Abstract
Inhibitors that bind to the paclitaxel- or vinblastine-binding sites of tubulin have been part of the pharmacopoeia of anticancer therapy for decades. However, tubulin inhibitors that bind to the colchicine-binding site are not used in clinical cancer therapy, because of their low therapeutic index. To address multidrug resistance to many conventional tubulin-binding agents, numerous efforts have attempted to clinically develop inhibitors that bind the colchicine-binding site. Previously, we have found that millepachine (MIL), a natural chalcone-type small molecule extracted from the plant Millettia pachycarpa, and its two derivatives (MDs) SKLB028 and SKLB050 have potential antitumor activities both in vitro and in vivo. However, their cellular targets and mechanisms are unclear. Here, biochemical and cellular experiments revealed that the MDs directly and irreversibly bind β-tubulin. X-ray crystallography of the tubulin–MD structures disclosed that the MDs bind at the tubulin intradimer interface and to the same site as colchicine and that their binding mode is similar to that of colchicine. Of note, MDs inhibited tubulin polymerization and caused G2/M cell-cycle arrest. Comprehensive analysis further revealed that free MIL exhibits an s-cis conformation, whereas MIL in the colchicine-binding site in tubulin adopts an s-trans conformation. Moreover, introducing an α-methyl to MDs to increase the proportion of s-trans conformations augmented MDs' tubulin inhibition activity. Our study uncovers a new class of chalcone-type tubulin inhibitors that bind the colchicine-binding site in β-tubulin and suggests that the s-trans conformation of these compounds may make them more active anticancer agents.
Keywords: microtubule, cancer prevention, cancer, natural product, tubulin, X-ray crystallography, drug resistance, Chalcone, Colchicine, Millepachine, s-trans conformation, Tubulin
Introduction
Microtubules are formed by noncovalent tubulin heterodimers (α- and β-tubulin) in a process that involves polymerization and depolymerization (1). Microtubules play important roles in key cell events especially in mitosis, and this makes them an attractive target for anti-cancer drug design (2). Currently, many microtubule inhibitors such as taxanes, Vinca alkaloids, and epothilones are used against many solid and hematologic malignancies (3–5). Although the microtubule inhibitors have been demonstrated to exert a high level of anti-cancer activity in clinical treatment or preclinical testing, their effectiveness is limited by multidrug resistance, peripheral neuropathy, and insolubility (6, 7). Membrane efflux pumps of the ATP-binding cassette family are the primary resistance mechanism developed by tumor cells exposed to microtubule-binding agents (8, 9).
Colchicine was the first identified tubulin destabilizing agent (10) and is commonly used as an unapproved drug to treat gout, familial Mediterranean fever, pericarditis, and Behçet's disease (11–14). In 2009, the Food and Drug Administration approved colchicine to treat acute gout flares and familial Mediterranean fever. Although colchicine was not approved to treat cancer, because of its low therapeutic index, numerous efforts were made to clinically develop colchicine-binding site inhibitors, because the favorable factors of most colchicine-binding site inhibitors are that they have no multidrug resistance (MDR)2 issues, they have a simple structure, and they are easy to synthesize (15).
Recently research revealed that another possible way to overcome MDR is to develop irreversible binding agents as suggested by Buey et al., who showed that cyclostreptin, a natural small molecule, retains its activity in cells that are resistant to paclitaxel by overexpression of P-glycoprotein through covalently (irreversibly) binding to Thr-220 and Asn-228 of β-tubulin (16). The authors proposed the idea that resistant tumor cells cannot escape the effect of compounds irreversibly binding to them by reducing their affinity for the target or by enhancing drug efflux, which suggests that the design of ligands that irreversibly react with tubulin might be an effective way to address drug resistance. For instance, zampanolide and (+)-dactylolide, new microtubule-stabilizing agents, retain full activity in MDR cell lines by covalently (irreversibly) binding to the taxane luminal site in β-tubulin (17, 18). T138067, a tubulin inhibitor that acts by covalent modification of β-tubulin residue Cys-239, retains full activity in MDR cells both in vitro and in vivo (19). Recently, Towle et al. (20) showed that irreversible mitotic blockade and persistent drug retention served as a good explanation for the excellent anti-cancer activity of eribulin on drug-resistant cell lines. All these findings proved that irreversible modification of tubulin may overcome MDR driven by ATP-binding cassette pumps.
Millepachine (MIL) was first isolated from Millettia pachycarpa Benth by our group (21). Our following study showed this novel chalcone possessed anti-cancer activity by inducing cell-cycle G2/M arrest and apoptosis in human hepatocarcinoma cells (22). Through modification of MIL, we achieved the derivatives SKLB028 and SKLB050 with low nanomolar IC50 values to various tumor cells including drug-resistant cell lines (see Fig. 1A), with obvious improved anti-cancer activities compared with MIL (23, 24). SKLB050 could circumvent multidrug resistance in a A2780CP (P-glycoprotein overexpression) model (24). In our present study, through biotinylation of MIL and structural biology studies, we found that MIL and its derivatives (MDs) could irreversibly bind to the colchicine site of β-tubulin and retained full activity toward MDR cells. Deeper mechanistic study revealed that free MIL adopts an s-cis conformation but is in an s-trans conformation in the colchicine site and that introducing an α-methyl to MDs to increase the proportion of s-trans conformations could clearly augment the activity of MDs.
Figure 1.
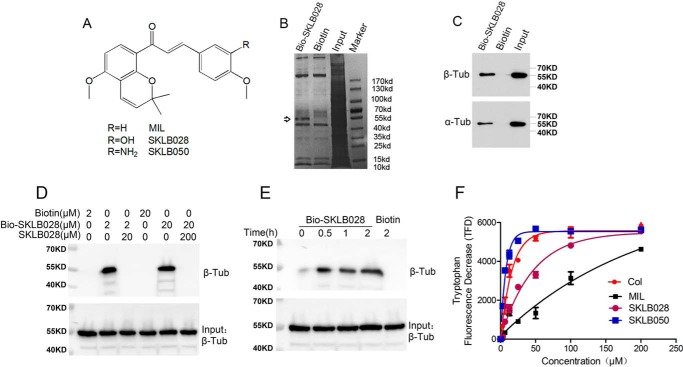
MIL directly targets β-tubulin. A, chemical structures of MIL. B, HepG2 cell lysates were incubated with Bio-SKLB028 or biotin, followed by pulldown with streptavidin–agarose. The precipitates were resolved by SDS-PAGE, and the gel was stained with Coomassie staining. C, HepG2 cell lysates were incubated with Bio-SKLB028 or biotin, followed by pulldown with streptavidin–agarose. The precipitates were detected by Western blotting by α- and β-tubulin. D, porcine brain tubulin (1 μm) were preincubated with or without indicated concentrations of SKLB028 for 2 h before incubation with lower concentration of Bio-SKLB028 for another 2 h, and different concentrations of biotin were incubated with tubulin for 2 h as control groups. Then all the samples were pulled down by streptavidin–agarose beads, and the eluted samples were bolted for β-tubulin. Also, total protein for each sample was detected by Western blotting for β-tubulin as a loading control. E, porcine brain tubulin (1 μm) was incubated with Bio-SKLB028 for different times and then pulled down by a streptavidin–agarose beads, and the eluted samples were bolted for β-tubulin. Also, total protein for each sample was detected by Western blotting for β-tubulin as a loading control. F, tryptophan-based binding assay to detect the Kd value of indicated compounds binding to tubulin. The indicated compounds at different concentrations (3.125, 6.25, 12.5, 25, 50, 100, and 200 μm) were incubated with tubulin for 30 min and then monitored at 295 nm (excitation) and 335 nm (emission) using Biotech Gen5 spectrophotometer. The dissociation constants were calculated from fitting curves of decreasing fluorescence using the GraphPad Prism software. Col, colchicine; Tub, tubulin; Col, colchicine.
