peptides. In the active agonist-bound calcitonin receptor, there is a high degree of overlap in the structural organization of ECL2 despite considerable sequence variation; Arg-281 that is positionally equivalent to Lys-288 of the GLP-1R appears to play a similar coordinating role in maintenance of this structure (19, 25). The organization of the ECL2 network is also required for calcium signaling, although it does not appear as important for peptide-mediated pERK (Figs. 10 and 11).
Figure 11.
Reorganization of the extracellular surface of the GLP-1R is critical to propagation of signaling. Amino acids involved in efficacy across cAMP (upper panels), [Ca2+]i (middle panels), and pERK (lower panels) for GLP-1 (left-hand panels) and exendin-4 (right-hand panels) were mapped onto the inactive GLP-1R model and the fully active exendin-P5 (ExP5)-bound GLP-1R structure. Displayed in surface representation are mutated amino acids that affect efficacy: yellow (2–5-fold reduction in affinity); light orange (5–10-fold reduction in affinity); red (>30-fold reduction in affinity); or green (increased affinity). Mutated residues not affected are displayed as gray (inactive receptor) or blue (active receptor).
The second network involves residues in TM1 and the TM7 proximal residues of ECL3 (Figs. 10 and 11). This network exhibits a higher degree of peptide-specific effects that are likely related to both differences in the peptide sequences and orientation of the peptides in the active structures. As noted above, this region of the receptor is involved in coordination of TM1 in the inactive structure and the reorganization and packing of TM1 with TM7 in the active structures (Fig. 9). Unlike GLP-1 and exendin-4, oxyntomodulin-mediated cAMP production is also weakly attenuated by mutation to amino acids in ECL1 that sit within the short α-helix formed in the active structure that extends to the ECL2 network. This is likely due to the predicted distinct positioning of oxyntomodulin when bound to the receptor (Fig. 10). The boundary of ECL1 at the top of TM3 is covalently linked to ECL2 by disulfide linkage of Cys-226 (TM3) and Cys-296 (ECL2), whereas Arg-227 in inactive/partially active structures may also stabilize ECL2. R227A mutation decreases affinity of all three peptides, but with minimal impact on signaling efficacy (32). As such, the extent to which the oxyntomodulin-specific effects are due to unique direct interactions with ECL1 versus potential differences in Cys-296 and Trp-297 is not clear.
Overall, the pattern of effect of mutation was similar for calcium and cAMP signaling across GLP-1 and exendin-4 where efficacy effects could be quantified, although there was generally a greater magnitude of effect on calcium signaling (Figs. 10 and 11). Previous pharmacological inhibitor studies revealed that both these pathways were regulated by G protein interaction at the WT receptor in the CHO–Flp-In cell background, although Gi and Gq interactions had more prominent roles in calcium mobilization (11); this is indicative of broad similarities in changes required to enable G protein coupling. Exendin-4–mediated signaling is also generally more sensitive to mutation than that mediated by GLP-1, and the required ECL2 network extends to the top of TM4 for this peptide (Fig. 10).
Peptide-mediated pERK was least sensitive to GLP-1R surface mutations, being principally confined to mutations of the distal ECL3/TM7 boundary, and residues of TM1 and the linker extension that provides physical connection to the ECD, which were revealed in this study (Figs. 10 and 11). This was particularly evident for exendin-4, which was least affected by mutation (Fig. 11). Intriguingly, there was effectively no involvement of the ECL2 residues that were absolutely required for cAMP and calcium signaling. Inhibitor studies at the WT receptor indicated that Gs and Gq have limited contribution to exendin-4–mediated pERK, with signaling principally driven by Gi, Gβγ, and arrestin interactions, although those are likely to be at least partly interdependent (11). For GLP-1, and more prominently oxyntomodulin, selective mutations in ECL2 also impacted pERK, and this could relate to greater contribution of Gq (GLP-1) and Gs (oxyntomodulin) in pERK at the WT receptor. Also of interest, for TM1 and ECL3/TM7, the effects of individual mutation were highly peptide-specific (Fig. 10). The data are consistent with a model whereby selective and peptide-specific interactions alter the TM1/TM7 interface linked to Gi/Gβγ/arrestin coupling to pERK. Moreover, our mutational data suggest these changes can occur relatively independent of the reorganization of TMs 5 and 6 that are critical for cAMP formation and [Ca2+]i mobilization, events that are Gs/Gq-dependent at the WT receptor. Exendin-P5 is a G protein-biased peptide agonist that exhibits bias toward cAMP relative to arrestin interaction, compared with the related exendin-4 peptide and GLP-1 (10, 17). It has distinct actions in vivo compared with exendin-4 (10). As noted above, comparison of the exendin-P5– and GLP-1–bound active GLP-1R structures reveals major differences in ECL3 and the upper TM boundaries of TMs 6 and 7 that are linked to distinct rotational differences in the upper region of TM1. Mutation in these domains has confirmed peptide-specific differences in the engagement of GLP-1 versus exendin-P5 with amino acids in TM1, as well as the TM6 proximal region of ECL3 (17), thereby providing additional structural evidence for distinctions in the role of these domains in propagation of signaling. An important caveat for extrapolation of our observations to more proximal measures of signaling is that they are based on inferences from WT receptor signaling. Direct measurement of proximal transducer engagement for mutant receptors will be required to validate hypotheses.
