Abstract
Introduction
Mutations in USH2A cause both isolated Retinitis Pigmentosa (RP) and Usher syndrome (that implies RP and hearing impairment). One of the most frequent variants identified in this gene and among these patients is the p.(Cys759Phe) change. However, the pathogenic role of this allele has been questioned since it was found in homozygosity in two healthy siblings of a Spanish family. To assess the causative role of USH2A p.(Cys759Phe) in autosomal recessive RP (ARRP) and Usher syndrome type II (USH2) and to establish possible genotype-phenotype correlations associated with p.(Cys759Phe), we performed a comprehensive genetic and clinical study in patients suffering from any of the two above-mentioned diseases and carrying at least one p.(Cys759Phe) allele.
Materials and methods
Diagnosis was set according to previously reported protocols. Genetic analyses were performed by using classical molecular and Next-Generation Sequencing approaches. Probands of 57 unrelated families were molecularly studied and 63 patients belonging to these families were phenotypically evaluated.
Results
Molecular analysis characterized 100% of the cases, identifying: 11 homozygous patients for USH2A p.(Cys759Phe), 42 compound heterozygous patients (12 of them with another missense USH2A pathogenic variant and 30 with a truncating USH2A variant), and 4 patients carrying the p.(Cys759Phe) allele and a pathogenic variant in another RP gene (PROM1, CNGB1 or RP1). No additional causative variants were identified in symptomatic homozygous patients. Statistical analysis of clinical differences between zygosity states yielded differences (p≤0.05) in age at diagnosis of RP and hypoacusis, and progression of visual field loss. Homozygosity of p.(Cys759Phe) and compound heterozygosity with another USH2A missense variant is associated with ARRP or ARRP plus late onset hypoacusis (OR = 20.62, CI = 95%, p = 0.041).
Conclusions
The present study supports the role of USH2A p.(Cys759Phe) in ARRP and USH2 pathogenesis, and demonstrates the clinical differences between different zygosity states. Phenotype-genotype correlations may guide the genetic characterization based upon specific clinical signs and may advise on the clinical management and prognosis based upon a specific genotype.
Introduction
Retinitis Pigmentosa (RP; MIM#268000) is the most common form of Inherited Retinal Dystrophies (IRD),with a prevalence of approximately 1 in 4000 [1]. It is characterized by primary degeneration of the rods in the early stage of the disease, with progressive evolution and, currently, without a treatment, leading to visual impairment or blindness [2]. Night blindness is the first symptom, followed by constriction of the peripheral visual field, and slow and progressive decrease of central vision [3]. RP is highly heterogeneous, both clinically and genetically. RP can be a non-syndromic disease which represents 70–80% of RP cases or it can be associated with other systemic alterations (syndromic RP; 20–30%) [4]. Usher syndrome (USH), the most frequent IRD syndromic disorder, is defined by sensorineural hearing loss together with RP. The three clinical subtypes: Usher syndrome type I (USH1) (MIM#276901), type II (USH2) (MIM#276902) and type III (USH3) (MIM#276903) are distinguished depending on the severity and onset of visual impairment and hearing loss, and on the presence of vestibular impairment [5].
The USH2A gene encodes for a transmembrane protein with a large extracellular portion containing 10 laminin EGF-like domains, 35 fibronectine type-III motifs as well as two laminin G domains [6,7] and is expressed in adult human retina, specially localized to the photoreceptor cells and in fetal human cochlea and eye [8–10]. Pathogenic variants in USH2A have been associated with both non-syndromic autosomal recessive RP (ARRP, 10–15% of the characterized cases) and USH2 (80%) [11–13], being the p.(Cys759Phe) one of the most frequent pathogenic variants in the Spanish population. This variant accounts for 4.5% of the RP cases [12,14] and for 8.1% of the USH2 patients [15]. In most cases it has been observed as an autosomal recessive inherited condition, although a rare case of uniparental isodisomy has been described [16], demonstrating that other mechanisms are possible.
The high prevalence of the p.(Cys759Phe) variant prompted us to determine the implication of this variant and to elucidate whether it is the cause of syndromic and non-syndromic RP in our cohort or otherwise it is a random association or a modifier variant of RP and USH2 [17].
Results
Spanish cohort carrying p.(Cys759Phe) pathogenic variant
A total of 57 probands belonging to Spanish families affected with non-syndromic ARRP or USH2, carrying the p.(Cys759Phe) variant either in homozygosis (11/57) or heterozygosis (46/57), were selected. These last included both patients in which only one p.(Cys759Phe) allele was found (17) and 29 patients previously characterized with a second allele using classical molecular techniques (ARRP/Usher genotyping microarray, Sanger sequencing or MLPA analysis).
In order to find the second mutated allele or to discard the implication of other causative genes in the pathology, we analyzed, by different targeted Next-Generation Sequencing (NGS) approaches, not only the non-fully characterized heterozygous families (17/57) but also the probands of the p.(Cys759Phe) homozygous families (11/57) and 24 of the 29 compound heterozygous probands previously characterized by classical molecular techniques. The non-analyzed heterozygotes by NGS were 3 compound heterozygous patients with no further sample available (RP-1574, RP-0391 and RP-1016/982) and 2 carriers of p.(Cys759Phe) allele characterized with other RD genes by classical molecular tests (RP-1899 and RP-0551).
After both classical and NGS studies, we could characterize 53 families with two mutations in the USH2A gene, including 11 homozygous and 42 compound heterozygous. Whenever co-segregation studies were possible (46 out of 53 families) USH2A variants co-segregated with the disease (pedigrees of homozygous and compound heterozygous families are shown in S1A and S1B Fig, respectively). A second USH2A mutated allele was found by NGS in 15 heterozygous cases, being 3 of them novel variants (Table 1). No other causative gene among the homozygous cases was found. In 4 additional cases, out of 46 heterozygous cases, pathogenic variants in RP1(2), PROM1 and CNGB1 genes were identified as cause of the disease, co-segregating in the families with RP (S1C Fig). Therefore, these patients are only carriers for USH2A p.(Cys759Phe) (Table 1). Among the compound heterozygous patients initially characterized by classical molecular techniques and re-analyzed using NGS, we found additional variants in the following families:
Table 1. Genotype of 46 Spanish families carrying the p.(Cys759Phe) pathogenic variant in compound heterozygous state or heterozygous carriers with other causative genes.