Results
MIL directly binds to tubulin
In our previous study, MIL and its derivatives, SKLB028 and SKLB050 (Fig. 1A), showed an effect on tubulin polymerization, and this prompted us to identify the potential targets of these compounds. For this purpose, the chemical probe biotin-tagged SKLB028 (hereafter named Bio-SKLB028) was synthesized (Fig. S1). Bio-SKLB028 retained full activity toward cancer cells compared with free SKLB028 (Table 1). Next, HepG2 cell lysates were incubated with Bio-SKLB028 or free biotin, and the mixtures were precipitated with streptavidin-coated agarose beads, followed by gel electrophoresis and Coomassie Blue staining. We observed one major protein band that co-precipitated specifically with Bio-SKLB028 beads, with an apparent molecular size of 55 kDa, whereas the biotin beads produced no such bands (Fig. 1B). MS data revealed that the specific band is mostly like β-tubulin (Fig. S2). Also, the specific band co-migrated with α- and β-tubulin, as determined by Western blotting analysis (Fig. 1C). Bio-SKLB028 also bound the in vitro porcine tubulin proteins in a concentration-dependent manner, but this binding was competitively inhibited by higher concentrations of unlabeled SKLB028 (Fig. 1D). Also, this binding of SKLB028 to tubulin was time-dependent (Fig. 1E). Changes in tryptophan fluorescence of tubulin were used as a test for drug binding (25). Using this tryptophan-based binding assay, as shown in Fig. 1F, we identified the Kd (dissociation constant) values of compounds binding to tubulin were 5.13 ± 0.62 μm (SKLB050), 11.03 ± 2.57 μm (colchicine), 31.69 ± 5.26 μm (SKLB028), and 139.3 ± 34.76 μm (MIL). All these results together proved that MIL directly binds to tubulin.
Table 1.
IC50 values of MDs, Bio-SKLB028 on HepG2, and A2780S cells
MTT assay was used to detect the IC50 (half-maximal inhibitory concentration) values of MDs, Bio-SKLB028 on HepG2, and A2780S cells. The data are expressed as means ± S.D.
| IC50 |
||||
|---|---|---|---|---|
| MIL | SKLB028 | SKLB050 | Bio-SKLB028 | |
| n m | ||||
| HepG2 | 2300 ± 220 | 120 ± 12 | 31 ± 2 | 298 ± 31 |
| A2780S | 3509 ± 572 | 89 ± 7 | 60 ± 4 | 159 ± 12 |
MDs inhibit tubulin polymerization and induce G2/M phase cell-cycle arrest
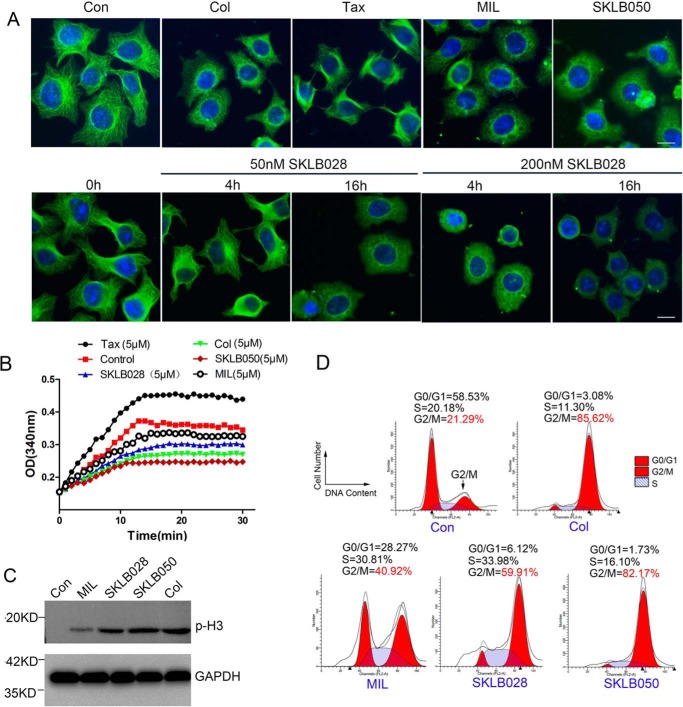
We further investigated the effect of MIL, SKLB028, and SKLB050 on tubulin inside the cells by immunofluorescence staining. As shown in Fig. 2A, colchicine inhibited tubulin polymerization, whereas paclitaxel obviously promoted tubulin polymerization in HCT-8/V cells. MDs inhibited tubulin polymerization in a colchicine-like manner. Furthermore, SKLB028 inhibited tubulin polymerization in a time- and dose-dependent manner. We further evaluated the effects of MDs on tubulin in vitro. The effects on the assembly of purified porcine tubulin were evaluated by measuring an increase in absorbance at 340 nm at 37 °C, by using colchicine and paclitaxel as comparative agents. Similar to colchicine, we found that SKLB028 and SKLB050 inhibited tubulin polymerization, and MIL showed a weak tubulin polymerization inhibition effect (Fig. 2B). We then investigated the effect of MDs on the cell cycle. As shown in Fig. 2C, we found that MDs could obviously increase the expression level of phosphorylation H3 (p-H3; a G2/M phase marker), and the flow cytometry results also showed that MDs could cause obvious G2/M phase arrest (Fig. 2D and Fig. S3). In all, MDs could inhibit tubulin polymerization and cause subsequent G2/M cell-cycle arrest.
Figure 2.
MDs inhibit tubulin polymerization and cause G2/M phase cell-cycle arrest. A, HCT-8/V cells were treated with 1 μm MIL, 50 nm SKLB050, 50 nm colchicine, or 50 nm paclitaxel for 16 h or treated with 50 nm or 200 nm SKLB028 for 4 or 16 h. Then tubulin morphology was detected by immunofluorescence with anti-β-tubulin and DAPI staining. Bar, 10 μm. B, indicated concentrations of compounds were co-incubated with tubulin (3 mg/ml) at 37 °C. Absorbance at 340 nm was detected every 1 min for 30 min. C, HCT-8/V cells were incubated with 1 μm MIL, 50 nm SKLB050, 50 nm colchicine, or 100 nm SKLB028 for 16 h; then cell lysates were collected and subjected to Western blotting for p-H3 detection. GAPDH was used as a loading control. D, HCT-8/V cells were incubated with 1 μm MIL, 50 nm SKLB050, 50 nm colchicine, and 100 nm for 16 h; subjected to PI staining; and analyzed by a flow cytometer for cell-cycle analysis. Col, colchicine; Con, control; Tax, paclitaxel.
MDs bind to the colchicine site of β-tubulin
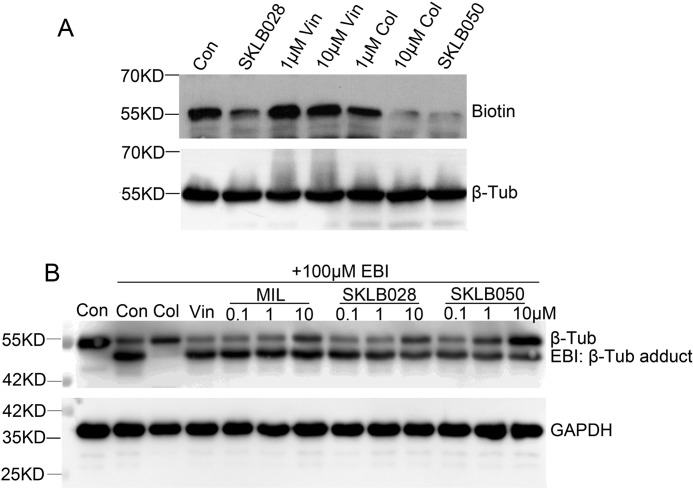
To identify the binding site of MDs on tubulin, a competition experiment between Bio-SKLB028 with SKLB028, vinblastine, colchicine, and SLB050 was performed. Porcine tubulin (1 μm) was preincubated with SKLB028 (10 μm), vinblastine (1 or 10 μm), colchicine (1 or 10 μm), or SKLB050 (10 μm) for 1 h before treating with Bio-SKLB028 (10 μm) for 2 h. Then the proteins were thoroughly washed and detected by Western blotting for biotin. As shown in Fig. 3A, SKLB028 and SKLB050 could inhibit the binding of Bio-SKLB028 to tubulin, which indicated that SKLB028, SKLB050, and Bio-SKLB028 bind to the same site on tubulin. Vinblastine showed no inhibition of the binding of SKLB028 to β-tubulin, whereas 1 μm colchicine weakly inhibited the binding of SKLB028 to β-tubulin and 10 μm colchicine strongly inhibited the binding of SKLB028 to tubulin. These data proved that MDs bind to the colchicine site of β-tubulin. Next, N,N′-ethylene-bis(iodoacetamide) (EBI), a homobifunctional thioalkylating agent, was used to cross-link the Cys-239 and the Cys-354 residues of β-tubulin involved in the colchicine-binding site (26). The covalent binding of EBI to β-tubulin forms an adduct that is easily detected by Western blotting as a second immunoreactive band of β-tubulin that migrates faster than the native β-tubulin band on SDS-PAGE (26). To comprehensively validate the binding of MDs to the colchicine site on β-tubulin, we detected whether MDs could inhibit the binding of EBI to β-tubulin. Our results showed that MIL and SKLB028 at high doses and SKLB050 at moderate and high doses could inhibit the formation of EBI–β-tubulin adducts (Fig. 3B). These results also indicated that MDs bind to the colchicine site of β-tubulin.