The novel structures for the GLP-1R are enabling us to begin to unravel the complexities associated with receptor activation and biased agonism. Combining new data from this study with our previous work on GLP-1R ECLs in the context of inactive and active structures has advanced our understanding of receptor domains that control signaling. Importantly, the work provides evidence for two, at least partially independent, structural domains linked to signaling. The first involves the interface between TMs 5 and 6 and is linked to reorganization of ECL2 into a structured network that is required for propagation of signaling linked to Gs and Gq-dependent pathways at the WT receptor. The second is the interface between TMs 1 and 7 that, although important for at least Gs-dependent cAMP signaling, may be independently linked to Gi/Gβγ/arrestin-mediated signaling that is the key driver of pERK at the WT receptor. Our data support a model where distinct peptide–receptor interactions can provide selective control of how these different networks are engaged.
Experimental procedures
Mutagenesis
Desired mutations were introduced to the N-terminal double c-Myc–labeled human GLP-1R gene in pDONR201 (Invitrogen) via the Muta-directTM kit (Beijing SBS Genetech Co., Ltd., China), and then LR recombination reactions were conducted to transfer the N-terminal double c-Myc–labeled human GLP-1R gene into the pEF5/FRT/V5-DEST destination vector using Gateway Technology (Invitrogen). The oligonucleotides for mutagenesis were purchased from GeneWorks (Thebarton, SA, Australia), and mutants were confirmed by automated DNA sequencing.
Stable cell line generation and cell culture
The mutant or wildtype (WT) receptor genes were integrated into the Flp-In-Chinese hamster ovary (Flp-In-CHO) cells, passage 4 (Invitrogen), using the Flp-InTM system. Stable Flp-In expression cell lines were generated through polyclonal selection, screened, and maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FBS, 600 μg/ml hygromycin B at 37 °C in 5% CO2. The WT receptor is expressed at ∼170,000 receptors/cell. Cell lines were routinely tested for mycoplasma and were mycoplasma free. Stable cells were frozen at passage 14, and all assays were performed with cells between passage 14 and 25.
Heterologous whole-cell competitive binding assay
Competition of 125I–exendin-4(9–39) binding to hGLP-1R was performed as described previously (13) on whole cells in 96-well plates using the radiolabeled antagonist 125I–exendin-4(9–39) (∼0.1 nm) and increasing concentrations of unlabeled peptide. Nonspecific binding was defined by co-incubation with 1 μm unlabeled exendin-4(9–39). Following overnight incubation, nonbound ligand was removed, and radioactivity was determined using a gamma counter.
Cell-surface expression by enzyme-linked immunosorbent assay
1.5 × 105 cells/well were seeded into 24-well culture plates and incubated overnight. Expression was determined through detection of N-terminal double c-Myc of GLP-1R by ELISA as described previously (13). Data were normalized to WT GLP-1R and Flp-In CHO parental cell lines.
Calcium mobilization
3 × 104 cells/well were seeded into 96-well culture plates and incubated overnight. Cells were incubated with Fluo4-AM for 45 min and stimulated with different concentrations of peptides, and fluorescence was determined in a FlexStation® plate reader every 1.36 s for 120 s after ligand addition as described previously (13). Data were normalized to the maximal response elicited by 100 μm ATP.
cAMP accumulation
3 × 104 cells/well were seeded into 96-well culture plates and incubated overnight. Cells were stimulated with increasing concentrations of ligands for 30 min in the presence of isobutylmethylxanthine. The liquid was discarded, changed to absolute ethanol, and volatilized to dry in room temperature. cAMP was detected using a Lance kit (PerkinElmer Life Sciences), as described previously (11). Data were normalized to the response of 100 μm forskolin.