| Categorya | Family ID | Patient ID | Technique (A2) | Gene | Nucleotide change (A2) | Amino acid change (A2) | Zygosis | Reference |
| Category B (Cys759Phe+USH2A missense) | RP-0366 | 96/0881 | Targeted-NGS | USH2A | c.754G>T | p.(Gly252Cys) | Heterozygous | [20] |
| RP-1979 | 12/1337 | Targeted-NGS | USH2A | c.1606T>C | p.(Cys536Arg) | Heterozygous | [21] | |
| RP-1053 | 06/0127 | Targeted-NGS | USH2A | c.3507G>C | p.(Trp1169Cys) | Heterozygous | This study | |
| RP-0721 | 02/0555 | Sanger | USH2A | c.3713C>G | p.(Thr1238Arg) | Heterozygous | [11] | |
| RP-2113 | 13/0464 | Targeted-NGS | USH2A | c.5462A>G | p.(Lys1821Arg) | Heterozygous | [22] | |
| RP-2504 | 15/2443 | Clinical exome | USH2A | c.9389G>T | p.(Trp3130Leu) | Heterozygous | This study | |
| RP-0752 | 02/1128 | Usher microarray | USH2A | c.9799T>C | p.(Cys3267Arg) | Heterozygous | [23] | |
| RP-2156 | 13/1196 | ARRP microarray | USH2A | c.9799T>C | p.(Cys3267Arg) | Heterozygous | [23] | |
| RP-2494 | 15/2242 | Clinical exome | USH2A | c.9799T>C | p.(Cys3267Arg) | Heterozygous | [23] | |
| RP-2372 | 14/1933 | ARRP microarray | USH2A | c.11156G>A | p.(Arg3719His) | Heterozygous | [24] | |
| RP-0653 | 01/0385 | Sanger | USH2A | c.12575G>A | p.(Arg4192His) | Heterozygous | [11] | |
| RP-1574 | 10/0653 | ARRP microarray | USH2A | c.13010C>T | p.(Thr4337Met) | Heterozygous | [23] | |
| Category C (Cys759Phe+USH2A truncating) | RP-1525 | 09/2102 | ARRP microarray | USH2A | c.100C>T | p.(Arg34*) | Heterozygous | [21] |
| RP-0391 | 97/0318 | ARRP microarray | USH2A | c.187C>T | p.(Arg63*) | Heterozygous | [21] | |
| RP-1802 | 11/1105 | Targeted-NGS | USH2A | c.920_923dupGCAA | p.(His308Glnfs*16) | Heterozygous | [6] | |
| RP-0016 | 934 | Sanger | USH2A | c.944_951dupCACAGCGG | p. (Cys318Hisfs*21) | Heterozygous | [25] | |
| RP-1412 | 09/0426 | Targeted-NGS | USH2A | c.1214delA | p.(Asn405Ilefs*3) | Heterozygous | [26] | |
| RP-0004 | 0729 | Sanger | USH2A | c.2135delC | p.(Ser712*) | Heterozygous | [25] | |
| RP-0879 | 04/0740 | ARRP microarray | USH2A | c.2135delC | p.(Ser712*) | Heterozygous | [25] | |
| RP-1104 | 06/0998 | ARRP microarray | USH2A | c.2299delG | p.(Glu767Serfs*21) | Heterozygous | [8] | |
| Categorya | Family ID | Patient ID | Technique (A2) | Gene | Nucleotide change (A2) | Amino acid change (A2) | Zygosis | Reference |
| Category C (Cys759Phe+USH2A truncating) | RP-1590 | 10/0779 | ARRP microarray | USH2A | c.2299delG | p.(Glu767Serfs*21) | Heterozygous | [8] |
| RP-1810 | 11/1176 | ARRP microarray | USH2A | c.2299delG | p.(Glu767Serfs*21) | Heterozygous | [8] | |
| RP-1858 | 11/1929 | ARRP microarray | USH2A | c.2299delG | p.(Glu767Serfs*21) | Heterozygous | [8] | |
| RP-2130 | 13/0792 | ARRP microarray | USH2A | c.2299delG | p.(Glu767Serfs*21) | Heterozygous | [8] | |
| RP-0605b | 00/0554 | ARRP microarray | USH2A | c.2431_2432delAA | p.(Lys811Aspfs*11) | Heterozygous | [27] | |
| RP-0610 | 00/0505 | ARRP microarray | USH2A | c.2431_2432delAA | p.(Lys811Aspfs*11) | Heterozygous | [27] | |
| RP-1817 | 11/1316 | ARRP microarray | USH2A | c.2431_2432delAA | p.(Lys811Aspfs*11) | Heterozygous | [27] | |
| RP-1016/RP-982 | 05/1231 | MLPA | USH2A | del Ex.22-29 | Heterozygous | This study | ||
| RP-0467 | 05/0084 | Sanger | USH2A | c.7595-2144A>G | p.(Lys2532Thrfs*56) | Heterozygous | [28] | |
| RP-1031 | 05/1440 | Sanger | USH2A | c.7595-2144A>G | p.(Lys2532Thrfs*56) | Heterozygous | [28] | |
| RP-1776 | 11/0774 | Sanger | USH2A | c.7595-2144A>G | p.(Lys2532Thrfs*56) | Heterozygous | [28] | |
| RP-2262 | 14/0248 | Targeted-NGS | USH2A | c.7595-2144A>G | p.(Lys2532Thrfs*56) | Heterozygous | [28] | |
| RP-0810c | 03/0809 | Sanger | USH2A | c.8435_8438delCCTA | p.(Thr2812Metfs*17) | Heterozygous | [23] | |
| RP-0385 | 10/0930 | MLPA | USH2A | del Ex.44 | Heterozygous | [29] | ||
| RP-2424 | 15/0499 | Clinical exome | USH2A | c.10759C>T | p.(Gln3587*) | Heterozygous | [30] | |
| RP-0061 | 05/0540 | Targeted-NGS | USH2A | c.11548+2T>G | Splicing defect | Heterozygous | [30] | |
| RP-2089 | 13/0144 | ARRP microarray | USH2A | c.11864G>A | p.(Trp3955*) | Heterozygous | [7] | |
| RP-2529 | 15/1890 | Clinical exome | USH2A | c.12457delG | p.(Ala4153Profs*14) | Heterozygous | This study | |
| RP-1059 | 06/0896 | Sanger | USH2A | c.13745delT | p.(Ile4582Lysfs*14) | Heterozygous | [11] | |
| RP-1422 | 09/0610 | Clinical exome | USH2A | c.13811+2T>G | splicing defect | Heterozygous | [31] | |
| RP-2396 | 14/2336 | Targeted-NGS | USH2A | c.14091delT | p.(Phe4697Leufs*2) | Heterozygous | [32] | |
| RP-0784 | 03/0735 | Clinical exome | USH2A | c.14180G>A | p.(Trp4727*) | Heterozygous | [13] | |
| Categorya | Family ID | Patient ID | Technique (A2) | Gene | Nucleotide change (A2) | Amino acid change (A2) | Zygosis | Reference |
| Other genes | RP-1914 | 12/0131 | Targeted-NGS | CNGB1 | c.2957A>T | p.(Asn986Ile) | Homozygous | [33] |
| RP-1899 | 11/2421 | ARRP microarray | PROM1 | c.1354dupT | p.(Tyr452Leufs*13) | Homozygous | [34] | |
| RP-0551 | 05/1342 | Sanger | RP1 | c.1625C>G | p.(Ser542*) | Homozygous | [35] | |
| RP-1772d | 11/0727 | Targeted-NGS | RP1 | c.2431delA | p.(Ser812Valfs*36) | Heterozygous | [22] |
aPatients were organized into different categories: compound heterozygous p.(Cys759Phe) + USH2A missense mutation (Category B), compound heterozygous p.(Cys759Phe) + USH2A truncating mutation (Category C), and "Other genes", carriers for p.(Cys759Phe) allele + causative mutation(s) in other RP gene.
bA splicing variant in CNGB3 (c.852+1G>C) in homozygosity was detected in the proband after NGS-reanalysis.
cA third USH2A variant [c.12574C>T; p.(Arg4192Cys] was detected in cis with p.Cys759Phe, after NGS re-analysis.
dThe RP-1772 family was re-classified as autosomal dominant RP.
Abbreviations: A2, second allele detected in USH2A or other RP gene; Technique (A2), technique by which the second mutation was detected; ARRP microarray, genotyping microarray for autosomal recessive retinitis pigmentosa.
Novel variants in USH2A are displayed in bold.
-
i)
In family RP-0004 an unreported likely pathogenic variant in PCDH15 was found: c.124G>T; p.(Gly42*). No second allele was detected by NGS analysis (Clinical Exome Solution; Sophia Genetics), which also includes copy number variations (CNV) detection.
-
ii)
RP-0810 also carried the USH2A variant c.12574C>T; p.(Arg4192Cys).This variant has been previously reported as pathogenic in a family with ARRP [18]. Segregation analysis in family RP-0810 showed that the variant was in cis with the p.(Cys759Phe) allele (S1B Fig). Both missense USH2A variants -p.(Cys759Phe) and p.(Arg4192Cys)- were in trans with a third USH2A truncating variant p.(Thr2812Metfs*17).
-
iii)
RP-0605 was a compound heterozygous family for USH2A variants p.(Cys759Phe) and p.(Lys811Aspfs*11). Using NGS, we additionally detected the CNGB3 c.852+1G>C variant in homozygous state. This variant was previously reported by Mayer et al. [19] in two families suffering from achromatopsia. Re-evaluation of the clinical history of our patient suggested the presence of two concurrent retinal pathologies. On the one hand, proband of the family RP-0605 is affected by RP+ hypoacusis. On the other hand, patient suffered from congenital nystagmus and reported photophobia and low VA since 3 years old. These latter symptoms and signs may be in keeping with a concurrent diagnosis of a cone dystrophy or an achromatopsia.
Variants found in both genes (USH2A and CNGB3) segregated in the family; therefore suggesting a RP diagnosis due to USH2A, and a likely cone dystrophy/achromatopsia diagnosis due to CNGB3 in the proband; and a cone dystrophy/achromatopsia diagnosis in his brother (sibling II:2, S1B Fig).
In silico predictions of mutated Cys759Phe Usherin
The Cys759 is a highly conserved position (98.16% in an alignment of 489 proteins, according to USMA tool) located inside a EGF laminin, a globular domain described to enhance USH2A stability in the basement membrane by prompting its interaction with collagen IV [36]. The substitution of the cysteine at position 759 by a phenylalanine would also disrupt a predicted disulfide bridge with Cys477 [12] as a source of erroneous protein folding and instability, as it is reported by USMA and Uniprot database.
In addition to the idea of the variant affecting a functional residue, the contiguous residue is glycosylated in a usherin orthologous protein (from fruit-fly) [37]. This has been used to predict a glycosylation site in USH2A Asn760 residue, based on residue conservation [38]. N-linked glycosylations are known to play a role in protein correct folding and cell-extracellular matrix attachment. The potential glycosylation at that position could be lost in the variant p.(Cys759Phe).
We also explored the functions that would be affected by the alteration of USH2A functionality. Thus, using the STRING database [39], we found 48 Gene Ontology terms (biological process) over-represented (False Discovery Rate, FDR<0.05), mainly related to ear and eye morphogenesis in the proteins interacting with USH2A (S1 Table).