Figure 3.
MDs bind to colchicine site of β-tubulin. A, porcine brain tubulin (1 μm) were treated with 1 μm SKLB028, 1 μm vinblastine, 10 μm vinblastine, 1 μm colchicine, 10 μm colchicine, or 1 μm SKLB050 for 1 h before treated with 1 μm Bio-SKLB028 for another 2 h. After totally wash of unbounded compounds and cross-linked by a UV hybridization incubator for 10 min, the samples were blotted for biotin. Total proteins were blotted for β-tubulin as a loading control. B, HCT-8/V cells were incubated with or without indicated concentrations of MDs for 2 h, and then 100 μm EBI was added to cells and incubated for another 2 h. Total protein was lysed with RIPA lysis buffer and subjected to Western blotting analysis for β-tubulin and GAPDH. Vin, vinblastine; Col, colchicine; Con, control; Tub, tubulin.
SKLB028 and SKLB050 were effective on MDR tumor cells
Next, we tested the anti-tumor activity of SKLB028 and SKLB050 on drug-resistant cells. A2780S, HCT-8 tumor cells, and three related drug-resistant cell lines expressing high levels of P-Pg170 or MRP1 (Fig. S4) were treated with SKLB028 or SKLB050 for 48 h. As shown in Table 2, SKLB028 and SKLB050 showed strong and similar growth inhibition against both sensitive and MDR cell lines, with a resistance factor ranging from 1.5 to 2.5, as compared with resistance factors of 186–733 for paclitaxel and vinblastine.
Table 2.
Verification of activities of taxol, vinblastine, SKLB028, SKLB050 on A2780S, A2780/T, HCT-8, HCT-8/V, and HCT-8/T cells
MTT assay was used to detect the IC50 (half-maximal inhibitory concentration) values. The data are expressed as means ± S.D.
| IC50 | |||||
|---|---|---|---|---|---|
| n m | |||||
| Cell lines | A2780S | A2780/T | HCT-8 | HCT-8/V | HCT-8/T |
| Paclitaxel | 46 ± 5 | 8600 ± 1000 (186)a | 36 ± 6 | 9200 ± 430 (255) | 10,800 ± 670 (300) |
| Vinblastine | 33 ± 7 | 7540 ± 876 (225) | 27 ± 3 | 9800 ± 670 (362) | 19,800 ± 1200 (733) |
| SKLB028 | 72 ± 8 | 150 ± 19 (2.1) | 98 ± 11 | 134 ± 13 (1.4) | 201 ± 56 (2.1) |
| SKLB050 | 23 ± 4 | 35 ± 5 (1.5) | 13 ± 2 | 19 ± 1 (1.5) | 33 ± 5 (2.5) |
a The numbers in parentheses are the calculated relative resistance values, obtained by dividing the IC50 value of the resistant line by the IC50 value of the parental line.
SKLB028 binds irreversibly to tubulin
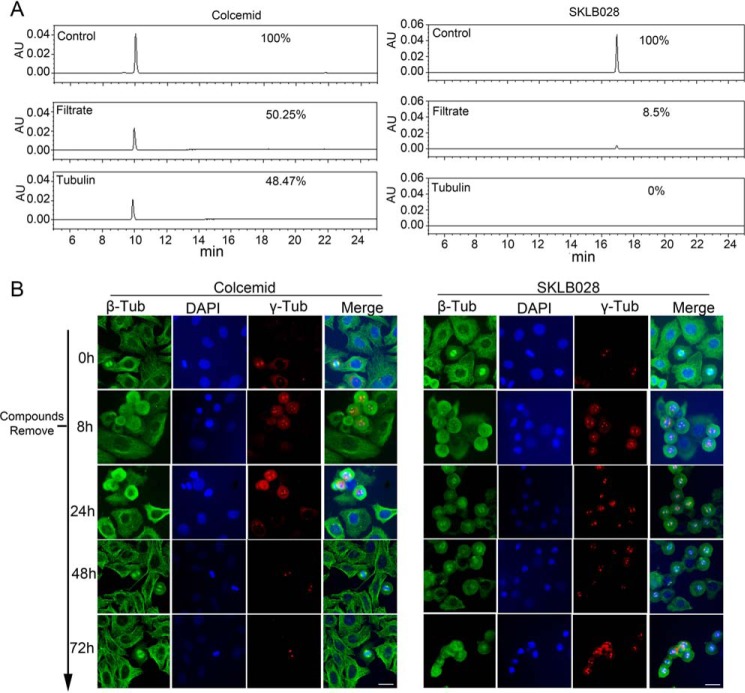
Because MIL and its derivatives strongly bind to β-tubulin, we tried to detect whether these compounds bind irreversibly to tubulin. Tubulin was incubated with SKLB028 or colcemid (a reversible tubulin inhibitor) and then separated by an ultrafiltration method. Compounds in the tubulin and filtrate fractions were detected by HPLC. Controls showed that SKLB028 and colcemid should have been detectable. In the colcemid-treated group, colcemid was detected in both the tubulin fraction and the filtrate fraction. In contrast, in the SKLB028-treated group, the tubulin fraction was devoid of SKLB028, and SKLB028 in the filtrate fraction was greatly reduced following incubation of SKLB028 with tubulin (Fig. 4A). In the absence of degradation products, SKLB028 must have reacted irreversibly with tubulin.
Figure 4.
MDs bind irreversibly to β-tubulin. A, tubulin (3 mg/ml) incubated with 30 μm colcemid or 30 μm SKLB028 for 1 h. Then tubulin part and filtrate part were separated with an ultrafiltration method. Compounds in tubulin part and filtrate part were detected by HPLC. Colcemid (30 μm) or SKLB028 (30 μm) dissolved in 200 μl of CH3OH were also detected by HPLC as controls. This experiment was repeated three times. B, high concentrations (10 μm) of SKLB028 or colcemid were co-incubated with HCT-8/V cells for 8 h, and then compounds were extensively washed for three times. The cells were further cultured to 72 h. At 0-, 8-, 24-, 48-, and 72-h points, tubulin morphology was analyzed by immunofluorescence. Bar, 20 μm. Tub, tubulin; AU, absorbance unit.
Similarly, there were differences in the effects of SKLB028 and colcemid on cellular tubulin. HCT-8/V cells were treated with high concentrations (10 μm) of SKLB028 or colcemid for 8 h; then the cells were extensively washed to remove compounds and were further cultured to 72 h. At 0-, 8-, 24-, 48-, and 72-h points, tubulin morphology and cell cycle were analyzed by immunofluorescence and flow cytometry, respectively. We observed recovery of the tubulin network in colcemid-treated cells. However, the effects of SKLB028 were irreversible. As shown in Fig. 4B, cells had typical tubulin networks at 0 h. After incubation with SKLB028 or colcemid for 8 h, the tubulin began to depolymerize, and a small portion of cells was arrested at G2/M phase, as indicated by abnormal (asymmetric and multipolar) spindles marked by γ-tubulin. After washing and another 16 h in culture, the depolymerization and G2/M phase arrest effect of SKLB028 had progressed, resulting in almost complete loss of the tubulin network. Also, more cells with abnormal spindles were observed. With colcemid, the tubulin network began to revert to a normal appearance. After a 40-h recovery, SKLB028-treated cells remained the same, with depolymerized tubulin and abnormal spindles. After 40 h without drug exposition, colcemid-treated cells had almost recovered to normal. After recovery for 64 h, SKLB028-treated cells showed the same morphology as those at 40 h, but the colcemid-treated cells showed typical microtubule networks again.