ERK1/2 phosphorylation
3 × 104 cells/well were seeded into 96-well culture plates and incubated overnight. Initially, pERK1/2 time-course experiments were performed over 1 h to identify the time point when the pERK1/2 response is maximal. Subsequently, dose responses for different agonists were determined at this peak time point with stimulation performed after serum starvation overnight. pERK1/2 was detected using an AlphaScreen assay as described previously (13). Data were normalized to the maximal response elicited by 10% FBS determined at 6 min.
Data analysis
IC50 values were estimated from competitive inhibition of 125I-exendin(9–39) binding using a three-parameter logistic equation (log(inhibitor versus response)) in Prism (version 7, Graphpad). In all cases, the concentration of the radioligand was ≤1% of the Kd values. Under these conditions, the IC50 approximates Ki, and such data are reported as pKi. Emax and EC50 were estimated from concentration-response curves using with a three-parameter logistic equation in Prism (version 7). These values are a composite of affinity, efficacy, and stimulus response coupling. The Black and Leff operational model of agonism (26) was applied to separate effects on pathway-specific signaling from those that modify ligand affinity. Derived values (τ) were normalized to experimentally determined levels of cell-surface expression to provide a measure of efficacy (τc) that is independent of affinity and altered cell-surface receptor expression (11). Log τc values for mutant receptors were statistically compared with those of the WT receptor using a one-way analysis of variance (ANOVA) and Dunnett's post-test. Significance was accepted at p < 0.05.
Molecular modeling and mapping of mutational effects
A homology model of the inactive GLP-1R TM domain was built using the minimally modified GCGR (PDB code 4L6R) (29), as described previously (28); the first amino acid in this model is Arg-134. The thermostabilized and full-length human GLP-1R bound to modified 11-mer peptide agonist (PDB code 5NX2) (15); the full-length and GLP-1 bound to rabbit GLP-1R in complex with Gs (PDB code 5VAI) (16), and the full-length and exendin-P5 bound to human GLP-1R in complex with Gs (PDB code 6B3J) (16) were used as deposited; the first amino acids in these structures are Thr-29, Thr-29, and Val-30, respectively.
Author contributions
S. L., D. Y., M.-W. W., D. W., and P. M. S. conceptualization; S. L., L. C., D. Y., Y.-L. L., C. K., A. C., and D. W. formal analysis; S. L., L. C., A. D., X. C., Y. F., C. K., P. Z., and T. C. investigation; S. L. methodology; S. L., M.-W. W., D. W., and P. M. S. writing-original draft; S. L., C. K., P. Z., A. C., M.-W. W., D. W., and P. M. S. writing-review and editing; D. Y., M.-W. W., D. W., and P. M. S. supervision; C. K., A. C., M.-W. W., D. W., and P. M. S. funding acquisition; P. M. S. project administration.
This work was supported in part by National Health and Medical Research Council of Australia Project Grants 1061044, 1065410, and 1126857; National Health and Medical Research Council Program Grant 1055134; Shanghai Science and Technology Development Fund Grant 15DZ2291600; National Natural Science Foundation of China Grant 81573479; and Strategic Priority Research Program of the Chinese Academy of Sciences Grants XDA12020347 and XDA12040308. The authors declare that they have no conflicts of interest with the contents of this article.
- GPCR
- G protein–coupled receptor
- GLP-1R
- glucagon-like peptide-1 receptor
- TM
- transmembrane
- ECL
- extracellular loop
- pERK
- phosphorylation of extracellular signal-regulated kinase 1/2
- ECD
- extracellular domain
- GCGR
- glucagon receptor
- NAM
- negative allosteric modulator
- PDB
- Protein Data Bank
- [Ca2+]i
- intracellular calcium
- ANOVA
- analysis of variance
- hGLP-1R
- human GLP-1R.