Genotype-phenotype correlation
In order to ease the comprehension of the p.(Cys759Phe) variant phenotype analysis, we divided those patients with the p.(Cys759Phe) allele into 3 genotype categories: i) Category A, all fourteen homozygous patients for the variant; ii) Category B, fourteen compound heterozygous patients with the p.(Cys759Phe) and a missense pathogenic variant; and iii) Category C, thirty-one patients with the p.(Cys759Phe) and, additionally, a truncating (nonsense, indels, deep-intronic and canonical splice site) variant. Patients presenting a causative variant in a different RP gene (not USH2A) were not included in these categories.
Based on the compiled clinical information, all the patients were assigned to one of the following three groups: ARRP or sporadic RP (ARRP/SRP), USH2 or RP + hypoacusis (when the available data were not sufficient for classification as USH2). Definition for this classification is detailed in "Clinical examination", in the "Material and methods" section. Ophthalmological data were available for 63 patients (56 families), including patients with mutations only in USH2A (59 cases from 52 families) and patients with mutations in other RP genes (4 cases from 4 families). Data on hearing loss were available for 52 patients (belonging to 45 families) with mutations only in USH2A and for 4 patients (4 families) with mutations in non-USH2A genes.
The number and percentage of patients classified according to the clinical subtype and genotype are shown in Table 2.
Table 2. Likelihood of presenting a specific clinical diagnosis for patients carrying p.(Cys759Phe) variant based on their genotype.
| Diagnosis | Category A: p.(Cys759Phe) Homozygous | Category B: Compound Heterozygous p.(Cys759Phe) +USH2A missense | Category C: Compound Heterozygous p.(Cys759Phe) +USH2A truncating | Category A vs Category B | Category A vs Category C | Category B vs Category C | |||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | likelihood of presenting Usher II diagnosis (p value, two-tailed Fisher´s test) | |||
| Usher syndrome type II | 0 | 0 | 0 | 0 | 8 | 25.8 | NA | 0.043 | 0.043 |
| ARRP/SRP | 9 | 64.3 | 9 | 64.3 | 19 | 61.3 | |||
| RP+hypoacusis | 5 | 35.7 | 5 | 35.7 | 4 | 12.9 | |||
| Total | 14 | 100 | 14 | 100 | 31 | 100 | |||
Fifty-nine cases belonging to 52 families were included in the analysis (4 families having causative mutations in other RP genes and presenting only the Cys759Phe allele in USH2A were excluded; also the proband of the family RP-2424, since only molecular information was available).
To facilitate the comprehension of the genotype-phenotype correlation analysis, the patients were classified into three different categories: homozygous patients for p.(Cys759Phe) variant (Category A); compound heterozygous patients carrying additionally other USH2A variant (missense, Category B and truncating, Category C).
The likelihood of presenting an USH2 diagnosis for each category was calculated by two-tailed Fisher´s test.
Abbreviations: RP, Retinitis Pigmentosa; AR, autosomal recessive; S, sporadic; NA, non applicable.
Complete clinical information of the studied patients is shown in S2 Table.
Patients presenting an USH2A truncating mutation in compound heterozygosity with p.(Cys759Phe) (Category C) were likely to present an USH2 diagnosis when compared with homozygous patients (Category A), or heterozygous patients from Category B (p = 0.043 in both cases, Table 2).
Additionally, there was more than a 20 fold increased chance (OR = 20.617 (1.130–376.212)) of presenting a non USH2 diagnosis when patients do not have a truncating variant in addition to p.(Cys759Phe) (p = 0.041, Table 3). The sensitivity and negative predictive value were both 100%.
Table 3. Likelihood of presenting a RP or RP + hypoacusis instead of a USH2 diagnosis in patients which carry a USH2A missense variant, comparing them with patients with an USH2A truncating variant, in addition to p.(Cys759Phe).
| Diagnosis | Category A+B | Category C | Category A+B vs Category C | ||
|---|---|---|---|---|---|
| N | % | N | % | likelihood of presenting a RP or RP+hypoacusis diagnosis (IC-95%) | |
| Usher syndrome type II | 0 | 0 | 8 | 25.8 | |
| ARRP/SRP + RP+hypoacusis | 28 | 100 | 23 | 74.2 | OR = 20.617 (1.130–376.212)[40]; p = 0.041 |
| Total | 28 | 100 | 31 | 100 | |
Fifty-nine cases belonging to 52 families were included in the analysis (4 families having causative mutations in other RP genes and presenting only the Cys759Phe allele in USH2A were excluded; also the proband of the family RP-2424, since only molecular information was available).
Abbreviations: IC, interval of confidence; RP, retinitis pigmentosa; AR, autosomal recessive; S, sporadic; O, odds ratio.
The mean and standard deviation (SD) of the analysed phenotype features for patients with RP, RP+hypoacusis or USH2, distributed by genotype categories, are shown in Table 4.
Table 4. Phenotypic findings in 59 patients from 52 families carrying the p.(Cys759Phe) variant, classified by their genotype.
| Age at diagnosis (yr) | NB onset (yr) | VF loss onset (yr) | VF, age at measurement (yr) | VF (degrees) | VA loss onset (yr) | VA, age at measurement (yr) | Best Eye Acuity (decimal) | Cataract, age at diagnosis (yr) | Hypoacusis age at diagnosis (yr) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Category A: p.(Cys759Phe) Homozygous | 43.1±10.7 (N = 7) | 29.5±10.0 (N = 11) | 31.0±10.0 (N = 12) | 49.9±19.9 (N = 11) | 12.1±7.3 (N = 12) | 41.5±11.0 (N = 6) | 46.1±20.6 (N = 12) | 1.0±1.3 (N = 12) | 54.2±10.3 (N = 5) | 72.5±3.5 (N = 6) |
| Category B: Compound Heterozygous p.(Cys759Phe)+USH2A missense | 43.9±10.7 (N = 8) | 25.8±13.5 (N = 13) | 27.8±11.4 (N = 12) | 42.9±8.9 (N = 13) | 13.0±10.8 (N = 13) | 37.8±13.3 (N = 6) | 49.1±9.9 (N = 9) | 0.5±3.4 (N = 10) | 45.2±10.6 (N = 9) | 31.3±20.1 (N = 4) |
| Category C: Compound Heterozygous p.(Cys759Phe)+USH2A truncating | 29.5±12.0 (N = 20) | 26.5±9.7 (N = 25) | 28.0±8.2 (N = 21) | 40.6±12.8 (N = 21) | 12.5±7.7 (N = 20) | 31.7±9.4 (N = 15) | 43.5±13.0 (N = 20) | 0.7±0.3 (N = 21) | 49.0±12.1 (N = 10) | 23.6±17.2 (N = 7) |
| Category A vs Category B+C | 0.069 | 0.362 | 0.365 | 0.205 | 0.922 | 0.149 | 0.896 | 0.373 | 0.230 | <0.001 |
| Category A vs Category C | 0.016 | 0.410 | 0.394 | 0.181 | 0.885 | 0.089 | 0.701 | 0.469 | 0.408 | <0.001 |
| Category A vs Category B | 0.897 | 0.442 | 0.465 | 0.295 | 0.417 | 0.614 | 0.661 | 0.232 | 0.159 | 0.023 |
| Category B vs Category C | 0.007 | 0.861 | 0. 938 | 0.554 | 0.523 | 0.334 | 0.215 | 0.151 | 0.479 | 0.547 |
| Category A+B vs Category C | 0.001 | 0.755 | 0.639 | 0.194 | 0.628 | 0.069 | 0.408 | 0.787 | 0.907 | 0.126 |
Families with mutations in other RD genes and patient with neither ophthalmological nor audiological available data (RP-2424) were excluded. Mean and SD values are displayed for each genetic category and phenotypic trait (Student’s t test). The number of patients included in the statistical analysis is indicated in brackets.
Abbreviations: NB, night blindness; VF, visual field; VA, visual acuity. Statistical significant differences between different categories are marked in bold (p≤ 0.05).
The analysis revealed statistically significant differences between patients belonging to Category C compared with the other groups (Categories A and/or B), for age at diagnosis (p = 0.001, Category A+B vs Category C). Carrying a truncating mutation was associated to an earlier age of diagnosis and age of onset of VA loss, and hypoacusis diagnosis tended to be earlier, although these differences did not become statistically significant (p = 0.069 and p = 0.126, Category A+B vs Category C, respectively). For all categories, VA loss appeared late in the disease´s evolution, being ≥0.4 (decimal) until the fifth decade of life.
The main noticed difference within patients was in age at diagnosis of hypoacusis. Patients from categories B and C are significantly more likely to present an earlier diagnosis of hearing loss than patients from category A (p = 0.023 and p<0.01, respectively), being the truncating group the one with an earlier diagnosis. Five of the patients presenting the p.(Cys759Phe) variant in homozygous state (5/14, 35.7%) referred hypoacusis at a relative old age (ranging from 50 to 76 years of age) and all of them reported mild hearing loss.