We also examined the HCT-8/V cells for DNA content by flow cytometry (Figs. S5 and S6), and these data also indicated a greater recovery after colcemid versus SKLB028 treatment, with a larger population of G2/M cells persisting after the SKLB028 treatment. All of these results proved that SKLB028 binds irreversibly to tubulin.
X-ray crystal structure of the tubulin complex with MIL, SKLB028, or SKLB050
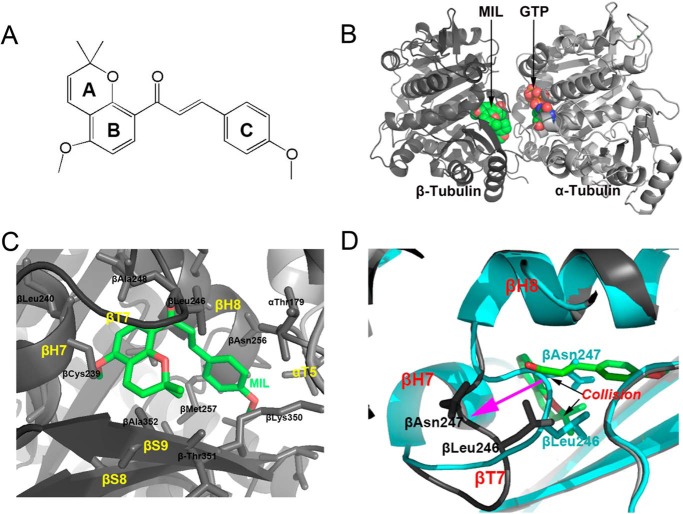
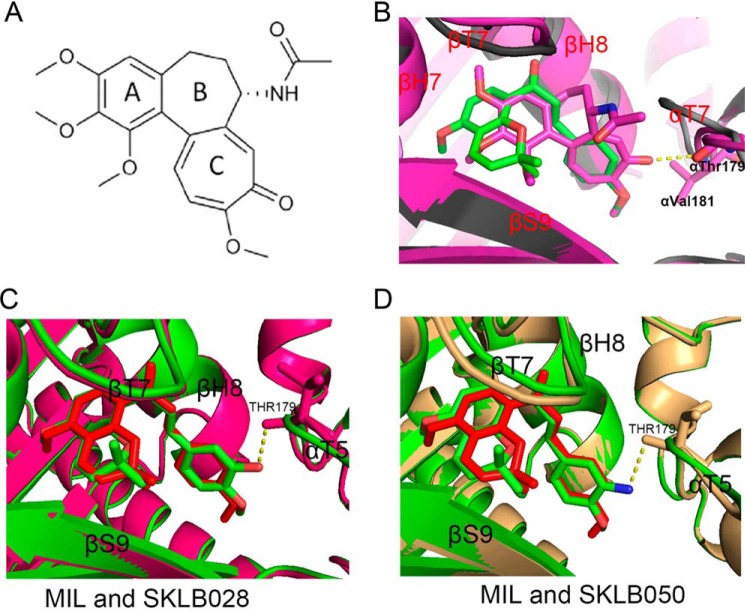
To obtain the detailed interaction of MIL and its derivatives with tubulin, we solved the crystal structure of tubulin–MIL, tubulin–SKLB028, and tubulin–SKLB050 complexes. Compounds were soaked into a well established crystallization system based on a protein complex composed of two αβ-tubulin heterodimers, one stathmin-like protein RB3, and one tubulin tyrosine ligase (T2R–TTL) (18, 27). With this method, we determined the tubulin–MIL (PDB code 5YLJ), tubulin–SKLB028 (PDB code 5YL2), and tubulin–SKLB050 (PDB code 5YLS) structures to resolutions of 2.70, 2.09, and 3.00 Å, respectively. The data collection and refinement statistics are given in Table 3. In general, there are no big changes in the overall structures of the three complexes when compared with the apo-tubulin structure (PDB code 5XP3). In the MIL–tubulin structure, MIL occupied a pocket formed by residues from helices H7 and H8, strands S8 and S9 and loop T7 of β-tubulin, and loop T5 of α-tubulin (Fig. 5); this binding site is the widely known colchicine site (28). In the MIL–tubulin structure, the A and B rings of MIL were buried deeply into the colchicine site, making key hydrophobic interactions with several residues of β-tubulin (Fig. 5C). Hydrogen-bond formation between MIL and tubulin was not observed. Through comparison of the apo– (PDB code 5XP3; 2.30 Å resolution) and MIL–tubulin structures, we found that the Asn-247 and Leu-248 residues of T7 loop on β-tubulin occupied the colchicine site. This observation suggested that the T7 loop must shift outward for the MIL binding, because the Asn-247 and Leu-248 residues in apo collide with MIL in the MIL–tubulin complex (Fig. 5D). Similar conformational changes were also observed in the colchicine–tubulin complex (28, 29). We also solved the structure of the colchicine–tubulin complex at 2.90 Å resolution (PDB code 5XIW). Superimposition of the MIL–tubulin structure onto the colchicine–tubulin structure revealed that the C ring of MIL superimposes well with the C rings of colchicine (Fig. 6, A and B). Similarly, the AB rings of MIL occupied the same pocket with the trimethoxybenzene of colchicine. However, colchicine interacts with α-tubulin with a hydrogen bond, whereas MIL shows no contacts with α-tubulin (Fig. 6B). We also compared the binding model of MIL with that of SKLB028 and SKLB050. As indicted in Fig. 6 (C and D), SKLB028 and SKLB050 adopted identical positions and conformations as MIL, except that SKLB028 and SKLB050 interact with α-tubulin through a hydrogen bond between the αThr-179 residue and hydroxyl or amino. These results implied that linkage between compounds (SKLB028 and SKLB050) with residues on α-tubulin may be the reason why SKLB028 and SKLB050 showed better activities than MIL.
Table 3.
Data collection and refinement statistics
| SKLB028 (PDB code 5YL2) | Colchicine (PDB code 5XIW) | MIL (PDB code 5YLJ) | SKLB050 (PDB code 5YLS) | Apo (PDB code 5XP3) | |
|---|---|---|---|---|---|
| Data collection statistics | |||||
| X-ray source | SSRF-BL17U1 | SSRF-BL17U1 | SSRF-BL17U1 | SSRF-BL17U1 | SSRF-BL19U1 |
| Wavelength (Å) | 0.97845 | 0.97845 | 0.97845 | 0.97845 | 0.97845 |
| Resolution range (Å) | 50.0–2.09 (2.15–2.09) | 50.0–2.90 (2.98–2.90) | 50–2.70 (2.77–2.70) | 50–3.00 (3.07–3.00) | 50.0–2.30 (2.37–2.30) |
| Space group | P 212121 | P 212121 | P 212121 | P 212121 | P 212121 |
| Unit cell: ambic (Å) | 105.542, 158.199, 181.797 | 105.491, 159.148, 181.702 | 105.231, 157.367, 182.684 | 105.767, 159.054, 183.151 | 105.168, 157.964, 181.408 |
| Total reflections | 1,034,053 | 441,140 | 507,371 | 415,606 | 881,784 |
| Unique reflections | 106,543 (7,060) | 65,289 (5,379) | 86,475 (6,924) | 62,464 (4,106) | 133,224 (10,957) |
| Redundancy | 6.7 (6.6) | 6.8 (7.0) | 6.8 (6.6) | 6.7 (6.8) | 6.6 (6.5) |
| Completeness (%) | 100 (100) | 100 (100) | 100 (99.9) | 100 (100) | 100 (100) |
| Mean I/σ(I) | 18.62 (2.25) | 14.4 (2.25) | 13.75 (2.25) | 15.90 (2.25) | 19.50 (2.31) |
| Remerge | 0.094 (0.588) | 0.164 (0.715) | 0.132 (0.695) | 0.120 (0.673) | 0.081 (0.662) |
|
| |||||
| Refinement statistics | |||||
| Rwork/Rfree | 0.208/0.253 | 0.204/0.253 | 0.216/0.256 | 0.208/0.252 | 0.189/0.24 |
| RMSD bond length (Å)a | 0.008 | 0.008 | 0.007 | 0.007 | 0.012 |
| RMSD bond angle (°) | 1.314 | 1.26 | 1.228 | 1.239 | 1.57 |
| Ramachandran plot statistics | |||||
| Favored regions (%) | 97.6 | 97 | 98.1 | 98 | 97.6 |
| Additional allowed regions (%) | 2.3 | 2.68 | 1.8 | 1.8 | 2.1 |
| Disallowed regions (%) | 0 | 0.32 | 0.1 | 0.2 | 0.3 |
a RMSD, root mean square deviation.