References
- 1. Santos, R., Ursu, O., Gaulton, A., Bento, A. P., Donadi, R. S., Bologa, C. G., Karlsson, A., Al-Lazikani, B., Hersey, A., Oprea, T. I., and Overington, J. P. (2017) A comprehensive map of molecular drug targets. Nat. Rev. Drug. Discov. 16, 19–34 10.1038/nrd.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luttrell, L. M., Maudsley, S., and Bohn, L. M. (2015) Fulfilling the promise of “Biased” G protein-coupled receptor agonism. Mol. Pharmacol. 88, 579–588 10.1124/mol.115.099630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Costa-Neto, C. M., Parreiras-E-Silva, L. T., and Bouvier, M. (2016) A pluridimensional view of biased agonism. Mol. Pharmacol. 90, 587–595 10.1124/mol.116.105940 [DOI] [PubMed] [Google Scholar]
- 4. Wootten, D., Miller, L. J., Koole, C., Christopoulos, A., and Sexton, P. M. (2017) Allostery and biased agonism at class B G protein-coupled receptors. Chem. Rev. 117, 111–138 10.1021/acs.chemrev.6b00049 [DOI] [PubMed] [Google Scholar]
- 5. Hollenstein, K., de Graaf, C., Bortolato, A., Wang, M. W., Marshall, F. H., and Stevens, R. C. (2014) Insights into the structure of class B GPCRs. Trends Pharmacol. Sci. 35, 12–22 10.1016/j.tips.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koole, C., Wootten, D., Simms, J., Valant, C., Sridhar, R., Woodman, O. L., Miller, L. J., Summers, R. J., Christopoulos, A., and Sexton, P. M. (2010) Allosteric ligands of the glucagon-like peptide 1 receptor (GLP-1R) differentially modulate endogenous and exogenous peptide responses in a pathway-selective manner: implications for drug screening. Mol. Pharmacol. 78, 456–465 10.1124/mol.110.065664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hager, M. V., Johnson, L. M., Wootten, D., Sexton, P. M., and Gellman, S. H. (2016) β-Arrestin-biased agonists of the GLP-1 receptor from β-amino acid residue incorporation into GLP-1 analogues. J. Am. Chem. Soc. 138, 14970–14979 10.1021/jacs.6b08323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hager, M. V., Clydesdale, L., Gellman, S. H., Sexton, P. M., and Wootten, D. (2017) Characterization of signal bias at the GLP-1 receptor induced by backbone modification of GLP-1. Biochem. Pharmacol. 136, 99–108 10.1016/j.bcp.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weston, C., Poyner, D., Patel, V., Dowell, S., and Ladds, G. (2014) Investigating G protein signalling bias at the glucagon-like peptide-1 receptor in yeast. Br. J. Pharmacol. 171, 3651–3665 10.1111/bph.12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang, H., Sturchler, E., Zhu, J., Nieto, A., Cistrone, P. A., Xie, J., He, L., Yea, K., Jones, T., Turn, R., Di Stefano, P. S., Griffin, P. R., Dawson, P. E., McDonald, P. H., and Lerner, R. A. (2015) Autocrine selection of a GLP-1R G-protein biased agonist with potent antidiabetic effects. Nat. Commun. 6, 8918 10.1038/ncomms9918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wootten, D., Reynolds, C. A., Smith, K. J., Mobarec, J. C., Koole, C., Savage, E. E., Pabreja, K., Simms, J., Sridhar, R., Furness, S. G. B., Liu, M., Thompson, P. E., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2016) The extracellular surface of the GLP-1 receptor is a molecular trigger for biased agonism. Cell 165, 1632–1643 10.1016/j.cell.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoare, S. R. (2005) Mechanisms of peptide and nonpeptide ligand binding to class B G-protein coupled receptors. Drug Discov. Today 10, 417–427 10.1016/S1359-6446(05)03370-2 [DOI] [PubMed] [Google Scholar]
- 13. Koole, C., Wootten, D., Simms, J., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2012) Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. J. Biol. Chem. 287, 3642–3658 10.1074/jbc.M111.309328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song, G., Yang, D., Wang, Y., de Graaf, C., Zhou, Q., Jiang, S., Liu, K., Cai, X., Dai, A., Lin, G., Liu, D., Wu, F., Wu, Y., Zhao, S., Ye, L., et al. (2017) Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature 546, 312–315 10.1038/nature22378 [DOI] [PubMed] [Google Scholar]