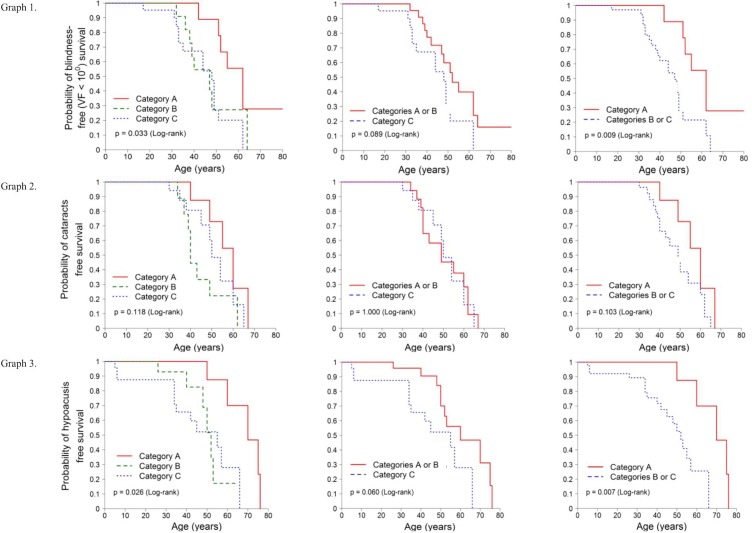
Survival analysis
The estimated survival curves for legal blindness (visual field, VF<10°), presence of cataracts and hypoacusis are shown in Fig 1.
Fig 1. Survival analysis.
Kaplan-Meier survival curves were estimated for each event and the curves of the different groups were compared using the log-rank test. The three categories of patients are considered separately, and then in two new regroupings (Category A + B) and (Category B + C). Category A: p.(Cys759Phe) homozygous, Category B: compound heterozygous p.(Cys759Phe) + USH2A missense variant, and Category C: compound heterozygous p.(Cys759Phe) + USH2A truncating variant. X axis: age in years. Y axis: probability of survival. Graph 1. Survival curve: fraction of patients free of legal blindness due to VF<10° over time. Graph 2. Survival curve: fraction of patients free of cataracts over time. Graph 3. Survival curve: fraction of patients free of hypoacusis over time.
In the case of legal blindness defined as VF<10°, the survival analysis revealed that there were differences between homozygous and heterozygous patients. Blindness takes less time to appear in heterozygous cases than in homozygous. Specifically, the median survival time is 62 years for homozygous compared to 49 years for heterozygous (Graph 1 in Fig 1).
There were no differences for cataracts (Graph 2 in Fig 1).
In the hypoacusis analysis, the difference also lied between homozygous and heterozygous patients, as it is shown in the Graph 3 (Fig 1). As for legal blindness due to VF loss, hearing loss takes less time to be perceived by compound heterozygous patients than by homozygous patients. Specifically, the median survival time was 70 years for homozygous and 53 years for heterozygous cases.
We found differences between genotype categories in the occurrence of legal blindness (VF) and hearing loss. By using the Cox proportional hazards models, we quantified how greater is the risk of presenting each of the two events in the heterozygous group compared to the homozygous patients. S3 Table shows the results of the Cox models for blindness (VF) and hearing loss. This analysis shows that the heterozygous group presents a statistically significant (three times, p = 0.014) higher risk of legal blindness (VF). In the case of hearing loss, the risk is >6 times higher for the heterozygous group (truncating + missense) than for the homozygous group (p = 0.017).
Discussion
In this work, we have analyzed by different molecular approaches a large cohort of non-syndromic ARRP and USH2 patients. All the fifty-seven probands studied were characterized, 53 of them presenting the p.(Cys759Phe) variant both in homozygous or compound heterozygous state. Even though in the literature this variant has been reported in over 90 RP patients and 40 USH2 patients [6,11–14,21,25,27,41–47], Gonzalez-del Pozo et al have questioned the pathogenic role of this variant, at least when it appears homozygously [17]. Herein we report a total of 11 homozygous families (14 patients) for the p.(Cys759Phe) variant (S1 Fig). In these families, any other candidate variants in the USH2A gene or other RP genes that could explain the RP or USH2 phenotype were not found by our NGS analysis. Nevertheless, variants in deep-intronic or regulatory regions and complex rearrangements cannot be discarded.
The p.(Cys759Phe) variant has been identified in a heterozygous state in 262 cases, presenting a population frequency of 0.09% in the Genome Sequencing Project (GnomAD) and, although this variant has been reported with a higher prevalence in Spanish population [48], there is no homozygous cases in control population even in Spanish population databases.
Furthermore, based on the in silico prediction, several arguments point to destabilization of Cys759Phe mutated USH2A protein in the extracellular matrix by: i) the change of the polar Cys by the hydrophobic Phe in position 759, ii) the disruption of a disulfide bond and, iii) a certain misfolding in a region with a role in promoting interactions. If USH2A interactome is altered, the functions affected are mainly related to the ear and eye morphogenesis, visual perception (phototransduction) or retina homeostasis, among others (S1 Table).
All these evidences suggest that this variant may play a specific role in the pathogenesis of non-syndromic RP or USH2. However, further functional and gene editing studies [49] could shed additional light on the role of this USH2A variant.
This is the first work analyzing phenotypic differences in patients presenting the p.(Cys759Phe) variant. Clinical differences between genotypes are not easy to assess. Nevertheless, large cohorts and systematic recording of phenotypic data across time are required to confirm a defined genotype-phenotype correlation, as we underline here [50–53]. In our analysis, it has been observed that visual impairment in p.(Cys759Phe) homozygous patients appears to be milder than in heterozygous patients, and that truncating variants seem to lead to a more severe visual alteration, with legal blindness due to VF loss occurring more than 10 years later in homozygous.
Hearing loss in patients with a truncating variant is also more severe when compared with both p.(Cys759Phe) homozygous patients and missense heterozygous patients, as already reported by Blanco-Kelly et al. [15].
Moreover, relative early onset (early 40´s to early 50´s) of cataracts (which is a relative frequent feature of RP) [54,55], with no differences between the three groups, but later than that reported for non-p.(Cys759Phe) USH2A patients (30´s) [15], again supports that p.(Cys759Phe) is responsible for a milder phenotype than other USH2A variants.
Given the statistical differences between homozygous and non-homozygous cases, we can say that the onset of visual symptoms and diagnosis of audiological impairment occur later in homozygous patients. High variability in sensorineural hearing loss for p.(Cys759Phe) has already been described [7,23]; however, hearing loss in homozygous patients is relatively infrequent (5/14). Regarding the patient-referred age at onset (7th decade of life) we cannot exclude that these patients suffered from age related hearing loss, rather than a hearing impairment due to USH2A defect.
Additionally, we observe that the p.(Cys759Phe) carrier patients with RP due to another RP gene have a visual phenotype characterized by a far earlier onset than the exhibited by our p.(Cys759Phe) cohort. None of them presented hypoacusis, and visual phenotype is coincident with that reported for their causative genes [22,33–35], thus suggesting that the p.(Cys759Phe) is not contributing to their RP. Besides, we believe that it is important to highlight that when coming across patients with the p.(Cys759Phe) variant and an early-adulthood onset of a RD phenotype, further genetic analysis of non-USH2A RP genes is recommended.
In conclusion, the p.(Cys759Phe) variant must be considered as pathogenic, since this variant, in coexistence with other pathogenic USH2A alleles, rendered in all cases a symptomatic phenotype, even though p.(Cys759Phe) variant might be related to a less severe ocular disease course than patients with other USH2A mutations. Moreover, the comprehensive molecular analysis of our homozygous and compound heterozygous p.(Cys759Phe) patients, did not reveal other candidate RP genes as responsible for their phenotype in most of the families analyzed by NGS means. Only in four out of 57 families with only one p.(Cys759Phe) allele, a different RP gene (not USH2A) was responsible for the disease, indicating that they were simply carriers of p.(Cys759Phe). Additionally, further findings only occurred in three of the NGS re-analyzed cases:
In family RP-0605, re-analysis with NGS uncovered the presence of two coexisting retinal diseases (RP and cone affectation), since biallelic pathogenic variants in two different RD-related genes (USH2A and CNGB3) have been identified. These facts bring into consideration the importance of, once the genotype is known, to go back to the phenotype, or curating the phenotype when performing, analysing and reporting molecular studies, and when considering the enrolment of patients into clinical trials [56,57]; moreover, when genetic diseases co-existence has been reported to be present up to 4.9% of cases with informative whole-exome sequencing [58].
Additionally, family RP-0810 carried three USH2A pathogenic changes. Unfortunately we cannot predict whether the presence of two in cis missense variants is having a more severe impact in USH2A function than their impact alone.