Figure 5.
Structure of the MIL–T2R–TTL complex. A, chemical structure of MIL. B, overall view of the complex formed between αβ-tubulin and MIL. C, close-up views of the interaction network observed between MIL (green sticks) and tubulin (black cartoon). Interacting residues of tubulin are shown in stick representation. Oxygen atoms are colored red and blue. D, superimposition of the MIL–T2R–TTL (cyan cartoon) and apo-T2R–TTL (black cartoon) structures. MIL is in green stick representation. Black arrows highlight regions of steric clashes between tubulin and MIL, purple arrow shows the large main chain changes between apo- from MIL complex.
Figure 6.
Comparison the binding modes of MIL– and colchicine–, MIL– and SKLB028–, MIL– and SKLB050–T2R–TTL complex. A, chemical structure of colchicine. B, superimposition of the MIL–T2R–TTL (gray cartoons) and colchicine–T2R–TTL (purple cartoons) structures. MIL and colchicine are in green and purple stick representation, respectively. C, superimposition of the MIL–T2R–TTL (green cartoons) and SKLB028–T2R–TTL (purple cartoons) structures. MIL and SKLB028 are in red and green stick representation, respectively. D, superimposition of the MIL–T2R–TTL (green cartoons) and SKLB050–T2R–TTL (pale yellow cartoons) structures. MIL and SKLB050 are in red and green stick representation, respectively.
Chalcones with s-trans conformation are more active
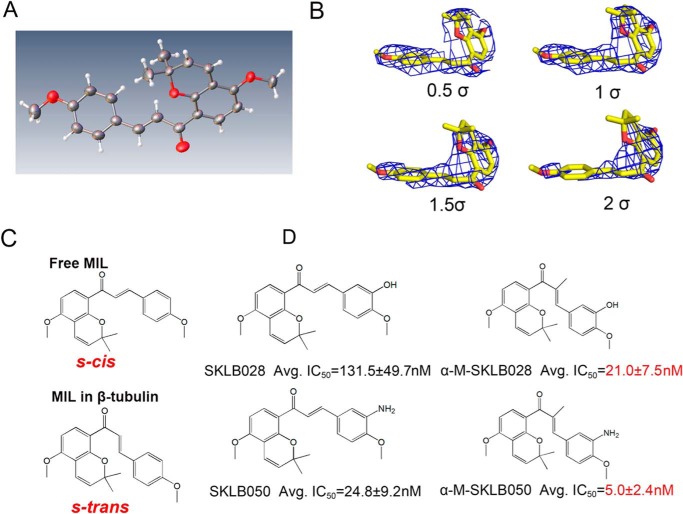
We compared the X-ray crystal structures of free MIL (CIF file of X-ray crystal structure of MIL is provided in the supporting information) with MIL in the tubulin complex. Interestingly, we found free MIL was in an s-cis conformation, whereas MIL in the tubulin complex adopts an s-trans conformation (Fig. 7, A–C), suggesting that the s-trans conformation of MIL might be the active component to bind to the colchicine site. To prove this hypothesis, we introduced an α-methyl to SKLB028 and SKLB050 to obtain α-M-SKLB028 and α-M-SKLB050 (synthesis of α-M-SKLB028 and α-M-SKLB050 is described in the supporting information), which may mainly adopt s-trans conformations (Fig. 7D); this structural change is thought to be a result of steric repulsion that would exist between the introduced α-methyl group and the A ring in the s-cis conformation. As expected, α-M-SKLB028 and α-M-SKLB050 were more active than SKLB028 and SKLB050 on five cancer cell lines (Fig. 7D and Table S1), respectively. Furthermore, we also observed that α-M-SKLB050 could induce cell-cycle arrest at a lower concentration (3 nm) compared with that of SKLB050 (30 nm) (Fig. S7). All these results implied that chalcones occupied the colchicine site in an s-trans conformational manner and that chalcones with s-trans conformation are more active.
Figure 7.
Chalcones with s-trans conformation are more active. A, X-ray structure of free MIL. B, stereo views of MIL in tubulin with the final refined 2Fo–Fc electron density contoured at 0.5, 1, 1.5, and 2 σ. C, structural formula of free MIL (s-cis) and MIL (s-trans) in tubulin complex. D, chemical structures of MDs and their average IC50s on five cancer cells (A2780S, A2780/T, HCT-8, HCT-8/V, and HCT-8/T; original data shown in Table S1).
Discussion
Tubulin inhibitors are commonly used chemotherapeutic agents for cancer therapy (3, 4, 30). Most tubulin inhibitors used in clinical were targeted to paclitaxel or vinblastine sites, such as paclitaxel, docetaxel, vinblastine, vincristine, vinorelbine, and eribulin (31–34). However, tubulin inhibitors binding to the colchicine site (the first discovered tubulin inhibitor site) (15, 30) are not used in clinical cancer therapy (15), although colchicine site tubulin inhibitors have obvious advantages over other tubulin inhibitors, such as having simple structure, being easy to synthesize, not being the substrate of multidrug-resistance protein, and not being easy to produce drug resistance (15). Therefore, the development of colchicine tubulin inhibitors has good prospects.
In our present study, we reported a new class of chalcone tubulin inhibitors. Through modification of millepachine (a novel natural compound), we generated SKLB028 and SKLB050, which possess better activity. Through biotinylation of MIL and pulldown experiments, we successfully determined that the target of MIL and its derivatives is β-tubulin. Considering that MIL and its derivatives are tubulin depolymerization agents, we competed MIL with conventional tubulin depolymerization agents, colchicine and vinblastine, respectively, and found that MIL inhibits the binding of colchicine to tubulin. Therefore, the binding site of MIL was the colchicine site. Importantly, the crystal structure of the tubulin–MIL complex also proved this. Because colchicine site tubulin inhibitors are not the substrate of multidrug-resistance proteins and not easy to produce drug resistance (15), we also tested the activity of MIL and its derivatives on three drug-resistant cell lines expressing high levels of MDR1 or MRP1 (A2780/T, HCT-8/T and HCT-8V). We found that SKLB028 and SKLB050 showed strong and similar growth inhibition against both sensitive and MDR cell lines, with a resistance factor ranging from 1.5 to 2.5, which supported the notion that colchicine site tubulin inhibitors are not the substrate of multidrug-resistance proteins.
Because the reversibility of the tubulin inhibitor binding to tubulin is an important parameter to determine the efficacy and toxicity of tubulin inhibitors in vivo (35), we also detected whether MIL and its derivatives bind irreversibly to tubulin. Through ultrafiltration and subsequent HPLC analysis, we found SKLB028 binds to tubulin in an irreversible way. Also, experiments at the cellular level showed that after a high dose of SKLB028 treatment for 8 h, the cellular morphology (indicated by tubulin staining) and cell-cycle arrest could not recover by 72 h. However, the colcemid-treated cells could recover after only 24 h. All these data indicated that SKLB028 binds to tubulin in an irreversible way. Of note, all of the assays here used to detect irreversible binding are indirect, because it is difficult to detect the irreversible binding directly when no covalent bond formed between MDs and tubulin.
Through comparison of the crystal structures of MIL–, SKLB028–, and SKLB050–tubulin complexes, we observed that SKLB028 and SKLB050 bind to the colchicine site in a position almost the same as MIL, except that SKLB028 and SKLB050 interact with α-tubulin through a hydrogen bond between the αThr-179 residue and hydroxyl or amino groups (in the same way as colchicine interacts with αVal-181). This implied that linkage between these compounds (SKLB028 and SKLB050) and residues on α-tubulin may be the reason why SKLB028 and SKLB050 showed better activities than MIL. Moreover, we also compared the X-ray crystal structures of free MIL with MIL in the tubulin complex. Interestingly, we found that free MIL was in an s-cis confromation, whereas MIL in the tubulin complex adopts an s-trans conformation, which suggested that the s-trans conformation was the active component. After the introduction of an α-methyl group to SKLB028 and SKLB050, the activities of these compounds were clearly improved. These results showed that chalcones occupied the colchicine site in an s-trans conformational manner and that naturally occurring chalcones that have an s-trans conformation are more active.