- 15. Jazayeri, A., Rappas, M., Brown, A. J. H., Kean, J., Errey, J. C., Robertson, N. J., Fiez-Vandal, C., Andrews, S. P., Congreve, M., Bortolato, A., Mason, J. S., Baig, A. H., Teobald, I., Doré, A. S., Weir, M., et al. (2017) Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546, 254–258 10.1038/nature22800 [DOI] [PubMed] [Google Scholar]
- 16. Zhang, Y., Sun, B., Feng, D., Hu, H., Chu, M., Qu, Q., Tarrasch, J. T., Li, S., Sun Kobilka, T., Kobilka, B. K., and Skiniotis, G. (2017) Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253 10.1038/nature22394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang, Y. L., Khoshouei, M., Glukhova, A., Furness, S. G. B., Zhao, P., Clydesdale, L., Koole, C., Truong, T. T., Thal, D. M., Lei, S., Radjainia, M., Danev, R., Baumeister, W., Wang, M. W., Miller, L. J., et al. (2018) Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555, 121–125 10.1038/nature25773 [DOI] [PubMed] [Google Scholar]
- 18. Zhang, H., Qiao, A., Yang, L., Van Eps, N., Frederiksen, K. S., Yang, D., Dai, A., Cai, X., Zhang, H., Yi, C., Cao, C., He, L., Yang, H., Lau, J., Ernst, O. P., et al. (2018) Structure of the glucagon receptor in complex with a glucagon analogue. Nature 553, 106–110 10.1038/nature25153 [DOI] [PubMed] [Google Scholar]
- 19. Liang, Y. L., Khoshouei, M., Radjainia, M., Zhang, Y., Glukhova, A., Tarrasch, J., Thal, D. M., Furness, S. G. B., Christopoulos, G., Coudrat, T., Danev, R., Baumeister, W., Miller, L. J., Christopoulos, A., Kobilka, B. K., et al. (2017) Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123 10.1038/nature22327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mukund, S., Shang, Y., Clarke, H. J., Madjidi, A., Corn, J. E., Kates, L., Kolumam, G., Chiang, V., Luis, E., Murray, J., Zhang, Y., Hötzel, I., Koth, C. M., and Allan, B. B. (2013) Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor. J. Biol. Chem. 288, 36168–36178 10.1074/jbc.M113.496984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao, L. H., Yin, Y., Yang, D., Liu, B., Hou, L., Wang, X., Pal, K., Jiang, Y., Feng, Y., Cai, X., Dai, A., Liu, M., Wang, M. W., Melcher, K., and Xu, H. E. (2016) Differential requirement of the extracellular domain in activation of class B G protein-coupled receptors. J. Biol. Chem. 291, 15119–15130 10.1074/jbc.M116.726620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin, Y., de Waal, P. W., He, Y., Zhao, L. H., Yang, D., Cai, X., Jiang, Y., Melcher, K., Wang, M. W., and Xu, H. E. (2017) Rearrangement of a polar core provides a conserved mechanism for constitutive activation of class B G protein-coupled receptors. J. Biol. Chem. 292, 9865–9881 10.1074/jbc.M117.782987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graaf, Cd., Donnelly, D., Wootten, D., Lau, J., Sexton, P. M., Miller, L. J., Ahn, J. M., Liao, J., Fletcher, M. M., Yang, D., Brown, A. J., Zhou, C., Deng, J., and Wang, M. W. (2016) Glucagon-like peptide-1 and its class B G protein-coupled receptors: a long march to therapeutic successes. Pharmacol. Rev. 68, 954–1013 10.1124/pr.115.011395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dods, R. L., and Donnelly, D. (2015) The peptide agonist-binding site of the glucagon-like peptide-1 (GLP-1) receptor based on site-directed mutagenesis and knowledge-based modelling. Biosci. Rep. 36, e00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dal Maso, E., Zhu, Y., Pham, V., Reynolds, C. A., Deganutti, G., Hick, C. A., Yang, D., Christopoulos, A., Hay, D. L., Wang, M. W., Sexton, P. M., Furness, S. G. B., and Wootten, D. (2018) Extracellular loops 2 and 3 of the calcitonin receptor selectively modify agonist binding and efficacy. Biochem. Pharmacol. 150, 214–244 10.1016/j.bcp.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Black, J. W., and Leff, P. (1983) Operational models of pharmacological agonism. Proc. R. Soc. Lond. B Biol. Sci. 220, 141–162 10.1098/rspb.1983.0093 [DOI] [PubMed] [Google Scholar]
- 27. Kenakin, T., and Christopoulos, A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 12, 205–216 [DOI] [PubMed] [Google Scholar]