NGS re-analysis of RP-0004 family has enabled us to detect one likely pathogenic allele in PCDH15. Nevertheless, we have not found a second allele in this gene. We would like to remark that our NGS analysis allows to us the study of CNV. Furthermore, mutations in PCDH15 are related to Usher syndrome type I and to non-syndromic hearing loss [59]; phenotypes that are not keeping with our patient clinical findings.
As regards genotype-phenotype correlation associated with USH2A p.(Cys759Phe) variant, the presence of a p.(Cys759Phe) allele in homozygous state or in combination with other USH2A missense mutation is associated with a RP or a RP with a late onset of hypoacusis clinical subtypes. This is in line with that reported by Lenassi et al. [13]. In their study they found that some missense USH2A alleles (among them, the p.(Cys759Phe) variant) were confined to nonsyndromic RP cases, being enriched in nonsyndromic RP compared to USH2 cases, whereas "null" variants were rare in nonsyndromic cases and common in USH2 (as in the present series). However, they proposed a model of allele hierarchy of variants affecting USH2A function that does not fit with the results obtained in our study. In the model proposed by Lenassi, retinal-specific alleles would yield a non-syndromic RP phenotype when they appear in homozygous state or in combination with other retinal-specific or USH2-specific alleles. In the present study, all patients diagnosed as USH2 carried a null USH2A variant in compound heterozygous state with p.(Cys759Phe), being the former variant allegedly confined to retinal disease.
Additionally, there is a phenotype associated to p.(Cys759Phe) homozygosity consisting on a later diagnosis of RP and slower progression of VF loss, with a very late hypoacusis diagnosis (around 7th decade).
In summary, our study objectively validates the pathogenicity of USH2A p.(Cys759Phe) and presents the clinical differences between p.(Cys759Phe) patients.
Materials and methods
Patients
Fifty-seven unrelated Spanish families diagnosed with RP or USH2 were recruited from the Biobank of the Fundación Jiménez Díaz Hospital (Madrid, Spain).
DNA was extracted from peripheral blood samples of index patients and their family members as described by Perez-Carro et al [22]. Informed consent was obtained from all subjects involved in this study. All procedures were reviewed and approved by the Ethics Committee of the hospital and adhered to the tenets of the Declaration of Helsinki and further reviews.
A total of 59 patients (from 52 families), presenting the p.(Cys759Phe) variant in homozygous (14/59) or in compound heterozygous (45/59) state were included in the phenotype analysis. Additionally, the phenotype of 4 patients (from 4 families) presenting the p.(Cys759Phe) variant in the USH2A gene and with pathogenic variants in the RP1(2), PROM1 or CNGB1 genes, are shown in S2 Table and they have not been statistically analyzed. Number of patients that underwent genetic and/or phenotype analysis are shown in S2 Fig.
Genetic analysis
All probands were previously screened for known mutations with classical molecular techniques: a specific ARRP/Usher genotyping microarray (AsperBiotech, Tartu, Estonia; http://www.asperbio.com/asper-ophthalmics) or Sanger sequencing. For those cases with the p.(Cys759Phe) variant heterozygously, Sanger sequencing to screen the deep intronic c.7595-2144A>G variant in USH2A and multiplex ligation-dependent probe amplification (MLPA probemixes P361 and P362, MRC-Holland), in order to find large deletions/duplications, were performed following the manufacturer’s instructions and analyzing the results with the Coffalyser software (MRC-Holland, Amsterdam, The Netherlands).
In order to characterize the incompletely characterized patients and to exclude the implication of additional variants in other RD and hypoacusis genes (rather than those identified in USH2A), homozygous and heterozygous p.(Cys759Phe) patients were analyzed with different NGS approaches based on: a) Targeted-NGS, different in-house gene panels were used along this research, containing genes previously associated with IRD (RetNet); and b) Clinical exome -TruSightOne (Illumina) or Clinical Exome Solution (Sophia Genetics)- containing more than 4.500 genes associated with known clinical phenotypes (OMIM Database). Genes included in each panel and those selected for the analysis of the clinical exome are detailed in S4 Table.
Sanger sequencing, as reported elsewhere [22], was performed to confirm pathogenic variants and to segregate them in the families.
Assessment of pathogenicity
Novel rare variants were checked in the 1000 Genomes Project, Exome Variant Server (EVS, version 0.0.30), Exome Aggregation Consortium (ExAC, version 0.3.1) and Genome Aggregation Database (gnomAD, version r2.0.2). Furthermore, 267 in-house whole exome from Spanish healthy individuals (CIBERER Collaborative Spanish Variant Server) were used to evaluate the frequency of the variants found in this study.
Four different predictive software programs were used to assess the pathogenesis of the missense variants: 1) Sorting Intolerant from Tolerant (SIFT), 2) Polymorphism Phenotyping v2 (Polyphen-2), 3) Align GVGD and 4) Mutation Taster. Those variants predicted as damaging by at least two different out of four prediction softwares were considered as a possible causative variant.
To check the effect of p.(Cys759Phe) in the USH2A structure and function, USMA (https://neuro-2.iurc.montp.inserm.fr/USMA/) and Uniprot, and PTMcode v.2 [38] databases were used, respectively.
For the prediction of the functional impact on the USH2A neighbourhood, we used STRING facility [39].
Clinical examination
All patients were classified according to their clinical history, pedigree data, results from ophthalmological studies [60–63], audiological test results (or self-reported hearing loss) and neurophysiological and vestibular test results [63–65].
A defined protocol, previously described by Blanco-Kelly et al. [15], was followed to collect the data for establishing the ophthalmological status.
The severity of visual impairment was established both for VA loss and VF loss, and classified following the WHO (World Health Organization) criteria, as detailed by Blanco-Kelly et al. [66].
Hearing loss severity was categorized according to audiological tests [63–65] as reported by Blanco-Kelly et al. [15].
Patients with mutations only in USH2A were classified in one of the three following groups: "ARRP", defined as RP and absence of hypoacusis (based on last audiogram or at clinical interrogation) at the time of assessment; "USH2", defined as RP plus hypoacusis with an USH2 audiogram and/or self-reported hypoacusis at early age of onset; and "RP+hypoacusis", defined as RP plus hypoacusis with non-USH2 audiogram and/or late age of onset. An USH2 audiogram is defined when it showed a neurosensorial and bilateral hearing loss, with mild-to-moderate loss at low and middle frequencies and moderate-to-profound loss at high frequencies. As an example, audiometries of two USH2 patients included in this study (RP-0061 and RP-1031) are depicted in S3 Fig.
Statistical analysis
The statistical analysis was performed for the patients presenting USH2A as responsible for their condition. The differences between values for the analyzed phenotypic aspects were tested by the Student´s t test. The differences in the frequency of the genotypes within the 3 types of phenotypes (ARRP/SRP, USH2 and RP+hypoacusis) were analysed by calculating odds ratio and χ2 or two-tailed Fisher’s test; when non-applicable, Sheskin D.J. (2004) [40] was applied.
Survival analysis for VF, cataracts and hypoacusis were estimated by the Kaplan-Meier method for each event, and the curves of the different groups were compared using the log-rank test. The three categories of patients were considered separately, and then in two new regroupings (Category A + B) and (Category B + C). Risk differences were estimated using Cox proportional hazards regression models.
Supporting information
(A) Pedigrees of homozygous p.(Cys759Phe) families. (B) Pedigrees of compound heterozygous families. (C) Pedigrees of families with causative mutations in other RP genes. Co-segregation analyses in family members are displayed when available. Abbreviations: m1, p.(Cys759Phe) allele; m2, second mutated allele in USH2A; m, mutated allele in other non-USH2A RD gene; wt, wild-type allele; NA, DNA not available.
(DOC)
*Three patients (RP-1574, RP-0391 and RP-1016/982) initially characterized using classical techniques were not re-analyzed by NGS due to lack of sample with enough quantity and/or quality. ** RP-2424 was excluded from phenotype studies, since no clinical information was available.
(DOC)
Audiograms show a typical Usher type II down-sloping pattern with bilateral hypoacusis from moderate to severe degree at high frequencies.
(DOC)
USH2A interacting proteins (with a STRING combined score > 0.400) compared to the whole genome. Abbreviations: GO, gene ontology; FDR, false discovery rate.
(DOC)
Proband from family RP-2424 was excluded from this table, since only molecular information was available. Families are organized in categories: category A, homozygous patients for p.(Cys759Phe) allele; category B, compound heterozygous patients p.(Cys759Phe) + USH2A missense mutation; category C, compound heterozygous patients p.(Cys759Phe) + USH2A truncating mutation; and families with pathogenic variants in other RP genes.