In conclusion, we identified β-tubulin as the cellular target of MIL and its derivative and showed that these compounds bind to the colchicine site in an irreversible manner. Importantly, we found that chalcone tubulin inhibitors with an s-trans dominant conformation were more active.
Experimental procedures
Reagents
Biotin, EBI, colchicine, paclitaxel, and colcemid were purchased from Selleck Chemicals. Penicillin, streptomycin, 4,6-diamidino-2-phenylindole (DAPI), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), DMSO, and propidium iodide (PI) were obtained from Sigma. Purified porcine brain tubulin was purchased from Cytoskeleton Inc. and supplied at 10 mg/ml in PEM buffer (80 mm PIPES, pH 6.9, 2 mm MgCl2, 0.5 mm EGTA, and 1 mm GTP) and preserved at −80 °C. Antibodies against α-tubulin, β-tubulin, biotin, p-H3, and GAPDH were obtained from Santa Cruz Biotechnology. Anti-γ-tubulin antibodies were obtained from Abcam. Anti-mouse secondary antibodies were from Zhongshanjinqiao Biotech. PIPES, EGTA, MgCl2, NaCl, glycerol, and other conventional reagents were purchased from Kelun Pharmaceutical or Sangon Biotech.
Cell lines and cell culture
Human ovarian carcinoma cells A2780/S and A2780/T (resistant to paclitaxel), human hepatoma cells HepG2, and human colon cancer cells HCT-116, HCT-8, and HCT-8/V (resistant to vinblastine) were obtained from the American Type Culture Collection. The cells were cultured in RPMI 1640 or DMEM containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in an atmosphere of 5% CO2. All cells used here were tested and authenticated every year in our laboratory by using an AmpFlSTR Identifiler PCR amplification kit (Applied Biosystems), and the cells were last tested in October 2017.
Pulldown of SKLB028-bound proteins
Biotin or Bio-SKLB028 (synthesis of Bio-SKLB028 described in supplementary information) was incubated with agarose beads (DynabeadsTM MyOneTM streptavidin T1; Invitrogen) to obtain biotin-conjugated or Bio-SKLB028–conjugated agarose beads. HepG2 cells were harvested and lysed for 30 min in RIPA buffer (Beyotime Biotech) supplemented with protease and phosphatase inhibitors (Beyotime Biotech). After centrifugation at 12,000 × g for 30 min, the supernatant (4 mg/ml) was collected and equally divided into two parts and then incubated with 0.1 ml of biotin or biotin–adenanthin beads in RIPA buffer overnight at 4 °C, respectively. After incubation, the beads were washed three times with RIPA buffer, and the bead-bound proteins were eluted, separated by SDS-PAGE, and visualized by Coomassie Brilliant Blue staining or subjected to Western blotting for β-tubulin and α-tubulin. Also, the specific bands in the Bio-SKLB028 group were further identified by MS, with the MS methods carried out rigorously following established laboratory protocols that were previously described (36).
Binding of SKLB028 to purified porcine tubulin
Porcine brain tubulin (2 μm) in PEM buffer (80 mm PIPES, pH 6.9, 0.5 mm EGTA, 2 mm MgCl2) was preincubated with or without 20 or 200 μm SKLB028 for 1 h before incubation with 2 or 20 μm Bio-SKLB028 for another 2 h; 2 or 20 μm biotin was incubated with tubulin for 2 h as control groups. Agarose beads were added to each sample and incubated for 15 min. The beads were then separated with centrifugation (12,000 r.p.m., 5 min) and washed with RIPA buffer three times. Bead-bound proteins were detected by Western blotting for β-tubulin. Also, total protein for each sample was detected by Western blotting for β-tubulin as a loading control. To do a time-dependent test, porcine brain tubulin (2 μm) was incubated with 2 μm Bio-SKLB028 for 0.5, 1, or 2 h. Biotin (2 μm) was incubated with tubulin for 2 h as a control. The same pulldown assay using agarose beads was performed, and the bead-bound proteins were detected by Western blotting for β-tubulin. Also, total protein for each sample was detected by Western blotting for β-tubulin as a loading control.
Tryptophan-based binding assay
The binding of small molecular to tubulin was detected using a tryptophan-based binding assay (25). Porcine brain tubulin (2 μm) in PEM buffer containing 15% glycerol was incubated with 3.125, 6.25, 12.5, 25, 50, 100, or 200 μm colchicine, MIL, SKLB028, or SKLB050 for 30 min. The tryptophan fluorescence of the protein was then monitored at 295 nm (excitation) and 335 nm (emission) using a Biotech Gen5 spectrophotometer. Dissociation constants were calculated from fitting curves of decreasing fluorescence using GraphPad Prism software.
Competition assay of SKLB028 with vinblastine or colchicine
Porcine brain tubulin (1 μm) in PEM buffer was preincubated with or without indicated concentrations of SKLB028, SKLB050, vinblastine, or colchicine for 1 h before incubation with Bio-SKLB028 for a further 2 h. All samples were then extensively washed with PEM buffer three times and then cross-linked by a UV hybridization incubator (360 nm) for 10 min. Next, samples were mixed with loading buffer and denatured at 100 °C in a water bath for 10 min. Finally, the samples were detected by Western blotting using a biotin antibody. Total proteins were loaded to detect β-tubulin as a control.
Immunofluorescence staining
HCT-8/V cells were seeded into 6-well plates and then treated with different compounds as indicated for 4 or 16 h. The cells were fixed with 4% paraformaldehyde and permeabilized with PBS containing 0.5% Triton X-100. After blocking for 30 min in 5% goat serum albumin at room temperature, the cells were incubated with monoclonal antibodies (anti-β-tubulin and anti-γ-tubulin) at 4 °C for 12 h. The cells were then washed three times with PBS followed by incubation with fluorescent secondary antibodies and labeling of the nuclei with DAPI. The cells were finally washed three times and visualized using a fluorescence microscope (Olympus, Tokyo, Japan).
In vitro tubulin polymerization assay
Porcine tubulin (3 mg/ml) was resuspended in PEM buffer with 15% glycerol and then preincubated with compounds or vehicle DMSO on ice. GTP was added to a final concentration of 1 mm before detecting the tubulin polymerization reaction. The reaction was monitored by measuring absorbance at 340 nm at 37 °C every minute in a Biotech Gen5 spectrophotometer.
Cell-cycle analysis
HCT-8/V cells were incubated with various concentrations of compounds for 16 h at 37 °C. The cells were then collected, washed with PBS, fixed in cold 70% ethanol overnight at 4 °C, and washed again with PBS, and the DNA was stained for a minimum of 10 min with 50 μg/ml PI containing 1 mg/ml of DNase-free RNase A. The samples were then analyzed using a flow cytometer (B FACSCalibur).
Western blotting detection of p-H3
HCT-8/V cells were incubated with various compounds for 16 h at 37 °C. The cells were then harvested and lysed in RIPA buffer. Equal amounts (30 μg) of total proteins were loaded on a gel for SDS-PAGE, and after separation, the proteins were transferred electrophoretically to polyvinylidene difluoride membranes, which were subsequently blocked in 5% skim milk. The p-H3 antibody was diluted in blocking buffer and incubated with the membranes overnight at 4 °C. The membranes were then washed with PBST and incubated with secondary antibodies for 1 h. Immunoreactivity detection was accomplished using enhanced chemiluminescence reagents (Millipore). GAPDH was used as a loading control.
EBI competition assay
HCT-8/V cells were incubated with different compounds for 2 h; then 100 μm EBI was added, and the cells were incubated for another 2 h. Total proteins were lysed with RIPA buffer and subjected to Western blotting analysis for β-tubulin. The detailed Western blotting method is as shown above for p-H3 or as our previous study (26). GAPDH was employed as a loading control.
HPLC detection of the binding of SKLB028 to tubulin
Purified porcine tubulin was resuspended in PEM buffer. Then equal volumes (100 μl) of tubulin solution (3 mg/ml) were incubated with 30 μm colcemid or 30 μm SKLB028 for 1 h at room temperature before being transferred to an ultrafiltration tube (Millipore, 10 kDa) and centrifuged at 12,000 rpm for 50 min at 4 °C. This ultrafiltration method could separate small molecule compounds from biomacromolecules (tubulin). After centrifugation, the filtrate was dried under vacuum and then dissolved in 200 μl of CH3OH to obtain a filtrate fraction. The tubulin (still in the ultrafiltration tube) was washed three times with 200 μl of PEM buffer (the PEM buffer was separated from tubulin also using an ultrafiltration method). The washed buffer was collected and dried under vacuum and then dissolved in 200 μl of CH3OH to get a tubulin fraction. The filtrate and tubulin fractions were then subjected to HPLC for detection of colcemid or SKLB028, respectively. Free colcemid (30 μm) or SKLB028 (30 μm) not incubated with tubulin were also dissolved in 200 μl of CH3OH and then detected by HPLC as controls, respectively.