- 28. Wootten, D., Reynolds, C. A., Koole, C., Smith, K. J., Mobarec, J. C., Simms, J., Quon, T., Coudrat, T., Furness, S. G., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2016) A hydrogen-bonded polar network in the core of the glucagon-like peptide-1 receptor is a fulcrum for biased agonism: lessons from class B crystal structures. Mol. Pharmacol. 89, 335–347 10.1124/mol.115.101246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siu, F. Y., He, M., de Graaf, C., Han, G. W., Yang, D., Zhang, Z., Zhou, C., Xu, Q., Wacker, D., Joseph, J. S., Liu, W., Lau, J., Cherezov, V., Katritch, V., Wang, M. W., and Stevens, R. C. (2013) Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449 10.1038/nature12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jazayeri, A., Doré, A. S., Lamb, D., Krishnamurthy, H., Southall, S. M., Baig, A. H., Bortolato, A., Koglin, M., Robertson, N. J., Errey, J. C., Andrews, S. P., Teobald, I., Brown, A. J., Cooke, R. M., Weir, M., and Marshall, F. H. (2016) Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–277 10.1038/nature17414 [DOI] [PubMed] [Google Scholar]
- 31. Yin, Y., Zhou, X. E., Hou, L., Zhao, L. H., Liu, B., Wang, G., Jiang, Y., Melcher, K., and Xu, H. E. (2016) An intrinsic agonist mechanism for activation of glucagon-like peptide-1 receptor by its extracellular domain. Cell Discov. 2, 16042 10.1038/celldisc.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wootten, D., Reynolds, C. A., Smith, K. J., Mobarec, J. C., Furness, S. G., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2016) Key interactions by conserved polar amino acids located at the transmembrane helical boundaries in class B GPCRs modulate activation, effector specificity and biased signalling in the glucagon-like peptide-1 receptor. Biochem. Pharmacol. 118, 68–87 10.1016/j.bcp.2016.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wootten, D., Simms, J., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2013) Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl. Acad. Sci. U.S.A. 110, 5211–5216 10.1073/pnas.1221585110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao, Q., Jeng, W., and Wheeler, M. B. (2000) Characterization of glucagon-like peptide-1 receptor-binding determinants. J. Mol. Endocrinol. 25, 321–335 10.1677/jme.0.0250321 [DOI] [PubMed] [Google Scholar]
- 35. López de Maturana, R., and Donnelly, D. (2002) The glucagon-like peptide-1 receptor binding site for the N terminus of GLP-1 requires polarity at Asp198 rather than negative charge. FEBS Lett. 530, 244–248 10.1016/S0014-5793(02)03492-0 [DOI] [PubMed] [Google Scholar]
- 36. Coopman, K., Wallis, R., Robb, G., Brown, A. J., Wilkinson, G. F., Timms, D., and Willars, G. B. (2011) Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor. Mol. Endocrinol. 25, 1804–1818 10.1210/me.2011-1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koole, C., Wootten, D., Simms, J., Valant, C., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2011) Polymorphism and ligand dependent changes in human glucagon-like peptide-1 receptor (GLP-1R) function: allosteric rescue of loss of function mutation. Mol. Pharmacol. 80, 486–497 10.1124/mol.111.072884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koole, C., Wootten, D., Simms, J., Miller, L. J., Christopoulos, A., and Sexton, P. M. (2015) Differential impact of amino acid substitutions on critical residues of the human glucagon-like peptide-1 receptor involved in peptide activity and small-molecule allostery. J. Pharmacol. Exp. Ther. 353, 52–63 10.1124/jpet.114.220913 [DOI] [PubMed] [Google Scholar]
- 39. Underwood, C. R., Garibay, P., Knudsen, L. B., Hastrup, S., Peters, G. H., Rudolph, R., and Reedtz-Runge, S. (2010) Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 285, 723–730 10.1074/jbc.M109.033829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Al-Sabah, S., and Donnelly, D. (2003) The positive charge at Lys-288 of the glucagon-like peptide-1 (GLP-1) receptor is important for binding the N-terminus of peptide agonists. FEBS Lett. 553, 342–346 10.1016/S0014-5793(03)01043-3 [DOI] [PubMed] [Google Scholar]