Abbreviations: Hom, homozygous; CH, compound heterozygous; Het, heterozygous; ARRP, autosomal recessive retinitis pigmentosa; SRP, sporadic retinitis pigmentosa; NB: night blindness; VF: visual field; VA: visual acuity; BCVA: best corrected visual acuity; CF: counting finger; LP: light perception; NLP: no light perception; NA: not availaible; y: years; IOP: intraocular pressure; EOG: electrooculagram; ERG: electroretinogram; MA: macular alteration; NR: non-recordable; RA: reduced amplitude; DL: delayed latencies; Sco: scotopic; Pho: photopic; OCT: optical coherence tomography; AR: Arden ratio; FA: fluorescent angiography; Typical RP fundus: pale optic disc, narrowed retina vessels and pigmentary changes (bone-spicules); OD: right eye; OS: left eye; BE: both eyes; RPE: Retinal Pigment Epithelium; —> progression; * Hypoacusis (age at last examination): audiogram and/or self-reported hypoacusis under clinical interrogation; RE: right ear; LE: left ear; Infancy# (≤6y): referred "infancy" was considered less than or equal to 6 years (for statistical analysis, infancy = 6 years); 1: mild hearing loss; 2: moderate hearing loss; 3: severe/profound hearing loss.
(DOC)
The table shows the results of the Cox models for the risk of presenting blindness and hearing loss, comparing p. (Cys759Phe) heterozygous patients (missense + truncating) with p. (Cys759Phe) homozygous patients. The models are summarized by the hazard ratio, its 95% confidence interval and the p value.
Abbreviations: VF, visual field; HR, hazard ratio; CI, confidence interval.
(DOC)
(A) In house IRD_NGS panel with 68 genes. (B) In house IRD_NGS panel with 75 genes. (C) Virtual panel with 205 genes selected for clinical exome analysis. (D) Virtual panel with 83 genes associated with deafness.
(DOC)
Acknowledgments
The authors are grateful to the families that participated in this study and to the colleagues who referred patients to us, specially to Dr. Encarna Guillén Navarro (Hospital Virgen de la Arrixaca, Murcia) and to Dr. Susanne Kohl (Centre for Ophthalmology-University Clinics Tübingen, Germany).
We also thank the Genetics and Ophthalmology Departments of the Fundación Jimenez Diaz University Hospital (FJD, Madrid), Asunción Giménez, Cristina Nieto, Cristina Villaverde, and Olga Zurita for their technical assistance.
This project was financially supported by the Center for Biomedical Network Research on Rare Diseases (CIBERER), FIS (PI16/00425 and PI16/00539) from Institute of Health Carlos III (ISCIII, Spanish Ministry of the Economy, Industry and Competitiveness), the European Regional Development Fund (ERDF), the Ramon Areces Foundation, the IIS-Fundación Jimenez Díaz-UAM Genome Medicine Chair, the Spanish National Organization of the Blind (ONCE) and the Spanish Fighting Blindness Foundation (FUNDALUCE). RP-C were sponsored by the Conchita Rábago Foundation, and LG-M and IM-M by the Río Hortega Programs (CM16/00126 and CM14/00079, respectively) from ISCIII.
Data Availability
All relevant data are available within the paper and its Supporting Information files.
Funding Statement
This project was financially supported by the Center for Biomedical Network Research on Rare Diseases (CIBERER), FIS (PI16/00425 and PI16/00539) from Institute of Health Carlos III (ISCIII, Spanish Ministry of the Economy, Industry and Competitiveness), the European Regional Development Fund (ERDF), the Ramon Areces Foundation, the IIS-Fundación Jimenez Díaz-UAM Genome Medicine Chair, the Spanish National Organization of the Blind (ONCE) and the Spanish Fighting Blindness Foundation (FUNDALUCE). RP-C were sponsored by the Conchita Rábago Foundation, and LG-M and IM-M by the Río Hortega Programs (CM16/00126 and CM14/00079, respectively) from ISCIII. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Den Hollander AI, Black A, Bennett J, Cremers FPM. Lighting a candle in the dark: Advances in genetics and gene therapy of recessive retinal dystrophies. J Clin Invest. 2010;120: 3042–3053. doi: 10.1172/JCI42258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivolta C, Sharon D, DeAngelis MM, Dryja TP. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum Mol Genet. 2002;11: 1219–1227. doi: 10.1093/hmg/11.10.1219 [DOI] [PubMed] [Google Scholar]
- 3.Ayuso C, Millan JM. Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Med. 2010;2: 34 doi: 10.1186/gm155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parmeggiani F S. Sorrentino F, Ponzin D, Barbaro V, Ferrari S, Di Iorio E. Retinitis Pigmentosa: Genes and Disease Mechanisms. Curr Genomics. 2011;12: 238–249. doi: 10.2174/138920211795860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith RJH, Berlin CI, Hejtmancik JF, Keats BJB, Kimberling WJ, Lewis R a, et al. Clinical diagnosis of the Usher syndromes. Am J Med Genet. 1994;50: 32–38. doi: 10.1002/ajmg.1320500107 [DOI] [PubMed] [Google Scholar]
- 6.Weston MD, Eudy JD, Fujita S, Yao S, Usami S, Cremers C, et al. Genomic structure and identification of novel mutations in usherin, the gene responsible for Usher syndrome type IIa. Am J Hum Genet. 2000;66: 1199–210. doi: 10.1086/302855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Wijk E, E Pennings RJ, te Brinke H, Claassen A, Yntema HG, Hoefsloot LH, et al. Report Identification of 51 Novel Exons of the Usher Syndrome Type 2A (USH2A) Gene That Encode Multiple Conserved Functional Domains and That Are Mutated in Patients with Usher Syndrome Type II. Am J Hum Genet. 2004;74: 738–744. doi: 10.1086/383096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eudy JD. Mutation of a Gene Encoding a Protein with Extracellular Matrix Motifs in Usher Syndrome Type IIa. Science (80-). 1998;280: 1753–1757. doi: 10.1126/science.280.5370.1753 [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Bulgakov O V., Darrow KN, Pawlyk B, Adamian M, Liberman MC, et al. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc Natl Acad Sci. 2007;104: 4413–4418. doi: 10.1073/pnas.0610950104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosgrove D and Zallocchi M. Usher protein functions in hair cells and photoreceptors. Int J Biochem Cell Biol. 2014;46: 80–89. doi: 10.1016/j.biocel.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ávila-Fernández A, Cantalapiedra D, Aller E, Vallespín E, Aguirre-Lambán J, Blanco-Kelly F, et al. Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol Vis. 2010;16: 2550–2558. doi:272 [pii] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Report Missense Mutation in the USH2A Gene: Association with Recessive Retinitis Pigmentosa without Hearing Loss. Am J Hum Genet. 2000;66: 1975–1978. doi: 10.1086/302926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenassi E, Vincent A, Li Z, Saihan Z, Coffey AJ, Steele-Stallard HB, et al. A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. Eur J Hum Genet. 2015;23: 1318–27. doi: 10.1038/ejhg.2014.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernal S, Ayuso C, Antinolo G, Gimenez A, Borrego S, Trujillo MJ, et al. Mutations in USH2A in Spanish patients with autosomal recessive retinitis pigmentosa: high prevalence and phenotypic variation. J Med Genet. 2003;40: e8(http://www.jmedgenet.com/cgi/content/full/40/1/. doi: 10.1136/jmg.40.1.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco-Kelly F, Jaijo T, Aller E, Avila-Fernandez A, López-Molina MI, Giménez A, et al. Clinical aspects of Usher syndrome and the USH2A gene in a cohort of 433 patients. JAMA Ophthalmol. 2015;133: 157–64. doi: 10.1001/jamaophthalmol.2014.4498 [DOI] [PubMed] [Google Scholar]
- 16.Rivolta C, Berson EL, Dryja TP. Paternal uniparental heterodisomy with partial isodisomy of chromosome 1 in a patient with retinitis pigmentosa without hearing loss and a missense mutation in the Usher syndrome type II gene USH2A. Arch Ophthalmol. 2002;120: 1566–1571. doi:eog10011 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Pozo MG del, Bravo-Gil N, Méndez-Vidal C, Montero-de-Espinosa I, Millán JM, Dopazo J, et al. Re-evaluation casts doubt on the pathogenicity of homozygous USH2A p.C759F. Am J Med Genet Part A. 2015;167: 1597–1600. doi: 10.1002/ajmg.a.37003 [DOI] [PubMed] [Google Scholar]