Reversibility detection
HCT-8/V cells were treated with high concentrations (10 μm) of SKLB028 or colcemid for 8 h, then were extensively washed to remove compounds, and were further cultured to 72 h. At 0, 8, 24, 48, and 72 h, tubulin morphology and cell cycle were analyzed by immunofluorescence and flow cytometry, respectively.
Structural biology
Preparation of the crystals of the T2R–TTL complex (where T2 is αβ-tubulin heterodimer, R is the stathmin-like protein RB3, and TTL is tubulin tyrosine ligase) were described in our previous study (36). To soak compounds into crystal, 0.1 μl of the compound solution (10 mm in DMSO) was added to the 2-μl crystal-containing drop for 24 h at 20 °C. Beamlines BL17U1 or BL19U1 at Shanghai Synchrotron Radiation Facility were used to obtain X-ray diffraction data. Structure determination and refinement was the same as our previous study (36). PyMOL was used to generate the figures.
Author contributions
J. Y., W. Y., T. Y., L. X., and L. C. conceptualization; J. Y., W. Y., Y. W., T. Y., and L. X. resources; J. Y., W. Y., Y. W., T. Y., L. X., X. Y., C. L., Z. L., X. C., M. H., L. Z., Q. Q., D. L., F. W., P. B., J. W., and L. C. data curation; J. Y., W. Y., Y. Y., Y. W., T. Y., L. X., L. Z., Q. Q., and H. P. software; J. Y., W. Y., Y. Y., T. Y., L. X., H. P., D. L., and F. W. formal analysis; J. Y., W. Y., C. L., Z. L., X. C., and L. C. supervision; J. Y., W. Y., and L. C. funding acquisition; J. Y., W. Y., Y. W., T. Y., and L. X. validation; J. Y., W. Y., Y. W., T. Y., L. X., Q. Q., H. P., and D. L. investigation; J. Y., W. Y., Y. W., T. Y., L. X., X. Y., J. W., H. Y., and L. C. methodology; J. Y., W. Y., T. Y., and L. X. writing-original draft; J. Y., W. Y., and L. C. project administration; J. Y., W. Y., and L. C. writing-review and editing.
Supplementary Material
Note added in proof
The 48- and 72-h time points in the colcemid group in Fig. S5 were inadvertently duplicated in the version of this paper that was published as a Paper in Press on April 24, 2018. This error has now been corrected and does not affect the results or conclusions of this work.
This work was supported by National Natural Science Foundation of China Grant U1402222 and Guangdong Innovative Research Team Program Grant 2011Y073. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1, Figs. S1–S7, Schemes S1–S3, and Methods.
- MDR
- multidrug resistance
- MD
- M. pachycarpa derivative
- MIL
- millepachine
- EBI
- N,N′-ethylene-bis(iodoacetamide)
- PDB
- Protein Data Bank
- DAPI
- 4,6-diamidino-2-phenylindole
- PI
- propidium iodide
- p-H3
- phosphorylation H3
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RIPA
- radioimmune precipitation assay
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Perez, E. A. (2009) Microtubule inhibitors: Differentiating tubulin-inhibiting agents based on mechanisms of action, clinical activity, and resistance. Mol. Cancer Ther. 8, 2086–2095 10.1158/1535-7163.MCT-09-0366 [DOI] [PubMed] [Google Scholar]
- 2. Jordan, M. A., and Kamath, K. (2007) How do microtubule-targeted drugs work? An overview. Curr. Cancer Drug Targets 7, 730–742 10.2174/156800907783220417 [DOI] [PubMed] [Google Scholar]
- 3. Bhalla, K. N. (2003) Microtubule-targeted anticancer agents and apoptosis. Oncogene 22, 9075–9086 10.1038/sj.onc.1207233 [DOI] [PubMed] [Google Scholar]
- 4. Michaud, L. B. (2009) The epothilones: how pharmacology relates to clinical utility. Ann. Pharmacother. 43, 1294–1309 10.1345/aph.1M005 [DOI] [PubMed] [Google Scholar]
- 5. Kim, J. S., Lee, Y. C., Nam, H. T., Li, G., Yun, E. J., Song, K. S., Seo, K. S., Park, J. H., Ahn, J. W., Zee, O., Park, J. I., Yoon, W. H., Lim, K., and Hwang, B. D. (2007) Apicularen A induces cell death through Fas ligand up-regulation and microtubule disruption by tubulin down-regulation in HM7 human colon cancer cells. Clin. Cancer Res. 13, 6509–6517 10.1158/1078-0432.CCR-07-1428 [DOI] [PubMed] [Google Scholar]
- 6. Kavallaris, M. (2010) Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer 10, 194–204 10.1038/nrc2803 [DOI] [PubMed] [Google Scholar]
- 7. Swain, S. M., and Arezzo, J. C. (2008) Neuropathy associated with microtubule inhibitors: diagnosis, incidence, and management. Clin. Adv. Hematol. Oncol. 6, 455–467 [PubMed] [Google Scholar]
- 8. Fojo, A. T., and Menefee, M. (2005) Microtubule targeting agents: basic mechanisms of multidrug resistance (MDR). Semin. Oncol. 32, S3–S8 [DOI] [PubMed] [Google Scholar]
- 9. Borst, P., Evers, R., Kool, M., and Wijnholds, J. (2000) A family of drug transporters: the multidrug resistance-associated proteins. J. Natl. Cancer Inst. 92, 1295–1302 10.1093/jnci/92.16.1295 [DOI] [PubMed] [Google Scholar]
- 10. Lockwood, A. H. (1979) Molecules in mammalian brain that interact with the colchicine site on tubulin. Proc. Natl. Acad. Sci. U.S.A. 76, 1184–1188 10.1073/pnas.76.3.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moreland, L. W., and Ball, G. V. (1991) Colchicine and gout. Arthritis Rheum. 34, 782–786 10.1002/art.1780340622 [DOI] [PubMed] [Google Scholar]
- 12. Sacks, S., Cordero, G., and Rojas, P. (1981) [Colchicine for familial Mediterranean fever (author's transl)]. Revista Médica De Chile 109, 239–241 [PubMed] [Google Scholar]
- 13. Imazio, M. (2015) Colchicine for pericarditis. Trends Cardiovasc. Med. 25, 129–136 10.1016/j.tcm.2014.09.011 [DOI] [PubMed] [Google Scholar]
- 14. Aktulga, E., Altaç, M., Müftüoglu, A., Ozyazgan, Y., Pazarli, H., Tüzün, Y., Yalçin, B., Yazici, H., and Yurdakul, S. (1980) A double blind study of colchicine in Behcet's disease. Haematologica 65, 399–402 [PubMed] [Google Scholar]
- 15. Lu, Y., Chen, J., Xiao, M., Li, W., and Miller, D. D. (2012) An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res. 29, 2943–2971 10.1007/s11095-012-0828-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buey, R. M., Calvo, E., Barasoain, I., Pineda, O., Edler, M. C., Matesanz, R., Cerezo, G., Vanderwal, C. D., Day, B. W., Sorensen, E. J., López, J. A., Andreu, J. M., Hamel, E., and Díaz, J. F. (2007) Cyclostreptin binds covalently to microtubule pores and lumenal taxoid binding sites. Nat. Chem. Biol. 3, 117–125 10.1038/nchembio853 [DOI] [PubMed] [Google Scholar]
- 17. Field, J. J., Pera, B., Calvo, E., Canales, A., Zurwerra, D., Trigili, C., Rodríguez-Salarichs, J., Matesanz, R., Kanakkanthara, A., Wakefield, S. J., Singh, A. J., Jiménez-Barbero, J., Northcote, P., Miller, J. H., López, J. A., et al. (2012) Zampanolide, a potent new microtubule stabilizing agent, covalently reacts with the taxane luminal site in both tubulin α,β-heterodimers and microtubules. Chem. Biol. 19, 686–698 10.1016/j.chembiol.2012.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prota, A. E., Bargsten, K., Zurwerra, D., Field, J. J., Díaz, J. F., Altmann, K. H., and Steinmetz, M. O. (2013) Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science 339, 587–590 10.1126/science.1230582 [DOI] [PubMed] [Google Scholar]