- 18.Corton M, Nishiguchi KM, Avila-Fernández A, Nikopoulos K, Riveiro-Alvarez R, Tatu SD, et al. Exome Sequencing of Index Patients with Retinal Dystrophies as a Tool for Molecular Diagnosis. PLoS One. 2013;8: 1–6. doi: 10.1371/journal.pone.0065574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer AK, Van Cauwenbergh C, Rother C, Baumann B, Reuter P, De Baere E, et al. CNGB3 mutation spectrum including copy number variations in 552 achromatopsia patients. Hum Mutat. 2017;38: 1579–1591. doi: 10.1002/humu.23311 [DOI] [PubMed] [Google Scholar]
- 20.Bravo-Gil N, Méndez-Vidal C, Romero-Pérez L, González-del Pozo M, Rodríguez-de la Rúa E, Dopazo J, et al. Improving the management of Inherited Retinal Dystrophies by targeted sequencing of a population-specific gene panel. Sci Rep. Nature Publishing Group; 2016;6: 23910 doi: 10.1038/srep23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreyer B, Tranebjaerg L, Rosenberg T, Weston MD, Kimberling WJ, Nilssen O. Identification of novel USH2A mutations: implications for the structure of USH2A protein. Eur J Hum Genet. 2000;8: 500–506. doi: 10.1038/sj.ejhg.5200491 [DOI] [PubMed] [Google Scholar]
- 22.Perez-Carro R, Corton M, Sánchez-Navarro I, Zurita O, Sanchez-Bolivar N, Sánchez-Alcudia R, et al. Panel-based NGS reveals novel pathogenic mutations in autosomal recessive retinitis pigmentosa. Sci Rep. Nature Publishing Group; 2016;6: 19531 doi: 10.1038/srep19531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aller E, Jaijo T, Beneyto M, Nájera C, Oltra S, Ayuso C, et al. Identification of 14 novel mutations in the long isoform of USH2A in Spanish patients with Usher syndrome type II. J Med Genet. 2006;43: e55 doi: 10.1136/jmg.2006.041764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Sheng X, Liu X, Li H, Liu Y, Rong W, et al. Targeted next-generation sequencing reveals novel USH2A mutations associated with diverse disease phenotypes: Implications for clinical and molecular diagnosis. PLoS One. 2014;9: 1–8. doi: 10.1371/journal.pone.0105439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernal S, Medà C, Solans T, Ayuso C, Garcia-Sandoval B, Valverde D, et al. Clinical and genetic studies in Spanish patients with Usher syndrome type II: Description of new mutations and evidence for a lack of genotype-phenotype correlation. Clin Genet. 2005;68: 204–214. doi: 10.1111/j.1399-0004.2005.00481.x [DOI] [PubMed] [Google Scholar]
- 26.Schwartz SB, Aleman TS, Cideciyan A V., Windsor EAM, Sumaroka A, Roman AJ, et al. Disease expression in Usher syndrome caused by VLGR1 gene mutation (USH2C) and comparison with USH2A phenotype. Investig Ophthalmol Vis Sci. 2005;46: 734–743. doi: 10.1167/iovs.04-1136 [DOI] [PubMed] [Google Scholar]
- 27.Nájera C, Beneyto M, Blanca J, Aller E, Fontcuberta A, Millán JM, et al. Mutations in myosin VIIA (MYO7A) and usherin (USH2A) in Spanish patients with Usher syndrome types I and II, respectively. Hum Mutat. 2002;20: 76–77. doi: 10.1002/humu.9042 [DOI] [PubMed] [Google Scholar]
- 28.Vaché C, Besnard T, le Berre P, García-García G, Baux D, Larrieu L, et al. Usher syndrome type 2 caused by activation of an USH2A pseudoexon: Implications for diagnosis and therapy. Hum Mutat. 2012;33: 104–108. doi: 10.1002/humu.21634 [DOI] [PubMed] [Google Scholar]
- 29.Glöckle N, Kohl S, Mohr J, Scheurenbrand T, Sprecher A, Weisschuh N, et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur J Hum Genet. 2014;22: 99–104. doi: 10.1038/ejhg.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Garcia G, Aparisi MJ, Jaijo T, Rodrigo R, Leon AM, Avila-Fernandez A, et al. Mutational screening of the USH2A gene in Spanish USH patients reveals 23 novel pathogenic mutations. Orphanet J Rare Dis. 2011;6: 65 doi: 10.1186/1750-1172-6-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besnard T, García-García G, Baux D, Vaché C, Faugère V, Larrieu L, et al. Experience of targeted Usher exome sequencing as a clinical test. Mol Genet genomic Med. 2014;2: 30–43. doi: 10.1002/mgg3.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonnet C, Riahi Z, Chantot-Bastaraud S, Smagghe L, Letexier M, Marcaillou C, et al. An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients. Eur J Hum Genet. 2016;24: 1730–1738. doi: 10.1038/ejhg.2016.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE. Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet. 2011;48: 145–151. doi: 10.1136/jmg.2010.083568 [DOI] [PubMed] [Google Scholar]
- 34.Pras E, Abu A, Rotenstreich Y, Avni I, Reish O, Morad Y, et al. Cone-rod dystrophy and a frameshift mutation in the PROM1 gene. Mol Vis. 2009;15: 1709–1716. [PMC free article] [PubMed] [Google Scholar]
- 35.Avila-Fernandez A, Corton M, Nishiguchi KM, Muñoz-Sanz N, Benavides-Mori B, Blanco-Kelly F, et al. Identification of an RP1 prevalent founder mutation and related phenotype in Spanish patients with early-onset autosomal recessive retinitis. Ophthalmology. Elsevier Inc.; 2012;119: 2616–2621. doi: 10.1016/j.ophtha.2012.06.033 [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharya G. A domain-specific usherin/collagen IV interaction may be required for stable integration into the basement membrane superstructure. J Cell Sci. 2004;117: 233–242. doi: 10.1242/jcs.00850 [DOI] [PubMed] [Google Scholar]
- 37.Zielinska DF, Gnad F, Schropp K, Wisniewski JR, Mann M. Mapping N-Glycosylation Sites across Seven Evolutionarily Distant Species Reveals a Divergent Substrate Proteome Despite a Common Core Machinery. Mol Cell. 2012;46: 542–548. doi: 10.1016/j.molcel.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 38.Minguez P, Letunic I, Parca L, Garcia-Alonso L, Dopazo J, Huerta-Cepas J, et al. PTMcode v2: A resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 2015;43: D494–D502. doi: 10.1093/nar/gku1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43: D447–D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. Technometrics. 2004;46: 1193 doi: 10.1198/tech.2004.s209 [Google Scholar]
- 41.Aller E, Nájera C, Millán JM, Oltra JS, Pérez-Garrigues H, Vilela C, et al. Genetic analysis of 2299delG and C759F mutations (USH2A) in patients with visual and/or auditory impairments. Eur J Hum Genet. 2004;12: 407–410. doi: 10.1038/sj.ejhg.5201138 [DOI] [PubMed] [Google Scholar]
- 42.Seyedahmadi BJ, Rivolta C, Keene JA, Berson EL, Dryja TP. Comprehensive screening of the USH2A gene in Usher syndrome type II and non-syndromic recessive retinitis pigmentosa. Exp Eye Res. 2004;79: 167–173. doi: 10.1016/j.exer.2004.03.005 [DOI] [PubMed] [Google Scholar]
- 43.Baux David, 1 Lise Larrieu, 1 Catherine Blanchet, 2 Christian Hamel, 2 Safouane Ben Salah, 2 Anne Vielle 1, Brigitte Gilbert-Dussardier, 3 Muriel Holder, 4 Patrick Calvas, 5 Nicole Philip, 6 Patrick Edery 7, Dominique Bonneau, 8 Mireille Claustres, 1, 9, 10 Sue Malcolm, 11 and Anne-Franc-oise Roux1 9. Molecular and In Silico Analyses of the Full-Length Isoform of Usherin Identify New Pathogenic Alleles in Usher Type II Patients. Hum Mutat. 2007;28: 781–789. doi: 10.1002/humu.20513 [DOI] [PubMed] [Google Scholar]
- 44.Dreyer B, Brox V, Tranebjærg L, Rosenberg T, Sadeghi AM, Möller C, et al. Spectrum of USH2A mutations in Scandinavian patients with Usher syndrome type II. Hum Mutat. 2008;29: 451–451. doi: 10.1002/humu.9524 [DOI] [PubMed] [Google Scholar]
- 45.Sandberg M a, Rosner B, Weigel-difranco C, Mcgee TL, Dryja TP, Berson EL. Disease Course in Patients with Autosomal Recessive Retinitis Pigmentosa due to the USH2A Gene. 2009;49: 5532–5539. doi: 10.1167/iovs.08-2009.Disease [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Zhang VW, Feng Y, Tian X, Li FY, Truong C, et al. Dependable and efficient clinical utility of target capture-based deep sequencing in molecular diagnosis of retinitis pigmentosa. Investig Ophthalmol Vis Sci. 2014;55: 6213–6223. doi: 10.1167/iovs.14-14936 [DOI] [PubMed] [Google Scholar]
- 47.Zhao L, Wang F, Wang H, Li Y, Alexander S, Wang K, et al. Next-generation sequencing-based molecular diagnosis of 82 retinitis pigmentosa probands from Northern Ireland. Hum Genet. 2015;134: 217–230. doi: 10.1007/s00439-014-1512-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dopazo J, Amadoz A, Bleda M, Garcia-Alonso L, Alemán A, García-García F, et al. 267 Spanish Exomes Reveal Population-Specific Differences in Disease-Related Genetic Variation. Mol Biol Evol. 2016;33: 1205–1218. doi: 10.1093/molbev/msw005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuster-García C, García-García G, González-Romero E, Jaijo T, Sequedo MD, Ayuso C, et al. USH2A Gene Editing Using the CRISPR System. Mol Ther—Nucleic Acids. Elsevier Ltd.; 2017;8: 529–541. doi: 10.1016/j.omtn.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang X-F, Huang F, Wu K-C, Wu J, Chen J, Pang C-P, et al. Genotype–phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet Med. 2015;17: 271–278. doi: 10.1038/gim.2014.138 [DOI] [PubMed] [Google Scholar]
- 51.Bernardis I, Chiesi L, Tenedini E, Artuso L, Percesepe A, Artusi V, et al. Unravelling the Complexity of Inherited Retinal Dystrophies Molecular Testing: Added Value of Targeted Next-Generation Sequencing. Biomed Res Int. 2016;2016 doi: 10.1155/2016/6341870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eandi CM, Dallorto L, Spinetta R, Micieli MP, Vanzetti M, Mariottini A, et al. Targeted next generation sequencing in Italian patients with Usher syndrome: Phenotype-genotype correlations. Sci Rep. Springer US; 2017;7: 1–8. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Testa F, Melillo P, Bonnet C, Marcelli V, de Benedictis A, Colucci R, Gallo B, Kurtenbach A, Rossi S, Marciano E, Auricchio A, Petit C, Zrenner E SS. Clinical presentation and disease course of Usher syndrome because of mutations in MYO7A or USH2A. Retina. 2017;37: 1581–1590. doi: 10.1097/IAE.0000000000001389 [DOI] [PubMed] [Google Scholar]
- 54.Auffarth GU, Nimsgern C, Tetz MR, Krastel H VH. Increased cataract rate and characteristics of Nd:YAG laser capsulotomy in retinitis pigmentosa. Ophthalmologe. 1997;94: 791–5. [DOI] [PubMed] [Google Scholar]
- 55.Pennings RJE, Huygen PLM, Orten DJ, Wagenaar M, van Aarem A, Kremer H, et al. Evaluation of visual impairment in Usher syndrome 1b and Usher syndrome 2a. Acta Ophthalmol Scand. 2004;82: 131–139. doi: 10.1111/j.1600-0420.2004.00234.x [DOI] [PubMed] [Google Scholar]
- 56.Magliulo G, Iannella G, Gagliardi S, Iozzo N, Plateroti R, Mariottini A TF. Usher’s Syndrome Type II: A Comparative Study of Genetic Mutations and Vestibular System Evaluation. Otolaryngol Head Neck Surg. 2017;157: 853–860. doi: 10.1177/0194599817715235 [DOI] [PubMed] [Google Scholar]
- 57.Sengillo JD, Cabral T, Schuerch K, Duong J, Lee W, Boudreault K, et al. Electroretinography Reveals Difference in Cone Function between Syndromic and Nonsyndromic USH2A Patients. Sci Rep. 2017;7: 1–11. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N Engl J Med. 2017;376: 21–31. doi: 10.1056/NEJMoa1516767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmed ZM, Riazuddin S, Ahmad J, Bernstein SL, Guo Y, Sabar MF, et al. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12: 3215–3223. doi: 10.1093/hmg/ddg358 [DOI] [PubMed] [Google Scholar]
- 60.Piazza L, Fishman GA, Farber M, Derlacki D, Anderson RJ. Visual acuity loss in patients with Usher’s syndrome. Arch Ophthalmol. 1986;104: 1336–9. doi: 10.1001/archopht.1986.01050210090031 [DOI] [PubMed] [Google Scholar]
- 61.Fishman GA; Robert J. Anderson MD LB and D, DJ. Prevalence of Foveal Lesions in Type 1 and Type 2 Usher’s Syndrome. Arch Ophthalmol. 1995;113: 770–773. Available: http://www.diabetes.org/diabetes-basics/type-2/ [DOI] [PubMed]
- 62.Edwards A, Fishman GA, Anderson RJ, Grover S, Derlacki DJ. Visual acuity and visual field impairment in Usher syndrome. Arch Ophthalmol. 1998;116: 165–8. doi: 10.1001/archopht.116.2.165 [DOI] [PubMed] [Google Scholar]
- 63.Seeliger M, Pfister M, Gendo K, Paasch S, Apfelstedt-Sylla E PP et al. Comparative study of visual, auditory, and olfactory function in Usher syndrome. Graefes Arch Clin Exp Ophthalmol. 1999;237: 301–7. [DOI] [PubMed] [Google Scholar]
- 64.Kumar A, Fishman G, Torok N. Vestibular and auditory function in Usher’s syndrome. Ann Otol Rhinol Laryngol. 1984;93: 600–608. doi: 10.1177/000348948409300613 [DOI] [PubMed] [Google Scholar]
- 65.Möller CG, Kimberling WJ, Davenport SL, Priluck I, White V, Biscone-Halterman K, et al. Usher syndrome: an otoneurologic study. The Laryngoscope. 1989. pp. 73–9. doi: 10.1288/00005537-198901000-00014 [DOI] [PubMed] [Google Scholar]
- 66.Blanco-Kelly F, García Hoyos M, Lopez Martinez MA, Lopez-Molina MI, Riveiro-Alvarez R, Fernandez-San Jose P, et al. Dominant retinitis pigmentosa, p.Gly56Arg mutation in NR2E3: Phenotype in a large cohort of 24 cases. PLoS One. 2016;11: 1–13. doi: 10.1371/journal.pone.0149473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Pedigrees of homozygous p.(Cys759Phe) families. (B) Pedigrees of compound heterozygous families. (C) Pedigrees of families with causative mutations in other RP genes. Co-segregation analyses in family members are displayed when available. Abbreviations: m1, p.(Cys759Phe) allele; m2, second mutated allele in USH2A; m, mutated allele in other non-USH2A RD gene; wt, wild-type allele; NA, DNA not available.
(DOC)
*Three patients (RP-1574, RP-0391 and RP-1016/982) initially characterized using classical techniques were not re-analyzed by NGS due to lack of sample with enough quantity and/or quality. ** RP-2424 was excluded from phenotype studies, since no clinical information was available.
(DOC)
Audiograms show a typical Usher type II down-sloping pattern with bilateral hypoacusis from moderate to severe degree at high frequencies.
(DOC)
USH2A interacting proteins (with a STRING combined score > 0.400) compared to the whole genome. Abbreviations: GO, gene ontology; FDR, false discovery rate.
(DOC)
Proband from family RP-2424 was excluded from this table, since only molecular information was available. Families are organized in categories: category A, homozygous patients for p.(Cys759Phe) allele; category B, compound heterozygous patients p.(Cys759Phe) + USH2A missense mutation; category C, compound heterozygous patients p.(Cys759Phe) + USH2A truncating mutation; and families with pathogenic variants in other RP genes.
Abbreviations: Hom, homozygous; CH, compound heterozygous; Het, heterozygous; ARRP, autosomal recessive retinitis pigmentosa; SRP, sporadic retinitis pigmentosa; NB: night blindness; VF: visual field; VA: visual acuity; BCVA: best corrected visual acuity; CF: counting finger; LP: light perception; NLP: no light perception; NA: not availaible; y: years; IOP: intraocular pressure; EOG: electrooculagram; ERG: electroretinogram; MA: macular alteration; NR: non-recordable; RA: reduced amplitude; DL: delayed latencies; Sco: scotopic; Pho: photopic; OCT: optical coherence tomography; AR: Arden ratio; FA: fluorescent angiography; Typical RP fundus: pale optic disc, narrowed retina vessels and pigmentary changes (bone-spicules); OD: right eye; OS: left eye; BE: both eyes; RPE: Retinal Pigment Epithelium; —> progression; * Hypoacusis (age at last examination): audiogram and/or self-reported hypoacusis under clinical interrogation; RE: right ear; LE: left ear; Infancy# (≤6y): referred "infancy" was considered less than or equal to 6 years (for statistical analysis, infancy = 6 years); 1: mild hearing loss; 2: moderate hearing loss; 3: severe/profound hearing loss.
(DOC)
The table shows the results of the Cox models for the risk of presenting blindness and hearing loss, comparing p. (Cys759Phe) heterozygous patients (missense + truncating) with p. (Cys759Phe) homozygous patients. The models are summarized by the hazard ratio, its 95% confidence interval and the p value.
Abbreviations: VF, visual field; HR, hazard ratio; CI, confidence interval.
(DOC)
(A) In house IRD_NGS panel with 68 genes. (B) In house IRD_NGS panel with 75 genes. (C) Virtual panel with 205 genes selected for clinical exome analysis. (D) Virtual panel with 83 genes associated with deafness.
(DOC)
Data Availability Statement
All relevant data are available within the paper and its Supporting Information files.