- 19. Shan, B., Medina, J. C., Santha, E., Frankmoelle, W. P., Chou, T. C., Learned, R. M., Narbut, M. R., Stott, D., Wu, P., Jaen, J. C., Rosen, T., Timmermans, P. B., and Beckmann, H. (1999) Selective, covalent modification of β-tubulin residue Cys-239 by T138067, an antitumor agent with in vivo efficacy against multidrug-resistant tumors. Proc. Natl. Acad. Sci. U.S.A. 96, 5686–5691 10.1073/pnas.96.10.5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Towle, M. J., Salvato, K. A., Wels, B. F., Aalfs, K. K., Zheng, W., Seletsky, B. M., Zhu, X., Lewis, B. M., Kishi, Y., Yu, M. J., and Littlefield, B. A. (2011) Eribulin induces irreversible mitotic blockade: implications of cell-based pharmacodynamics for in vivo efficacy under intermittent dosing conditions. Cancer Res. 71, 496–505 10.1158/1538-7445.AM2011-496,10.1158/0008-5472.CAN-10-1874 [DOI] [PubMed] [Google Scholar]
- 21. Ye, H., Fu, A., Wu, W., Li, Y., Wang, G., Tang, M., Li, S., He, S., Zhong, S., Lai, H., Yang, J., Xiang, M., Peng, A., and Chen, L. (2012) Cytotoxic and apoptotic effects of constituents from Millettia pachycarpa Benth. Fitoterapia 83, 1402–1408 10.1016/j.fitote.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 22. Wu, W., Ye, H., Wan, L., Han, X., Wang, G., Hu, J., Tang, M., Duan, X., Fan, Y., He, S., Huang, L., Pei, H., Wang, X., Li, X., Xie, C., et al. (2013) Millepachine, a novel chalcone, induces G2/M arrest by inhibiting CDK1 activity and causing apoptosis via ROS-mitochondrial apoptotic pathway in human hepatocarcinoma cells in vitro and in vivo. Carcinogenesis 34, 1636–1643 10.1093/carcin/bgt087 [DOI] [PubMed] [Google Scholar]
- 23. Cao, D., Han, X., Wang, G., Yang, Z., Peng, F., Ma, L., Zhang, R., Ye, H., Tang, M., Wu, W., Lei, K., Wen, J., Chen, J., Qiu, J., Liang, X., et al. (2013) Synthesis and biological evaluation of novel pyranochalcone derivatives as a new class of microtubule stabilizing agents. Eur. J. Med. Chem. 62, 579–589 10.1016/j.ejmech.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 24. Yang, Z., Wu, W., Wang, J., Liu, L., Li, L., Yang, J., Wang, G., Cao, D., Zhang, R., Tang, M., Wen, J., Zhu, J, Xiang, W., Wang, F., Ma, L., et al. (2014) Synthesis and biological evaluation of novel millepachine derivatives as a new class of tubulin polymerization inhibitors. J. Med. Chem. 57, 7977–7989 10.1021/jm500849z [DOI] [PubMed] [Google Scholar]
- 25. Chinen, T., Liu, P., Shioda, S., Pagel, J., Cerikan, B., Lin, T. C., Gruss, O., Hayashi, Y., Takeno, H., Shima, T., Okada, Y., Hayakawa, I., Hayashi, Y., Kigoshi, H., Usui, T., et al. (2015) The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nat. Commun. 6, 8722 10.1038/ncomms9722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao, D., Liu, Y., Yan, W., Wang, C., Bai, P., Wang, T., Tang, M., Wang, X., Yang, Z., Ma, B., Ma, L., Lei, L., Wang, F., Xu, B., Zhou, Y., et al. (2016) Design, synthesis, and evaluation of in vitro and in vivo anticancer activity of 4-substituted coumarins: a novel class of potent tubulin polymerization inhibitors. J. Med. Chem. 59, 5721–5739 10.1021/acs.jmedchem.6b00158 [DOI] [PubMed] [Google Scholar]
- 27. Prota, A. E., Bargsten, K., Northcote, P. T., Marsh, M., Altmann, K. H., Miller, J. H., Díaz, J. F., and Steinmetz, M. O. (2014) Structural basis of microtubule stabilization by laulimalide and peloruside A. Angewandte Chemie 53, 1621–1625 10.1002/anie.201307749 [DOI] [PubMed] [Google Scholar]
- 28. Wei, W., Ayad, N. G., Wan, Y., Zhang, G. J., Kirschner, M. W., and Kaelin, W. G., Jr. (2004) Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428, 194–198 10.1038/nature02381 [DOI] [PubMed] [Google Scholar]
- 29. Dorléans, A., Gigant, B., Ravelli, R. B., Mailliet, P., Mikol, V., and Knossow, M. (2009) Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc. Natl. Acad. Sci. U.S.A. 106, 13775–13779 10.1073/pnas.0904223106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakagawa-Goto, K., Oda, A., Hamel, E., Ohkoshi, E., Lee, K. H., and Goto, M. (2015) Development of a novel class of tubulin inhibitor from desmosdumotin B with a hydroxylated bicyclic B-ring. J. Med. Chem. 58, 2378–2389 10.1021/jm501859j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clegg, A., Scott, D. A., Hewitson, P., Sidhu, M., and Waugh, N. (2002) Clinical and cost effectiveness of paclitaxel, docetaxel, gemcitabine, and vinorelbine in non-small cell lung cancer: a systematic review. Thorax 57, 20–28 10.1136/thorax.57.1.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wieczorek, M., Tcherkezian, J., Bernier, C., Prota, A. E., Chaaban, S., Rolland, Y., Godbout, C., Hancock, M. A., Arezzo, J. C., Ocal, O., Rocha, C., Olieric, N., Hall, A., Ding, H., Bramoullé, A., et al. (2016) The synthetic diazonamide DZ-2384 has distinct effects on microtubule curvature and dynamics without neurotoxicity. Sci. Transl. Med. 8, 365ra159 10.1126/scitranslmed.aag1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cortes, J., O'Shaughnessy, J., Loesch, D., Blum, J. L., Vahdat, L. T., Petrakova, K., Chollet, P., Manikas, A., Diéras, V., Delozier, T., Vladimirov, V., Cardoso, F., Koh, H., Bougnoux, P., Dutcus, C. E., et al. (2011) Eribulin monotherapy versus treatment of physician's choice in patients with metastatic breast cancer (EMBRACE): a phase 3 open-label randomised study. Lancet 377, 914–923 10.1016/S0140-6736(11)60070-6 [DOI] [PubMed] [Google Scholar]
- 34. Doodhi, H., Prota, A. E., Rodríguez-García, R., Xiao, H., Custar, D. W., Bargsten, K., Katrukha, E. A., Hilbert, M., Hua, S., Jiang, K., Grigoriev, I., Yang, C. H., Cox, D., Horwitz, S. B., Kapitein, L. C., et al. (2016) Termination of protofilament elongation by eribulin induces lattice defects that promote microtubule catastrophes. Curr. Biol. 26, 1713–1721 10.1016/j.cub.2016.04.053 [DOI] [PubMed] [Google Scholar]
- 35. Thomas, N. E., Thamkachy, R., Sivakumar, K. C., Sreedevi, K. J., Louis, X. L., Thomas, S. A., Kumar, R., Rajasekharan, K. N., Cassimeris, L., and Sengupta, S. (2014) Reversible action of diaminothiazoles in cancer cells is implicated by the induction of a fast conformational change of tubulin and suppression of microtubule dynamics. Mol. Cancer Ther. 13, 179–189 10.1186/1476-4598-13-179,10.1158/1535-7163.MCT-13-0479 [DOI] [PubMed] [Google Scholar]
- 36. Yang, J., Wang, Y., Wang, T., Jiang, J., Botting, C. H., Liu, H., Chen, Q., Yang, J., Naismith, J. H., Zhu, X., and Chen, L. (2016) Pironetin reacts covalently with cysteine-316 of α-tubulin to destabilize microtubule. Nat. Commun. 7, 12103 10.1038/ncomms12103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.